Abstract
Ribulose 1,5-bisphosphate carboxylase/oxygenase (RubisCO) catalyzes the assimilation of atmospheric CO2 into organic matter and is thus central to the existence of life on earth. The beginning of the 2000s was marked by the discovery of a new family of proteins, the RubisCO-like proteins (RLPs), which are structural homologs of RubisCO. RLPs are unable to catalyze CO2 fixation. The RLPs from Chlorobaculum tepidum, Bacillus subtilis, Geobacillus kaustophilus, and Microcystis aeruginosa have been shown to participate in sulfur metabolism. Whereas the precise function of C. tepidum RLP is unknown, the B. subtilis, G. kaustophilus, and M. aeruginosa RLPs function as tautomerases/enolases in a methionine salvage pathway (MSP). Here, we show that the form II RubisCO enzyme from the nonsulfur purple bacterium Rhodospirillum rubrum is also able to function as an enolase in vivo as part of an MSP, but only under anaerobic conditions. However, unlike B. subtilis RLP, R. rubrum RLP does not catalyze the enolization of 2,3-diketo-5-methylthiopentyl-1-phosphate. Instead, under aerobic growth conditions, R. rubrum RLP employs another intermediate of the MSP, 5-methylthioribulose-1-phosphate, as a substrate, resulting in the formation of different products. To further determine the interrelationship between RubisCOs and RLPs (and the potential integration of cellular carbon and sulfur metabolism), the functional roles of both RubisCO and RLP have been examined in vivo via the use of specific knockout strains and complementation studies of R. rubrum. The presence of functional, yet separate, MSPs in R. rubrum under both aerobic (chemoheterotrophic) and anaerobic (photoheterotrophic) growth conditions has not been observed previously in any organism. Moreover, the aerobic and anaerobic sulfur salvage pathways appear to be differentially controlled, with novel and previously undescribed steps apparent for sulfur salvage in this organism.
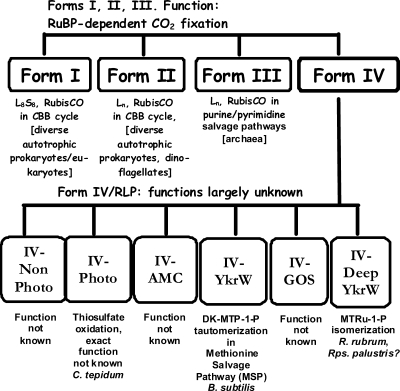
Ribulose 1,5-bisphosphate (RuBP) carboxylase/oxygenase (RubisCO) is the key enzyme of the Calvin-Benson-Bassham (CBB) reductive pentose phosphate pathway. This enzyme catalyzes the primary CO2 fixation reaction and is found in diverse organisms, including plants, most photosynthetic and chemoautotrophic microorganisms, and many archaea (25). On the basis of amino acid sequence similarities, the RubisCO family of proteins has been classified into four groups, i.e., form I, form II, form III, and form IV (Fig. 1). The enzymes classified under forms I, II, and III are all able to catalyze the RubisCO reaction, i.e., carboxylation/oxygenation of RuBP. The most recently discovered group of enzymes in the RubisCO family are the form IV or RubisCO-like proteins (RLPs). These proteins have thus far been identified in proteobacteria, cyanobacteria, archaea, and algae (2, 4, 8, 11, 12, 21, 25, 26). RLPs have been further divided into six different subgroups based on sequence similarities within the group: IV-Photo, IV-Nonphoto, IV-YkrW, IV-DeepYrkW, IV-GOS (Global Ocean Sequencing), and IV-AMC (Acid Mine Consortium) (25, 26). Despite sharing a level of sequence similarity with the bonafide RubisCOs, the RLPs are unable to carry out CO2/O2 fixation because their sequences contain dissimilar residues at positions analogous to RubisCO's active-site residues (25). The structures of the Geobacillus kaustophilus and Chlorobaculum tepidum RLPs have now been solved, and there are indeed differences between the tertiary structures of these two proteins and the bonafide RubisCO enzymes (14, 17, 25). Moreover, distinct patterns of active-site residue identities among the different clades of the RLP lineage suggest that these subgroups of RLPs are likely to utilize different substrates and perform dissimilar reactions (23, 25, 26).
FIG. 1.
Summary of the different classes of RubisCO found in nature so far (25). Forms I, II, and III catalyze bonafide CO2/O2 fixation reactions by using RuBP as the substrate. Form IV RubisCO (RLP) does not catalyze RuBP-dependent CO2/O2 fixation and is divided into six known clades (25), with only representatives of the type IV-YkrW and IV-DeepYkrW subgroups shown to catalyze defined, yet distinct, reactions (Fig. 2).
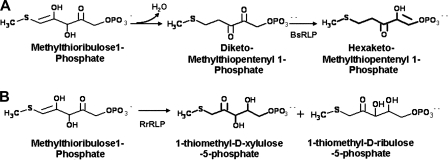
Previous studies performed with the Chlorobaculum tepidum RLP (of the IV-Photo group) gave the first indication that the RLPs may be involved in some aspect of sulfur metabolism (12, 13). This was later substantiated when the precise function was established for the Bacillus subtilis (2), Microcystis aeruginosa (4), and Geobacillus kaustophilus (14) RLPs of the IV-YkrW group. All three proteins catalyze a tautomerase/enolase reaction of a methionine salvage pathway (MSP) in which the substrate 2,3-diketo-5-methylthiopentanyl-1-phosphate (DK-MTP 1P) is converted to 2-hydroxy-3-keto-5-thiomethylpent-1-ene 1-phosphate (HK-MTP 1P) (Fig. 2). This reaction is very reminiscent of the enolization of RuBP catalyzed by RubisCO. Moreover, form II RubisCO from Rhodospirillum rubrum was shown to complement an RLP mutant strain of B. subtilis, with the ability to catalyze the identical tautomerase/enolase reaction (2). Interestingly, in addition to the presence of a form II RubisCO gene (cbbM), the genome of R. rubrum also encodes an RLP that clusters with the IV-DeepYkrW group (25). The function of this protein was recently determined, and it was shown to catalyze a distinct reaction that uses 5-methylthioribulose-1-phosphate as the substrate (15). Via an unprecedented 1,3-proton transfer, with two successive 1,2-proton transfers from its substrate, R. rubrum RLP catalyzes the formation of two products, i.e., 1-thiomethyl-d-xylulose-5-phosphate and 1-thiomethyl-d-ribulose-5-phosphate, at a 3:1 ratio (15) (Fig. 2). The novel reaction catalyzed by this RLP suggests that R. rubrum likely uses a different pathway to salvage sulfur compounds.
