Cytoskeletal proteins, including microfilaments, microtubules, and intermediate filaments, play a pivotal role in the treatment of cancer, as their regulation by small molecules arrests progression through the cell cycle.[1] Marine natural products contain a diversity of molecules that target the cytoskeleton. For instance, the cyclic peptide jasplakinolide induces assembly and stabilization of actin microfilaments.[2] The cytoskeleton is also accessed by other classes of natural products. Several polyketides, including halichondrin B and spongistatin, target microtubule stabilization,[3] while phor-boxazole B employs cytokeratin as a foundation to recruit critical cycle regulators.[4] Our interest focused on deep-sea actinomycetes[5] in an effort to identify metabolites that target other components of the cytoskeleton.
The uptake and localization of the previously described ammosamides A (1) and B (2) was first investigated.[6] Though brightly colored, the metabolites lacked sufficient fluorescence to be evaluated at physiologically relevant levels. Using methods developed through collaborative studies, we prepared an affinity probe to investigate these cellular events.[7] Specifically, a highly fluorescent 7-dimethylaminocoumarin-4-acetic acid derivative was chosen, since it lacked toxicity, offered synthetic flexibility, and served both as a fluorescent label and an epitope to a monoclonal antibody (mAb). This duality, referred to herein as an immunoaffinity fluorescence probe (IAF), was viewed as advantageous by allowing both molecular and cellular studies to be conducted with the same probe.
To prepare the IAF probe, ammosamide B (2) was coupled with acid 3 to afford adduct 4 in 10% yield after rigorous HPLC purification (Scheme 1).[8] Adduct 4 was the only product obtained in this reaction, and attempts at optimization proved unsuccessful, suggesting that the congested environment of the aromatic amines at C-6 and C-8 in 2 made efficient coupling difficult. As anticipated, adduct 4 contained 1H and 13C NMR spectroscopy resonances in accordance with the coupling of fragments 2 and 3 (see NMR spectroscopy table in the Supporting Information). Mass spectral analysis of 4 provided the molecular formula C25H21ClN6O5 (HR-ESI-FTMS-Orbitrap: m/z [M+H]+: 521.1338). The position of the label at C-8-N was determined by inspection of the NOESY spectrum of 4, as a key NOE correlation between the C-8 amide NH and the C-1a methyl group could be discerned.
Scheme 1.
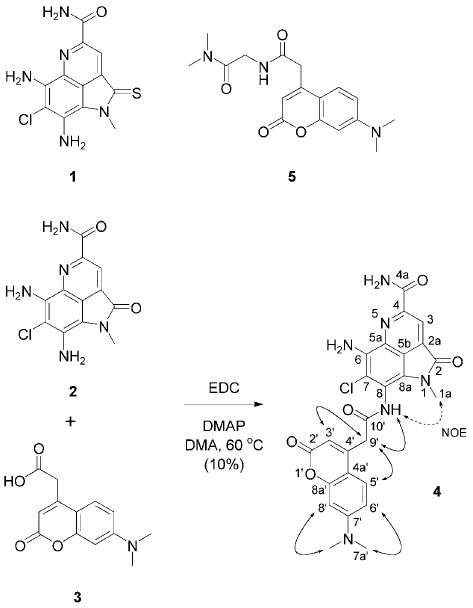
Structures of ammosamide A (1), ammosamide B (2), 7-dimethylaminocoumarin-4-acetic acid (3), ammosamide B probe 4 and control dye 5. Probe 4 was prepared by coupling 2 and 3. ROESY (bold arrows) and NOESY correlations (dotted arrows) used in the elucidation of the structure of probe 4 are shown. EDC=N′-(3-dimethylaminopropyl)-N-ethylcarbodiimide; DMAP=4-dimethylaminopyridine; DMA=N,N-dimethylacetamide.
Synthetic alkaloid 4 displayed properties desirable of an ammosamide B probe as it was cytotoxic, fluorescent, and selectively recognized by a monoclonal antibody. Cytotoxicity data collected with HCT-116 cells indicated that 4 (IC50 = 17 μm) maintained an essential proportion of the activity of 1 and 2 (IC50 = 320 nm). The probe contained three absorptions at λmax = 293, 353, 606 nm and fluoresced with an emission at λmax = 461 nm when excited at λ = 353 nm. The affinity of 4 to an antibody was also established, as microequilibrium dialysis (Harvard Apparatus) indicated that 4 bound to mouse immunoglobulin G (IgG), XRI-TF35, with a Kd of 1.1 ± 0.2 nm.
