Abstract
Anaerobic ethylbenzene metabolism in the betaproteobacterium Aromatoleum aromaticum is initiated by anaerobic oxidation to acetophenone via (S)-1-phenylethanol. The subsequent carboxylation of acetophenone to benzoylacetate is catalyzed by an acetophenone-induced enzyme, which has been purified and studied. The same enzyme is involved in acetophenone metabolism in the absence of ethylbenzene. Acetophenone carboxylase consists of five subunits with molecular masses of 70, 15, 87, 75, and 34 kDa, whose genes (apcABCDE) form an apparent operon. The enzyme is synthesized at high levels in cells grown on ethylbenzene or acetophenone, but not in cells grown on benzoate. During purification, acetophenone carboxylase dissociates into inactive subcomplexes consisting of the 70-, 15-, 87-, and 75-kDa subunits (apcABCD gene products) and the 34-kDa subunit (apcE gene product), respectively. Acetophenone carboxylase activity was restored by mixing the purified subcomplexes. The enzyme contains 1 Zn2+ ion per αβγδ core complex and is dependent on the presence of Mg2+ or Mn2+. In spite of the presence of Zn in the enzyme, it is strongly inhibited by Zn2+ ions. Carboxylation of acetophenone is dependent on ATP hydrolysis to ADP and Pi, exhibiting a stoichiometry of 2 mol ATP per mol acetophenone carboxylated. The enzyme shows uncoupled ATPase activity with either bicarbonate or acetophenone in the absence of the second substrate. These observations indicate that both substrates may be phosphorylated, which is consistent with isotope exchange activity observed with deuterated acetophenone and inhibition by carbamoylphosphate, a structural analogue of carboxyphosphate. A potential mechanism of ATP-dependent acetophenone carboxylation is suggested.
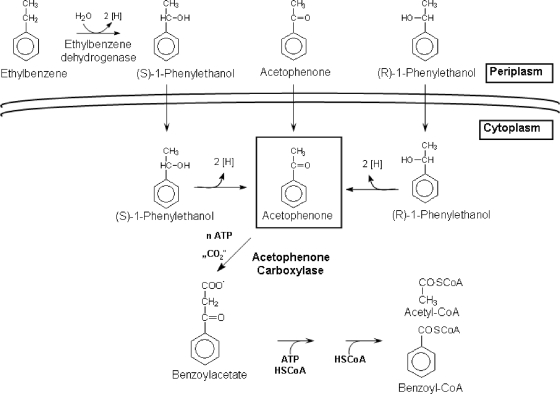
Ethylbenzene belongs to the BTEX (benzene, toluene, ethylbenzene, and xylene) group of petroleum-derived hydrocarbons with extensive industrial and ecological relevance. Anaerobic catabolism of ethylbenzene proceeds via different pathways in denitrifying and sulfate-reducing bacteria. The latter generate a succinate adduct of ethylbenzene as the first intermediate, probably by addition of fumarate to the methylene carbon atom (14). However, denitrifying bacteria are capable of oxygen-independent hydroxylation of the methylene group of ethylbenzene to yield (S)-1-phenylethanol (1, 12, 20), which is catalyzed by the molybdenum enzyme ethylbenzene dehydrogenase (11, 15, 21). The pathway continues with the oxidation of (S)-1-phenylethanol to acetophenone by an alcohol dehydrogenase (10, 16). Acetophenone may also be produced from (R)-1-phenylethanol or directly used as a substrate. The enzymes of further acetophenone catabolism are uncharacterized. From the observed CO2 dependence of ethylbenzene and acetophenone degradation, acetophenone was proposed to be carboxylated to benzoylacetate, which is then activated to the coenzyme A (CoA) thioester and thiolytically cleaved to benzoyl-CoA and acetyl-CoA (1, 6, 20, 22). On the level of benzoyl-CoA, the pathway of anaerobic ethylbenzene degradation flows into that of anaerobic benzoate degradation (for reviews, see references 2, 8, and 9) (Fig. 1).
FIG. 1.
Proposed catabolic pathway of ethylbenzene, phenylethanol, and acetophenone in A. aromaticum strain EbN1.
In this communication, we identify and characterize the postulated enzyme responsible for acetophenone carboxylation in Aromatoleum aromaticum strain EbN1. The enzyme is specifically induced in ethylbenzene- and acetophenone-grown cells. Acetophenone carboxylation is shown to be dependent on ATP hydrolysis, reminiscent of but distinct from the related carboxylation of acetone (25).
MATERIALS AND METHODS
Growth of bacteria and preparation of cell extract.
A. aromaticum strain EbN1 was grown on mineral medium with ethylbenzene or acetophenone as a carbon source and nitrate as an electron acceptor (22). Growth of precultures (1-liter scale) was performed as described previously (20, 22). Fermentor cultures (100 to 200 liters) were run in fed-batch mode with a growth-limiting and exponentially increasing feeding rate of nitrate and a discontinuous supply of ethylbenzene or acetopheneone, respectively. Cells of strain EbN1 from a typical fermentor were harvested during the exponential growth phase at an optical density of 4.0. The harvested cells were immediately frozen and stored in liquid nitrogen. Escherichia coli strain XL1-Blue MRF (Stratagene, Heidelberg, Germany) was used for overexpression experiments. Recombinant cells were grown at 37°C in Luria-Bertani (LB) medium. Ampicillin was added to the E. coli cultures to a final concentration of 100 μg ml−1.
Frozen cells of strain EbN1 (50 g [wet mass]) were suspended in 100 ml of 20% glycerol containing 0.5 mg DNase I. Extracts of E. coli strain XL1-Blue were prepared from 30 g (wet mass) of frozen cells suspended in 60 ml 125 mM Tris-HCl buffer (pH 8.3) containing 0.5 mg DNase I. The cell suspensions were passed through a French pressure cell at 137 MPa. Cell debris and membranes were removed by ultracentrifugation (100,000 × g) at 4°C for 1 h. The supernatant (cell extract) was used immediately or kept frozen at −70°C.
Molecular biological techniques.
Standard protocols were used for DNA isolation, cloning, transformation, amplification, and purification (23).
Heterologous expression of the apcEC-His6 subunit in E. coli.
The apcE gene was amplified (primers apcEfor, CAC CAT GGA AGC CGC TGG AGC, and apcErev, TTC GCC GCG CAC GCG CTC GAC CTG) and cloned into the pBAD-102 Topo cloning vector (Invitrogen, Germany). The N-terminally fused thioredoxin gene present in the initial plasmid was subsequently cut out by restriction with NcoI and religation, yielding a construct coding for ApcE with a C-terminal His tag (ApcEC-His6), which was transformed and expressed in E. coli XL1-Blue. The cells were grown in a 200-liter fermentor at 37°C in Luria-Bertani broth containing 100 μg of ampicillin ml−1 and induced at an optical density of 0.75 with 0.2% (vol/vol) arabinose as an inductor. After additional growth for 4 h, the cells were harvested and stored in liquid nitrogen until they were used.
