Abstract
RND (resistance-nodulation-division) family transporters in Gram-negative bacteria frequently pump out a wide range of inhibitors and often contribute to multidrug resistance to antibiotics and biocides. An archetypal RND pump of Escherichia coli, AcrB, is known to exist as a homotrimer, and this construction is essential for drug pumping through the functionally rotating mechanism. MdtBC, however, appears different because two pump genes coexist within a single operon, and genetic deletion data suggest that both pumps must be expressed in order for the drug efflux to occur. We have expressed the corresponding genes, with one of them in a His-tagged form. Copurification of MdtB and MdtC under these conditions showed that they form a complex, with an average stoichiometry of 2:1. Unequivocal evidence that only the trimer containing two B protomers and one C protomer is active was obtained by expressing all possible combinations of B and C in covalently linked forms. Finally, conversion into alanine of the residues, known to form a proton translocation pathway in AcrB, inactivated transport only when made in MdtB, not when made in MdtC, a result suggesting that MdtC plays a different role not directly involved in drug binding and extrusion.
Bacterial multidrug resistance is a major public health problem (10, 17). One widespread resistance mechanism involves the multidrug resistance (MDR) transporters. Among these, the resistance-nodulation-cell division (RND) family transporters, such as the AcrAB-TolC system in Escherichia coli, play a major role in drug resistance in Gram-negative bacteria because they allow the direct extrusion of drug molecules into extracellular space, and because they sometimes confer an increased level of tolerance to an astonishingly wide range of toxic compounds (18). In general, an RND-type exporter protein (such as AcrB), located in the inner membrane, forms a tripartite complex with a periplasmic adaptor protein, such as AcrA, and a homotrimeric outer membrane channel, such as TolC (18). The drug efflux process requires the presence of all three components. The crystallographic structures of AcrB (13, 14, 22, 24), AcrA (11, 27), and TolC (2, 8) are known, and models of the tripartite complex have been proposed (6, 27).
AcrB is a homotrimeric transporter (14) located in the inner membrane and uses the proton gradient as the energy source (31). The homotrimeric structure is thought to be functionally important, or even essential, as each protomer appears to undergo a series of mandatory conformational alterations during the process of drug export, often called “functionally rotating mechanism,” as deduced from the structure of the asymmetric trimers of AcrB (13, 22, 24). This mechanism was also supported by the observation that, in a trimer in which protomers were covalently linked to each other, inactivation of one protomer alone abolishes the activity of the entire trimeric complex (29).
Not all RND-type transporters, however, follow this homotrimeric organization. The mdtABC genes of E. coli encode an RND system that is unusual in that it contains two different RND pump genes, mdtB and mdtC, in addition to its own adaptor gene, mdtA. Previous genetic studies have demonstrated that the deletion of either of the two RND pump genes abolishes (1) the resistance to β-lactams, novobiocin, and bile salt derivatives, like deoxycholate, or narrows the range of pump substrates (15), a result suggesting that the functional unit is likely a heteromultimeric pump formed by MdtB/MdtC proteins. However, no direct data have so far been presented supporting the interaction between these proteins or the stoichiometry of the complex. Because the heterooligomeric composition of this pump was unexpected based on the accepted notion of how the homotrimeric pump functions by the functionally rotating mechanism, we examined here the nature of the MdtBC complex in detail.
In this study, we first purified the oligomeric transporter by labeling either MdtB or MdtC with a His tag. We obtained a trimeric complex(es) containing both MdtB and MdtC in an approximately 2:1 ratio. However, we could not rule out the possibility that there were mixtures of trimers containing different ratios of the B and C proteins. We therefore utilized the recently introduced technology of creating covalently linked trimers (29), and we show here that the only active trimers are those containing two units of MdtB and one unit of MdtC.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
E. coli strains and plasmids used in this study are listed in Table 1. E. coli DH5α (20) was used as a cloning host for plasmids. A protease- and recombinase-deficient E. coli B strain lacking both the AcrAB and MdtABC efflux systems, BL21KAMR [BL21 ΔacrAB ΔmdtABC Δ(srl-recA)306::Tn10(Tcr)], was constructed for the expression of a giant gene encoding a covalently linked Mdt complex. For this, acrAB and mdtABC mutations were introduced into E. coli BL21 (Novagen). First, the ΔacrAB::Kmr from RAM1337 (MC4100 Δara ΔacrAB::Kmr; a gift from R. Misra) was introduced by P1cml,clr100-mediated transduction (12), and the kanamycin resistance gene was removed by using FLP recombinase from the curable plasmid pCP20 (4). Second, the ΔmdtABC::Kmr from HS276 (26) was introduced into E. coli BL21 ΔacrAB by transduction, and the kanamycin resistance cassette was removed. Finally, the Δ(srl-recA)306::Tn10 allele from strain BLR (Novagen) was introduced into BL21 ΔacrAB ΔmdtABC by transduction to produce BL21KAMR. The presence of the ΔacrAB, ΔmdtABC, and ΔrecA::Tn10 mutations was confirmed by PCR.
TABLE 1.
E. coli strains and plasmids used in this study
| Strain or plasmid | Genotype and/or characteristicsb | Reference/source |
|---|---|---|
| Strains | ||
| DH5α | F−endA1 recA1 ϕ80lacZΔM15 Δ(lacZYA-argF)U169 hsdR17(rK− mK+) supE44 thi-1 gyrA96 relA1 phoA deoR λ− | 20 |
| AG100A | K-12 argE3 thi-1 rpslL xyl mtl Δ(gal-uvrB) supE44 ΔacrAB::Tn903(Kmr) | 19 |
| AG100A ΔmdtABC | AG100A derivative lacking mdtABC genes | This work |
| AG100A ΔmdtBCD | AG100A derivative lacking mdtBCD genes | 1 |
| BL21 | E. coli B F−ompT hsdSB(rB− mB−) gal dcm lon | Novagen |
| BLR | E. coli B F−ompT hsdSB(rB− mB−) gal dcm lon Δ(srl-recA)306::Tn10(Tcr) | Novagen |
| BL21KAMR | BL21 ΔacrAB ΔmdtABC Δ(srl-recA)306::Tn10(Tcr) | This study |
| Plasmids | ||
| pSPORT1 | pMB1 ori, medium-copy-no. cloning vector; lac-inducible expression; Apr | Gibco BRL |
| pSlinkXH | pSPORT1 derivative carrying linker sequence on an XbaI-HindIII fragment | This study |
| p10His | pSPORT1 derivative carrying a His10-tag sequence | This study |
| pACYC177 | p15a ori, low-copy-no. cloning vector; Apr, Kmr | 3 |
| pAMB10 | pACYC177 harboring mdtB under lac promoter with its own SD sequence and a His10-tag sequence at the 3′ end; Kmr | This study |
| pAMC10 | pACYC177 harboring mdtC under lac promoter with the engineered S.D. sequence and a His10-tag sequence at the 3′ end; Kmr | This study |
| pHSG576 | pSC101 ori, low-copy-no. cloning vector; lac-inducible expression; Cmr | 30 |
| pHSGS | pHSG576 containing pSPORT1-derived MCS sequence under lac promoter; Cmr | This study |
| pRARE2 | p15a ori, contains tRNA genes argU, argW, argX, ileX, glyT, leuW, proL, metT, thrT, tyrU, and thrU; Cmr | Novagen |
| pRARE2-km | pRARE2 containing kan (Kmr) instead of cat (Cmr) | This study |
| pAcrB | pSPORT1 derivative harboring acrB with a His6 tag sequence at the 3′ end | Y. Takatsuka |
| pSBHis | pSPORT1 derivative harboring mdtB with a His10 tag sequence at the 3′ end | This study |
| pSCHis | pSPORT1 derivative harboring mdtC with a His10 tag sequence at the 3′ end | This study |
| pSBCHis | pSPORT1 derivative harboring mdtB followed by mdtC with a His10 tag sequence at the 3′ end | This study |
| pSCBHis | pSPORT1 derivative harboring mdtC followed by mdtB with a His10 tag sequence at the 3′ end | This study |
| pSABCHis | pSPORT1 derivative harboring mdtA and mdtB followed by mdtC with a His10 tag sequence at the 3′ end | This study |
| pHMBC10 | pHSG576 derivative harboring mdtBC with a His10 tag sequence at the 3′ end | This study |
| pHMABC10 | pHSG576 derivative harboring mdtABC with a His10 tag sequence at the 3′ end | This study |
| pHMA-B10 | pHSG576 derivative harboring mdtA and mdtB with a His10 tag sequence at the 3′ end | This study |
| pHMA-C10 | pHSG576 derivative harboring mdtA and mdtC with a His10 tag sequence at the 3′ end | This study |
| pHMA-BB10a | pHSG576 harboring mdtA and an artificial gene for a covalently linked dimer of MdtB with a C-terminal His10 tag | This study |
pHSG576-based plasmids containing mdtA as well as artificial genes for covalently linked transporters are denoted pHMA-XX10 or pHMA-XXX10, where X is B (mdtB) or C (mdtC), with the artificial gene underlined. “10” indicates the His10 tag. Here only one example is shown. For other plasmids, see Fig. 3A.