We then turned to evaluate the uptake of probe 4 in live mammalian cells using fluorescence microscopy. Upon addition of 1 mL of a 50 μm solution, probe 4 concentrated into HeLa, HCT-116, and PC-3 cells within minutes (Figure 1). Remarkably, the entire fluorescence from this aliquot was taken up from solution and localized within the cell. Titration studies indicated that the limit of this uptake was 0.24 ± 0.03 pmolcell−1, 0.21 ±0.02 pmolcell−1, and 0.27 ± 0.04 pmol cell−1 for HCT-116, HeLa, and PC-3 cells, respectively.
Figure 1.
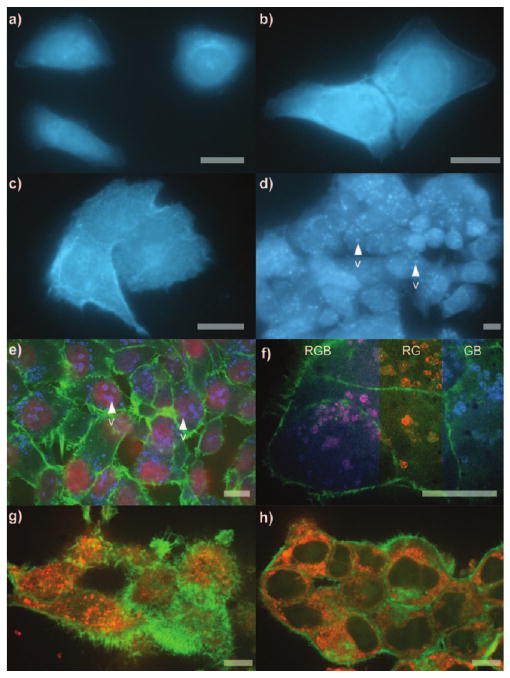
Cellular phenotype. a–c) Images from 106 cells incubated with 1 mL of 50 μm probe 4 in DMEM for 15 min. a) HeLa cells. b) HCT-116 cells. c) PC-3 cells. d) HeLa cells in (a) after incubation at 37°C with 5% CO2 for 12 h. e) Three-colored confocal micrographs of the cells in (d) after fixation and staining of the nucleus with Syto608[9a] (R), actin with FITC-phalloidin,[9b] (G) and probe 4 (B). f) Cells in (d) after staining the lysosomes with LysoTracker Red DND-99[9c] (R), actin with FITC-phalloidin[9b] (G) and probe 4 (B). Color mixing of R and B channels overlap to form magenta as depicted by three color mixing (RGB). Individual R and B channels are shown by RG and GB composites. g,h) HCT-116 cells treated for 5 h with 1 mL of 50 μm probe 4 in DMEM per 106 cells, fixed, and then stained. Microtubules were stained with BODIPY 564/570 paclitaxel[9d] (R) and actin with FITC-phalloidin (G). Colors are denoted as (R)=red, (B)=blue, (G)=green. Bar denotes 10 μm.
Fluorescence microscopy was again employed to detail the intracellular localization of probe 4. In HeLa cells, the probe was observed throughout the cytosol with modest localization apparent within small vesicles (Figure 1a). Comparable uptake was observed in HCT-116 cells (Figure 1b). In PC-3 cells, the localization of probe 4 appeared less specific, appearing throughout the cytosol. After treatment for 15 min, the fluorescence from probe 4 was retained in cells even after repetitive washing with media, though control dye 5 was completely washed from all three cell lines and remained indistinguishable from untreated cells.