Acetophenone carboxylase assays.
The activity of acetophenone carboxylase was measured either by (i) incorporation of [14C]bicarbonate into nonvolatile acid-stable products or (ii) acetophenone- and/or HCO3−-dependent ATP hydrolysis. Unless otherwise indicated, the assays were performed using a standard assay mixture containing 100 mM MOPS (morpholinepropanesulfonic acid)/KOH, pH 6.5, 10 mM MgCl2, 5 mM ATP, 20 mM NH4Cl, and 40 mM KHCO3.
(i) Substrate-dependent incorporation of [14C]bicarbonate.
Acetophenone carboxylase activity was measured via substrate-dependent incorporation of radioactivity from NaH14CO3 into acid-stable products. The assays were performed in 1-ml stoppered glass vials (0.3 to 0.5 ml standard assay mixture). In addition to the standard assay mixture, 10 kBq of NaH14CO3 (final specific radioactivity per assay, 0.25 Bq nmol−1) and enzyme (0.1 to 0.3 mg protein) were added. After 1 min of preincubation at 30°C, a 100-μl control sample was withdrawn and mixed with 30 μl of 5 M NaHSO4 to reach a final pH of 2.0 and to precipitate the protein. In the remaining assay mixture, the reaction was then initiated by adding acetophenone (1 mM end concentration) and incubating the mixture at 30°C. At various time points, 100-μl samples were withdrawn and acidified by adding 30 μl 5 M NaHSO4 to terminate the enzyme reaction. Volatile 14CO2 (nonfixed) was removed from the acidified samples by vigorously shaking the samples in open scintillation vials for 3 h. The remaining radioactivity in the samples was determined by liquid scintillation counting. Samples in which either the substrate or ATP was omitted served as controls.
(ii) Continuous, coupled spectrophotometric assay of ADP formation.
Acetophenone- and HCO3−-dependent ADP formation of purified acetophenone carboxylase was determined spectrophotometrically by coupling ADP formation to the oxidation of NADH, using a modified coupled enzyme assay (26). The assay mixture (0.5 to 1 ml) contained 2 mM phosphoenolpyruvate, 0.4 mM NADH, 1 U ml−1 pyruvate kinase, and 4 U ml−1 lactate dehydrogenase, in addition to the standard assay mixture described above. Cuvettes were preincubated at 30°C for 3 min, and then purified acetophenone carboxylase was added (0.3 mg), and after further incubation for 2 min, the reaction was initiated by adding 1 mM acetophenone. The assays were monitored by the decrease of absorbance at 365 nm. To determine ATP hydrolysis rates induced by only one of the two substrates, HCO3− was omitted from the standard assay mixture and the assay was initiated with either HCO3− (40 mM end concentration) or acetophenone (1 mM end concentration). Possible contamination of the enzyme fractions by myokinase was checked by adding AMP (2 mM end concentration) to the assay mixture. In inhibition experiments, ADP (2 mM end concentration) was added at the end of all assays to exclude inhibitory effects on the auxiliary enzymes of the coupled enzyme assay.
The pH optimum of the reaction was determined using a MES (morpholineethanesulfonic acid)-KOH buffer system in the range of pH 5.5 to 6.5 and a MOPS-KOH buffer system in the range of pH 6.5 to 8.0 (50 mM each). The residual activity of acetophenone carboxylase was determined using the first assay described above. Dependence on nucleoside triphosphates (NTPs) (5 mM each) and substrate (1 mM each) and the apparent Km values for acetophenone, ATP, and HCO3− were determined by the first assay described above. To study the inhibition of ATPase and carboxylase activities of acetophenone carboxylase, various concentrations of Me2+ ions (e.g., 0.05 to 1 mM Zn2+ or 0.2 to 4 mM Ni2+) and carbamoylphosphate (0 to 10 mM) were added to the standard assay mixtures. Residual activities for formation of 14C-labeled benzoylacetate or ATP hydrolysis were determined using both assays described above.
Purification of acetophenone carboxylase core enzyme (subunits ApcABCD).
Purification was performed at 4°C under oxic conditions. Extract from ethylbenzene-grown cells (50 g [wet mass]) was applied to a DEAE-Sepharose column (Amersham Biosciences; diameter, 26 mm; volume, 70 ml), which had been equilibrated with buffer A (10 mM Tris-Cl, pH 7.5, 2 mM MgCl2, 10% [vol/vol] glycerol). The column was washed with four column volumes of buffer A and eluted in step gradients with 100, 150, and 220 mM KCl in buffer A at a constant flow rate of 3 ml min−1. Most of subunit ApcE of acetophenone carboxylase eluted at 150 mM KCl (pool I), and the rest of the ApcE and the subunits ApcABCD eluted at 220 mM KCl (pool II). The fractions were pooled and concentrated 5-fold by ultrafiltration (Amicon concentrator with a YM-70 membrane). The concentrated pool II was diluted 2-fold with buffer B (20 mM MES-KOH, pH 6.5, 5 mM MgCl2, 10% [vol/vol] glycerol) and again applied to a DEAE-Sepharose column (diameter, 16 mm; volume, 16 ml) that had been equilibrated with buffer C (20 mM MES-KOH, pH 5.5, 5 mM MgCl2, 10% [vol/vol] glycerol). The column was washed with four column volumes of buffer B and eluted in a step gradient with 150 mM KCl in buffer B and a subsequent linear gradient from 150 to 500 mM KCl in buffer B over five column volumes. Fractions containing the ApcABCD subunits eluted from the column during the linear gradient and were pooled and concentrated by ultrafiltration (Amicon concentrator with a YM-30 membrane). The concentrated pool was further purified by gel filtration on a Superdex-200 high-load column (Amersham Biosciences; diameter, 26 mm; volume 320 ml), which had been equilibrated with buffer D (10 mM Tris-HCl, pH 7.5, 100 mM KCl). The fractions containing subunits ApcABCD of acetophenone carboxylase were pooled and concentrated by ultrafiltration (Amicon concentrator with a YM-30 membrane).
Purification of ApcE.