Apr, ampicillin resistant; Kmr, kanamycin resistant; Cm r, chloramphenicol resistant; Tcr, tetracycline resistant.
Cells were grown in LB broth (Difco tryptone, 1% [wt/vol]; Difco yeast extract, 1%; and NaCl, 0.5%) or on LB agar plates supplemented, when necessary, with the following concentrations of antibiotics: ampicillin, 100 μg/ml; kanamycin, 35 μg/ml; chloramphenicol, 7 μg/ml (for the maintenance of low-copy-number plasmids in a ΔacrAB ΔmdtABC strain) or 35 μg/ml (for the plasmid selection in the DH5α strain).
Construction of plasmids.
Chromosomal DNA from DH5α was used to amplify the genes mdtA, mdtB, and mdtC by PCR. All primer sequences are available from the authors upon request. The C-terminal His10-tagged sequence was introduced initially via the PCR primers into the vector pSPORT1 for the construction of the p10His vector plasmid and then was added from this plasmid to the 3′ terminus of various genes. To express the native (or His-tagged) transporter proteins MdtB and MdtC, the corresponding genes were amplified and sequentially cloned into the PstI/SacI and SacI/HindIII sites of vector p10His, producing pSBCHis or pSCBHis.
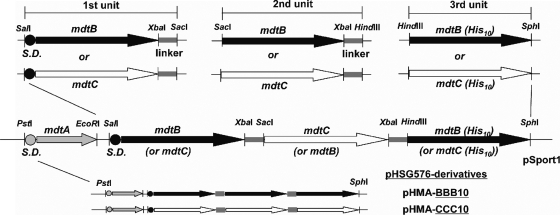
We created an artificial gene encoding a covalently linked Mdt complex, either as a dimer or a trimer, as described previously (29). Three protomer units (named first, second, and third unit) were constructed separately as a SalI-SacI fragment having the first subunit DNA sequence with its upstream Shine-Dalgarno (SD) sequence and a downstream linker sequence, as a SacI-HindIII fragment having the second subunit DNA sequence with a downstream linker sequence, and as a HindIII-SphI fragment having the third subunit DNA sequence with a His10-tagged sequence followed by a stop codon at its 3′ end, respectively (Fig. 1). PCR amplification of either the mdtB or mdtC sequence, together with a cognate or an engineered SD sequence, using appropriate primers as a SalI-XbaI fragment, and subsequent insertion of the resulting amplicon in frame with a linker sequence contained in a pUC19 derivative (29) as an XbaI-SacI fragment resulted in the first unit of an about 3.3-kbp SalI-SacI DNA fragment containing the first subunit sequence with an upstream SD sequence and a translation start cordon (ATG), followed by a linker sequence. We used as a linker sequence a 135-bp internal sequence of acrB, which encodes 45 amino acid residues from Met496 to Arg540 (29) corresponding to cytosolic α-helix (Iα) and its flanking regions (14). The linker sequence bridging the second and third subunit sequences was amplified using a forward primer linkXFw (29) and a reverse primer linkHinRv (5′-TGCATAAGCTTACGCCCCGTACTGCGCAG-3′; the underlined sequence represents the HindIII restriction site) as an XbaI-HindIII fragment and cloned into the appropriate sites of pSportI for the construction of pSlinkXH. Then either mdtB or mdtC was amplified as a SacI-XbaI fragment and cloned into pSlinkXH, resulting in the second unit of an about 3.2-kbp SacI-HindIII fragment containing the second subunit DNA sequence with a downstream linker sequence. For construction of the third unit, either mdtB or mdtC with a His10-tagged sequence followed by a stop codon at the 3′ end was amplified as a HindIII-SphI fragment and cloned into the respective site of a pSportI. Since each unit contained either mdtB or mdtC subunit sequences, we generated four dimeric intermediates (B-B-, B-C-, C-B-, and C-C-) of about 6.5 kbp by inserting the first unit of the SalI-SacI fragment in front of the second unit of the SacI-HindIII fragment cloned in pSPORT1. Finally, eight trimeric sequences with a His10-tagged sequence attached to the 3′ end (B-B-B, B-B-C, B-C-B, B-C-C, C-B-B, C-B-C, C-C-B, and C-C-C) were constructed by inserting the third unit of the HindIII-SphI fragment behind each dimeric intermediate SalI-HindIII fragment cloned in pSPORT1. To construct the second unit of a gene encoding a dimeric Mdt complex, either mdtB or mdtC with a His10-tagged sequence followed by a stop codon at the 3′ end was amplified as a SacI-HindIII fragment and cloned into the respective sites of pSPORTI. Four dimeric sequences with a His10-tagged sequence at the 3′ end (B-B, B-C, C-B, and C-C) were constructed by inserting the first unit of the SalI-SacI fragment in frame with the second unit of the SalI-HindIII fragment cloned in pSPORT1. To coexpress MdtA and a covalently linked Mdt complex from a single plasmid, mdtA with its cognate SD sequence was amplified as a PstI-EcoRI fragment and cloned into the corresponding sites of a pSPORT1 derivative.
FIG. 1.
Construction of pHSG576 plasmids containing artificial genes for the expression of covalently linked MdtB and/or MdtC transporters. The mdtB or mdtC gene, connected to a downstream linker sequence coding for the horizontal helix of AcrB (29), was made as a SalI-SacI, SacI-HindIII, or HindIII-SphI sequence depending on whether it was intended as the first, second, or third unit, respectively, and was cloned into the MCS site of pHSGS, the first unit together with the upstream mdtA gene. For the final constructs, only pHMA-BBB10 and pHMA-CCC10 are shown here as examples. The artificial genes for covalently linked dimers and trimers are underlined in the names of the plasmids. For other constructs, see Fig. 3A, and for details, see the text.