Pretreatment of cells with 1 mL of 10 μm ammosamide A (1) or 10 μm ammosamide B (2) in Dulbecco's modified Eagle medium (DMEM) reduced the uptake of probe 4, leading to images with approximately 10 % of the fluorescence intensity as those observed in Figure 1a–c (the relative effects of 1 and 2 were indistinguishable), thereby supporting the conclusion that 4 is a reliable mimetic of the ammosamides. Attempts to elute 4 from the cells in Figure 1a–c by the addition of 10–100 μm 1 or 2 failed, leaving cells with at least 90 % retention of their original fluorescence. Finally, the blue fluorescent stain in Figure 1a–c remained after formalin fixation and washing with 95% ethanol (conditions that typically elute small-molecule ligands from the cell). The combination of these observations suggests that the uptake of probe 4 was accompanied by either a very strong noncovalent or covalent interaction.
After 12 h of incubation at 37 °C under a 5 % CO2 atmosphere, the blue fluorescence from probe 4 was vesiculated (Figure 1d). This response occurred after a 15 min treatment with probe 4, followed immediately by washing and incubation of the cells in fresh media for 12 h. Confocal microscopy of cells costained with a panel of organelle stains (Figure 1e) indicated that that the localization occurred in the lysosomes. In particular, the overlap of red fluorescence from the red lysosome stain, LysoTracker Red DND-99,[9c] and the blue fluorescence from probe 4 provided compelling evidence in support of this observation (Figure 1f).
The effect of probe 4 on the cell cycle was next evaluated using fluorescence activated cell sorting (FACS) analysis. Unsynchronized cells were halted at G1, G2, and during mitosis (Figure 2a). Cells synchronized with l-mimosine and treated at G1-phase with probe 4 were halted at the G1/S progression (Figure 2b). Cells synchronized with thymidine and treated with probe 4 during S-phase were inhibited during G2 and mitosis (Figure 2c). Comparable cell cycle data was also obtained from studies on ammosamide A (1; Figure 2a–c), indicating that probe 4 provided an accurate representation of the bioactivity. The complexity of this inhibition suggested the involvement of either multiple cell cycle regulators or a regulator that was involved in multiple stages of the cell cycle.
Figure 2.
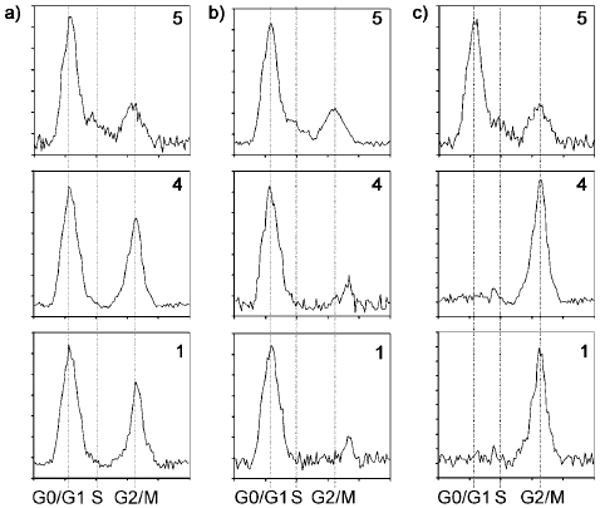
Cell cycle response. HCT-116 cells were treated with either 50 μm control dye 5 (normal cell proliferation), probe 4 or ammosamide A (1) in DMEM per 106 cells. a) Unsynchronized HCT-116 cells. b) HCT-116 cells synchronized and treated at G0 and incubated for 12 h. c) HCT-116 cells synchronized and treated at S and incubated for 12 h.
Taking advantage of the IAF tag's dual functionality, probe 4 was also applied to screen lysates of HCT-116 cells for protein targets. Co-immunoprecipitation (co-IP) was conducted from the lysate of 108 cells treated with 5 μm 4 for 12 h. The cells were scraped from the plate, concentrated by centrifugation at 300 rpm at 4°C for 5 min, washed three times with PBS pH 7.2 (5 mL), and lysed in PBS pH 7.2 (0.5 mL) containing protease inhibitor cocktail (Roche) by agitation through a 30 gauge needle. The crude lysate was centrifuged at 13000 rpm for 5 min at 4°C to remove insoluble matter and subjected to spin dialysis on a 9 kDa spin filter (iCON, Pierce Biotechnology) to concentrate the lysate to approximately 1 mgmL−1 total protein content. Samples of the resulting lysate were precipitated with Affigel Hz containing 12.5 mgmL−1 of the anti-dye XRI-TF35 mAb. After binding, the resin was washed repetitively with PBS pH 7.2 at 4 °C, and protein was then eluted by treatment with 0.1m Tris-Cl pH 6.8 containing 5 μm (lane L2, Figure 3) or 50 μm 3 (lane L3, Figure 3). Over five repetitions, a band at approximately 220 kDa appeared after eluting the resin with media containing 3 as a vehicle to release bound protein. Western blot analysis with an alkaline phosphatase conjugated anti-mouse polyclonal antibody (Pierce) indicated that this band did not contain fragments of the XRI-TF35 mAb. The band in lane L3 (Figure 3) was excised and submitted to LC/MS/MS protein ID analysis, which revealed a protein most similar to those in the myosin family, with 22–28% coverage of the amino acid residues.