FIG. 2.
Distinct reactions catalyzed by type IV-YkrW (A) and type IV-DeepYkrW (B) classes of form IV RubisCO/RLP, exemplified by the proteins from B. subtilis and R. rubrum, respectively.
The presence of an RLP-encoding gene triggered the search for additional genes in the R. rubrum genome that might be homologs of known enzymes that participate in a conventional MSP. Several genes were indeed identified to encode homologs of MSP enzymes. However, to this point there is no experimental evidence for the existence of a functional MSP (21) in R. rubrum. Thus, in this study, we sought to determine the role of the RLP and RubisCO protein in sulfur salvage since each protein catalyzes different reactions and RubisCO is known to be synthesized only under anaerobic conditions (6, 7). Moreover, it is well appreciated that R. rubrum possesses a versatile metabolic capacity and is able to grow under both anaerobic and aerobic conditions, using a variety of carbon sources. The involvement of RLP and RubisCO in sulfur salvage was thus determined and found to be associated with aerobic and anaerobic metabolism, respectively.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
R. rubrum strains used in the current study are Str-2 (wild type; a spontaneous streptomycin-resistant derivative of strain S1 [ATCC 11170]) and I19A (cbbM mutant; a form II RubisCO disruption strain [10]). PYE complex medium consisting of 0.3% peptone, 0.3% yeast extract, 10% Ormerod's basal salts (19), and 15 μg of biotin per liter was used for aerobic chemoheterotrophic growth of R. rubrum in conjugation experiments. Ormerod's medium (OM) (19), containing dl-malate as the carbon source, was used for all photoheterotrophic growth experiments and was also used as the defined medium under aerobic chemoheterotrophic growth conditions. MTA (5-methylthioadenosine)-dependent growth was achieved with sulfur-depleted OM, prepared by replacing the sulfate salts with equimolar amounts of chloride salts. Antibiotics used for selection of R. rubrum mutants and transconjugants were kanamycin (50 μg ml−1), gentamicin (10 μg ml−1), tetracycline (36 μg ml−1), and streptomycin (50 μg ml−1).
Escherichia coli strain DH5α (Invitrogen) was used as the host strain for all the cloning procedures; strain SM-10 was used as the donor strain in the conjugation experiments (22). E. coli cultures were grown in Luria-Bertani (LB) medium containing 1% tryptone, 0.5% yeast extract, and 1% NaCl (wt/vol). Antibiotics used for plasmid selection in E. coli were ampicillin at 100 μg ml−1, kanamycin at 50 μg ml−1, gentamicin at 15 μg ml−1, erythromycin at 30 μg ml−1, and chloramphenicol at 30 μg ml−1. Antibiotics and media components were purchased from either Sigma or Fisher. A list of all strains and plasmids used in this study is provided (Table 1).
TABLE 1.
Bacterial strains and plasmids used in this study
| Bacterial strain or plasmid | Characteristic(s) | Reference |
|---|---|---|
| Strains | ||
| E. coli DH5α | Common cloning strain | 9 |
| E. coli SM-10 | Donor strain in conjugation experiments | 22 |
| E. coli BL21(DE3) | High-level expression by IPTG induction of T7 RNA polymerase from lacUV5 promoter | 27 |
| R. rubrum Wt (Str-2) | Wild-type strain, streptomycin resistant | 10 |
| R. rubrum cbbM (I19A) mutant | RubisCO disruption strain | 10 |
| R. rubrum rlpA mutant | RLP disruption strain | This study |
| R. rubrum cbbM rlpA mutant | RubisCO/RLP double disruption strain | This study |
| Plasmids | ||
| pTBRrRLP | R. rubrum rlpA gene cloned into pCRTOPOBLUNT | This study |
| pTBRLPGm | Gentamicin gene inserted into StuI site of pTBRrRLP | This study |
| pTB-MPI | R. rubrum mtrI gene cloned into pCRTOPOBLUNT | This study |
| pTB-MPIGm | Gentamicin gene inserted into mtrI gene in pTB-MPI | This study |
| pUC1318Gm | Source of Gm cartridge | 1 |
| pSUP202 | Suicide vector | 22 |
| pSUP-RLPGm | Disrupted rlpA gene from pTBRLPGm cloned into PstI site of pSUP202 | This study |
| pSUP-MPIGm | Disrupted mtrI gene from pTB-MPIGm cloned into EcoRI site of pSUP202 | This study |
| pRK415 | Broad-host-range vector | 16 |
| pRPR | pRK415 vector containing R. rubrum RLP promoter region | This study |
| pRP-RRLP | R. rubrum rlpA gene cloned into pRPR | This study |
| pRP-CRLP | C. tepidum RLP gene cloned into pRPR | This study |
| pRP-BRLP | B. subtilis RLP gene cloned into pRPR | This study |
| pRP-PRLP1 | R. palustris RLP1 gene cloned into pRPR | This study |
| pRP-PRLP2 | R. palustris RLP2 gene cloned into pRPR | This study |
| pRPS-MCS3 | Broad-host-range plasmid containing R. rubrum cbbM promoter and cbbR gene | 24 |
| pRPS-RrcbbM | R. rubrum cbbM gene cloned into pRPS-MCS3 | This study |
| pRPS-RpcbbM | R. palustris cbbM gene cloned into pRPS-MCS3 | Satagopan and Tabita, unpublished |
MTA-dependent growth of R. rubrum.
