Abstract
Little is known about how differentiating cells reorganize their cellular structure to perform specialized physiological functions. MIST1, an evolutionarily conserved transcription factor, is required for the formation of large, specialized secretory vesicles in gastric zymogenic (chief) cells (ZCs) as they differentiate from their mucous neck cell progenitors. Here, we show that MIST1 binds to highly conserved CATATG E-boxes to directly activate transcription of 6 genes, including those encoding the small GTPases RAB26 and RAB3D. We next show that RAB26 and RAB3D expression is significantly downregulated in Mist1−/− ZCs, suggesting that MIST1 establishes large secretory granules by inducing RAB transcription. To test this hypothesis, we transfected human gastric cancer cell lines stably expressing MIST1 with red fluorescent protein (RFP)-tagged pepsinogen C, a key secretory product of ZCs. Those cells upregulate expression of RAB26 and RAB3D to form large secretory granules, whereas control, non-MIST1-expressing cells do not. Moreover, granule formation in MIST1-expressing cells requires RAB activity because treatment with a RAB prenylation inhibitor and transfection of dominant negative RAB26 abrogate granule formation. Together, our data establish the molecular process by which a transcription factor can directly induce fundamental cellular architecture changes by increasing transcription of specific cellular effectors that act to organize a unique subcellular compartment.
Developmentally regulated transcription factors (TFs) play central roles in cell fate specification and differentiation of all cell types. During the complex process of specified cellular differentiation, certain TFs are turned on only at the final steps of the transcription regulation hierarchy. Presumably, those TFs directly activate major cell effector genes and in turn govern the establishment of a differentiated cell's designated morphology and function. There is emerging evidence that there might indeed be a limited number of TFs that are employed in diverse tissues to induce specific gene cassettes that regulate cell structure. For example, it was recently shown that differentiation in general, no matter the tissue, induced preferential expression of the same types of genes that govern secretion and communication with extracellular space (13). Further, it has been shown that X-box binding protein 1 (XBP1), a transcription factor that is also developmentally regulated and required for wholesale changes in cell structure in certain cell lineages, directly activates multiple secretory pathway genes that help establish the abundant rough endoplasmic reticulum (rER) and mitochondria needed by specialized secretory cells, such as plasma cells, pancreatic acinar cells, and intestinal Paneth cells (30, 33, 56, 59). Another example of a developmentally important TF that regulates cell structure is TFEB, a basic helix-loop-helix (bHLH) recently shown to upregulate a cohort of target genes that specifically regulate lysosome function and biogenesis (54).
The corpus of the adult murine stomach is a useful system for studying the role of developmentally regulated TFs in coordinating cell structural changes because the gastric epithelium comprises multiple secretory cell lineages that are constantly renewed in a spatiotemporally organized fashion (6, 28, 39). The zymogenic (also known as chief) cell (ZC) lineage occupies the base of the gastric unit, migrating from the stem cell zone and passing first through a distinct mucous neck cell progenitor phase before rapid terminal differentiation, including dramatic expansion of the rER and apical accumulation of large secretory vesicles filled with pepsinogen and other digestive enzymes (5, 29).
The bHLH transcription factor MIST1 is critical for ZC architectural maturation (see Fig. 1) (50). MIST1 is a developmentally regulated and highly cell lineage-specific TF, with an onset of expression only during terminal differentiation of—along with ZCs—a few secretory cells in mammals (26, 48, 49). MIST1-expressing cells, ranging from immunoglobulin-secreting plasma cells (4) to alveolar breast lobular cells (68), are scattered in diverse tissues and have little in common either in their developmental origins or in the specific substances they elaborate. However, they do share a specialized physiological function, high-capacity secretion of proteins, indicating that MIST1 may be the transcriptional architect initiating the subset of structural changes required for such functionality. Indeed, the loss of MIST1 leads to smaller cell cytoplasms, with smaller secretory granules, reduced secretory protein stores, and a decrease in secretory capacity (5, 34, 50). Modulating cell architecture is likely a critical feature of normal development because MIST1 is a highly conserved gene in organisms from Drosophila melanogaster to zebrafish to mammals, reflected not only by the high degree of homology among MIST1 orthologs but also the specific sequences of the genomic cis elements they bind, CATATG-type E-boxes (18-20, 45, 46).
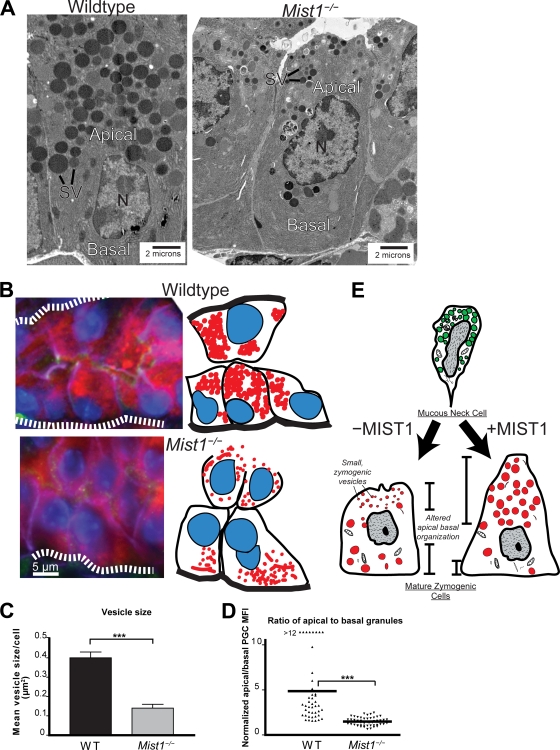
FIG. 1.