Figure 3.
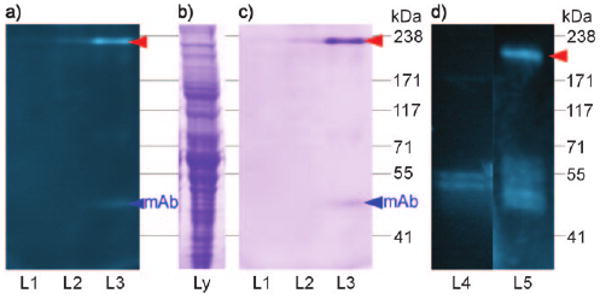
Co-immunoprecipitation (co-IP) studies. a) A 3–8% Trisacetate SDS-PAGE gel depicting fluorescent bands arising from the co-IP of lysate from 108 HCT-116 cells treated with probe 4 and Affigel Hz resin containing 12.5 mg mL−1 of XRI-TF35 mAb. After incubation for 12 h and multiple washings with PBS pH 7.2 at 4°C, the bound protein was eluted from XRI-TF35-Affigel Hz resin with 0.1 m Tris-Cl pH 6.8 (L1), 5 μm 3 in 0.1 m Tris-Cl pH 6.8 (L2), or 50 μm 3 in 0.1 m Tris-Cl pH 6.8 (L3) at 23 °C. b) HCT-116 lysate stained with GelCode blue. c) GelCode blue staining of the gel in (a). d) A 4% Tris-glycine SDS-PAGE gel depicting fluorescent bands from the co-IP of a 50 μg mL−1 sample of rabbit skeletal myosin that was incubated with 10 μm control 5 in PBS pH 7.2 (L4) or 10 μm probe 4 in PBS pH 7.2 (L5) and Affigel Hz resin containing 12.5 mg mL−1 of XRI-TF35 mAb. The bands in L4 and L5 were obtained after multiple washings of the resin with PBS pH 7.2 at 4°C and subsequent elution of the bound protein with 50 μm 3 in PBS pH 7.2 at 23 °C. Red arrow denotes bands of interest.
The immunoprecipitation experiment was repeated using rabbit skeletal muscle myosin. A fluorescent band at approximately 200 kDa corresponding to the heavy chain of myosin was co-immunoprecipitated from these solutions (L5, Figure 3d), employing the same methods used to isolate myosin from HCT-116 cells (Figure 3a–c). In fact, probe 4 was effective at fluorescently labeling this protein with an yield of 35 ± 5% and 82 ± 3% following treatment with 100 μL of 10 μm 4 and 100 μL of 50 μm 4 in DMEM for 2 h at room temperature.[10] The yield of this labeling remained within 5% deviation after spin dialysis on a 5 kDa molecular-weight cutoff filter (Centricon) or by microequilibrium dialysis (Harvard Apparatus), indicating that probe 4 did not diffuse from the protein. Furthermore, control experimentation with 5 did not return myosin, as determined by fluorescent gel analysis (L4, Figure 3d). The fluorescence from 4 was retained in the myosin band after SDS-PAGE gel analysis. Probe 4 (or the coumarin dye in probe 4) appeared to have attached to myosin. Ammosamide A (1) and ammosamide B (2) also stained rabbit muscle myosin as determined by the uptake of absorption at λmax = 580 nm for protein treated with 1 and λmax = 520 nm for protein treated with 2. Combined with the fact that neither dye 3 nor control dye 5 yielded a myosin conjugate, these data suggest that functionality within the ammosamide structure alone was responsible for targeting myosin.