Single colonies were used to inoculate culture tubes containing Ormerod's malate minimal medium under aerobic conditions at 30°C with shaking at 200 rpm. Growth proceeded until the mid-exponential phase (A660, ∼0.6 to 0.8). Cells were harvested by centrifuging cultures at 12,000 × g for 3 min; cell pellets were washed three times with sulfur-depleted medium and then resuspended in the same medium. Washed cells were inoculated into sulfur-depleted malate minimal medium supplemented with MTA. As a negative control in all the experiments, cells were also inoculated into sulfur-depleted medium lacking any exogenous sulfur source. Anaerobic photoheterotrophic MTA-dependent growth was accomplished by performing the same procedure described above, using cells grown chemoheterotrophically and then made anaerobic inside an anaerobic chamber (Coy Labs, Grass Lake, MI) that maintained an atmosphere of 2.5 to 3% hydrogen and balance nitrogen. Anaerobic media were prepared under a 100% nitrogen atmosphere and dispensed (10 ml per tube) in 25-ml tubes fitted with butyl rubber stoppers with an aluminum seal crimped over the stopper (Bellco Glass Inc., Vineland, NJ). Anaerobic cultures were grown in the light at 27°C in a growth chamber (Environment Growth Chambers, Chagrin Falls, OH). In the experiments testing for MTA-dependent growth, all strains were cultured in defined media with MTA or methionine (positive control) as the sole sulfur source, or without any sulfur source (negative control). The concentration of MTA and methionine used in the media was 1 mM. The antibiotics kanamycin and gentamicin, which are commercially available as sulfate salts, were not used for MTA-dependent growth because they would have introduced sulfate into the medium. All the growth experiments were performed using three independent clonal isolates of each strain; the results shown are data from at least two independent growth experiments.
Molecular biology protocols.
Genomic DNA was purified using the Wizard genomic DNA purification kit (Promega). Southern blot analysis and PCRs were carried out using standard protocols (3, 18). All the genes were amplified from the genomic DNA of the respective organisms by performing PCR using either Taq DNA polymerase (Invitrogen) or Pfu DNA polymerase (Stratagene). Plasmid DNA from both E. coli and R. rubrum cells was isolated using a plasmid miniprep kit (Qiagen).
Inactivation of the rlpA (RLP) gene.
The R. rubrum rlpA gene was amplified from genomic DNA by PCR with Pfu DNA polymerase (Invitrogen) by using primers 5′-CGAGGACAGGATCCGCGCCATCGG-3′ and 5′-GCGCCCCTGCAGGCGATCGTCTCC-3′. The PCR-amplified rlpA region was cloned into the pCR-Blunt II-TOPO vector (Invitrogen), resulting in plasmid pTBRrRLP. The rlpA gene was disrupted after insertion of an XmnI- and AfeI-digested gentamicin cassette from pUC1318Gm (Table 1) into StuI-digested pTBRrRLP. This resulted in plasmid pTBRLPGm. The disrupted rlpA gene was then subcloned into suicide vector pSUP202 (Table 1). Plasmid pTBRLPGm was digested with NsiI, and the fragment containing the disrupted rlpA gene was ligated into PstI-digested pSUP202. This resulted in the formation of plasmid pSUP-RLPGm, which was transferred to R. rubrum by conjugation. Transconjugants were selected for gentamicin resistance and tetracycline sensitivity. An RLP and form II RubisCO double disruption strain was created by disrupting rlpA in form II RubisCO disruption strain I19A (cbbM mutant). In the current study, I19A is referred to as the cbbM mutant. The rlpA disruption was confirmed after performing a Southern blot analysis using the rlpA gene as a probe.
Cloning for complementation studies.
Plasmid pRPR was constructed by cloning the putative promoter region of the R. rubrum rlpA gene into pRK415 (Table 1). The promoter region was amplified by PCR using a forward primer incorporating the NdeI site (5′-GGCGTGGATCATATGACGGTGCGCCTGG-3′) and a reverse primer incorporating the AseI site (5′-CAGTCTGTCCGTATTAATATGTCTCCCGCGGC-3′). The PCR product was digested with NdeI and AseI and cloned into the corresponding sites in pRK415. This resulted in plasmid pRPR, which was used for expressing various genes under the direction of the RLP promoter for all complementation studies. All the RubisCO and RLP genes, except the B. subtilis RLP gene, were amplified from the genomic DNA of their respective organisms. The B. subtilis RLP gene was amplified from plasmid pUC19ykrW (S. S. Scott and F. R. Tabita, unpublished data). All the forward and reverse primers introduced NdeI and BamHI sites, respectively (see Table S1 in the supplemental material). The PCR products were digested with NdeI and BamHI restriction enzymes and cloned into AseI- and BamHI-digested pRPR.
Inactivation of the 5-methylthioribose-1-phosphate isomerase (mtrI) gene.
The region surrounding the mtrI gene was amplified using the primers 5′-GGGGAACATATGTCCGAGGCGTATCGGC-3′ and 5′-GCGACCGCGGATCCGGTCGGGAAACGAGGCG-3′ and cloned into the pCR-Blunt II-TOPO vector (Invitrogen). This resulted in plasmid pTB-MPI. The mtrI gene was disrupted by inserting an AccI- and AfeI-digested gentamicin cassette into AccI- and SrfI-digested pTB-MPI. This deleted 245 bp of the mtrI gene. The disrupted gene was subcloned into the suicide vector pSUP202. The disrupted gene fragment was digested with EcoRI and inserted into the EcoRI site of pSUP202; this resulted in the formation of plasmid pSUP-MPIGm. Plasmid pSUP-MPIGm was transferred to wild-type R. rubrum by conjugation; transconjugants were selected for gentamicin resistance. The genotype of the mtrI disruption strain was confirmed by Southern blot analysis.
Bacterial conjugation and selection of transconjugants.