The pSPORT1 derivatives, however, were not suitable for assays of efflux activity, as described in Results. These assays, therefore, were carried out by using a low-copy-number vector, pHSG576. To facilitate transfer of cloned genes from a pSPORT1 derivative to pHSG576, a multiple cloning site (MCS) sequence from the PstI to AatII sites in pSPORT1 was introduced into pHSG576 as follows. pSPORT1 was digested with AatII, rendered blunt ended using T4 DNA polymerase, and cut with PstI. The resulting 89-bp fragment (for handling purposes, a several-hundred-bp BamHI fragment was inserted into the MCS to increase the size of this fragment), having one blunt end, was ligated into the PstI/FspI sites of pHSG576, resulting in pHSGS. The mdtA and the artificial genes constructed on pSPORT1 were transferred as the PstI-SphI fragment into the corresponding sites of pHSGS.
In some experiments, MdtB or MdtC monomers were expressed together with a covalently linked Mdt complex. For this, the mdtB or mdtC sequence with its cognate or engineered SD sequence and a His10-tagged sequence at the 3′ end was amplified and cloned initially into the SalI/HindIII sites of pSPORT1. Then, the AatII-EcoRV fragment containing the mdtB or mdtC gene along with the lac promoter was cut from the resulting pSPORT1 derivative and cloned into the AatII-FspI sites of pACYC177 for the construction of pAMB10 or pAMC10. All constructs were verified by sequencing.
Site-directed mutagenesis.
In order to examine whether inactivation of one protomer inactivates the entire linked trimer, mutations were initially introduced into the mdtB or mdtC genes harbored in pHMABC10 by using the QuikChange site-directed mutagenesis protocol (Stratagene). To introduce the mutant sequences into the covalently linked trimers, the mutant mdtB or mdtC sequences were amplified by PCR as a SalI-XbaI fragment or a SacI-XbaI fragment, respectively, and then were used to replace the wild-type sequences in the pHMA-BCB10 plasmid (see Fig. 3 and 5). In addition, the third unit containing a mutant mdtB with a His10-tagged sequence was created by PCR amplification as a HindIII-SphI fragment and was used to replace the last BHis unit in the linked trimer. All mutations were confirmed by sequencing.
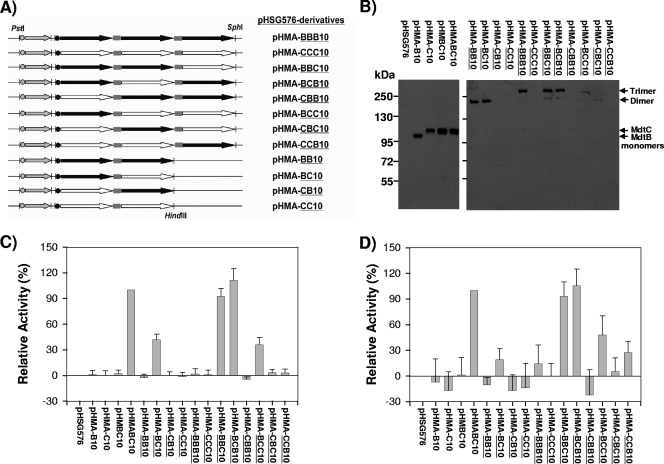
FIG. 3.
Covalently linked Mdt complexes. (A) Schematic representation of the plasmids containing mdtA and artificial genes for a covalently linked Mdt dimer or trimer. (B) Expression of a covalently linked Mdt complex in E. coli. A covalently linked Mdt complex was expressed as a C-terminal His10-tagged fusion protein from a pHSG576-based plasmid in E. coli BL21KAMR, from which total membrane fractions were isolated. Inner membrane proteins (20 μg per sample) solubilized with N-lauroylsarcosine were resolved by SDS-PAGE and then electroblotted onto a PVDF membrane for Western analysis. Detection was performed by an anti-His tag antibody. (C and D) In vivo activity of a covalently linked Mdt complex. The ability of plasmids to confer cloxacillin (C) or deoxycholate (D) resistance to the E. coli BL21KAMR was measured by the gradient plate method as described in Materials and Methods. Each bar represents the mean ± standard deviation of data obtained from three separate experiments.
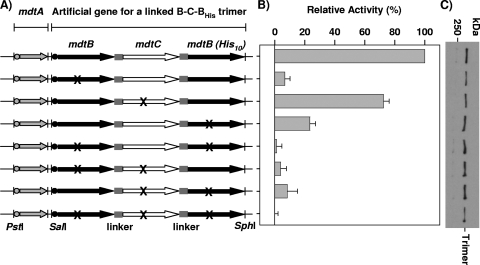
FIG. 5.
Proton translocation mutants in a linked Mdt complex. (A) Introduction of a proton translocation mutation into an artificial gene for a linked MdtB-C-BHis complex harbored in pHMA-BCB10. To express a linked Mdt complex having only one mutant MdtB subunit, D410A mutation was introduced in either one of the two mdtB subunit sequences of the artificial gene. Similarly, D401A mutation was introduced into the mdtC sequence. X represents the location of these mutations in the proton translocation pathway. (B) Activities of proton translocation mutants of a linked Mdt B-C-BHis trimer in the BL21KAMR host. Efflux activities were estimated from the levels of resistance to cloxacillin by using the gradient plate method. Error bars show standard deviations. (C) Expression level of proton translocation mutants of a linked Mdt B-C-BHis trimer. The linked Mdt trimers were analyzed by Western blotting using an anti-His-tag antibody.
Western blot analysis.
For the expression analysis of MdtB, MdtC, and a covalently linked Mdt complex, each of three colonies of the fresh transformant of BL21KAMR carrying a pHSG576 derivative was inoculated into 5 ml of LB medium with chloramphenicol and grown to an optical density at 600 nm (OD600) of ca. 1.0, with shaking at 30°C without isopropyl-β-d-thiogalactopyranoside (IPTG) induction. The cultured bacteria were harvested, resuspended in 10 mM HEPES-KOH (pH 7.5) buffer at an OD600 of 30, and lysed by sonication in the presence of a complete, EDTA-free protease inhibitor cocktail (Roche). Unbroken cells were removed by low-speed centrifugation. The supernatant was then centrifuged at 150,000 × g for 1 h at 4°C, and the resulting pellets were resuspended in 10 mM HEPES-KOH (pH 7.5) buffer containing 1.5% (wt/vol) N-lauroylsarcosine. Solubilized inner membrane protein fractions were obtained as supernatants after centrifugation at 150,000 × g for 1 h at 4°C. Proteins were resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) on 7.5% gels and transferred onto a polyvinyldifluoride (PVDF) membrane (Immobilon-P; Millipore Corp.). The membranes were incubated with affinity-purified anti-MdtB antibody, affinity-purified anti-MdtC antibody, or anti-tetra-His antibody (Qiagen). Western blots were developed by using the peroxidase-conjugated secondary antibody and Western Lightening chemiluminescence reagent plus (PerkinElmer Life and Analytical Sciences), and exposure to the X-ray film followed.
Drug susceptibility.
Drug susceptibilities of E. coli cells were tested usually at 30°C without IPTG induction by two different methods. Susceptibilities of BL21KAMR containing a pHSG576 derivative, to cloxacillin and deoxycholate, were determined by a gradient plate method (5, 28), in which a linear concentration gradient of cloxacillin (Sigma) or deoxycholate (Sigma) was prepared in square agar plates containing 500 μg/ml of sodium deoxycholate or 0.40 μg/ml of cloxacillin in the lower layer. The medium was a slightly modified LB, with the yeast extract decreased to 0.5% and NaCl content increased to 1.0%. Mid-exponential-phase culture from a single colony of the fresh transformant, grown in LB broth with chloramphenicol, was harvested and resuspended in fresh LB broth. The cell suspension, after the cell density was adjusted to an OD600 of 0.1, was streaked as a line across the plate, in parallel with the direction of drug gradient. After 24 h of incubation at 30°C, bacterial growth across the plates was measured. The relative activity of each construct was calculated as described previously (28) using pHSG576 (arbitrarily assigned as 0%) and pHMABC10 (arbitrarily assigned as 100%) as negative and positive controls. The twofold serial dilution method was used to determine the MIC of novobiocin for E. coli BL21KAMR carrying two compatible plasmids, a pHSG576 derivative and a pACYC177 derivative. Mid-exponential-phase culture from a single colony of the fresh transformant, grown in LB broth with chloramphenicol and kanamycin, was harvested and resuspended in fresh LB broth. The cell suspension, after cell density was adjusted to an OD600 of 0.05, was used to inoculate LB broth tubes (final OD600 = 0.025) containing twofold serial dilutions of novobiocin and incubated at 30°C without shaking. After 24 h of incubation, OD600 readings below 0.15 were read as indicating the absence of bacterial growth.