The wild-type ApcE subunit was partially purified by four chromatographic steps at 4°C under aerobic conditions. Combined ApcE-containing fractions of three runs on DEAE-Sepharose (pool I [see above]) were applied to a Macroprep ceramic hydroxapatite column (Bio-Rad; diameter, 16 mm; volume, 7 ml), which had been equilibrated with buffer E (20 mM MOPS-KOH, pH 7.0, 5 mM MgCl2, 10% [vol/vol] glycerol). The column was washed with four column volumes of buffer E and eluted in a linear gradient from 0 to 50 mM K-phosphate in buffer E (pH 7.0) over 10 column volumes. Fractions containing ApcE were pooled and concentrated by ultrafiltration (Amicon concentrator with a YM-30 membrane). The ApcE pool was then mixed with ammonium sulfate to a final concentration of 1 M (NH4)2SO4 and applied to a phenyl-Sepharose column (Amersham Biosciences; diameter, 16 mm; volume, 10 ml) that had been equilibrated with buffer F [20 mM MOPS-KOH, pH 7.0, 5 mM MgCl2, 1 M (NH4)2SO4]. The column was washed with three column volumes of buffer F and eluted in a step gradient to 500 mM (NH4)2SO4 in buffer F. The fractions containing ApcE were concentrated and dialyzed against 1 liter buffer G (10 mM Tris-HCl, pH 7.5) for 12 to 16 h. The dialyzed sample was further purified by gel filtration on a Superdex-200 high-load column (Amersham Biosciences; diameter, 16 mm; volume 120 ml), which had been equilibrated with buffer D. Fractions containing subunit ApcE of acetophenone carboxylase were pooled and concentrated by ultrafiltration (Amicon concentrator with a YM-30 membrane).
Purification of heterologously expressed ApcEC-His6.
Heterologously expressed ApcEC-His6 was purified from 30 g of E. coli XL1-Blue cells under aerobic conditions via two chromatographic steps at 4°C. A chelating Sepharose column (Amersham Biosciences; diameter, 26 mm; volume, 80 ml), which was loaded with nickel ions according to the manufacturers' instructions, was used first. After the column was equilibrated with buffer H (20 mM Tris-HCl, pH 7.5, 250 mM KCl), an extract of overproducing E. coli cells was applied, and the column was washed with two column volumes of buffer H containing 40 mM imidazole at a flow rate of 4 ml min−1. ApcEC-His6 was then eluted by applying buffer H containing 150 mM imidazole. The sample was concentrated 10-fold by ultrafiltration and dialyzed against 1 liter buffer G for 12 to 16 h. The dialyzed pool was further purified by gel filtration on a Superdex-200 high-load column (Amersham Biosciences; diameter, 26 mm; volume, 320 ml), which had been equilibrated with buffer D. The fractions containing ApcEC-His6 were pooled and concentrated by ultrafiltration.
Reconstitution experiments.
The inactive core complex of acetophenone carboxylase obtained after gel filtration chromatography (ApcABCD) was reconstituted by adding either native or recombinant subunit ApcE. Stoichiometries were determined by mixing constant amounts of the core complex ApcABCD (140 μg; 0.29 nmol) with increasing amounts of purified recombinant subunit ApcEC-His6 (1.25 to 15 μg; 0.04 to 0.44 nmol) by incubating the mixture at 30°C for 20 min and then measuring acetophenone carboxylase activity using the first assay described above. Optimal activity was reconstituted with a stoichiometry of 2 or 3 ApcABCD heterotetramers per ApcE subunit.
Analytical methods.
14C-labeled benzoylacetate generated by the first assay described above was analyzed by high-performance liquid chromatography (HPLC) on a reversed-phase C18 column (LiChrosphere 100; endcapped; 5 μm; 125 by 4 mm; Merck, Darmstadt, Germany) equilibrated with buffer J (50 mM formic acid, 20 mM K2HPO4, pH 3.0, 5% acetonitrile) at a flow rate of 1 ml min−1. The column was washed for 5 min with buffer J and then developed by applying a gradient from 5 to 20% acetonitrile in buffer H over 10 min, followed by another 15-min elution with 20% acetonitrile in buffer J. The eluate was monitored simultaneously for absorption at 260 nm and radioactivity, using a photo diode array detector and a radioactivity flowthrough analyzer in series. The retention times were 16 min for benzoylacetate and 23 min for acetophenone. The products formed from ATP during the carboxylation of acetophenone were analyzed by HPLC using a Mono-Q column (1 ml; Pharmacia) at a flow rate of 1 ml min−1. Acid-precipitated enzyme assays were applied to the column and developed by using a linear gradient from 0 to 500 mM KCl in buffer K (50 mM K-phosphate, pH 6.7) over 25 min. The retention times were as follows: AMP, 2.7 min; ADP, 5.5 min; ATP, 7.8 min.
GC-MS analysis of isotopic exchange.
Assays (1 ml) for analysis of isotopic exchange were performed essentially as described for the spectrophotometric assay of ADP formation (ii), using 0.5 mM NADH and omitting HCO3−. All buffers were flushed with nitrogen gas to exclude traces of dissolved oxygen and CO2. Purified acetophenone carboxylase (0 to 0.4 mg/ml) was added, and the reaction was started by the addition of 1 mM [methyl-2H3]acetophenone. At various time points (0, 10, and 30 min), 200-μl samples were withdrawn, and the reaction was stopped by adding 12.5 μl of NaHSO4 (2.5 M). The reaction products were extracted with 200 μl dichloromethane and analyzed via gas chromatography-mass spectrometry (GC-MS). The samples were analyzed with a Hewlett-Packard 5890 Series II Plus GCMSD on a 30m DB-5HS column (J & W Scientific) at a constant pressure of 20 lb/in2; 1 μl of sample was injected via a splitless-type injector onto the column by an autosampler. Elution started at 40°C, followed by an increase to 150°C at 10°C/min and a further increase to 230°C at 25°C/min. Mass detection was in the range of 40 to 450 m/z.
Other methods.
Protein concentrations were determined by Coomassie dye binding with bovine serum albumin as a standard (5). Discontinuous SDS-PAGE was performed in 12.5% (wt/vol) polyacrylamide gels as described previously (17). Native molecular masses were estimated by gel filtration from the retention volumes of standard proteins eluted under the same conditions. Purified core enzyme (1.7 mg ml−1) and purified ApcEC-His6 subunit (0.5 mg ml−1) were analyzed for the presence of metals by inductively coupled plasma emission spectroscopy (ICP-OES). The N termini of the subunits were sequenced by automated Edman degradation. The acetophenone concentrations used for enzymatic assays were verified by UV spectra of stock solutions (ɛ245 nm = 16.3 mM−1 cm−1). The spectra of the acetophenone carboxylase subcomplexes were recorded under aerobic conditions. The formation of a potential enzyme-phosphate intermediate of acetophenone carboxylase was investigated by incubation with either 0.1 mM [γ-32P]ATP or [β-32P]ATP (specific radioactivities, 1,500 Bq/nmol) in standard assay mixture for 30 min at 30°C, followed by SDS-PAGE and analysis by phosphorimager (Fujix BAS-IP MP 2040; Fuji, Tokyo, Japan). Benzoylacetate was always prepared fresh from its ethyl ester (5 mg), which was dissolved in 1 ml of 1 M NaOH, hydrolyzed by incubation for 30 min, and neutralized with HCl.
RESULTS
Activity of acetophenone carboxylase.