MIST1 is required for development and maintenance of large zymogenic secretory granules and apical-basal orientation. (A) TEM of ZCs from wild-type and Mist1−/− mice. Note the central position of the nucleus, along with reduced sizes of mature apical secretory vesicles in the Mist1−/− ZC. SV, mature secretory vesicles; N, nucleus. (B) The left panels show epifluorescence of wild-type and Mist1−/− ZCs in the base of a gastric unit (oriented with the stomach lumen to the left and the base to the right). Also shown is the cell and granule orientation observed by immunolabeling, with anti-GIF (red) labeling granules, anti-β-actin (green) labeling mesenchymal cells at base of cells and gastric unit lumen, anti-E-cadherin (purple) labeling basolateral cell-cell borders, and bisbenzimide (blue) labeling nuclei. The dashed white lines represent basement membranes. The right panels are cartoons of fluorescently labeled cells. Note that Mist1−/− ZCs have nuclei closer to the lumen, with fewer, more basally localized secretory granules. (C) Secretory vesicle areas from multiple ZCs were quantified from TEM photomicrographs. For all figures, ** represents a P value of <0.01 and *** represents a P value of <0.001. WT, wild type. (D) Multiple ZCs from multiple mice were quantified for intracellular apical (lumen to nucleus) and basal (base to nucleus) secretory granule immunofluorescence in immunolabeling experiments like those described for panel B. Each point is the ratio of apical to basal mean fluorescence intensity determined by anti-PGC (PGC being a principal component of zymogenic granules) in an individual ZC. Note that wild-type ZCs have predominately apical distribution of granules, whereas apical and basal granule distributions are nearly equal in Mist1−/− ZCs. (E) Cartoon showing ZCs arising from mucous neck cell progenitors in the neck of the gastric unit. In the absence of MIST1, ZCs migrating into the base exhibit apical-basal organization and granule formation/maintenance deficits (see also reference 50).
To coordinate specific changes in cell structure, MIST1 must directly activate transcription of a group of cellular effector genes that coordinate structural changes. This cassette of structure-modifying genes would likely be conserved across tissues and species. However, no MIST1 targets that might coordinate such structural changes have yet been identified. Ras-like small GTPases are important regulators of vesicular formation and transport in eukaryotic cells, making them potential structure-regulating gene products (15, 17). It has been shown that Rab3d expression is decreased in Mist1−/− pancreatic acinar cells, but Rab3d has not yet been shown to be a direct target of MIST1, and Rab3d is expressed in non-MIST1-expressing cells as well. Furthermore, transgenic expression of Rab3d was insufficient to restore the defects caused by the loss of MIST1 function in pancreatic acinar cells (26), indicating that other, as yet unidentified MIST1 targets must also play a role. A RAB paralogous to RAB3D, RAB26, has not been extensively studied, but its expression is restricted to a limited number of highly secretory cells, for which it has been proposed to be involved in regulated vesicular secretion (40, 64, 67).
Here, we show that MIST1 is sufficient to upregulate 16 genes in 2 different gastric cell lines. We further show that it directly associates with CATATG E-boxes in 6 of those genes that have highly evolutionarily conserved CATATG E-boxes within the first intron. Two MIST1-induced genes, RAB3D and RAB26, stood out as representing a possible mechanism by which MIST1, acting at the level of transcription, could regulate formation of exocrine secretory granules in differentiating cells. We show that RAB3D and RAB26 are expressed in gastric ZCs in the corpus of the mouse stomach in a MIST1-dependent manner. We next developed a system to test the functional importance of MIST1 induction of these RABs. We show that gastric epithelial cells stably expressing MIST1, but not control cell lines, produce large ZC-like secretory granules when they are transfected with red fluorescent protein (RFP)-tagged pepsinogen C (PGC). Expression levels of RAB26 and RAB3D were also specifically induced in those cell lines. Finally, we show that formation of large granules in these cells is dependent on RAB activity because granule formation is abrogated by (i) inhibition of RAB26 and RAB3D function by disrupting RAB prenylation and (ii) transfection with a dominant negative RAB26 construct. We therefore propose that MIST1 regulates large-secretory-granule formation, a key aspect of differentiated secretory cell function, via activating transcription of RAB26 and RAB3D. Together, our data demonstrate how a developmentally regulated TF can induce secretory-cell architectural changes by transcriptionally regulating specific membrane trafficking genes.
MATERIALS AND METHODS
Mice.
All experiments involving animals were performed according to protocols approved by the Washington University School of Medicine Animal Studies Committee. Germ line Mist1−/− mice were generated as described previously (49) and maintained in a specific-pathogen-free barrier facility. Control mice were the littermates of Mist1−/− mice resulting from heterozygote-by-heterozygote crosses.
Immunofluorescence.
Stomachs were prepared and stained as described previously (50). Briefly, stomachs were inflated with freshly prepared methacarn fixative and suspended in fixative for 15 to 30 min at room temperature, followed by multiple rinses in 70% ethyl alcohol (EtOH), arrangement in 2% agar in a tissue cassette, and routine paraffin processing. Sections (5 μm) were deparaffinized and rehydrated, and then antigen retrieval was performed by boiling in 50 mM Tris-HCl, pH 9.0. Slides were blocked in 1% bovine serum albumin (BSA) and 0.3% Triton X-100 in phosphate-buffered saline (PBS) and then incubated in primary antibodies followed by secondary antibodies (see below). Slides were incubated for 5 min in 1 μg/ml bisbenzimide (Invitrogen, Carlsbad, CA) prior to mounting in 1:1 glycerol-PBS.
For immunofluorescence analysis of cultured cells, cells were grown in Lab-Tek Chamber Slide 4-well Permanox slides (Thermo Fisher Scientific, Rochester, NY) and fixed in 4% paraformaldehyde for 10 min at room temperature. For antibody staining, slides were permeabilized by 0.1% Triton X-100 in PBS and blocked in 2% BSA and 0.05% Triton X-100 in PBS. Fluorescence microscopy and imaging were performed using a Zeiss Axiovert 200 microscope with Axiocam MRM camera at room temperature. Optical sectioning was performed using the Apotome adaptor following calibration. In most cases, 1-μm sections were taken from the cell-substrate interface to the full height of the cell. Contrast (maximal, minimal, and midtone) adjustment and fluorescent channel overlay and pseudocoloring were performed with Photoshop (Adobe Systems, San Jose, CA). All adjustments were performed on the entire image equally. Cartoon cell traces and illustrations were produced using Illustrator (Adobe Systems, San Jose, CA).