Purification of MdtBC complex.
An overnight culture of E. coli AG100A ΔmdtABC cells carrying either the plasmid pSBCHis or pSCBHis was inoculated into 1 liter of LB medium with ampicillin. Expression of both proteins was induced by adding IPTG to a final concentration of 0.4 mM when the OD600 reached 0.6. After 4 h of induction, cells were harvested and resuspended in buffer A (20 mM Tris-HCl, pH 7.0; 200 mM KCl; and 5 mM imidazole) containing 100 μg/ml DNase I. Cells were lysed by two passages through a French pressure cell at 15,000 lb/in2 in the presence of 1 mM phenylmethanesulfonyl fluoride and EDTA-free protease inhibitor cocktail at 4°C. Lysate was centrifuged at low speed to remove unbroken cells, and the supernatant was centrifuged for 1 h at 161,000 × g at 4°C. Pelleted membranes were dissolved in 20 ml of buffer B (20 mM Tris-HCl [pH 7.0], 200 mM KCl, 5 mM imidazole, 10% glycerol, and 2% n-dodecyl-β-d-maltose [DM]) at 4°C. Solubilized membrane proteins, recovered as supernatant after ultracentrifugation for 1 h at 161,000 × g at 4°C, were loaded onto a Talon His-Bind cobalt chelating column (BD Biosciences) equilibrated with buffer C (20 mM Tris-HCl [pH 7.0], 200 mM KCl, 5 mM imidazole, 10% glycerol, and 0.02% DM). The flowthrough fraction was reloaded onto the column twice, in order to improve the recovery of the MdtBC complex. Washing was carried out with 20-column volumes of buffer C. MdtBC was eluted with the same buffer containing increasingly higher concentrations of imidazole, as specified. The fractions containing pure MdtBC complexes were pooled and loaded onto a HiPrep 26/10 desalting column (Amersham) equilibrated with a buffer containing 20 mM HEPES-KOH [pH 7.0], 200 mM KCl, 10% glycerol, and 0.02% DM. MdtBC was eluted in the same buffer, concentrated in a 50,0000-kDa cutoff ultrafiltration device (Vivaspin; Sartorius) to a protein concentration of 2 to 3 mg/ml and stored at 4°C.
BN-PAGE.
Blue native-PAGE (BN-PAGE) was performed according to the method described by Schägger et al. (21), with the following modifications. Bio-Rad gradient gels (4 to 15% acrylamide) were used to resolve the complexes in a buffer containing 25 mM Tris and 0.2 mM glycine (pH 8.3) without SDS. Coomasie G-250 was added to the samples but omitted from the cathode buffer.
Anti-peptide polyclonal antibodies.
Rabbit polyclonal antibodies were raised against the synthetic peptides after conjugation to keyhole limpet hemocyanin. The peptide fragment of MdtB used corresponded to residues 45 to 60, and that of MdtC to residues 674 to 692. These antibodies were affinity purified against the corresponding peptide coupled to SulfoLink resins (Pierce) according to the manufacturer's instructions. For the Western blot analysis, 1.08 μg/ml and 0.05 μg/ml of affinity-purified anti-MdtB and anti-MdtC antibody, respectively, was used.
RESULTS
MdtB and MdtC form heterotrimeric complex(es).
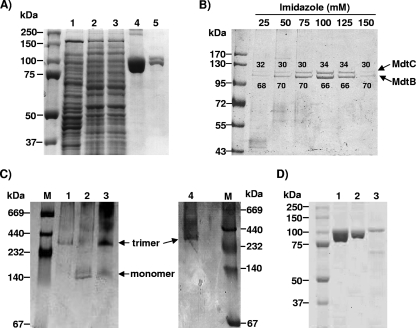
The first biochemical evidence of the interaction between MdtB and MdtC occurred with the copurification of MdtB and MdtC, in which both of the two transporters were expressed from plasmids, with only one of them His tagged at the C terminus. Purification on the Talon Co2+ column led to the copurification of other, untagged protein, regardless of whether MdtB or MdtC was His tagged (Fig. 2A, lanes 4 and 5). The apparent molecular mass of these bands matched the theoretical values calculated from the deduced amino acids sequences. Antibodies specific for MdtB or for MdtC specifically detected the recombinant proteins (data not shown). The identities of these proteins were further confirmed by tryptic digestion followed by mass spectroscopy analysis, which yielded most of the fragments expected for MdtB or MdtC (data not shown).
FIG. 2.
Copurification analysis showed that MdtB and MdtC form a heterotrimeric complex. (A) Copurification of MdtBHis and MdtC from E. coli AG100A ΔmdtABC-pSCBHis. The gels were stained with Coomassie blue. Lane 1, crude extract after French pressure cell lysis; lane 2, membrane protein extract after solubilization with DM; lane 3, Co2+ column flowthrough; lane 4, dissociated protomers from copurified MdtBHisC complexes eluted with 100 mM imidazole; lane 5, dissociated protomers from MdtB2C complex isolated by the stepwise gradient elution with imidazole, as specified in Materials and Methods. (B) SDS-PAGE analysis of the components of copurified MdtBHisC from stepwise imidazole gradient elution fractions. Proteins were expressed from plasmid pSCBHis. The gels were stained with Coomassie blue. The ratio of MdtB (lower band) to MdtC (upper band) within the copurified MdtBC complex, quantified by densitometry with Image J software, is shown as percentage values above or below each protein band. (C) BN-PAGE analysis of MdtBC complex reveals a trimeric aggregation state. The gels were stained with Coomassie blue. Affinity-purified MdtB (lane 1), MdtC (lane 2), and MdtBC (lane 3) were analyzed and compared with purified AcrB trimer (lane 4). High-molecular-weight standards for native electrophoresis (Amersham Biociences) are shown in lane M. (D) SDS-PAGE analysis of MdtB and MdtC monomers, produced by the dissociation of oligomers in SDS-containing sample buffer at room temperature. Lane 1, copurified MdtBC complex after dissociation into subunits; lane 2, His-tagged MdtB after affinity purification on Co2+ column; lane 3, His-tagged MdtC after affinity purification on Co2+ column. MdtC preparation shows several low-molecular-mass bands corresponding to degradation products of the protein.
To determine the mass of the complex containing MdtB and MdtC, we analyzed the purified complex by BN-PAGE (21). The MdtBC oligomer showed a major band of approximately 300 kDa corresponding to a trimer, as well as a minor band of approximately 100 kDa corresponding to dissociated, monomeric MdtB and MdtC (Fig. 2C, lane 3). The mobility of the 300-kDa band was very similar to that of the purified AcrB trimer (Fig. 2C, lane 4).