Acetophenone has been assumed to be carboxylated to benzoylacetate during anaerobic ethylbenzene metabolism, because ethylbenzene and acetophenone are degraded only in the presence of CO2 or HCO3− in the medium (1, 6, 22). Therefore, extracts of cells of strain EbN1 grown on ethylbenzene, acetophenone, or benzoate under denitrifying conditions were assayed for the formation of products from H14CO3− and acetophenone. Fixation of H14CO3− into acid-stable products was indeed detected with extracts of cells grown on ethylbenzene or acetophenone, but not those of control cells grown on benzoate. The specific activities of 9 to 21 nmol bicarbonate fixed min−1 mg of protein−1 were recorded, depending on the cell batch, which is close to the minimum expected value for an enzyme involved in ethylbenzene catabolism in the analyzed cell cultures: a value of 12.7 nmol min−1 mg of protein−1 was derived from a typical growth rate of the culture of 0.024 h−1 and an experimentally determined molar growth yield of 29 g dry cell mass formed per mol of ethylbenzene consumed. Acetophenone carboxylation activity in cell extracts was not sensitive to avidin, indicating that the enzyme is independent of biotin. Moreover, oxygen did not affect the enzyme activity; therefore, all further experiments were carried out under aerobic conditions. Activity was detected only in assay mixtures containing acetophenone, HCO3−, ATP, and additional divalent metal ions (e.g., Mg2+). Mn2+ could be substituted for Mg2+. Additionally, monovalent cations, e.g., K+ or NH4+, at concentrations of 20 to 40 mM were required for optimal activity. The carboxylation reaction was linearly time dependent within the first 5 min and protein dependent between 0 and 1.0 mg protein from cell extracts. The pH optimum for this reaction was determined to be pH 6.5, using different buffer systems covering a range of pH 5.5 to 8.0. Different nucleoside triphosphates were tested for the ability to support acetophenone carboxylase activity. In preliminary experiments with cell extracts, GTP, CTP, UTP, or ITP yielded 30 to 100% of the activity observed with ATP, but only ATP supported activity with purified acetophenone carboxylase (see below), indicating that the extract contained NTP-dependent kinases converting residual ADP to ATP. The formation of benzoylacetate as a product of acetophenone carboxylation was confirmed by comigration of 14C-labeled benzoylacetate with the unlabeled standard in HPLC analysis of the assay mixture. Benzoylacetate was formed only in extracts of cells grown on ethylbenzene or acetophenone, but not in extracts of cells grown on benzoate (data not shown), and the formation was strictly dependent on the addition of ATP.
Purification of acetophenone carboxylase.
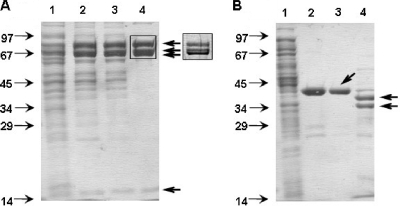
Acetophenone carboxylase was purified chromatographically from cell extracts of acetophenone- or ethylbenzene-grown cells. During the first two steps of purification by DEAE-Sepharose chromatography, activity was completely lost (Table 1). This could be ascribed to the dissociation of active acetophenone carboxylase into two inactive subcomplexes. Both subcomplexes of the enzyme were enriched and purified separately, and activity could be reconstituted by recombining the two subcomplexes. We succeeded in purifying the larger “core” complex of acetophenone carboxylase from cell extracts of strain EbN1 to apparent homogeneity. It contained four subunits, which were confirmed as ApcABCD (21) by N-terminal sequencing (Fig. 2 A, lane 4). The purified complex showed a specific activity of about 50 to 60 nmol (min mg)−1 when combined with purified recombinant ApcE (Table 1).
TABLE 1.
Purification of acetophenone carboxylase subcomplex Ia
| Step | Vol (ml) | Protein (mg) | Total activity (μmol min−1) | Sp act [nmol (min mg)−1] | Purification (fold) | Yield (%) |
|---|---|---|---|---|---|---|
| Extraction | 100 | 3,000 | 27 (24) | 9 (8) | 1 | 100 |
| DEAE, pH 7.5 | 25 | 300 | 6.9 (0.6) | 23 (2) | 2.6 (0.25) | 26 (2.5) |
| DEAE, pH 6.5 | 13 | 143 | 5.4 (0) | 38 (0) | 4.2 (−) | 20 (−) |
| Gel filtration | 3 | 42 | 2.1 (0) | 51 (0) | 5.7 (−) | 8 (−) |
Activity was measured by the determination of acid-stable 14C using a standard assay mixture. The values in parentheses were determined in the absence of exogenous ApcEC-His6. Minuses indicate nonaccessible values due to loss of activity.
FIG. 2.
Purification of the subcomplexes of acetophenone carboxylase. Fractions were analyzed by SDS-PAGE (12.5% polyacrylamide). (A) Purification of the “core” enzyme composed of a 70-kDa subunit (ApcA), a 15-kDa subunit (ApcB), an 87-kDa subunit (ApcC), and a 75-kDa subunit (ApcD). Lanes: 1, extract of A. aromaticum strain EbN1 grown on ethylbenzene (20 μg protein); 2, DEAE-Sepharose fraction, pH 7.5 (15 μg protein); 3, DEAE-Sepharose fraction, pH 6.5 (12 μg protein); 4, Superdex-200 gel filtration fraction (8 μg protein). The inset gives a closer look at the subunits ApcACD as separated by a 9% SDS-polyacrylamide gel. The arrows point to the subunits ApcABCD. (B) Purification of subunit ApcEC-His6. Lanes: 1, extract of E. coli strain XL1-Blue after 4 h of induction (20 μg protein); 2, Ni2+-chelating Sepharose fraction (10 μg protein); 3, Superdex-200 gel filtration fraction (8 μg protein); 4, Superdex-200 gel filtration fraction of enriched wild-type ApcE of A. aromaticum strain EbN1 (8 μg protein). Arrows: lane 3, heterologously expressed ApcEC-His6; lane 4, two bands of enriched ApcE from A. aromaticum strain EbN1.
Dependence of acetophenone carboxylation activity on the AcpE subunit.
Loss of acetophenone carboxylation activity during purification was consistently associated with the loss of an ethylbenzene-induced protein of 34 kDa from the fractions containing the “core” complex. This lost protein was enriched separately over four chromatographic steps. After the last chromatographic step, two main protein bands with apparent masses of 34 and 38 kDa were visible on SDS-PAGE (Fig. 2B, lane 4). The expected molecular mass of ApcE as calculated from the apcE gene sequence is 32.1 kDa. The N-terminal sequences of both proteins were identical with that of the apcE gene product. Thus, the double bands may have originated from posttranslational modification or from partial proteolytic cleavage of the protein close to the C-terminal end. The main peak of the enriched protein showed a native mass of 40 ± 10 kDa, as determined by gel filtration, suggesting that the protein is monomeric. Adding this protein to the purified inactive core complex reconstituted acetophenone carboxylation activity; therefore, it is regarded as a functional subunit (ɛ) of the enzyme. However, the ApcE subunit could not be purified to homogeneity from cells of strain EbN1, and the protein may have been partially degraded or otherwise damaged during purification.