Conjugation was performed by biparental matings. R. rubrum recipient strains were grown for 3 to 4 days in PYE (complex) medium to the late exponential or early stationary phase (optical density at 660 nm [OD660], ∼1.2 to 1.5); the cells were then diluted 1:10 and grown for 1 to 2 days until mid- to late exponential phase (OD660, ∼0.9 to 1.2). E. coli strain SM-10 was used as the donor strain for the matings. Overnight cultures of E. coli grown in LB medium with the appropriate antibiotics were diluted 1:10 in LB (without antibiotic) and incubated at 37°C with shaking at 220 rpm for 2 h. Matings were set up by combining recipient cells (1.0 ml) with donor cells (1.0 ml) in an Eppendorf tube and centrifuging the cells for 4 min at 13,600 × g in a microcentrifuge. This mating mixture pellet was resuspended in 30 μl of PYE medium, and the resuspension was spotted onto a PYE medium plate. Control plates containing either recipient cells only or donor cells with an empty plasmid (pRPR) without any insert were prepared as described above and included in each conjugation experiment. The mating PYE plates were incubated in the dark at 30°C overnight.
Following mating, cells from each plate were resuspended in 1 ml of PYE medium. Dilutions of 10−1 to 10−4 were plated onto PYE medium plates containing the appropriate antibiotics. The R. rubrum wild-type strain is resistant to streptomycin. Streptomycin was used as a counterselection for E. coli whenever wild-type R. rubrum was the recipient strain. Kanamycin and gentamicin were also used for counterselection when the RLP/RubisCO double disruption strain was used as the recipient. Selection was accomplished in all experiments by incubating plates in the dark at 30°C until colonies appeared (6 to 10 days). Colonies were grown in PYE or Ormerod's malate medium broth, supplemented with the appropriate antibiotics, and used for further manipulations.
RESULTS
Correlation between the presence of RLP and a functional MSP.
If an organism is grown on MTA as the sole sulfur source, needed sulfur-containing amino acids must be synthesized as a result of MTA metabolism via some type of sulfur salvage pathway or MSP. It was previously shown that both R. rubrum and Rhodopseudomonas palustris are capable of growth by using MTA as the sole sulfur source under aerobic chemoheterotrophic growth conditions, whereas neither Rhodobacter sphaeroides nor Rhodobacter capsulatus was able to metabolize MTA (25). All four organisms were able to grow on media containing methionine as the sole sulfur source, showing that R. capsulatus and R. sphaeroides do not lack the ability to metabolize methionine. Based on the abilities to metabolize MTA, it could thus be concluded that R. rubrum and R. palustris must have a mechanism to salvage sulfur from MTA, presumably via some form of MSP. Using B. subtilis as a paradigm, in order for R. rubrum and R. palustris to grow using MTA as the sole sulfur source, an enolase/tautomerase that would catalyze the conversion of DK-MTP 1P to HK-MTP 1P would be required as part of the MSP (21). This ability to use MTA correlated with the presence of one or more RLP genes in the genome of each of these organisms, genes which are not found in R. capsulatus or R. sphaeroides (25). Thus, our working hypothesis was that R. capsulatus and R. sphaeroides lacked any means to convert MTA to methionine, while both R. rubrum and R. palustris possess this metabolic capability by virtue of possessing RLP genes.
Role of RLP and RubisCO in MTA-dependent growth of R. rubrum.
The RLP gene was disrupted after insertion of a gentamicin gene cassette within the coding sequence via homologous recombination; the genotype of this strain was confirmed after Southern blot analysis (data not shown). The rlpA gene was disrupted both in the wild type as well as in a form II RubisCO disruption (cbbM mutant) background. Compared to their respective parent strains, single rlpA disruption and RLP/RubisCO double-disruption strains did not show any apparent phenotypic difference when grown under either photoheterotrophic or chemoheterotrophic conditions by using ammonium sulfate, a readily assimilable sulfur source.
The rlpA disruption strain of R. rubrum was incapable of using MTA as the sole sulfur source under aerobic growth conditions (Fig. 3A), indicating that RLP is required for metabolizing MTA. The form II RubisCO disruption strain (cbbM mutant) was able to grow using MTA as the sole sulfur source under these conditions. As expected, the RubisCO/RLP double disruption strain (cbbM rlpA mutant) was unable to carry out aerobic MTA-dependent growth (Fig. 3A).
FIG. 3.
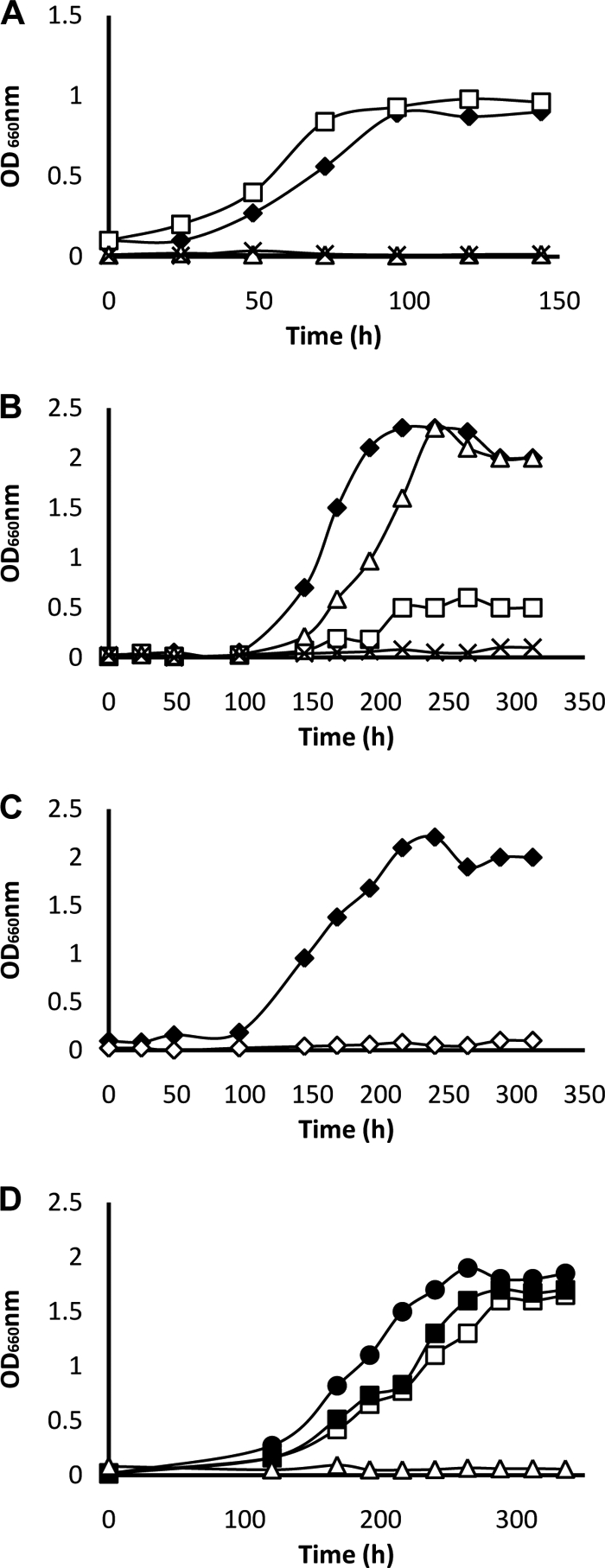
(A) Growth of R. rubrum strains under aerobic conditions with MTA as the sole sulfur source. Malate was used as the carbon source. (B) Growth of R. rubrum strains under anaerobic conditions with MTA as the sole sulfur source. In all cases, a defined malate minimal medium was utilized. ♦, wild type; ▵, rlpA mutant; □, cbbM mutant; ×, cbbM rlpA mutant. (C) Complementation of the R. rubrum cbbM rlpA mutant strain under anaerobic photoheterotrophic conditions in malate minimal medium containing MTA as the sole sulfur source. ⋄, cbbM rlpA mutant with no plasmid; ♦, cbbM rlpA mutant containing the R. rubrum cbbM gene, plasmid RrcbbM. (D) cbbM rlpA mutant containing R. palustris wild-type and mutant cbbM genes. •, RpcbbM; ▪, RpcbbMI165T; □, RpcbbMI165V; ▵, no plasmid. The R. rubrum rlpA gene was not able to complement the cbbM rlpA mutant strain under anaerobic growth conditions.