Given the role of myosin in cytoskeletal structuring,[11] the effect of 4 on actin and microtubule assembly was explored. HCT-116 cells treated with probe 4 were fibrotic and contained greater than a ten-fold increase in actin filaments near their plasma membrane (Figure 1g). Marked microtubule depolymerization, apparent from the formation of aggregates throughout the cell, was observed as well (Figure 1g,h). The combination of these effects could arise from a lack in geometrical assembly of both actin and/or microtubule fragments resulting from the modification of myosin.[12]
Histological studies were pursued to evaluate whether binding of probe 4 was restricted to myosin II in skeletal muscle. A procedure was developed that allowed mouse tissue to be stained with 4 under conditions in which unbound materials, such as control 5, could be washed from the tissue (Figure 4o). Muscle was indeed a primary target, as evidenced by staining of smooth muscle (m, Figure 4g,n), skeletal muscle (Figure 4k), cardiac muscle (my, Figure 4h), and smooth muscle in blood vessels (s in Figure 4a, e in Figure 4f, pv in Figure 4j). However, muscle cells were not the only cell type that was stained with 4. High concentrations of 4 were found in epithelial cells (te, Figure 4b), erythro-blasts (ei, Figure 4c), adipocytes (a, Figure 4f), nerve cells (py, Figure 4d,e), dermal cells (de and ed, Figure 4m), as well as in the lamina propria (lp, Figure 4g), villi (v, Figure 4g), intestinal crypts (cp, Figure 4g), connecting tubules (ct, Figure 4i), bronchioles (br, Figure 4j) and islets of Langerhans (iL, Figure 4l).
Figure 4.
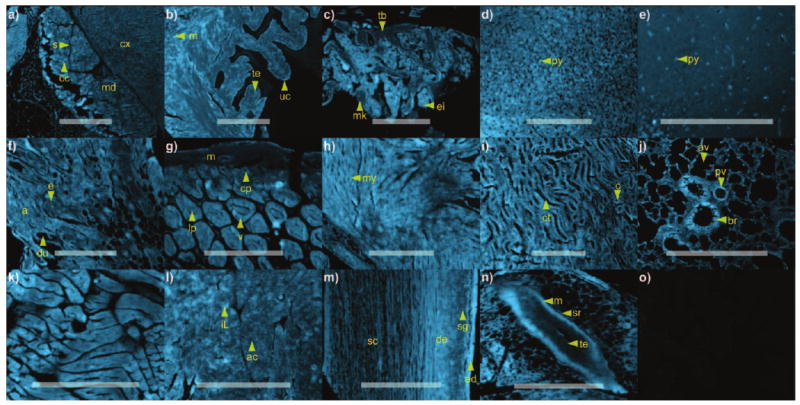
Histological analysis. A microarray containing select Mus musculus tissues[13] was treated for 4 h with 2 mL of 1 μm probe 4 in PBS pH 7.2. Images from each tissue section were collected using an identical exposure, thereby allowing the intensity to be compared between each image. Tissues stained with probe 4: a) adrenal gland, b) bladder, c) bone marrow, d) cerebellum, e) cerebral cortex, f) breast, g) intestine, h) heart, i) kidney, j) lung, k) skeletal muscle, l) pancreas, m) skin, and n) spleen. o) Control experimentation was conducted in parallel by treating the tissue microarray under the identical conditions used to collect the images in (a–m) with 1 μm control dye 5 in PBS pH 7.2. For all panels, the background from 5 was not visible under the exposure time used for image collection, as demonstrated by the uptake control probe 5 in heart tissue. Anatomical denominations: a=adipose cells, ac=acini, av=alveoli, br=bronchiole, c=capillaries, cc=chrimaffin cells, cp=crypts, ct=connecting tubules, cx=cortex, de=dermis, du=ducts, e=epithelial cells, ed=epidermis, ei=erythroid island, iL=islet of Langerhans, lp=lamina propria, m=muscularis, md=medulla, mk=megakaryocyte, my=myocytes, pv=pulminary vetricle, py=pyramidal cells, s=sinusoids, sc=subcutis, sg=sebaceous glands, sr=serosa, tb=trabecular bone, te=transitional epithelium, uc=umbrella cells, v=villus. Bars denote 1 mm.