Purification of recombinant ApcE as a His-tagged protein.
Because of the apparent heterogeneity and low recovery rate of ApcE from wild-type cells, we cloned and expressed the apcE gene separately in E. coli and added a C-terminal His tag for easy purification. The level of synthesis of recombinant ApcEC-His6 in E. coli was rather low and was not increased by using various concentrations of the inductor arabinose. However, the recombinant protein could be easily purified by affinity chromatography and gel filtration, with a yield of 8 mg protein per 30 g wet cell mass. This protein was added to the core complex and indeed reconstituted acetophenone carboxylase activity, as previously observed with enriched wild-type ApcE (Table 1). Therefore, recombinant His-tagged ApcE seems to be fully functional and to retain the catalytic competence of wild-type ApcE. These results also exclude the possibility that any other protein of strain EbN1 is required to reconstitute acetophenone carboxylase activity.
Molecular properties of acetophenone carboxylase.
The core complex of acetophenone carboxylase consists of four subunits with apparent molecular masses of 87, 75, 70, and 15 kDa that correspond to the apcABCD gene products (Fig. 2A, lane 4). The native molecular mass of the complex was determined to be 485 ± 15 kDa by gel filtration chromatography. Therefore, the core complex most probably forms an (αβγδ)2-hetero-octamer. The three larger subunits of the complex were previously described as acetophenone-induced proteins in strain EbN1 (6, 15) (Fig. 2A, lane 1), which are lacking in extracts from cells grown on benzoate. It was evident that acetophenone carboxylase was highly induced and represented a sizable fraction of the total soluble protein in cultures grown on ethylbenzene or acetophenone, consistent with the rather low enrichment factor of purified protein (Table 1). The high level of expression of acetophenone carboxylase presumably compensated for the relatively low specific activity of the enzyme, i.e., high levels of acetophenone carboxylase were required to support cell growth at the observed rates. Elemental analysis by ICP-OES revealed that the purified acetophenone carboxylase core complex contained zinc in stoichiometric amounts (1.1 mol Zn per mol of αβγδ heterotetramer). No further metals or selenium was detected. The UV-visible (UV-Vis) spectrum of the enzyme exhibited an absorption maximum at 280 nm without additional absorbance peaks in the wavelength range from 300 to 600 nm, indicating that no UV-Vis-absorbing cofactors were present.
Catalytic properties of acetophenone carboxylase.
Purified acetophenone carboxylase catalyzed the HCO3−- and ATP-dependent carboxylation of acetophenone to benzoylacetate. The reaction followed Michaelis-Menten kinetics with the following apparent Km values (± standard deviations [SD]; from at least 3 parallel measurements): 33 ± 15 μM for acetophenone, 0.54 ± 0.17 mM for HCO3−, and 0.5 ± 0.1 mM for ATP. The substrate specificity of acetophenone carboxylase was tested in enzyme assays with a number of different ketones, including aliphatic ketones, such as acetone and 2-butanone; aromatic ketones (propiophenone, 4-hydroxy-acetophenone, and 4-amino-acetophenone); and the heterocyclic aromatic ketone 4-acetyl-pyridine. Of these substrates, only the aromatic ketones acetophenone, propiophenone, and 4-acetyl-pyridine were converted by the enzyme. Propiophenone was carboxylated at approximately the same specific rate as acetophenone, whereas the activity detected with 4-acetyl-pyridine was lower (108% and 22% of the rate with acetophenone, respectively). None of the ring-substituted acetophenone derivatives, 4-hydroxy-acetophenone and 4-amino-acetophenone, was used as a substrate, and neither were any of the aliphatic ketones.
ATPase activity of acetophenone carboxylase.
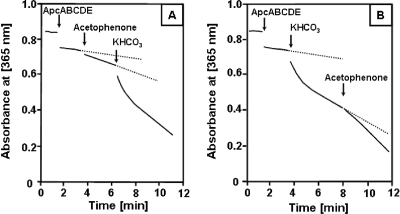
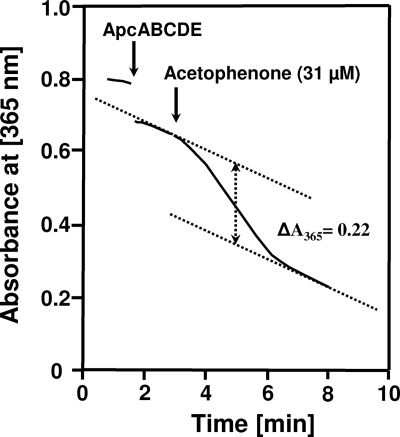
A continuous spectrophotometric assay was developed that relied on coupling acetophenone carboxylase-catalyzed ATP hydrolysis to NADH oxidation, using the coupling enzymes pyruvate kinase and lactate dehydrogenase. This assay was only possible with purified enzyme, because the 1-phenylethanol dehydrogenase isoenzymes present in cell extracts also catalyze acetophenone-dependent oxidation of NADH (16). The assay was independent of added myokinase as an additional auxiliary enzyme, suggesting hydrolysis of ATP exclusively to ADP and no formation of AMP during the carboxylation reaction. The exclusive generation of ADP from ATP during carboxylation of acetophenone was confirmed by HPLC analysis (data not shown). Small amounts of AMP occurred only after long incubation periods and may be explained by chemical hydrolysis of enzymatically formed ADP, as suggested by similar amounts of ATP hydrolyzed to ADP in controls without enzyme. The separate subcomplexes of acetophenone carboxylase (ApcABCD or ApcEC-His6) alone did not hydrolyze ATP at high rates (data not shown). However, after acetophenone carboxylase was reconstituted by mixing the two subcomplexes, slow ATP hydrolysis in the absence of substrates started immediately (Fig. 3). Thus, all five subunits are required for the acetophenone-carboxylating activity and the ATP-hydrolytic activity of the enzyme. The specific activity of reconstituted acetophenone carboxylase for ATP hydrolysis in the absence of acetophenone and HCO3− was 6 nmol (min mg)−1. The rate of ATP hydrolysis was 2- to 3-fold increased when either one of the substrates, HCO3− or acetophenone, was added in the absence of the second (Fig. 3), with specific activities of 16 nmol (min mg)−1 in assays started with HCO3− and 11 nmol (min mg)−1 in those started with acetophenone. Adding the second substrate to these assays resulted in further acceleration of the ATP hydrolysis rate, which reached specific activities of 34 nmol (min mg)−1 with the enzyme batch used, independently of whether the reaction was first started with acetophenone followed by HCO3− or vice versa. In assays with limited concentrations of acetophenone as the only substrate, the rate of ATP hydrolysis did not decrease after consumption of 2-fold stoichiometric amounts of NADH (data not shown), revealing the reaction to be uncoupled. However, assays containing limiting amounts of acetophenone in the presence of HCO3− allowed us to determine the stoichiometry of ATP hydrolysis relative to acetophenone carboxylation. From the measured value of 2.1 mol NADH oxidized per mol acetophenone added to the assay, hydrolysis of 2 ATP per carboxylated acetophenone was inferred (Fig. 4).