The following primary antibodies were used for immunostaining: goat anti-human gastric intrinsic factor (GIF) (1:2,000; gift of David Alpers, Washington University), sheep anti-PGC (1:10,000; Abcam), and rabbit antiactin (1:200; gift of Thaddeus Stappenbeck [clone AC-40; Sigma, St. Louis, MO]). Secondary antibodies used were Alexa Fluor 488-, 594-, and 647-conjugated donkey anti-goat, anti-rabbit, anti-sheep, and anti-mouse antibodies (1:500; Invitrogen).
Immunofluorescence quantification.
Immunofluorescence quantification to determine cytoplasmic fluorescence intensity was performed with ImageJ software. For tissue sections, the cytoplasm was divided into regions fully apical and fully basal to the nucleus. Mean fluorescence intensity (MFI) was determined in each region after subtraction of background; background was determined by averaging PGC fluorescence in 4 parietal cells—which do not express PGC—in the same unit. Results from 51 cells from 7 gastric units in two separate knockout mice and 50 cells from 6 units in two control mice were compiled.
For cell line vesicle quantification, lines stably expressing MIST1 were transfected with PGC-RFP and given the appropriate treatment. Large-vesicle cells (defined as cells containing 3 or more vesicles, each ≥1.5 μm) and diffuse-vesicle cells (cells with no visible vesicles) and total PGC-RFP-transfected cells were scored. For enhanced green fluorescent protein (eGFP)-human T77N mutant RAB26 (hRAB26T77N) fluorescence intensity quantification, MIST1-stable cell lines were cotransfected with PGC-RFP and eGFP-hRAB26T77N. Sixteen-bit images captured in Zeiss Axiovision software were analyzed in ImageJ as follows. We determined mean green and red cytoplasmic fluorescence intensities for large-vesicle cells (as defined above) and diffuse-vesicle cells (defined by ≥135 intensity units above background and no vesicles ≥1 μm) in each region after subtraction from the median background (green and red fluorescence in an area with no cells) and normalization to average nuclear green fluorescence (nuclei were green due to expression of MIST1-eGFP) for ≥10 cell nuclei/field. A total of 245 cells were scored across 3 transfections.
EM.
For transmission electron microscopy (TEM) studies, stomachs were fixed, sectioned, stained, and imaged as described previously (50). For TEM of cell lines, cells were plated on Lab-Tek Chamber Slide 4-well Permanox slides (Thermo Fisher Scientific). After being rinsed in PBS, they were fixed in modified Karnovsky's fixative (2.5% glutaraldehyde, 2% paraformaldehyde in 0.1 mmol/liter cacodylate buffer). TEM thin sections were cut directly from cell cultures embedded on the original Permanox substrate. To quantify vesicle size, fields of MIST1-stable and eGFP control cells were randomly selected, digital images were captured at ×4,000 magnification, and ImageJ was used to determine areas of all vesicle profiles in all cells in the field in each TIFF image. Twenty cells were analyzed from each cell line (∼500 total vesicles scored).
Cell lines and transient transfection.
HGC-27 cells (HPACC, Porton Down, United Kingdom) were maintained at 37°C in 5% CO2 in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum, 1% nonessential amino acids, 1% glutamine, and 100 ng/ml each of penicillin and streptomycin. AGS cells (from ATCC, Manassas, VA) were grown in RPMI 1640 supplemented with 10% fetal bovine serum, 0.9% glutamine, 0.4% HEPES, 1% Na pyruvate, 2.5% glucose, and 100 ng/ml each of penicillin and streptomycin. Cells were passaged every 3 days by using trypsin-EDTA. For transient transfection, 1 × 106 AGS or HGC-27 cells were transfected via electroporation using Nucleofector I (Amaxa-Lonza, Basel, Switzerland), program B-023, and cell line transfection solution V. For each electroporation, 3 μg PGC-RFP (see below), 5 μg eGFP-hRAB26T77N (see below), or 3 μg pmaxGFP (Amaxa-Lonza) plasmid was used. Cells were replated on Lab-Tek Chamber Slide 4-well Permanox slides and routinely analyzed 24 h posttransfection.
GeneChip analysis.
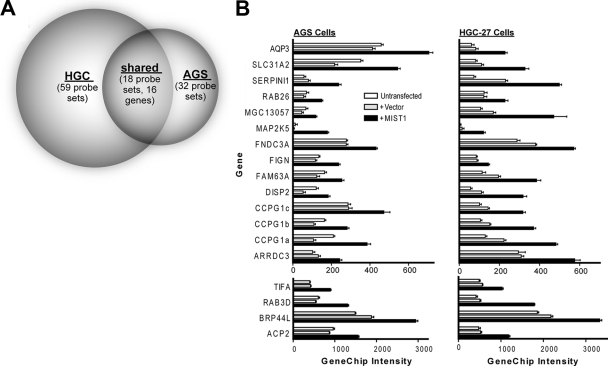
Untransfected HGC-27 and AGS control plates were grown to 70 to 80% confluence on 10-cm dishes. For cells transfected with either MIST1-eGFP or control eGFP vector, 1 × 106 AGS or HGC-27 cells were transfected with 1.5 μg pmaxGFP plus either 3 μg MIST1 cDNA or 3 μg empty vector by using the transfection conditions described above. The GFP-positive population of each transfectant was isolated by flow cytometric cell sorting using a FACSVantage (Becton-Dickinson, San Jose, CA) at 24 h posttransfection and then replated for an additional 24 h before harvesting. Cells were harvested with trypsin-EDTA. RNA was extracted by using a Qiagen RNeasy kit, and RNA quality was verified by visualization on an agarose gel. Five micrograms of total RNA was used to prepare labeled target by using Affymetrix GeneChip one-cycle target labeling reagents. Ten micrograms of labeled target cRNA was hybridized to HGU133_Plus_2 GeneChips. For both AGS and HGC-27 cells, expression profiles of MIST1-plus-pmaxGFP-transfected cells were compared with profiles of empty-vector-plus-pmaxGFP-transfected cells. An initial inclusive candidate MIST1 target list was generated by determining by dChip analysis (69) the transcripts that had a 90% confidence lower-bound fold-change increase of ≥1.2 and an intensity greater than 100 in 4 independent comparisons, combined as either MIST1-expressing AGS cells versus vector-transfected cells and versus untransfected cells or MIST1-expressing HGC-27 cells versus vector-transfected cells and versus untransfected cells. This list of 211 genes was then filtered to generate AGS and HGC-27 MIST1-induced transcripts by filtering genes whose expression intensity was <100 in MIST1-transfected cells and whose mean fold change in MIST1-transfected cells relative to either control population was <1.5. Thirty-two AGS probe sets and 59 HGC-27 probe sets met those criteria, and 18 of those, representing 16 individual genes, were shared (see Fig. 2A).