To summarize, a dual affinity-fluorescent label was used to prepare a single analogue of ammosamide B (2). This IAF-labeled analogue 4 was used to identify cell uptake and localization of its conjugated natural product, as verified by blocking and control experimentation. Subsequent cell cycle studies and activity assays indicated that probe 4 and the parent ammosamides display comparable bioactivity. A 220 kDa protein from HCT-116 cell lysates was identified by means of co-immunoprecipitation. LC/MS/MS protein analysis suggested that this protein arose from myosin, which was subsequently confirmed by in vitro labeling of an authentic sample of myosin II with probe 4. Further confocal microscopic studies showed that exposure to 4 delivered hyperfragmented actin fibers and unassembled clusters of microtubules. These observations, as well as the translation of 4 over time into lysosomes, were consistent with a loss of myosin function.[14] Histological analyses demonstrated that 4 was not limited to skeletal muscle, indicating that it likely interacts with several of the myosin families.
This work has revealed a novel means in which a marine natural product interacts with the cytoskeleton by targeting the motor protein myosin. Recent studies have shown that screening of small-molecule libraries can identify synthetic materials, such as blebbistatin, that display high affinity to select isoforms of myosin II.[15] Using a cocrystal structure of (S)-(−)-blebbistatin bound to myosin II,[16] ammosamide A (1), ammosamide B (2), and probe 4 docked into the blebbistatin binding pocket. Probe 4 was capable of forming interactions with key residues Tyr261, Ser456, and Gln634 within the (S)-(−)-blebbistatin binding pocket (Figure 5a). The pendant coumarin label adopted a position comparable to the phenyl residue in (S)-(−)-blebbistatin, extending into a channel with the potential to form hydrogen-bonding interactions with Lys 265 and Asp 590 (Figure 5b). Though probe 4 may fit within the (S)-(−)-blebbistatin binding pocket, detailed studies are required to determine the actual mechanism by which the probe binds to and labels myosin.
Figure 5.
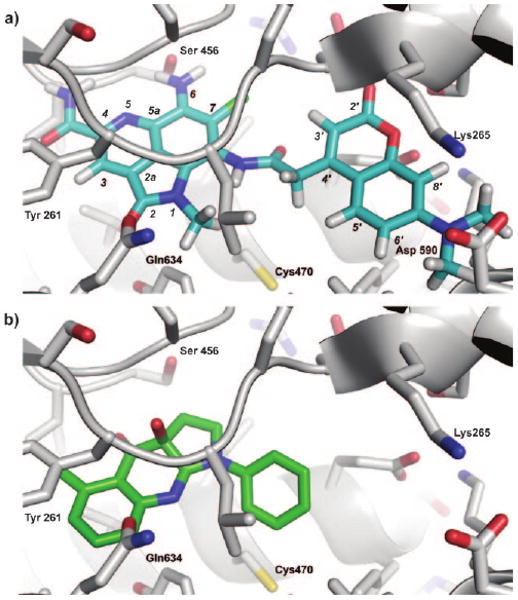
Docking studies. a) A depiction of ammosamide probe 4 (blue) docked in Dictyostelium discoideum myosin II. b) The binding of (S)-(−)-blebbistatin (green) within the same pocket of Dictyostelium discoideum myosin II. Docking was conducted with Autodock 3 using the coordinates from PDB accession number 3bz7.[16]
Studies are underway to evaluate the role that the ammosamides play in cell cycle regulation, cytokinesis, and cell migration. Like blebbistatin and its analogues,[17] the ammosamides may serve as tools to probe a variety of processes regulated by myosin. Fluorescent probe 4 itself, which binds to myosin, could find use in certain physiological and chemical studies.[18] In the context of cancer research, the ability to prepare an active fluorescent probe from the ammosamides offers a unique opportunity to visualize the downstream cytoskeletal regulatory events that arise from the modification of myosin. In this way, the suitability of members of the myosin family as chemotherapeutic targets can be further validated.[15b]
Supplementary Material
Footnotes
Supporting information for this article is available on the WWW under http://dx.doi.org/10.1002/anie.200804107.