FIG. 3.
Substrate dependence of ATP hydrolysis catalyzed by acetophenone carboxylase. (A) Assay started first by the addition of acetophenone (1 mM) and then supplied with HCO3− (40 mM). (B) Assay started with the substrates in the reverse order. The time points when the compounds were added are indicated by arrows.
FIG. 4.
Stoichiometry of ATP hydrolysis. A coupled enzyme assay was started with a limiting amount of acetophenone (31 μM). The corresponding amount of oxidized NADH was calculated from the absorbance difference (ΔA365) between the start of the reaction and its leveling off. The time points when the compounds were added are indicated by arrows.
Inhibition of acetophenone carboxylase.
For all tested inhibitors, effects on either the carboxylating activity or the ATPase activity of acetophenone carboxylase were studied separately. Both activities were inhibited by Zn2+ and Ni2+ ions, but only inhibition by Zn2+ was also observed in crude extracts of strain EbN1. Inhibition to half-maximal activities of ATPase and carboxylation in the assays were observed in the presence of 0.2 mM ZnCl2, whereas NiCl2 affected the two activities differently (50% inhibited ATPase activity at 1 mM and carboxylation activity at 2.5 mM, respectively). Additional inhibitory effects were observed in the presence of high concentrations of Mg2+ or Mn2+ ions, which are required as essential cosubstrates by the enzyme. The dependence of activity on the concentrations of these metals followed optimum curves, with activity optima close to the concentration of ATP added (5 mM). Concentrations of >5 mM inhibited the enzyme (50% inhibition at 10 mM Mn2+ and 20 mM Mg2+, respectively), indicating that Mg2+ or Mn2+ is required only for complexing and not for the reaction itself.
Interestingly, carbamoylphosphate was identified as a further inhibitor of ATPase and carboxylating activity of acetophenone carboxylase (50% inhibition at 2 and 8 mM carbamoylphosphate, respectively). This is especially remarkable because carbamoylphosphate can be regarded as a structural analogue of carboxyphosphate, a probable intermediate of the reaction cycle. Finally, inhibitory effects of nonhydrolyzable ATP analogues, like AMP-CPP (α,β-methylene-ATP), AMP-PCP (β,γ-methylene-ATP), and AMP-PNP (β,γ-imido-ATP), were studied. None of these supported acetophenone carboxylase activity. After preincubation of enzyme assay mixtures with AMP-CPP and AMP-PCP (2 mM each) for 5 min and initiation with 10 mM ATP, the acetophenone carboxylase activity was not affected, whereas preincubation with AMP-PNP (2 mM) completely inhibited acetophenone carboxylase activity. The inhibition by AMP-PNP was not competitive, as the addition of a 5-fold surplus of ATP did not restore enzyme activity. Therefore, AMP-CPP and AMP-PCP apparently do not bind to the enzyme, whereas AMP-PNP seems to bind tightly enough to prevent its displacement by ATP.
Proton-deuteron exchange activity of acetophenone carboxylase.
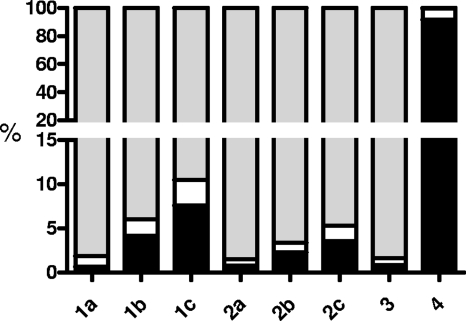
The catalytic properties of acetophenone carboxylase indicate that phospho-enol-acetophenone may be formed as an activated intermediate from acetophenone, concurrent with the abstraction of a proton from the methyl group of the side chain (Fig. 6, reaction 2). In an uncoupled assay containing only acetophenone but no HCO3−, ATPase activity was reduced to 65% when [methyl-2H3]acetophenone was used, indicating a kinetic isotope effect of H+ release from the methyl group in the course of the reaction. Interestingly, the rate of activity in the presence of HCO3− was not influenced (data not shown). This indicates that phosphorylation of acetophenone is not rate limiting in the overall reaction. Proton abstraction from Cα was confirmed by assaying for proton-deuteron exchange in [methyl-2H3]acetophenone during the uncoupled enzyme reaction via GC-MS analysis. A time- and enzyme-dependent isotope exchange reaction was indeed detected, but only with the fully reconstituted enzyme with both subcomplexes. The product was a mixture of predominantly nonlabeled acetophenone and minor amounts of [methyl-2H]acetophenone (Fig. 5), while the amount of [methyl-2H2]acetophenone could not be quantified due to an interfering MS peak. The data indicate that multiple exchange events occur while acetophenone is bound at the active site. This is also supported by the finding that the total number of deuterons exchanged was greater than the amount of ATP simultaneously consumed. Therefore, proton-deuteron exchange in the methyl group appears to be dependent on ATP hydrolysis, whereas the higher stoichiometry may indicate a reversible exchange reaction of the phosphate of acetophenone-enol-phosphate and an amino acid, such as His, Asp, or Arg. However, no P-labeled enzyme was determined in SDS-PAGE after treatment with either [γ-32P]ATP or [β-32P]ATP.
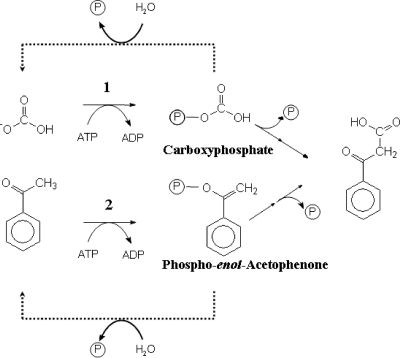
FIG. 6.
Proposed mechanism for acetophenone carboxylation by acetophenone carboxylase.
FIG. 5.
GC-MS analysis of isotope exchange products during uncoupled ATPase activity. [methyl-2H3]acetophenone is converted to nonlabeled acetophenone and minor amounts of [methyl-2H]acetophenone. [methyl-2H2]acetophenone could not be detected because of an interfering MS peak. Lanes: 1, assay containing 0.4 mg of Apc after 0 (a), 5 (b), and 10 min (c) of reaction time; 2, assay containing 0.2 mg of Apc after 0 (a), 5 (b), and 10 min (c) of reaction time; 3 and 4, controls with [methyl-2H3]acetophenone and nonlabeled acetophenone, respectively, in the absence of enzyme, after 10 min of reaction time. Black, nonlabeled acetophenone; white, [methyl-2H]acetophenone; gray, [methyl-2H3]acetophenone.