If MdtB and MdtC assembled into trimers in a more or less random manner, we would expect complexes of different compositions, such as B3, B2C, BC2, and C3. When either MdtB or MdtC carries a His tag fused to the C terminus, each of these possible trimers would be expected to contain 0, 1, 2, or 3 tags and may bind to the Ni/Co-chelating matrix with different affinities. It may then be possible to separate these species by elution using a gradient or steps of imidazole. When the proteins were expressed from pSCBHis and the His-tagged complexes were eluted using a stepwise gradient of imidazole at 25, 50, 75, 100, 125, and 150 mM, complexes were eluted at several of these steps. Although in some early experiments the B-to-C ratios appeared to show some variation, this was not reproducible, and in the majority of experiments (an example is shown in Fig. 2B) each of the complexes seemed to contain similar, roughly 2:1 ratios of MdtB (lower band) to MdtC (upper band). The same 2:1 ratio of MdtB/MdtC was also observed when the His tag was fused to MdtC (data not shown). Thus, it seems likely that the assembly is not random, and most complexes contain the favored ratio of two molecules of MdtB and one molecule of MdtC, although small fractions with different stoichiometric ratios could have escaped detection due to the limited resolution of the stepwise elution procedure.
To examine whether MdtB or MdtC alone is able to form homotrimeric complexes, we independently expressed either MdtB or MdtC alone in its His-tagged form. Both proteins were present exclusively in the cytoplasmic membrane fraction. His-tagged MdtB was highly expressed in the absence of MdtC from the plasmid pSBHis and was readily purified following the same protocol as that for the MdtBC complex (Fig. 2D, lane 2). However, MdtC appeared to be unstable when expressed alone from the plasmid pSCHis. Purification of MdtC yielded a partially degraded preparation, and the expression levels were much lower than those obtained for MdtB (Fig. 2D, lane 3). When purified MdtB was analyzed by BN-PAGE, mobility similar to the MdtB2C and AcrB trimers was observed (Fig. 2C, lane 1). A faint band corresponding to the trimeric MdtC was also observed by BN-PAGE (Fig. 2C, lane 2). Results thus indicate that MdtB (and possibly MdtC) may be assembled as a homotrimer when expressed individually.
Creation of an artificial gene encoding a covalently linked Mdt complex.
The results described above suggested that MdtB and MdtC tended to become assembled as a B2C trimer. However, it was unclear whether the isolated trimer was completely homogeneous. Furthermore, the formation of B3 and C3 trimers under extreme conditions suggested that the assembly does not always proceed with the 2:1 stoichiometric ratio, and we could not rule out the possibility that different ratios also gave rise to functional trimers. Indeed, Nagakubo et al. (15) reported that expression of MdtC alone produced significant levels of resistance to bile salts. Thus, in order to find out the composition of the functional trimer in an unequivocal manner, we produced artificial proteins in which the component protomers were covalently linked via cytosolic linkers so that one subunit's C terminus was connected to the next subunit's N terminus, in a way similar to that of linked AcrB trimers recently described (29). The major advantage of this approach is that composition and activity of the artificial protein complex can be determined simultaneously for intact cells. We thus created artificial genes encoding covalently linked Mdt complexes, either as a dimer or a trimer (Fig. 1). Since two different SD sequences provided by the mdtB and mdtC genes can give rise to translational signals of different strengths, the upstream SD sequence and GTG start codon of the first subunit mdtC sequence were replaced by the corresponding sequences of mdtB to enable direct comparison of expression levels and activities of the artificial gene constructs.
Initially we made all artificial gene constructs using a medium-copy-number vector, pSPORT1. Our preliminary attempt to assay the novobiocin efflux activity of a pSPORT1 derivative containing the artificial gene constructs in a ΔacrAB ΔmdtBCD host strain was unsuccessful, presumably because of the insufficient cellular MdtA level expressed from a single chromosomal copy of mdtA. In one experiment conducted at 37°C, pSBCHis containing mdtBC showed less than a twofold increase in the novobiocin MIC when introduced into the AG100A ΔmdtBCD strain. In contrast, a pSABCHis plasmid, containing mdtA in addition to mdtBC, conferred a 16-fold increase in the MIC (data not shown). We therefore cloned the mdtA gene with its cognate SD sequence into the PstI/EcoRI sites of a pSPORT1 derivative containing an artificial gene construct to coexpress MdtA and a covalently linked Mdt complex from a single recombinant plasmid. However, these constructs were still less than ideal, because the high levels of expression of the linked proteins apparently led to their degradation and the reassembly of the resultant protomers, as judged from the results of the MIC assay (data not shown).
For these reasons, our experiments described below were performed by using the pHSG576 vector, a lower-copy-number plasmid. The PstI-SphI fragment containing mdtA and an artificial gene for a linked Mdt complex was transferred from the pSPORT1 derivative to pHSG576. In addition, a pHSG576 derivative allowed for the susceptibility test against β-lactams.
Expression of a covalently linked Mdt complex.
To examine expression levels and activities of covalently linked Mdt complexes from the low-copy-number pHSG576 derivatives, we took several measures to reduce proteolytic degradation of the expressed full-length product. We used a protease-deficient (lon and ompT) BL21KAMR strain as an expression host (see also reference 29), we relied on the baseline expression from the pLac promoter without IPTG induction, and we used a low growth temperature (30°C). Homologous recombination between tandemly repeated mdtB or mdtC sequences was prevented by the ΔrecA background of BL21KAMR. Western blot analysis of the N-lauroylsarcosine-solubilized inner membrane proteins from the exponential phase cells using anti-His4 antibody showed that a full-length product, either as a dimer or a trimer, was expressed only from an artificial gene having the mdtB sequence at the 5′ end (Fig. 3B). The poor expression of linked products having mdtC at the 5′ end was surprising, especially because expression of the monomeric mdtC gene in pHMA-C10 was at a level comparable to that of the monomeric mdtB gene in pHMA-B10 (Fig. 3B). The most likely explanation, at this point, is that the presence of the covalently linked sequence downstream interferes with the correct folding of MdtC, unless the trimeric assembly is initiated from MdtB. This idea is also consistent with the finding that MdtC that was overproduced alone in the presence of IPTG was degraded extensively in the absence of MdtB (Fig. 2D, lane 3).
Activity of a covalently linked Mdt complex.
The activity of the linked Mdt complexes was measured for intact cells by their ability to increase cloxacillin and deoxycholate resistance in BL21KAMR, using the gradient plate method (Fig. 3C and D). The highest drug efflux activities were observed with constructs B-C-B and B-B-C, expected to produce a linked B2C complex, and they were comparable to that of the natural unlinked trimer expressed from pHMABC10 harboring a chromosomal sequence of mdtABC. The lack of activity of another construct (C-B-B) was due apparently to the absence of appreciable expression of this full-length complex in the host cell (Fig. 3B). BL21KAMR expressed a linked B-B-B homotrimer at a high level (Fig. 3B), but the transporter showed no activity (Fig. 3C and D). Cells could not express a linked C-C-C homotrimer (Fig. 3B), and no appreciable activity was seen (Fig. 3C and D).