This work was the result of financial support from the US National Cancer Institute (CA44848 to W.F.). S.P.G. is grateful to the Fundação para a Ciência e Tecnologia, Portugal, for a postdoc fellowship. The authors thank Sara Kelly (Scripps Institution of Oceanography) for cell culturing, Timothy Foley and Michael D. Burkart (University of California at San Diego) for assistance with the production of the XRI-TF35 mAb resin, and Qishan Lin (CFG at University of Albany) for protein ID analyses.
References
- 1.a) Reayi A, Arya P. Curr Opin Chem Biol. 2005;9:240–247. doi: 10.1016/j.cbpa.2005.04.007. [DOI] [PubMed] [Google Scholar]; b) Sudakin V, Yen TJ. BioDrugs. 2007;21:225. doi: 10.2165/00063030-200721040-00003. [DOI] [PubMed] [Google Scholar]; c) Liaw TY, Chang MH, Kavallaris M. Curr Drug Targets. 2007;8:739. doi: 10.2174/138945007780830836. [DOI] [PubMed] [Google Scholar]
- 2.a) Christian OE, Compton J, Christian KR, Mooberry SL, Valeriote FA, Crews P. J Nat Prod. 2005;68:1592. doi: 10.1021/np050293f. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Senderowicz AM, Kaur G, Sainz E, Laing C, Inman WD, Rodriguez J, Crews P, Malspeis L, Grever MR, Sausville EA, Duncan KKL. Natl Cancer Inst J. 1995;87:46. doi: 10.1093/jnci/87.1.46. [DOI] [PubMed] [Google Scholar]
- 3.a) Simmons TL, Andrianasolo E, McPhail K, Flatt P, Gerwick WH. Mol Cancer Ther. 2005;4:333. [PubMed] [Google Scholar]; b) Hamel E. Pharmacol Ther. 1992;55:31. doi: 10.1016/0163-7258(92)90028-x. [DOI] [PubMed] [Google Scholar]
- 4.Forsyth CJ, Ying L, Chen J, La Clair JJ. J Am Chem Soc. 2006;128:3858. doi: 10.1021/ja057087e. [DOI] [PubMed] [Google Scholar]
- 5.Fenical W, Jensen PR. Nat Chem Biol. 2006;2:666. doi: 10.1038/nchembio841. [DOI] [PubMed] [Google Scholar]
- 6.Hughes CC, MacMillan JB, Gaudêncio SP, Jensen PR, Fenical W. Angew Chem. 2009;121:739. doi: 10.1002/anie.200804890. [DOI] [PMC free article] [PubMed] [Google Scholar]; Angew Chem Int Ed. 2009;48:725. [Google Scholar]
- 7.Alexander MD, Burkart MD, Leonard MS, Portonovo P, Liang B, Ding X, Joullie MM, Gulledge BM, Aggen JB, Chamberlin AR, Sandler J, Fenical W, Cui J, Gharpure SJ, Polosukhin A, Zhang HR, Evans PA, Richardson AD, Harper MK, Ireland CM, Vong BG, Brady TP, Theodorakis EA, La Clair JJ. ChemBioChem. 2006;7:409. doi: 10.1002/cbic.200500466. [DOI] [PubMed] [Google Scholar]
- 8.Given the instability of the thiolactam in ammosamide A (1), the probe was constructed from ammosamide B (2).
- 9.a) Antinozzi PA, Garcia-Diaz A, Hu C, Rothman JE. Proc Natl Acad Sci USA. 2006;103:3698. doi: 10.1073/pnas.0510521103. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Wulf E, Deboben A, Bautz FA, Faulstich H, Wieland T. Proc Natl Acad Sci USA. 1979;76:4498. doi: 10.1073/pnas.76.9.4498. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Sorensen CE, Novak I. J Biol Chem. 2001;276:32925. doi: 10.1074/jbc.M103313200. [DOI] [PubMed] [Google Scholar]; d) Bicamumpaka C, Page M. Int J Mol Med. 1998;2:161. doi: 10.3892/ijmm.2.2.161. [DOI] [PubMed] [Google Scholar]
- 10.The yield of this reaction was based on the relative fluorescent uptake of 4 after repetitive removal by dialysis on a 9 kDa spin filter (iCON, Pierce Biotechnology).