FIG. 2.
Transient transfection with MIST1 in AGS and HGC-27 gastric cell lines increases expression of a common cohort of genes. (A) Venn diagram showing the number of Affymetrix HGU133_Plus_2 GeneChip probe sets increased in two different gastric epithelial cell lines (HGC-27 and AGS) following transient transfection with MIST1. (B) The expression intensities on the GeneChips for the 18 probe sets (16 genes) that were increased in both cell lines relative to those of vector-transfected and untransfected controls.
Bioinformatic analysis of MIST1 targets.
The ECR browser (44) was used to analyze the human genomic sequence for each gene 5′ to the transcription start. Sequence was analyzed up to 50 kb or until an exon from the neighboring gene was reached. The first intron up to 50 kb was also analyzed. When alternate splice forms existed, the farthest 5′ exon documented was used and the intron following it was considered the first intron. CATATG sequences within analyzed sequences were identified using the dreg application in the EMBOSS suite. Conservation among species aligned in the ECR browser was then determined at each of these CATATG sites. Note that only human, chimp, cow, dog, rat, mouse, opossum, and chicken data were available in the ECR browser at the time of the analysis, but conservation undoubtedly extends across numerous species not diagrammed in Fig. 3. All 16 putative MIST1 targets were analyzed in this way. Six genes showed the conserved intronic pattern depicted in Fig. 3A and B. Seven genes showed scattered CATATG sequences or none, and any observed conservation was restricted to primates. Two genes (MAP2K5 and FIGN) were slight variants. The MAP2K5 gene was highly conserved throughout and showed sporadic CATATG sequences that were conserved with no single predominant site and no particular pattern to the species conservation (not shown). The FIGN gene has a short intron 1 and has two highly conserved CATATG sequences in the second intron (not shown). In addition to the 16 putative targets, we also analyzed the first introns of the following RAB family members for conserved CATATG sequences: RAB3A, -B, and -C; RAB27A and -B; RAB37; and RAB9A and -B. Many of those had no CATATG sites; none had sites conserved outside primates.
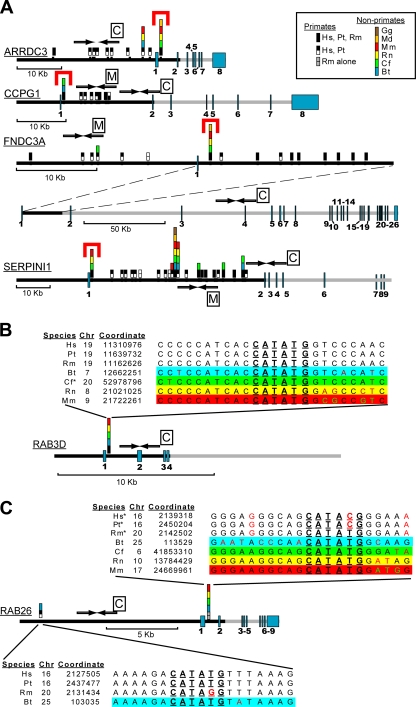
FIG. 3.
Several genes activated by MIST1 overexpression have evolutionarily conserved, first intronic canonical MIST1 E-box sequences. (A) All MIST1-type E-boxes (CATATG) were identified in the first introns and 5′ to transcription start for the 16 human genes from Fig. 2 (see Materials and Methods). Evolutionary conservation relative to the other vertebrate species listed in legend (species identified by 2-letter abbreviations of species names) was then determined. Genes (gray bars) are schematized to scale with numbered exons in blue and regions of genes analyzed in black. Note the dramatic conservation of CATATG sequences located within 1 kb of the end of the first exon in these 4 genes (red brackets). Primers for each of these conserved intronic sequences were analyzed for MIST1 binding (Fig. 4); control primers were generated to assess MIST1 binding to regions with no CATATG (amplicons indicated by arrows with boxed C's). In some cases, MIST1 binding to regions with nonconserved CATATG sequences (denoted by arrows with boxed M's) was also assessed. (B and C) Schematic as shown in panel A for the two RAB genes induced by MIST1 transfection but with detailed sequence alignment at each CATATG. *, consensus sequence runs on an antiparallel strand for the given species. Note that the conserved intronic CATATG in RAB26 in humans and chimpanzees has a single point mutation, destroying the E-box. The genomes of those species have another MIST1 binding site ∼11.5 kb upstream of the transcription start; bovine genomes have both intronic and 5′ sites. Gg, Gallus gallus; Hs, Homo sapiens; Pt, Pan troglodytes; Rm, Rhesus macaque; Md, Monodelphis domestica; Mm, Mus musculus; Rn, Rattus norvegicus; Cf, Canis lupus familiaris; Bt, Bos taurus.
Establishment of lines stably expressing MIST1-eGFP and eGFP.
A total of 1 × 106 HGC-27 cells were transfected with 3 μg MIST1-eGFP cDNA or 3 μg eGFP cDNA by using the transfection conditions described above. After 24 h, transfectants were replated at a limiting dilution under selection with 500 ng/μl Geneticin (Gibco-Invitrogen). Wells that grew out over 14 to 21 days were analyzed by flow cytometry for the level and homogeneity of GFP signal.
ChIP.