We also observed some marginal activity in BL21KAMR expressing a linked B-C dimeric complex or a linked B-C-C trimeric complex (Fig. 3C and D). Since monomeric fragments generated by the proteolytic degradation or premature termination of the linked proteins can associate with an expressed linked complex or other monomeric fragments and can give rise to a functional oligomer, we wanted to determine the actual oligomeric entity responsible for the activity associated with a linked B-C dimer generated by pHMA-BC10 and a linked B-C-C trimer generated by pHMA-BCC10. (The covalently linked genes are underlined.) For this purpose, a linked B-C dimer was coexpressed with an MdtB or an MdtC monomer in BL21KAMR bearing two compatible plasmids, pHMA-BC10 of pSC101 origin and pAMB10 or pAMC10 of p15a origin. If the linked B-C dimer is the stable and active species, the dimer will not associate with the coexpressed MdtB or MdtC monomer, and the coexpression of these monomers would produce no changes in the efflux activity. However, if the activity (shown in Fig. 3C and D) was due to the B2C trimer produced by the association of a linked B-C dimer with an MdtB monomer (that may be produced by degradation or termination of a B-C dimer), then the coexpression of additional MdtB monomers (but not that of MdtC monomers) will help in the production of a B2C complex, causing an increase in activity. This was clearly the case, as seen with data shown in boldface in Table 2. On the other hand, if the associated trimeric product of a linked B-C dimer and an MdtC monomer (that is, BC2 complex) is the active form, the coexpression of MdtC monomers should increase the activity; this was clearly not the case (Table 2). Thus, the actual oligomeric complex for the activity associated with pHMA-BC10 is not the linked B-C dimer but the trimeric association product of a linked B-C dimer and an MdtB monomer generated by proteolytic degradation or premature termination of B-C.
TABLE 2.
Novobiocin susceptibility of the BL21KAMR strains coexpressing a linked Mdt complex and a monomeric MdtB or MdtCa
| Plasmid combinations | MIC (μg/ml) of novobiocin |
|---|---|
| pHSG576 + pACYC177 | 1.56 |
| pHSG576 + pAMB10 | 1.56-3.13 |
| pHSG576 + pAMC10 | 1.56 |
| pHMA-B10 + pACYC177 | 1.56 |
| pHMA-B10 + pAMC10 | 12.5 |
| pHMA-C10 + pACYC177 | 1.56 |
| pHMA-C10 + pAMB10 | 25.0 |
| pHMA-BC10 + pACYC177 | 6.25 |
| pHMA-BC10 + pAMB10 | 25.0 |
| pHMA-BC10 + pAMC10 | 6.25 |
| pHMA-BCC10 + pACYC177 | 6.25 |
| pHMA-BCC10 + pAMB10 | 25.0 |
| pHMA-BCC10 + pAMC10 | 6.25 |
| pHMA-BCB10 + pACYC177 | 12.5 |
| pHMA-BCB10 + pAMB10 | 12.5 |
| pHMA-BCB10 + pAMC10 | 12.5 |
For the boldface data, the B2C trimer was presumably produced by the association of a linked B-C dimer with an MdtB monomer, and the coexpression of additional MdtB monomers helped in the production of a B2C complex, causing an increase in activity.
A similar coexpression experiment was conducted with a linked B-C-C trimer. Table 2 shows that coexpression of a linked B-C-C trimer with an MdtB monomer, but not with an MdtC monomer, led to a significant increase in activity, implying that the marginal activity associated with a B-C-C trimer plasmid pHMA-BCC10 was not the activity of a B-C-C trimer per se but rather the result of the reassembling of degraded (or prematurely terminated) fragments to a stable and functional B2C complex. Consistent with this notion, activity was nearly unchanged when a linked B-C-B trimer was coexpressed with either MdtB or MdtC monomer (Table 2).
Rare codons and their effect on the expression of MdtB and MdtC.
The genes mdtB and mdtC are rich in rare codons in E. coli. The mdtB transcript contains 5 CCC, 1 AGA/AGG, 2 GGA, 6 CGG/CGA, 1 AUA, and 5 CUA, for a total of 20 rare codons, while the mdtC transcript contains 8 CCC, 10 GGA, 11 CGG/CGA, 3 AUA, and 5 CUA, for a total of 37 rare codons. (For comparison, the acrB transcript contains a total of 7 rare codons.) In order to rule out the possibility that the poor expression of some mdtC-containing constructs was caused by the presence of rare codons, we expressed the artificial genes in BL21KAMR cells cotransformed with a derivative (pRARE2-km) of plasmid pRARE2 (Novagen) supplying tRNAs for seven rare codons (AUA, AGG, AGA, CUA, CCC, CGG, and GGA). We measured the novobiocin efflux activity of BL21KAMR/pRARE2-km/pHMA-CBB10 as a measure of expression of a full-length C-B-B product because BL21KAMR harboring pHMA-CBB10 showed no activity due to the absence of appreciable expression of a full-length C-B-B (Fig. 3B). Supply of rare tRNAs by pRARE2-km had little effect on the efflux activity of BL21KAMR/pRARE2-km/pHMA-CBB10 (data not shown). Thus, the low expression of a linked complex having MdtC at the N terminus cannot be explained by the presence of rare codons in the mdtC gene. Possibly the correct folding of the nascent MdtC protein may require the help of MdtB that is folded first.
Proton translocation mutants of MdtB-C complex.
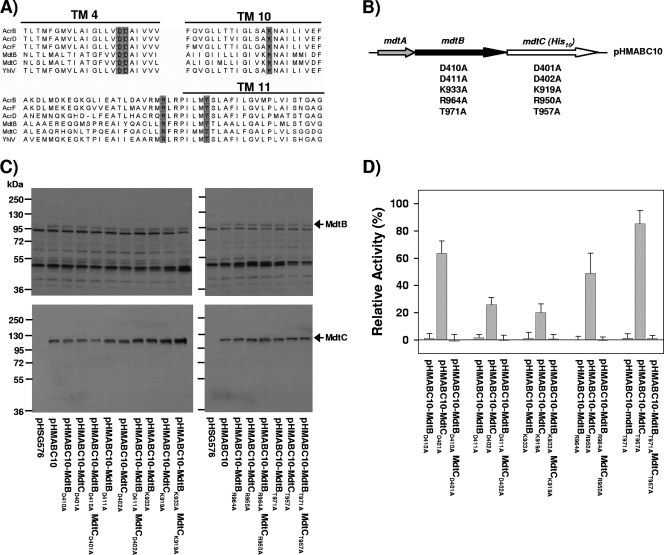
Sequence alignment with other E. coli RND transporters showed that five charged and polar amino acid residues that are involved in the proton translocation pathway in AcrB (7, 28) are conserved in MdtB and MdtC (Fig. 4A). In AcrB, these five conserved amino acid residues are essential for its activity (28), consistent with the notion of proton motive force-driven drug transport mechanism of AcrB. Thus, it is reasonable to assume that MdtB or MdtC may also use proton motive force to energize substrate transport. To address the question of whether MdtB and MdtC subunits play a role in pump function similar to that of the protomers of the homotrimeric AcrB, we replaced each of the five predicted proton translocation residues with alanine in the unlinked MdtB or MdtC expressed from pHMABC10 (Fig. 4B). The expression level of mutant MdtB and MdtC proteins was examined by Western blot analysis using affinity-purified anti-MdtB and anti-MdtC antibodies (Fig. 4C). Due to the low binding affinity between MdtB and anti-MdtB antibody, the concentration of anti-MdtB antibody required to detect the MdtB protein was 20-fold-higher than the concentration of anti-MdtC antibody needed to detect MdtC protein. This resulted in several nonspecific bands on Western blots. However, it is clear that all mutant MdtB and MdtC proteins were expressed in the inner membrane at levels that are similar to levels of wild-type MdtB and MdtC (Fig. 4C). When any of the single proton translocation mutations was introduced into MdtB, the activity of the Mdt complex formed thereof was nearly abolished, as judged by the ability to increase cloxacillin resistance in BL21KAMR cells (Fig. 4D). On the other hand, conversion to alanine of any putative proton translocation residues in MdtC had only modest effects on resistance: 20 to 85% of the activity remained depending on the altered residue.
FIG. 4.