Because RubisCO expression and function in R. rubrum require anaerobiosis (6), we decided to first test for the presence of a functional MSP under anaerobic photoheterotrophic growth conditions with the wild-type strain. As shown in Fig. 3B, the wild type is able to utilize MTA (Fig. 3B). This was surprising because the known MSP of B. subtilis has an oxygen-requiring enzymatic step (21). Further analysis showed that the R. rubrum rlpA mutant strain, which cannot metabolize MTA as a sulfur source under aerobic chemoheterotrophic growth conditions (Fig. 3A), was able to grow in the same exact medium under anaerobic photoheterotrophic growth conditions (Fig. 3B). It was also observed that the R. rubrum cbbM mutant (form II RubisCO deletion) strain, which still had a functional RLP, was barely able to grow on MTA media under anaerobic conditions, indicating the potential involvement of RubisCO (instead of RLP) in MTA metabolism under these growth conditions (Fig. 3B). The RubisCO/RLP double disruption strain (cbbM rlpA mutant) was unable to metabolize MTA under anaerobic photoheterotrophic conditions, further confirming the requirement of a functional RubisCO under these growth conditions (Fig. 3B).
Complementation of R. rubrum cbbM rlpA strain under aerobic growth conditions.
Because of its inability to metabolize MTA under either aerobic or anaerobic growth conditions, the cbbM rlpA strain was used for complementation experiments. Different RLPs and RubisCOs were tested for their ability to support MTA-dependent growth in the cbbM rlpA strain by expressing them on plasmid pRPR, which is a derivative of pRK415 (see Materials and Methods). Plasmid pRPR was constructed by cloning the presumptive upstream promoter region of the rlpA gene from R. rubrum into pRK415 (10) (Table 1); if this sequence did, in fact, contain the R. rubrum rlpA promoter, it would ensure similar levels of transcription for all the different genes. Genes encoding the form I and form II RubisCOs from R. palustris and the RLPs from R. rubrum (RrRLP), C. tepidum (CtRLP), B. subtilis (BsRLP), and R. palustris (RpRLP1) were tested for their ability to support MTA-dependent growth under aerobic growth conditions. As expected, the R. rubrum rlpA gene, when expressed on the plasmid, was able to complement for MTA-dependent growth of the cbbM rlpA mutant strain, indicating that a promoter sequence was present within the upstream region that was used to construct the expression plasmid. B. subtilis ykrW (mtnW) was able to partially rescue the MTA-dependent growth phenotype, whereas none of the other genes were able to complement for MTA-dependent growth (Table 2; see also Fig. S1 in the supplemental material).
TABLE 2.
Growth of R. rubrum strains under different growth conditionsa
| Strain genotype | Aerobic growth |
Anaerobic growth |
||
|---|---|---|---|---|
| Methionine | MTA | Methionine | MTA | |
| Wt | + | + | + | + |
| cbbM | + | + | + | − |
| rlpA | + | − | + | + |
| cbbM rlpA | + | − | + | − |
| cbbM rlpA/Rrrlp+ | + | + | ND | ND |
| cbbM rlpA/RpcbbM+ | + | + | ||
| cbbM rlpA/RrcbbM+ | + | − | + | + |
| cbbM rlpA/Ctrlp+ | + | − | ND | ND |
| cbbM rlpA/Bsrlp+ | + | + | ND | ND |
| cbbM rlpA/Rprlp2+ | + | − | ND | ND |
+ growth; −, no growth; ND, not determined. The genes expressed on plasmids for complementation are underlined.
Complementation of the cbbM rlpA disruption strain under anaerobic growth conditions.
When expressed on a plasmid (pRPS-RrcbbM), the R. rubrum form II RubisCO (cbbM) gene was able to rescue the MTA-dependent growth phenotype of the cbbM rlpA strain under anaerobic growth conditions (Fig. 3C), further confirming the results observed with the rlpA mutant strain. The R. palustris form II RubisCO (RpCbbM), which resembles RrCbbM, was also able to support MTA-dependent growth under anaerobic conditions in the cbbM rlpA strain of R. rubrum (Fig. 3D).