DISCUSSION
In this study, a soluble enzyme consisting of five subunits that catalyzes the so far unproven carboxylation of acetophenone to benzoylacetate, the third step of anaerobic catabolism of ethylbenzene, was purified from A. aromaticum and characterized. The reaction catalyzed is as follows: acetophenone + 2 ATP + HCO3−→benzoylacetate + 2 ADP + 2 Pi. Mono- and divalent cations are required for optimal activity. Acetophenone carboxylase is induced specifically in cells grown on ethylbenzene or acetophenone, but not in cells grown on benzoate. Four of the five subunits (87, 75, 70, and 15 kDa) form an inactive subcomplex (core enzyme), and addition of the fifth (34-kDa) subunit is required for activity. The natural substrates of the enzyme are acetophenone (apparent Km, 33 μM), MgATP (apparent Km, 0.46 mM), and HCO3− (apparent Km, 0.54 mM). It also accepts a few other aromatic and heterocyclic ketones without substituents. The low apparent Km value for acetophenone fits well with the observed Km values of the enzymes from the previous steps in the pathway, ethylbenzene dehydrogenase (0.5 μM) and (S)-1-phenylethanol dehydrogenase (6 μM). The high apparent Km value determined for HCO3− is consistent with the requirement for high concentrations of HCO3− in the medium (30 mM) during growth on ethylbenzene or acetophenone (6). No nucleotides other than ATP were capable of supporting acetophenone carboxylation with purified acetophenone carboxylase. The carboxylation of acetophenone is a thermodynamically unfavorable process (ΔG°′ at pH 7 for acetone carboxylation with HCO3−, +11.8 kJ/mol), but the hydrolysis of ATP to ADP (ΔG°′ = −31 kJ/mol) would theoretically provide sufficient energy to drive the carboxylation reaction. Interestingly, during the carboxylation of acetophenone, two molecules of ATP are hydrolyzed, forming ADP and Pi. The hydrolysis of the ATP of acetophenone carboxylase occurs with the addition of only one of the substrates, acetophenone or HCO3− (uncoupled reaction). The HCO3−-dependent ATPase activity can be explained by assuming nonproductive formation and hydrolysis of a carboxyphosphate intermediate (13, 19). This behavior has also been reported for carbamoylphosphate synthase, which involves the formation of an unstable carboxyphosphate intermediate. Further evidence for carboxyphosphate as a possible intermediate is provided by the inhibition of ATPase and carboxylating activity of the enzyme by carbamoylphosphate, a structural analogue of carboxyphosphate. Carbamoylphosphate is not accepted as an alternative substrate, indicating that the carboxyphosphate needs to be cleaved prior to forming the new C-C bond. The observed acetophenone-dependent ATPase activity suggests the transfer of a second phosphoryl group to acetophenone, either directly or via a phosphoryl-enzyme intermediate. Proton-deuteron exchange studies of Cα-proton abstraction indicate that several exchange events must occur per hydrolyzed ATP. This may be explained by assuming a rapid exchange of the high-energy phosphoryl group of acetophenone-enol-phosphate with an amino acid of the enzyme. However, such a phosphorylated enzyme intermediate would be quite unstable, as we did not detect labeling in any subunit by SDS-PAGE after treatment with either [γ-32P]ATP or [β-32P]ATP.
Acetophenone carboxylation probably involves an initial hydrolysis of the phosphodiester of phospho-enol-acetophenone, which isomerizes to yield the carbanion of acetophenone, which then acts as a nucleophile in attacking the activated CO2. The stoichiometrically bound zinc atom may have a role in correctly orienting the activated intermediates. More specifically, it may stabilize either the enol state of acetophenone for initial phosphorylation or the acetophenone carbanion intermediate of the proposed mechanism.
Acetophenone carboxylase is unique in its biochemical details. Close analogues of the apcABCDE genes apparently coding for the same type of enzyme are found in only two other sequenced genomes (Rubrobacter xylanophilus [NC_008148] and Rhodococcus jostii [NC_008270]). The subunits of the core complex show similarities to N-methyl-hydantoinases, which catalyze the ATP-dependent hydrolysis of cyclic amide bonds (18). The initial step for these enzymes is the activation of the keto group via phosphoryl group transfer from ATP, forming the activated enol intermediate, ADP, and inorganic phosphate, similar to what has been proposed for acetophenone carboxylase.
The only known paralogous enzymes with similar biochemical functions are acetone carboxylases from several aerobic and anaerobic bacteria, including A. aromaticum, which catalyze the ATP-dependent carboxylation of acetone to acetoacetate (7, 25). However, these enzymes consist of only three subunits in an α2β2γ2 composition (24), encoded by the acxABC genes. The three subunits of acetone carboxylase show low sequence similarities (20 to 31% identity) to those of the acetophenone carboxylase core complex (ApcABCD), which harbors one paralogue of AcxA (ApcD) and AcxC (ApcB) and two paralogues of AcxB (ApcAC). However, acetone carboxylase does not contain a paralogue of ApcE. Purified acetone carboxylase also contains Mn2+ instead of Zn2+ and forms AMP during the reaction (3). We exclude a similar role for Mn in acetophenone carboxylase for several reasons: (i) cultures of A. aromaticum strain EbN1 showed equal growth rates on acetophenone regardless of whether Mn2+ was added to or depleted from the growth media; (ii) ICP-OES analysis of purified acetophenone carboxylase revealed 2.2 mol Zn/α2β2γ2δ2 core enzyme, but no significant amounts of Mn in either subcomplex; and (iii) purified acetophenone carboxylase was silent in electron paramagnetic resonance (EPR) spectroscopy (data not shown).
Studies of the Cα-proton abstraction from acetone showed that acetone carboxylase exhibits a proton-deuteron exchange pattern similar to that observed for acetophenone carboxylase (4). Furthermore, acetone carboxylase also shows an uncoupled ATPase reaction when bicarbonate is omitted, under which conditions ATP is predominantly hydrolyzed to ADP. Therefore, activation of both acetone and acetophenone may be accomplished by phosphorylation via the γ-phosphoryl group of an ATP cosubstrate, while the activation of the second substrate, HCO3−, might be brought about either via a second ATP or via the β-phosphoryl group of the initially formed ADP (4).
The identification and characterization of acetophenone carboxylase fills a gap in the proposed ethylbenzene metabolic pathway and proves the conversion of acetophenone to benzoylacetate. The properties of the enzyme reported in this study suggest a novel mechanism for ATP-dependent acetophenone carboxylation (Fig. 6). Based on the data presented, we suggest that one ATP is used for the activation of HCO3− to carboxyphosphate (as in carbamoylphosphate synthase) and a second ATP for the formation of phospho-enol-acetophenone (as in ATP-dependent N-methyl-hydantoinases or acetone carboxylase).