- 11.a) Hehnly H, Stamnes M. FEBS Lett. 2007;581:2112. doi: 10.1016/j.febslet.2007.01.094. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Sweeney HL, Houdusse A. Curr Opin Cell Biol. 2007;19:57. doi: 10.1016/j.ceb.2006.12.005. [DOI] [PubMed] [Google Scholar]
- 12.a) Wu X, Xiang X, Hammer JA. Trends Cell Biol. 2006;16:135. doi: 10.1016/j.tcb.2006.01.004. [DOI] [PubMed] [Google Scholar]; b) Bement WM, Yu HY, Burkel BM, Vaughan EM, Clark AG. Curr Opin Cell Biol. 2007;19:95. doi: 10.1016/j.ceb.2006.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.a) Brown LA, Huntsman DJ. J Mol Histol. 2007;38:151. doi: 10.1007/s10735-006-9069-y. [DOI] [PubMed] [Google Scholar]; b) Hewitt SM. Methods Enzymol. 2006;410:400. doi: 10.1016/S0076-6879(06)10020-8. [DOI] [PubMed] [Google Scholar]
- 14.a) McNeil PL. J Cell Sci. 2002;115:873. doi: 10.1242/jcs.115.5.873. [DOI] [PubMed] [Google Scholar]; b) Apodaca G. Traffic. 2001;2:149. doi: 10.1034/j.1600-0854.2001.020301.x. [DOI] [PubMed] [Google Scholar]; c) Araki N. Front Biosci. 2006;11:1479. doi: 10.2741/1897. [DOI] [PubMed] [Google Scholar]
- 15.a) Cheung A, Dantzig JA, Hollingworth S, Baylor SM, Goldman YE, Mitchison TJ, Straight AF. Nat Cell Biol. 2002;4:83. doi: 10.1038/ncb734. [DOI] [PubMed] [Google Scholar]; b) Ramamurthy B, Yengo CM, Straight AF, Mitchison TJ, Sweeney HL. Biochemistry. 2004;43:14832. doi: 10.1021/bi0490284. [DOI] [PubMed] [Google Scholar]
- 16.Lucas-Lopez C, Allingham JS, Lebl T, Lawson CP, Brenk R, Sellers JR, Rayment I, Westwood NJ. Org Biomol Chem. 2008;6:2076. doi: 10.1039/b801223g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.a) Haviv L, Gillo D, Backouche F, Bernheim-Groswasser A. J Mol Biol. 2008;375:325. doi: 10.1016/j.jmb.2007.09.066. [DOI] [PubMed] [Google Scholar]; b) Wang HH, Tanaka H, Qin X, Zhao T, Ye LH, Okagaki T, Katayama T, Nakamura A, Ishikawa R, Thatcher SE, Wright GL, Kohama K. Am J Physiol Heart Circ Physiol. 2008;294:H2060. doi: 10.1152/ajpheart.00970.2007. [DOI] [PubMed] [Google Scholar]; c) Martens JC, Radmacher M. Pfluegers Arch. 2008;456:95. doi: 10.1007/s00424-007-0419-8. [DOI] [PubMed] [Google Scholar]; d) Eddinger TJ, Meer DP, Miner AS, Meehl J, Rovner AS, Ratz PH. J Pharmacol Exp Ther. 2007;320:865. doi: 10.1124/jpet.106.109363. [DOI] [PubMed] [Google Scholar]; e) Dou Y, Arlock P, Arner A. Am J Physiol Cell Physiol. 2007;293:C1148. doi: 10.1152/ajpcell.00551.2006. [DOI] [PubMed] [Google Scholar]
- 18.a) Farman GP, Tachampa K, Mateja R, Cazorla O, Lacampagne A, de Tombe PP. Pflugers Arch. 2008;445:995. doi: 10.1007/s00424-007-0375-3. [DOI] [PubMed] [Google Scholar]; b) Rehfeldt E, Engler AJ, Eckhardt A, Ahmed F, Discher DE. Adv Drug Deliv Rev. 2007;10:1329. doi: 10.1016/j.addr.2007.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