Chromatin immunoprecipitation (ChIP) was performed as described by Im and coworkers (22). Approximately 108 HGC-27 cells that stably express MIST1-GFP fusion proteins were harvested for one ChIP experiment. Fifteen microliters of MIST1 antiserum (rabbit polyclonal anti-human MIST1) or serum from the rabbit prior to immunization (preimmune control) together with protein A/G plus agarose (Santa Cruz Biotechnology, Santa Cruz, CA) was added to the cell lysate for immunoprecipitation. Regular PCR and quantitative real-time PCR (qRT-PCR) were performed (the sequences used and all other primer sequences are available upon request) to assess the quantity of genomic sequences immunoprecipitated by either preimmune control or MIST1 antiserum, as well as a 1:10 dilution of the cell extract prior to immunoprecipitation.
RAB inhibitor studies.
3-Pyridinyl ethylidene hydroxyl phosphonocarboxylate (3-PEHPC), an analog of the bisphosphonate drug risedronate, was generated as previously described (10). 3-PEHPC was stored in a stock solution of 106 mM at 4°C in aqueous solution at pH 6.3. For cell culture experiments, 3-PEHPC was added directly to the appropriate dilution in tissue culture media. Multiple concentrations were tested up to 5 mM, and effectiveness was determined by dispersion of transfected eGFP-RAB26 plasmid. Based on these experiments, a final concentration of 5 mM was used in all experiments described in the manuscript.
In situ hybridization.
Rab26 cDNA was obtained from OpenBiosystems (catalog no. MMM1013-98478848). Rab3d cDNA was obtained via amplification from a mouse stomach cDNA library and cloned into T-Easy vector (Promega, Madison, WI). Digoxigenin (DIG)-labeled RNA probes were synthesized via in vitro transcription following plasmid linearization (DIG RNA labeling mix, product no. 11277073910; Roche Applied Sciences, Indianapolis, IN). The antisense RNA probe was synthesized with T7 RNA polymerase, and the sense RNA probe was synthesized using Sp6 RNA polymerase. Adult B6 strain mice were perfused with 0.9% NaCl and 4% paraformaldehyde (PFA) sequentially, and the stomachs were removed and further fixed in 4% PFA at 4°C for 8 h or more. Tissue was blocked, frozen, and sectioned as previously described (50). Sections (20 μm) were dipped in water several times to remove OCT compound (Sakura Finetek, Torrance, CA), fixed in 4% PFA, treated with proteinase K, refixed in 4% PFA, and then treated with 0.1 M triethanolamine before incubation in hybridization buffer (50% formamide, 5× SSC [1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate], 1× Denhardt's solution, 250 μg/ml carrier yeast tRNA, 500 μg/ml salmon sperm DNA, 100 μg/ml heparin, 5 mM EDTA, 0.1% Tween 20) for 1 to 2 h at 65°C. The tissue sections were incubated with DIG-labeled RNA probes in hybridization buffer overnight at 65°C. After RNA hybridization, slides were washed in 2× SSC at 65°C for 15 min and then at room temperature for 5 min, followed by RNase (1 μg/ml in 2× SSC) incubation at 37°C for 30 min and washes with 2× SSC (room temperature) for 5 min twice and 0.2× SSC (60°C) for 30 min twice. DIG-RNA hybridized to tissue sections was visualized by incubating the slides first in blocking solution (1% BSA in PBS) for 1 h and with antidigoxigenin antibody (1:2,000; Roche) at 4°C overnight, followed by washing in PBT (PBS-0.1% Tween 20), equilibrating in alkaline phosphatase (AP) staining buffer (100 mM Tris, pH 9.5, 50 mM MgCl2, 100 mM NaCl, 0.1% Tween 20), and color development with nitroblue tetrazolium (NBT)-BCIP (5-bromo-4-chloro-3-indolylphosphate) (Roche) until a strong signal was detected in the sections hybridized to antisense probes; specificity was confirmed by lack of signal in sense-hybridized sections developed in the same experiments for the same length of time. Reactions were stopped by PBT, and sections were mounted in 1:1 glycerol-PBS.
Graphing and statistics.
All graphs and statistics were determined with GraphPad Prism, except the Venn diagram, which was generated using BioVenn (21), and then traced using Adobe Illustrator. Statistical analysis was, in the case of simple control-versus-experimental condition comparison, by Student's t test. Otherwise, significances were determined by a one-way analysis of variance (ANOVA) test with Dunnett's multiple-comparison correction.
LCM and qRT-PCR.
Preparation of stomach frozen sections for laser-capture microdissection (LCM) has been described previously (50). Briefly, stomachs were excised immediately following sacrifice, quickly flushed with room-temperature PBS, inflated by duodenal injection of OCT compound (Sakura Finetek, Torrance, CA), frozen in Cytocool II (Richard-Allen Scientific, Kalamazoo, MI), and cut into serial 7-μm-thick cryosections, which were mounted on Superfrost slides (Fisher Scientific), fixed in 70% EtOH, rehydrated in nuclease-free water (nuclease-free solutions from Ambion, Austin, TX), and then incubated in Alexa Fluor 488-conjugated Griffonia simplicifolia GS-II (diluted 1:500 in nuclease-free water) for 15 min. Sections were washed in nuclease-free water and dehydrated in graded ethanol followed by xylene. ZCs were identified as corpus cells that were basal to GS-II labeling and which did not show the dark silhouettes and characteristic shape of parietal cells following xylene dehydration. Four wild-type mice and 5 Mist1−/− mice were used for dissection (PixCell II LCM apparatus [7.5-μm spot diameter] and CapSure HS LCM caps; Arcturus, Mountain View, CA) to generate two caps per mouse. RNA was purified by PicoPure kit (Arcturus), and RNA integrity was confirmed by an Agilent 2100 Bioanalyzer (Palo Alto, CA). qRT-PCR was performed only on RNA that had sharp 18S and 28S bands. All RNA from each cap was treated with DNase I (Invitrogen) and then reverse transcribed using the SuperScript III (Invitrogen) standard protocol (most cDNA syntheses started with 10 ng of total RNA). Measurements of cDNA levels were performed by qRT-PCR using a Stratagene (La Jolla, CA) MX3000P detection system, and Absolute QPCR SYBR green mix (Thermo Scientific) fluorescence was used to quantify relative amplicon amounts of mouse RAB3D, RAB26, CCPG1, and 18S rRNA. Data analysis to allow expression of qRT-PCR data in terms of cycles above the background, normalized to 18S rRNA amplicon intensity, was performed as described in detail previously (5).