Previously, a mutation in an essential residue (Ile-164) of R. rubrum CbbM was shown to retain only 1% of the wild-type level of activity (5). Mutant R. palustris CbbM constructs which had point mutations in the same RubisCO active-site residue were available in the laboratory. This residue, Ile-165 in R. palustris CbbM, was substituted with different residues, and some of the resultant mutants were severely compromised with regard to their ability to fix CO2. When introduced into a RubisCO deletion strain of Rhodobacter capsulatus (20), the wild-type RpCbbM was able to complement for RubisCO function, whereas an I165T mutant was unable to do so (S. Satagopan and F. R. Tabita, unpublished results). In order to determine if this residue, which is critical for RubisCO function, is also required for alleviating the MTA phenotype of the cbbM rlpA strain, the pRPS-MCS-I165T construct was conjugated into the cbbM rlpA strain of R. rubrum. The resultant strain was able to grow on MTA medium under anaerobic photoheterotrophic conditions (Fig. 3D), indicating that this RubisCO-compromised mutant protein was functional with regard to MTA metabolism (summarized in Table S2 in the supplemental material).
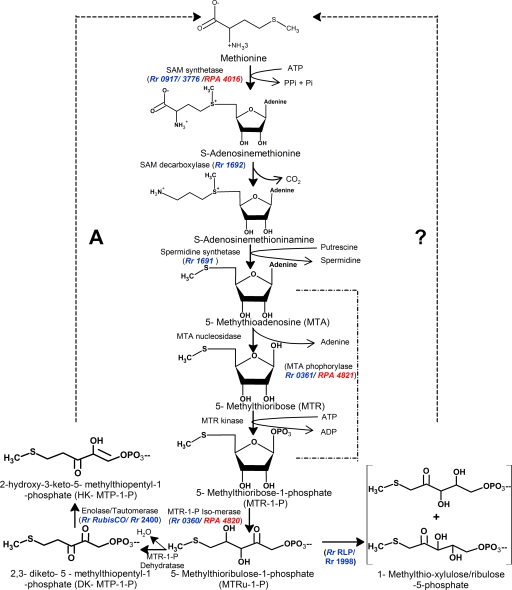
Disruption of the R. rubrum 5-methylthioribose-1-phosphate isomerase (mtrI) gene.
The mtrI gene that encodes 5-methylthioribose-1-phosphate isomerase catalyzes the reaction that results in the formation of 5-methylthio-ribulose-1-phosphate (MTRu 1P). Homologs of this enzyme are present in both R. rubrum (Rr 0360) and R. palustris (RPA 4820) (Fig. 4). Although R. rubrum's RLP and RubisCO appear to have functional differences when grown under aerobic and anaerobic conditions, it is possible that the mtrI homolog catalyzes a reaction that would be critical for MTA metabolism under both aerobic and anaerobic conditions. To test this possibility, the mtrI gene was disrupted by insertion of a gentamicin cassette into the open reading frame, using a strategy that is similar to the generation of the rlpA disruption strain. The resultant mutant was tested for its ability to metabolize MTA. It was unable to support MTA-dependent growth under either aerobic chemoheterotrophic or anaerobic photoheterotrophic conditions (see Fig. S2A and S2B, respectively, in the supplemental material). These results provided further evidence for the necessity of some form of MTA metabolic pathway under both aerobic and anaerobic growth conditions. Furthermore, the mtrI gene product catalyzes a reaction that appears to be critical to this pathway under both aerobic and anaerobic conditions.
FIG. 4.
MSP. In B. subtilis and related organisms, the IV-YkrW RLP, encoded by the mtnW (ykrW) gene, participates in an enolase/tautomerase reaction of the MSP whereby DK-MTP 1P is converted to 2-hydroxy-3-keto-5-methylthiopentenyl-1-phosphate (HK-MTPenyl-1P), as shown on the left side of the scheme (21). In R. rubrum, RubisCO (CbbM) catalyzes this reaction to allow for anaerobic MTA-dependent growth; however, it is not clear at this time how the product of this reaction is further metabolized to methionine, indicated by the dashed line on the left and in panel A. Under aerobic growth conditions, R. rubrum RLP catalyzes a novel reaction whereby 5-methylthioribulose-1-phosphate is converted to a racemic mixture of 1-methylthioxylulose-5-phosphate or 1-methylythioribulose-5-phosphate (15) (shown in large parentheses on the right side). However, there is no known means by which the products of this reaction may be converted to sulfur-containing amino acids to allow for aerobic growth on MTA as the sole sulfur source (indicated by the dashed line and question mark on the right). Homologs of the B. subtilis MSP genes found in the genomes of R. rubrum and R. palustris CGA 009 are shown in blue and red, respectively. The absence of blue or red gene designations indicates that known genes of the B. subtilis MSP paradigm are not found or are not apparent in the genomes of R. rubrum or R. palustris, respectively.
DISCUSSION
MTA is a by-product of spermidine biosynthesis, as well as acyl homoserine lactone and ethylene biosynthesis. In most organisms, including plants and humans, MTA is converted back to methionine by an MSP (21). Whereas the RLPs of the IV-YkrW group catalyze enolization (tautomerization) of DK-MTP 1P as part of the MSP (2, 4, 11, 14) (Fig. 4), it is surprising that the form II RubisCO (CbbM) from R. rubrum could also function as a DK-MTP 1P tautomerase/enolase in vitro and could complement for the loss of RLP function in B. subtilis (2). Curiously, R. rubrum RLP uses a different substrate and was shown to catalyze a novel isomerization reaction whereby 5-methylthio-d-ribulose 1-phosphate is converted to a 3:1 mixture of 1-methylthio-xylulose 5-phosphate and 1-methylthioribulose 5-phosphate (15) (Fig. 2). These findings suggested that both RLP and RubisCO may have different physiological roles relative to MTA metabolism in R. rubrum. We show here that MTA may serve as the sole sulfur source under both aerobic and anaerobic growth conditions. Although the presence of an MSP under anaerobic growth conditions has long been speculated, there has been no evidence reported thus far. Because of the presence of an oxygen-requiring dioxygenase step in currently constituted MSP schemes (Fig. 4) (21), it is not possible for this traditional pathway to function in the absence of oxygen in either R. rubrum or R. palustris, both of which have been shown to metabolize MTA under these conditions (23). Moreover, it appears that the mtrI gene is functional and required for both aerobic and anaerobic MTA metabolism. It is thus possible that the product of this reaction is ultimately metabolized into methionine via a mechanism which bypasses the dioxygenase step of the current MSP paradigm (21). It is interesting that homologs of genes whose products would catalyze the reactions beyond the RLP-catalyzed step are absent within the genomes of both R. rubrum and R. palustris, suggesting that the aerobic part of this pathway may also function differently from the current B. subtilis MSP paradigm (Fig. 4).