Proton translocation mutants of MdtB and MdtC. (A) Sequence alignment of MdtB and MdtC with other E. coli RND transporters. The gray boxes indicate the conserved charged and polar residues thought to be involved in the proton translocation pathway. (B) Construction of a proton translocation mutant of the Mdt BC complex. To produce a complex having a single mutation only in the subunit of one type, each of the five conserved charged and polar residues was mutated into alanine either in MdtB or MdtC gene harbored in pHMABC10. In addition, equivalent sites in both the MdtB and MdtC genes were mutated simultaneously into alanine to produce a complex with a single mutation in all subunits. (C) Expression levels of the mutant MdtB and MdtC proteins. N-lauroylsarcosine-solubilized inner membrane fraction was prepared from BL21KAMR cells containing a mutant plasmid and analyzed by Western blotting using affinity-purified polyclonal anti-MdtB (top) and anti-MdtC (bottom) antibodies. In panels C and D, plasmids containing mutated sequences are shown by indicating after the hyphen the mutant protein: for example, pHMABC10-MdtBD410A means the pHMABC10 plasmid whose mdtB gene was replaced by the mutant gene coding for the MdtB protein with the D410A mutation. (D) Activity of a proton translocation mutant of MdtBC complex. Efflux activity of the BL21KAMR cells harboring the mutant plasmid was estimated from its level of resistance to cloxacillin by using the gradient plate method and is represented as a percentage of the activity of the wild-type MdtBC complex expressed from pHMABC10. Error bars show standard deviations.
The different effects of mutations in MdtB versus MdtC could not be interpreted in a straightforward manner, however, because in the expected Mdt B2C complex they contribute two versus one subunit(s). Thus, it is conceivable that the effect of mutations in MdtC is less because the complex still has two functional subunits, whereas that of mutations in MdtB is more severe because two out of the three protomers are altered. In order to avoid such complications, we constructed linked Mdt B-C-BHis complexes containing the D410A (or D401A) mutation in only one MdtB (or MdtC) subunit (Fig. 5A). When the activity of the linked Mdt B-C-BHis complex having a mutated MdtB subunit was measured by using cloxacillin as a substrate, only 6.8 to 23.5% of activity was observed compared to the linked B-C-BHis complex having all functional subunits (Fig. 5B). On the other hand, the activity of the linked B-C-BHis complex having a mutated MdtC subunit showed 72.6% of activity, which is similar to the activity observed with the unlinked complex formed by two wild-type MdtB and one MdtC with the D401A mutation (MdtCD401A). The results indicate that MdtB and MdtC behave differently in relation to mutations in proton translocation residues, a finding that will be analyzed in Discussion.
DISCUSSION
The prototype multidrug efflux pump of the RND family, AcrB, exists as a homotrimer, and this homotrimeric structure is intimately connected to the mode of action of this pump, the functionally rotating mechanism of drug export (13, 22, 24, 29). Thus, the presence in E. coli of MdtBC, an RND pump complex with two similar but different component proteins, was rather unexpected.
In the current study, we first showed, by adding His tags only to MdtB or MdtC and by coexpressing it with untagged partners, that trimeric proteins with the approximate composition of B2C were eluted from the cobalt column. We tried to resolve trimers of differing compositions by eluting with a stepwise gradient of imidazole but could not obtain trimers of other compositions in a reproducible manner. Nevertheless, we could not completely rule out the possibility that the obtained trimer was actually a mixture of different trimers, which could not be resolved because of the ineffectiveness of the separation method. Furthermore, at least mdtB was expressed well in the absence of mdtC, and the proteins apparently formed a B3 homooligomer. Because of these uncertainties, we decided to get an unequivocal answer on the composition of the trimer by producing covalently linked trimers, as was recently done with AcrB (29).
The results showed that covalently linked B-B-C and B-C-B constructs showed high activity, as expected from the hypothesis that B2C is the active trimer (Fig. 3C and D). However, there were two unexpected complications to this approach. First, the linked B-C dimer and the linked B-C-C trimer showed modest activity. A linked dimer of AcrB was earlier shown to produce some efflux activity, and this was ascribed to the generation of monomeric proteins, either by premature termination of transcription or translation or by proteolytic degradation of the linked products (see reference 9), followed finally by the reassembly of these monomeric proteins to generate the active trimer (29). A similar explanation likely applies here, and we confirmed this interpretation experimentally by coexpressing either the MdtC monomer or the MdtB monomer together with linked oligomers (Table 2). The second complication was the lack of expression of the linked dimer and trimers beginning with MdtC. Since monomeric MdtC is expressed reasonably well (Fig. 3B), this phenomenon was unexpected. Possibly the presence, at its C terminus, of the linker sequence and/or another transporter sequence affects adversely the folding of the first MdtC protomer with the linked constructs, and this effect is avoided if MdtB is expressed and folded ahead of MdtC.
Our results thus show that the active transporter is the Mdt B2C complex. The crucial question now is why this heterotrimer is required for drug pumping activity. This structure is especially unexpected because the idea of the functionally rotating mechanism, proposed (13, 22, 24) and supported (29) for the homotrimeric AcrB, assumes that each of the protomeric units plays an equal role by successively going through a three-stage cycle of conformational changes. In this connection, our results of the mutant MdtB or MdtC unit within the linked trimer (Fig. 5) seem quite relevant. Since the protonation of one or both of the Asp residues in transmembrane segment 4 (TM4) (23) appears to produce the “extrusion” conformer of AcrB that is needed for the pumping out of the drug (13, 22, 24), the observation that MdtB cannot tolerate the conversion of these Asp residues to Ala (Fig. 4D and 5) suggests strongly that MdtB participates in the binding and the proton motive force-driven extrusion of the drug. Furthermore, the observation that the inactivation of only one of the two MdtB protomeric units suffices to inactivate the entire complex indicates that there is a strong cooperative interaction between these two units. This is consistent with the prediction that conformational changes in one protomer must be accommodated by the compensatory conformational alterations in other protomers in AcrB (13, 22, 24) and with the observation that a trimer consisting of mutant proteins alone cannot produce a conformation expected for the extrusion protomer (25). It is also consistent with the positive cooperativity kinetics of AcrB with some substrates (16). MdtC, in contrast, seems to tolerate the alterations of residues involved in proton translocation (Fig. 4D and 5). This suggests that it plays a function different from that of MdtB, perhaps a function not involving the direct extrusion of the drug. This conclusion is consistent with our previous finding that the presence of the chromosomal mdtC gene alone did not preserve any intrinsic resistance to deoxycholate, even in the presence of the transcriptional activator BaeR expressed from a multicopy plasmid (1), and with the finding in this study that the expression of MdtC alone from plasmid pHMA-C10 did not produce any resistance to deoxycholate (Fig. 3D), although Nagakubo et al. (15) reported that MdtC alone confers resistance to bile salts. Thus, the precise function of MdtC is currently unclear and awaits further study. Of this connection, we note that MdtB resembles AcrB and other RND drug transporters in some aspects, but MdtC does not. For example, the calculated pI values for MdtB and AcrB are very similar (5.3 and 5.4, respectively), but that for MdtC is very different (value of 8.4). Finally, it is unclear at present what advantages this structurally asymmetric trimer confers over the functionally rotating mechanism of the AcrB homotrimer.
A search for homologs of MdtC with the program BLAST, as well as for pairs of RND transporters coded by related genes at the website www.membranetransport.org, showed that homologs of MdtBC are widespread among Enterobacteriaceae. Interestingly, a homolog even exists in Salmonella enterica serovar Typhi CT18, which appears to have lost many transporters, leaving behind only the homologs of AcrB, AcrD, and MdtBC among multidrug efflux RND pumps; this suggests that MdtBC homologs play an important role in the physiology or ecology of Enterobacteriaceae. Outside Enterobacteriaceae, however, MdtBC homologs are not always found in bacterial genomes, although it appears widespread among pseudomonads (Pseudomonas putida, P. aeruginosa, P. fluorescens, and P. syringae). It is found in Rhodopseudomonas palustris, but not in Rhodobacter sphaeroides or in Sinorhizobium meliloti, Rhizobium leguminosarum, or Agrobacterium tumefaciens (which contains 10 multidrug efflux RND pumps). What this uneven distribution of MdtBC homologs means in terms of its physiological role is also a subject for future study.