qRT-PCR with stable MIST1-expressing and eGFP control HGC-27 cells was performed after RNA isolation by using TRIzol-solubilized tissue extract (Invitrogen) following the manufacturer's protocol, quantification using the Beckman Coulter DU 640 spectrophotometer (Fullerton, CA), and cDNA synthesis as described previously (5) (1 μg of starting total RNA was used). Measurements of RAB3D, RAB26, and 18S rRNA cDNA levels were performed and analyzed as described above.
Plasmid preparation.
The phMist1-EGFP-C1 expression plasmid was constructed with the coding region of human MIST1 cDNA (IMAGE identification number 8322448; Open Biosystems, Huntsville, AL) followed by a 30-aa peptide linker added in frame to the amino terminus of eGFP in pEGFP-C1 (Clontech Laboratories, Inc., Mountain View, CA) by the restriction site-free PCR method of ribocloning (1; http://barnes1.wustl.edu/∼wayne/riboclon.htm) using Klentaq-LA enzyme (2) and accurate PCR conditions. The riboprimers used did not match the vector or target products, so they were first elongated on longer Band-Aid primer templates. The vector region was amplified by PCR using primers DNA3as and pEGFP-DNA3sb with V5s and EGFP-V5asb on pEGFP-C1 DNA. The hMIST1 target was PCR amplified using DNA3s and hMIST-DNA3b with V5as after sequential elongations on bandaids hMIST-V5b3, hMIST-V5b2, and hMIST-V5b1. The construct coding region pMT6-5 (5,416 bp) was verified correct by DNA sequencing.
The pcDNA3.1 PGCRFP fusion construct was generated using a modified sequence overlap extension PCR strategy. The PGC fragment was amplified by PCR from a pcDNA3.1PGC plasmid (a kind gift of Susan Guttentag, University of Pennsylvania [16]) with the T7 primer (forward) and a reverse primer that contained both the PGC 3′ end and the RFP 5′-end coding sequence, separated by a small linker sequence. The RFP sequence was amplified by PCR from a pCS2 Notch-RFP plasmid (kind gift of Rafi Kopan, Washington University), a forward primer (complementary to the reverse primer used to amplify the PGC fragment), and a reverse primer. The PGC and RFP fragments were then purified and mixed together with T7 and RFP reverse primer to amplify the PGC-RFP fusion fragment. This PCR fragment was then digested with EcoRI and XhoI and cloned into pcDNA3.1 vector, which was linearized with the same pair of restriction enzymes.
pEGFP-hMist1-C1 (designed by a method similar to that used for phMist1-EGFP-C1) was monomerized to pmEGFP-hMist1-C1 by site-directed mutagenesis using reverse and forward primers (58) to convert leucine 222 to lysine and introduce a new restriction site, that for AflII. The monomeric eGFP (meGFP)-hRAB26 expression plasmid was constructed with the coding region of hRAB26 cDNA (IMAGE identification number 5262795; Open Biosystems) added in frame to the carboxyl terminus of eGFP, replacing the hMIST1 coding region in pEGFP-hMist1-C1 by ribocloning (1, 2). PCR was performed on the vector region by using primers DNA3as and pcD3LKs on pmEGFP-hMist1-C1 DNA. The hRAB26 target was PCR amplified using DNA3s and hRAB26-DNA3b with pcD3LKas and hRAB26-pcD3LKs. The construct coding region (5,583 bp) was verified to be correct by DNA sequencing.
meGFP-hRAB26 was mutagenized to meGFP-hRAB26T77N by site-directed mutagenesis to convert the threonine at amino acid position 77 to an asparagine, and a new BbsI restriction site was introduced. The construct coding region was verified to be correct by DNA sequencing.
Microarray data accession number.
GeneChip data sets have been deposited in GEO (accession number GSE16924).
RESULTS
Identifying MIST1 direct targets that might regulate secretory-cell granule formation and establishment of apical-basal polarity.
During maturation, ZCs undergo three dramatic changes in cell biology: they (i) greatly upregulate their rER network, (ii) form large exocrine secretory granules, and (iii) expand their apical cytoplasm, with their nuclei moving basally. Mist1 expression begins as ZCs start to differentiate from their mucous neck cell precursors (50); it is involved in both formation/maintenance of the secretory granules and the apical-basal reorganization. In the absence of Mist1, ZCs exhibit markedly smaller zymogen-containing, secretory vesicles than the wild type (Fig. 1A, C, and E) . Vesicle sizes, as measured by TEM of multiple ZCs in multiple gastric units, differed, with means of 0.40 ± 0.03 μm2 (wild type) and 0.14 ± 0.02 μm2 (Mist1−/− mice) (P < 0.001; Fig. 1B and C). The defects in apical-basal organization are illustrated in Fig. 1B and D. The ratio of supranuclear to infranuclear secretory vesicle fluorescence decreased from an apical predominance of (4.2 ± 0.7):1 to an essentially equal distribution of apical and basal granules (ratio of [1.49 ± 0.06]:1) in Mist1−/− ZCs (Fig. 1D). The changes reflect inappropriate basolateral trafficking of granules and/or overly apical positioning of the nucleus. MIST1 loss-of-function defects are summarized in the diagram in Fig. 1E. The loss of MIST1 does not result in diminished rER (50).