We have shown that the RLP disruption strain of R. rubrum is incapable of MTA-dependent growth under aerobic growth conditions. Although an intact cbbM gene is present in this strain, it is barely expressed under aerobic growth conditions (6). Further, R. rubrum cells that do contain exogenously expressed RubisCO were found to oxidatively inactivate and then subsequently degrade this protein under aerobic conditions (6, 7). It was thus not surprising that the rlpA-disruption strain falied to metabolize MTA in the presence of oxygen. The inability of form I (CbbLS) and form II (CbbM) RubisCOs from R. palustris to support aerobic MTA-dependent growth of the cbbM rlpA mutant strain of R. rubrum (Table 2; see also Fig. S1 in the supplementary material) may also be attributed to either poor gene expression or oxidative inactivation of the proteins or may perhaps be due to the fact that these proteins are inherently unable to catalyze the RLP-type reaction. In contrast, the results from complementation to aerobic MTA-dependent growth with different RLP genes in the cbbM rlpA strain (Table 2) seem to be reflective of the differences between the putative active-site residue sets used by different subgroups of RLPs. It may also be attributable to the differences in other structural features (17, 25). Only B. subtilis ykrW (mtnW) showed some ability to complement the cbbM rlpA strain under these conditions. Inasmuch as B. subtilis and R. rubrum RLPs catalyze reactions that involve different substrates, it is apparent that the products of both reactions are somehow incorporated into a pathway that allows for aerobic MTA-dependent growth.
Both the R. rubrum and R. palustris cbbM genes were able to support MTA-dependent growth of the cbbM rlpA strain when expressed on a broad-host-range plasmid under anaerobic photoheterotrophic growth conditions. This is undoubtedly due to the fact that R. rubrum form II RubisCO (CbbM) can catalyze the DK-MTP 1P reaction (2) (and by extension, so does R. palustris CbbM). Based on knowledge of the structure and reaction mechanism employed by RubisCO, it is apparent that both substrates interact at the same active site (2, 14, 17, 25). Interestingly, the RubisCO active-site mutant (R. palustris cbbM I165T mutant), which is compromised in its ability to fix CO2, is able to complement for the MTA phenotype. This indicates that active-site residues required for RubisCO function are not necessarily equivalent for the enolase/tautomerase reaction of the anaerobic MSP catalyzed by form II RubisCO. Residue Ile-165 is in van der Waals contact with two other RubisCO active-site residues, K191 and D193, and magnesium ions (5). Clearly, as shown by the in vivo complementation studies, the interaction of these residues may not be critical for binding of the substrate DK-MTP 1P used in MTA metabolism.
Most importantly, it appears that RubisCO catalyzes two separate reactions in R. rubrum and thus appears to participate in both carbon (via the CBB CO2 assimilatory cycle) and sulfur (via an MSP) metabolism. Clearly, MTA-dependent growth in R. rubrum requires RLP under aerobic growth conditions, but the organism obligatorily requires RubisCO to grow with MTA as the sole sulfur source under anaerobic growth conditions. These findings suggest that the genes and at least one of the proteins required for MTA-dependent growth are likely to be differentially regulated in R. rubrum, and this scenario is probably the same for R. palustris as well. Clearly, these studies are indicative of the plasticity of RubisCO's active site to function in a physiologically relevant fashion in two separate and important pathways. Further work on the mechanism of the reaction catalyzed by RubisCO in anaerobic MTA metabolism will help in understanding how the different residues interact with diverse substrates that are turned over by the RubisCO active site.
Supplementary Material
Acknowledgments
This work was initially supported by grant GM24497 from the National Institutes of Health and by grant DE-FG02-91ER20033 from the Office of Biological and Environmental Research (Genomics:GTL Program) of the U.S. Department of Energy.
We acknowledge Sriram Satagopan for providing the pRPS-MCS-I165T construct for carrying out complementation studies of R. rubrum.
Footnotes
Published ahead of print on 28 December 2009.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Arsene, F., P. A. Kaminski, and C. Elmerich. 1996. Modulation of NifA activity by PII in Azospirillum brasilense: evidence for a regulatory role of the NifA N-terminal domain. J. Bacteriol. 178:4830-4838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ashida, H., Y. Saito, C. Kojima, K. Kobayashi, N. Ogasawara, and A. Yokota. 2003. A functional link between RuBisCO-like protein of Bacillus and photosynthetic RuBisCO. Science 302:286-290. [DOI] [PubMed] [Google Scholar]
- 3.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl. 1987. Current protocols in molecular biology. Green & Wiley, New York, NY.