Acknowledgments
This work was supported by the Deutsche Forschungsgemeinschaft, Bonn, Germany.
We thank G. Fuchs and N. Gad'on, Universität Freiburg, for their help with fermentor cultures. We also thank R. Auxier, Chemical Analysis Facility, University of Georgia, for ICP-OES analysis and R. Brunisholz, ETH Zürich, for N-terminal sequencing.
Footnotes
Published ahead of print on 4 January 2010.
REFERENCES
- 1.Ball, H. A., H. A. Johnson, M. Reinhard, and A. M. Spormann. 1996. Initial reactions in anaerobic ethylbenzene oxidation by a denitrifying bacterium, strain EB1. J. Bacteriol. 178:5755-5761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Boll, M., G. Fuchs, and J. Heider. 2002. Anaerobic oxidation of aromatic compounds and hydrocarbons. Curr. Opin. Chem. Biol. 6:604-611. [DOI] [PubMed] [Google Scholar]
- 3.Boyd, J. M., H. Ellsworth, and S. A. Ensign. 2004. Bacterial acetone carboxylase is a manganese-dependent metalloenzyme. J. Biol. Chem. 279:46644-46651. [DOI] [PubMed] [Google Scholar]
- 4.Boyd, J. M., and S. A. Ensign. 2005. ATP-dependent enolization of acetone by acetone carboxylase from Rhodobacter capsulatus. Biochemistry 44:8543-8553. [DOI] [PubMed] [Google Scholar]
- 5.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 6.Champion, K. M., K. Zengler, and R. Rabus. 1999. Anaerobic degradation of ethylbenzene and toluene in denitrifying strain EbN1 proceeds via independent substrate-induced pathways. J. Mol. Microbiol. Biotechnol. 1:157-164. [PubMed] [Google Scholar]
- 7.Clark, D. D., and S. A. Ensign. 1999. Evidence for an inducible nucleotide-dependent acetone carboxylase in Rhodococcus rhodochrous B276. J. Bacteriol. 181:2752-2758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Heider, J., and G. Fuchs. 1997. Microbial anaerobic aromatic metabolism. Anaerobe 3:1-22. [DOI] [PubMed] [Google Scholar]
- 9.Heider, J., A. M. Spormann, H. R. Beller, and F. Widdel. 1999. Anaerobic bacterial metabolism of hydrocarbons. FEMS Microbiol. Rev. 22:459-473. [Google Scholar]
- 10.Höffken, H. W., M. Duong, T. Friedrich, M. Breuer, B. Hauer, R. Reinhardt, R. Rabus, and J. Heider. 2006. Crystal structure and enzyme kinetics of the (S)-specific 1-phenylethanol dehydrogenase of the denitrifying bacterium strain EbN1. Biochemistry 45:82-93. [DOI] [PubMed] [Google Scholar]
- 11.Johnson, H. A., D. A. Pelletier, and A. M. Spormann. 2001. Isolation and characterization of anaerobic ethylbenzene dehydrogenase, a novel Mo-Fe-S enzyme. J. Bacteriol. 183:4536-4542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Johnson, H. A., and A. M. Spormann. 1999. In vitro studies on the initial reactions of anaerobic ethylbenzene mineralization. J. Bacteriol. 181:5662-5668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kazlauskas, R. J., and M. Whitesides. 1985. Synthesis of methoxycarbonyl phosphate, a new reagent having high phosphoryl donor potential for use in ATP cofactor regeneration. J. Org. Chem. 50:1069-1076. [Google Scholar]
- 14.Kniemeyer, O., T. Fischer, H. Wilkes, F. O. Glockner, and F. Widdel. 2003. Anaerobic degradation of ethylbenzene by a new type of marine sulfate-reducing bacterium. Appl. Environ. Microbiol. 69:760-768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kniemeyer, O., and J. Heider. 2001. Ethylbenzene dehydrogenase, a novel hydrocarbon-oxidising molybdenum/iron-sulfur/heme enzyme. J. Biol. Chem. 276:21381-21386. [DOI] [PubMed] [Google Scholar]
- 16.Kniemeyer, O., and J. Heider. 2001. (S)-1-Phenylethanol dehydrogenase of Azoarcus sp. strain EbN1, an enzyme of anaerobic ethylbenzene catabolism. Arch. Microbiol. 176:129-135. [DOI] [PubMed] [Google Scholar]
- 17.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 18.Ogawa, J., J. M. Kim, W. Nirdnoy, Y. Amano, H. Yamada, and S. Shimizu. 1995. Purification and characterization of an ATP-dependent amidohydrolase, N-methylhydantoin amidohydrolase, from Pseudomonas putida 77. Eur. J. Biochem. 229:284-290. [DOI] [PubMed] [Google Scholar]
- 19.Powers, S. G., and A. Meister. 1978. Carbonic-phosphoric anhydride (carboxy phosphate). Significance in catalysis and regulation of glutamine-dependent carbamyl phosphate synthetase. J. Biol. Chem. 253:1258-1265. [PubMed] [Google Scholar]
- 20.Rabus, R., and J. Heider. 1998. Initial reactions of anaerobic metabolism of alkylbenzenes in denitrifying and sulfate-reducing bacteria. Arch. Microbiol. 170:377-384. [Google Scholar]
- 21.Rabus, R., M. Kube, A. Beck, F. Widdel, and R. Reinhardt. 2002. Genes involved in the anaerobic degradation of ethylbenzene in a denitrifying bacterium, strain EbN1. Arch. Microbiol. 178:506-516. [DOI] [PubMed] [Google Scholar]
- 22.Rabus, R., and F. Widdel. 1995. Anaerobic degradation of ethylbenzene and other aromatic hydrocarbons by new denitrifying bacteria. Arch. Microbiol. 163:96-103. [DOI] [PubMed] [Google Scholar]
- 23.Sambrook, T., E. F. Frisch, and J. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- 24.Sluis, M. K., and S. A. Ensign. 1997. Purification and characterization of acetone carboxylase from Xanthobacter strain Py2. Proc. Natl. Acad. Sci. U. S. A. 94:8456-8461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sluis, M. K., F. J. Small, J. R. Allen, and S. A. Ensign. 1996. Involvement of an ATP-dependent carboxylase in a CO2-dependent pathway of acetone metabolism by Xanthobacter strain Py2. J. Bacteriol. 178:4020-4026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ziegler, K., K. Braun, A. Böckler, and G. Fuchs. 1987. Studies on the anaerobic degradation of benzoic acid and 2-aminobenzoic acid by a denitrifying Pseudomonas strain. Arch. Microbiol. 149:62-69. [Google Scholar]