To determine the genes that might be activated by MIST1 to regulate cellular structural changes, we transiently transfected either MIST1 or empty vector control plasmid into two human gastric cell lines (AGS and HGC-27). eGFP plasmid was cotransfected under both conditions, and cells expressing high levels of GFP (i.e., highly transfected cells) were isolated by fluorescence-activated cell sorting (FACS). Global gene expression changes specific to MIST1 transfection were assayed by use of Affymetrix HGU133_Plus_2 GeneChips. A total of 16 genes exhibited increased expression (90% confidence interval, ≥1.2-fold change; intensity difference, ≥100) in both MIST1-transfected cell lines relative to respective parental and empty-vector-transfected control populations (Fig. 2 and Table 1).
TABLE 1.
Functions of gene products upregulated by MIST1 transfection
| Gene product | Potential function(s)/characterization | Authors (reference[s]) |
|---|---|---|
| Products of genes that directly regulate vesicular formation and transport | ||
| RAB3D | Regulates vesicular docking and fusion during regulated exocytosis | Millar et al. (38) |
| RAB26 | Involved in regulated vesicular secretion | Yoshie et al. (67), Wagner et al. (64), Nashida et al. (40) |
| Factors that are secreted or associate with specialized membrane compartments | ||
| ACP2 (acid phosphatase 2, lysosomal) | Regulates lysosomal function; its deficiency causes abnormal lysosomal storage and inclusions | Saftig et al. (53), Mannan et al. (36) |
| SLC31A2 (solute carrier family 31 [copper transporters], member 2) | Cellular copper uptake | Zhou and Gitschier (70) |
| SERPINI1 (serpin peptidase inhibitor, clade I [neuroserpin], member 1) | Secreted protein; mutations associate with two families with FENIB, an autosomal dominant form of dementia | Stoeckli et al. (60), Davis et al. (11, 12) |
| DISP1 (dispatched homolog 1) | Multitransmembrane protein, hedgehog transporter | Ma et al. (35), Cohen (8) |
| Other products of genes that may potentially regulate vesicular transportation and cellular architecture | ||
| CCPG1 (cell cycle progression gene 1) | Novel scaffolding protein that regulates guanine nucleotide factor Dbs and promotes CDC42 activation by Dbs | Kostenko et al. (31) |
| FNDC3A (fibronectin type III domain-containing 3A) | Cell adhesion and migration | Obholz et al. (41), Shan et al. (57) |
| AQP3 (aquaporin 3) | Water channel in many cell types | Inase et al. (23) |
| ARRDC3 (arresting domain-containing 3) | Associated with the plasma membrane, endosomes, and lysosomes during endocytosis; plays a regulatory role in cell proliferation | Oka et al. (43) |
| FIGN (fidgetin) | One of the “meiotic” or subfamily-7 group of AAA proteins; AAA proteins are molecular chaperones involved in a variety of activities like proteolysis, peroxisome biogenesis, membrane fusion, endosome sorting, and meiotic spindle formation | Patel and Latterich (47), Cox et al. (9) |
| Signaling molecules | ||
| TIFA (TRAF-interacting protein with forkhead-associated domain) | Associates with TRAF and regulates tumor necrosis factor receptor signaling | Ea et al. (14), Kanamori et al. (27), Takatsuna et al. (61) |
| MAP2K5 (mitogen-activated protein kinase kinase 5) | Interacts with MAPK7; MAP2K5/MAPK7 protein cascade is a novel signaling pathway | Zhou et al. (71) |
| Products of genes whose molecular function is unclear | ||
| BRP44L (brain protein 44-like) | Unknown | Jiang et al. (25) |
| MGC13057 (chromosome 2 open reading frame 88) | Unknown | |
| FAM63A (family with sequence similarity 63, member A) | Unknown |
MIST1 directly associates with conserved CATATG E-boxes in 6 MIST1-induced genes.
To determine which MIST1-induced genes were likely direct targets of MIST1, we first examined the entire genomic region upstream of the transcription start sites and the entire first intron of each gene for conserved E-box sequences of the CATATG type, shown previously to be the binding site for both mammalian MIST1 (62) and DIMMED, its fly ortholog (19, 45, 46). ARRDC3, CCPG1, FNDC3A, and SERPINI1 all contain remarkably conserved CATATG sequences within the first intron (Fig. 3A). The RAB3D gene has the canonical MIST1 binding site in a region between 167 and 205 bp downstream of the end of the first exon in multiple species (Fig. 3B). RAB26 has a similarly located CATATG that is conserved in multiple mammalian genomes except human; however, the only CATATG in the 5′ upstream and intronic human genomic DNA we examined is in a region 11.3 kb upstream of the RAB26 transcription start (chromosome 16 [Chr16], coordinate 2127505), a location also conserved in chimpanzees (Fig. 3C). The remainder of the 16 MIST1-activated genes showed no conserved CATATG sites in the first intron.
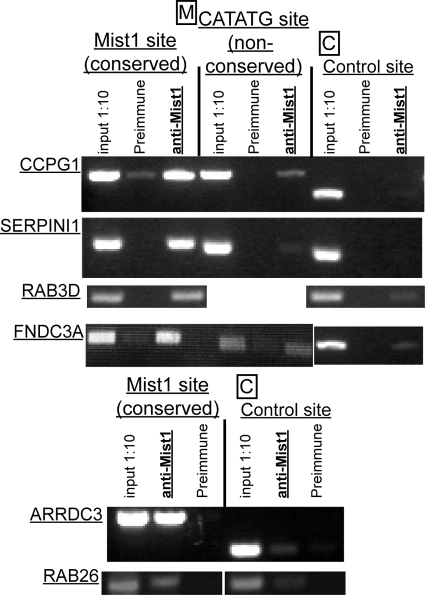
To confirm direct MIST1 binding to the conserved CATATG sites, we performed ChIP on HGC-27 gastric cells stably expressing a MIST1-eGFP plasmid (henceforth called HGC-MIST1). Primers closely flanking the conserved CATATG sites shown in Fig. 3A to C generated amplicons in the MIST1-immunoprecipitated genomic DNA fragments with greater efficiency than in preimmune control-precipitated DNA in all 6 genes (Fig. 4). In contrast, primers for control regions without CATATG sequences generated little to no amplicon in the MIST1-immunoprecipitated DNA. We also examined nonconserved CATATG sites in CCPG1, SERPINI1, and FNDC3A (Fig. 4). Only the site in CCPG1 showed possible occupation by MIST1, though this site was within 5 kb of the conserved CATATG, and genomic DNA was fragmented to an average size of 1 kb in these experiments. Hence, we cannot rule out the possibility that a small population of longer genomic DNA fragments derived from precipitation of MIST1 at the conserved site might account for amplification at this region. Overall, the results indicate that MIST1 occupies specific transcription regulatory elements in ARRDC3, CCPG1, FNDC3A, SERPINI1, RAB26, and RAB3D genes.