Acknowledgments
This study was supported in part by U.S. Public Health Service grant AI-09644 from National Institute of Allergy and Infectious Diseases.
We thank Rajeev Misra and H. C. Sulavik for strains and Yumiko Takatsuka for her advice on construction of linked oligomer genes.
Footnotes
Published ahead of print on 28 December 2009.
REFERENCES
- 1.Baranova, N., and H. Nikaido. 2002. The baeSR two-component system activates transcription of the yegMNOB (mdtABCD) transporter gene cluster in Escherichia coli and increases its resistance to novobiocin and deoxycholate. J. Bacteriol. 184:4168-4176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bavro, V. N., Z. Pietras, N. Furnham, L. Perez-Cano, J. Fernandez-Recio, R. Misra, and B. Luisi. 2008. Assembly and channel opening in a bacterial drug efflux machine. Mol. Cell 30:114-121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chang, A. C., and S. N. Cohen. 1978. Construction and characterization of amplifiable multicopy DNA cloning vehicles derived from the P15A cryptic miniplasmid. J. Bacteriol. 134:1141-1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U. S. A. 97:6640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Elkins, C. A., and H. Nikaido. 2002. Substrate specificity of the RND-type multidrug efflux pumps AcrB and AcrD of Escherichia coli is determined predominately by two large periplasmic loops. J. Bacteriol. 184:6490-6498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Eswaran, J., E. Koronakis, M. K. Higgins, C. Hughes, and V. Koronakis. 2004. Three's company. Component structures bring a closer view of tripartite drug efflux pumps. Curr. Opin. Struct. Biol. 14:741-747. [DOI] [PubMed] [Google Scholar]
- 7.Guan, L., and T. Nakae. 2001. Identification of essential charged residues in transmembrane segments of the multidrug transporter MexB of Pseudomonas aeruginosa. J. Bacteriol. 183:1734-1739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Koronakis, V., A. Sharff, E. Koronakis, B. Luisi, and C. Hughes. 2002. Crystal structure of the bacterial membrane protein TolC central to multidrug efflux and protein export. Nature 405:914-919. [DOI] [PubMed] [Google Scholar]
- 9.Kurland, C., and J. Gallant. 1996. Errors of heterologous protein expression. Curr. Opin. Biotechnol. 7:489-493. [DOI] [PubMed] [Google Scholar]
- 10.Li, X.-Z., and H. Nikaido. 2004. Efflux-mediated drug resistance in bacteria. Drugs 64:159-204. [DOI] [PubMed] [Google Scholar]
- 11.Mikolosko, J., K. Bobyk, H. I. Zgurskaya, and P. Ghosh. 2006. Conformational flexibility in the multidrug efflux system protein AcrA. Structure 14:577-587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- 13.Murakami, S., R. Nakashima, E. Yamashita, T. Matsumoto, and A. Yamaguchi. 2006. Crystal structures of a multidrug transporter reveal a functionally rotating mechanism. Nature 443:173-179. [DOI] [PubMed] [Google Scholar]
- 14.Murakami, S., R. Nakashima, E. Yamashita, and A. Yamaguchi. 2002. Crystal structure of bacterial multidrug efflux transporter AcrB. Nature 419:587-593. [DOI] [PubMed] [Google Scholar]
- 15.Nagakubo, S., K. Nishino, T. Hirata, and A. Yamaguchi. 2002. The putative response regulator BaeR stimulates multidrug resistance of Escherichia coli via a novel multidrug exporter system, MdtABC. J. Bacteriol. 184:4161-4167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nagano, K., and H. Nikaido. 2009. Kinetic behavior of the major multidrug efflux pump AcrB of Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 106:5854-5858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nikaido, H. 2009. Multidrug resistance in bacteria. Annu. Rev. Biochem. 78:119-146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nikaido, H. 1996. Multidrug efflux pumps of gram-negative bacteria. J. Bacteriol. 178:5853-5859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Okusu, H., D. Ma, and H. Nikaido. 1996. AcrAB efflux pump plays a major role in the antibiotic resistance phenotype of Escherichia coli multiple-antibiotic-resistance (Mar) mutants. J. Bacteriol. 178:306-308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 21.Schägger, H., and G. von Jagow. 1991. Blue native electrophoresis for isolation of membrane protein complexes in enzymatically active form. Anal. Biochem. 199:223-231. [DOI] [PubMed] [Google Scholar]
- 22.Seeger, M. A., A. Schiefner, T. Eicher, F. Verrey, K. Diedrichs, and K. M. Pos. 2006. Structural asymmetry of AcrB trimer suggests a peristaltic pump mechanism. Science 313:1295-1298. [DOI] [PubMed] [Google Scholar]
- 23.Seeger, M. A., C. von Ballmoos, F. Verrey, and K. M. Pos. 2009. Crucial role of Asp408 in the proton translocation pathway of multidrug transporter AcrB: evidence from site-directed mutagenesis and carbodiimide labeling. Biochemistry 48:5801-5812. [DOI] [PubMed] [Google Scholar]
- 24.Sennhauser, G., P. Amstutz, C. Briand, O. Storchenegger, and M. G. Grütter. 2007. Drug export pathway of multidrug exporter AcrB revealed by DARPin inhibitors. PLos Biol. 5:e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Su, C. C., M. Li, R. Gu, Y. Takatsuka, G. McDermott, H. Nikaido, and E. W. Yu. 2006. Conformation of the AcrB multidrug efflux pump in mutants of the putative proton-relay pathway. J. Bacteriol. 188:7290-7296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sulavik, M. C., C. Houseweart, C. Cramer, N. Jiwani, N. Murgolo, J. Greene, B. DiDomenico, K. J. Shaw, G. H. Miller, R. Hare, and G. Shimer. 2001. Antibiotic susceptibility profiles of Escherichia coli strains lacking multidrug efflux pump genes. Antimicrob. Agents Chemother. 45:1126-1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Symmons, M. F., E. Bokma, E. Koronakis, C. Hughes, and V. Koronakis. 2009. The assembled structure of a complete tripartite bacterial multidrug efflux pump. Proc. Natl. Acad. Sci. U. S. A. 106:7173-7178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Takatsuka, Y., and H. Nikaido. 2006. Threonine-978 in the transmembrane segment of the multidrug efflux pump AcrB of Escherichia coli is crucial for drug transport as a probable component of the proton relay network. J. Bacteriol. 188:7284-7289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Takatsuka, Y., and H. Nikaido. 2009. Covalently linked trimer of the AcrB multidrug efflux pump provides support for the functional rotating mechanism. J. Bacteriol. 191:1729-1737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Takeshita, S., M. Sato, M. Toba, W. Masahashi, and T. Hashimoto-Gotoh. 1987. High-copy-number and low-copy-number plasmid vectors for lacZ alpha-complementation and chloramphenicol- or kanamycin-resistance selection. Gene 61:63-74. [DOI] [PubMed] [Google Scholar]
- 31.Zgurskaya, H. I., and H. Nikaido. 1999. Bypassing the periplasm: reconstitution of the AcrAB multidrug efflux pump of Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 96:7190-7195. [DOI] [PMC free article] [PubMed] [Google Scholar]