- 4.Carré-Mlouka, A., A. Méjean, P. Quillardet, H. Ashida, Y. Saito, A. Yokota, I. Callebaut, A. Sekowska, E. Dittmann, C. Bouchier, and N. T. de Marsac. 2006. A new rubisco-like protein coexists with a photosynthetic rubisco in the planktonic cyanobacteria Microcystis. J. Biol. Chem. 281:24462-24471. [DOI] [PubMed] [Google Scholar]
- 5.Chène, P., A. G. Day, and A. R. Fersht. 1997. Role of isoleucine-164 at the active site of rubisco from Rhodospirillum rubrum. Biochem. Biophys. Res. Commun. 232:482-486. [DOI] [PubMed] [Google Scholar]
- 6.Cook, L. S., H. Im, and F. R. Tabita. 1988. Oxygen-dependent inactivation of ribulose 1,5-bisphosphate carboxylase/oxygenase in crude extracts of Rhodospirillum rubrum and establishment of a model inactivation system with purified enzyme. J. Bacteriol. 170:5473-5478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cook, L. S., and F. R. Tabita. 1988. Oxygen regulation of ribulose 1,5-bisphosphate carboxylase activity in Rhodospirillum rubrum. J. Bacteriol. 170:5468-5472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Derelle, E., C. Ferraz, S. Rombauts, P. Rouzé, A. Z. Worden, S. Robbens, F. Partensky, S. Degroeve, S. Echeynié, R. Cooke, Y. Saeys, J. Wuyts, K. Jabbari, C. Bowler, O. Panaud, B. Piégu, S. G. Ball, J. P. Ral, F. Y. Bouget, G. Piganeau, B. De Baets, A. Picard, M. Delseny, J. Demaille, Y. Van de Peer, and H. Moreau. 2006. Genome analysis of the smallest free-living eukaryote Ostreococcus tauri unveils many unique features. Proc. Natl. Acad. Sci. U. S. A. 103:11647-11652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dower, W. J., J. F. Miller, and C. W. Ragsdale. 1988. High efficiency transformation of E. coli by high voltage electroporation. Nucleic Acids Res. 16:6127-6145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Falcone, D. L., and F. R. Tabita. 1993. Complementation analysis and regulation of CO2 fixation gene expression in a ribulose 1,5-bisphosphate carboxylase-oxygenase deletion strain of Rhodospirillum rubrum. J. Bacteriol. 175:5066-5077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Frangeul, L., P. Quillardet, A. M. Castets, J. F. Humbert, H. C. Matthijs, D. Cortez, A. Tolonen, C. C. Zhang, S. Gribaldo, J. C. Kehr, Y. Zilliges, N. Ziemert, S. Becker, E. Talla, A. Latifi, A. Billault, A. Lepelletier, E. Dittmann, C. Bouchier, and N. T. de Marsac. 2008. Highly plastic genome of Microcystis aeruginosa PCC 7806, a ubiquitous toxic freshwater cyanobacterium. BMC Genomics 9:274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hanson, T. E., and F. R. Tabita. 2001. A ribulose-1,5-bisphosphate carboxylase/oxygenase (RubisCO)-like protein from Chlorobium tepidum that is involved with sulfur metabolism and the response to oxidative stress. Proc. Natl. Acad. Sci. U. S. A. 98:4397-4402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hanson, T. E., and F. R. Tabita. 2003. Insights into the stress response and sulfur metabolism revealed by proteome analysis of a Chlorobium tepidum mutant lacking the Rubisco-like protein. Photosyn. Res. 78:231-248. [DOI] [PubMed] [Google Scholar]
- 14.Imker, H. J., A. A. Fedorov, E. V. Fedorov, S. C. Almo, and J. A. Gerlt. 2007. Mechanistic diversity in the RuBisCO superfamily: the “enolase” in the methionine salvage pathway in Geobacillus kaustophilus. Biochemistry 46:4077-4089. [DOI] [PubMed] [Google Scholar]
- 15.Imker, H. J., J. Singh, B. P Warlick, F. R. Tabita, and J. A. Gerlt. 2008. Mechanistic diversity in the RuBisCO superfamily: a novel isomerization reaction catalyzed by the RuBisCO-like protein from Rhodospirillum rubrum. Biochemistry 47:11171-11173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Keen, N. T., S. Tamaki, D. Kobayashi, and D. Trollinger. 1988. Improved broad-host-range plasmids for DNA cloning in Gram negative bacteria. Gene 70:191-197. [DOI] [PubMed] [Google Scholar]
- 17.Li, H., M. R. Sawaya, F. R. Tabita, and D. Eisenberg. 2005. Crystal structure of a RuBisCO-like protein from the green sulfur bacterium Chlorobium tepidum. Structure 13:779-789. [DOI] [PubMed] [Google Scholar]
- 18.Mukhopadhyay, B., E. F. Johnson, and M. J. Ascano. 1999. Conditions for vigorous growth on sulfide and reactor-scale cultivation protocols for the thermophilic green sulfur bacterium Chlorobium tepidum. Appl. Environ. Microbiol. 65:301-306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ormerod, J. G., K. S. Ormerod, and H. Gest. 1961. Light-dependent utilization of organic compounds and photoproduction of molecular hydrogen by the photosynthetic bacteria; relationships with nitrogen metabolism. Arch. Biochem. Biophys. 94:449-463. [DOI] [PubMed] [Google Scholar]
- 20.Paoli, G. C., P. Vichivanives, and F. R. Tabita. 1998. Physiological control and regulation of the Rhodobacter capsulatus cbb operons. J. Bacteriol. 180:4258-4269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sekowska, A., V. Dénervaud, H. Ashida, K. Michoud, D. Haas, A. Yokota, and A. Danchin. 2004. Bacterial variations on the methionine salvage pathway. BMC Microbiol. 4:4-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Simon, R., U. Priefer, and A. Puhler. 1983. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in gram negative bacteria. Biotechnology 1:784-791. [Google Scholar]
- 23.Singh, J. 2008. Functional relationships among Rubisco family members. Ph.D. thesis. The Ohio State University, Columbus, OH.
- 24.Smith, S., and F. R. Tabita. 2003. Positive and negative selection of mutant forms of prokaryotic (cyanobacterial) ribulose-1,5-bisphosphate carboxylase/oxygenase. J. Mol. Biol. 331:557-569. [DOI] [PubMed] [Google Scholar]
- 25.Tabita, F. R., T. E. Hanson, H. Li, S. Satagopan, J. Singh, and S. Chan. 2007. Function, structure, and evolution of the RubisCO-like proteins and their RubisCO homologs. Microbiol. Mol. Biol. Rev. 71:576-599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tabita, F. R., T. E. Hanson, S. Satagopan, B. H. Witte, and N. E. Kreel. 2008. Phylogenetic and evolutionary relationships of RubisCO and the RubisCO-like proteins and the functional lessons provided by diverse molecular forms. Philos. Trans. R. Soc. Lond. B Biol. Sci. 363:2629-2640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Weiner, M. P., C. Anderson, B. Jerpseth, S. Wells, B. Johnson-Browne, and P. Vaillancourt. 1994. Studier pET system vectors and hosts. Strateg. Mol. Biol. 7:41-43. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.