FIG. 4.
Conserved MIST1 E-box sequences are directly bound by MIST1. Gel electrophoresis images of PCR amplicons representative of ≥3 independent ChIP experiments using anti-MIST1 antibody followed by primers either to the conserved intronic CATATG [Mist1 site (conserved)] or to nonconserved CATATG and control regions denoted in Fig. 3. Input 1:10, genomic DNA prior to immunoprecipitation diluted 1:10; preimmune, immunoprecipitation from nonspecific rabbit antiserum. All images taken from PCRs run for 35 cycles, though each reaction volume was sampled at multiple cycles to ensure amplification had not plateaued. Note that conserved CATATG sequences show much stronger amplification of MIST1-bound DNA than preimmune control-bound DNA, and no MIST1-bound DNA was detectable at control sites without CATATG sequences. Nonconserved CATATG sites showed little to no amplifiable MIST1-bound DNA.
RAB3D and RAB26 are ZC specific and MIST1 dependent in vivo.
Of the 6 MIST1-regulated genes, two, RAB3D and RAB26, have been previously proposed to play a role in regulated secretion (15). Thus, we reasoned that these two RABs might be the cellular effectors that MIST1 induces to govern maintenance and formation of large secretory granules in exocrine secretory cells. We examined other related RAB genes purported to be involved in regulated exocytosis (RAB3A, RAB3B, RAB3C, RAB27A, RAB27B, and RAB37) and did not find similarly conserved intronic CATATG sequences (not shown), nor was expression of these other RAB family members affected by forced MIST1 expression (Fig. 2B). Thus, RAB3D and RAB26 appear to be specific and direct targets of MIST1 transcriptional regulation.
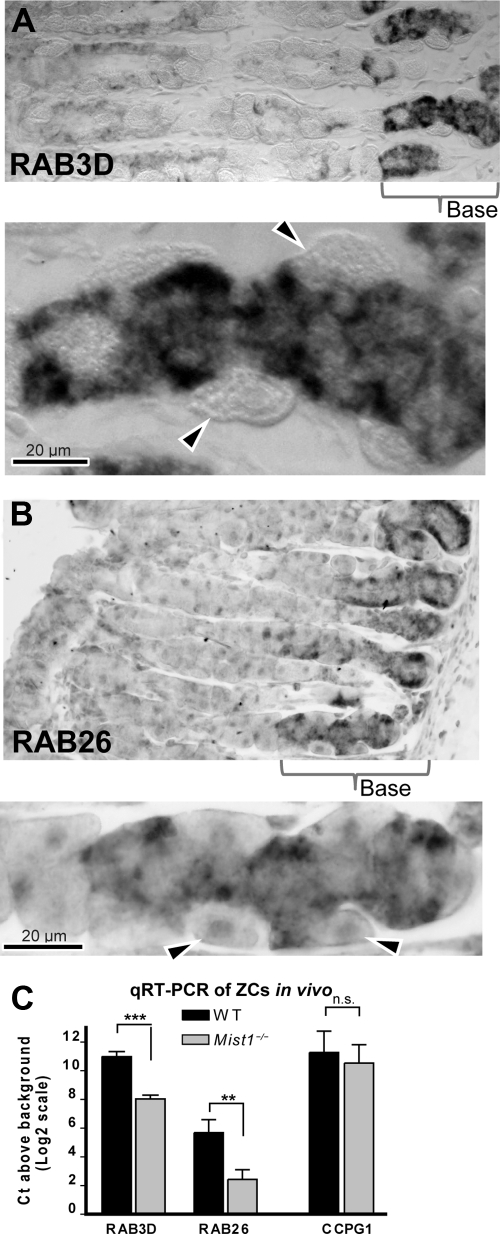
If RAB3D and RAB26 regulate secretory granule formation, they should be expressed in ZCs and be MIST1 dependent in vivo. In situ hybridization showed that RAB26 and RAB3D expression in mouse stomachs was confined to MIST1-expressing ZCs in wild-type mice (Fig. 5A and B). To determine whether either RAB depended on MIST1 expression, we used LCM on ZCs from the bases of Mist1−/− mice and their wild-type littermates. In Mist1−/− mice, RAB26 message was barely detectable above the background signal (1.9 cycles above water; 3 independent mice and dissections), whereas it was expressed at 11-fold-higher levels in wild-type mice (P < 0.01; Fig. 5C). Mist1−/− mice also showed a significant decrease (8-fold; P < 0.001) in RAB3D expression (Fig. 5C). Hence, Rab26 and Rab3d are transcriptionally regulated by MIST1 in vivo as well.
FIG. 5.
RAB3D and RAB26 expression are zymogenic cell specific and MIST1 dependent in vivo. Images of in situ hybridization of mouse fundic gastric gland sections showing antisense RAB3D (A) and RAB26 (B) signals confined to the ZCs at the base of the glands are shown. Note the absence of signal in parietal cells (arrowheads in the higher-magnification view below each main panel). (C) Results of quantitative RT-PCR of ZCs dissected by LCM from Mist1−/− mice. RAB3D and RAB26 expression levels are significantly decreased in these mice. Note that the y axis values are expressed as log2 values relative to levels in wells containing no cDNA following 18S rRNA normalization; hence, zero indicates essentially no detectable levels of given amplicon. RAB26 expression in the Mist1−/− mice is nearly completely abrogated. Primers for another potential MIST1 target, CCPG1, indicate no significant change in expression (n.s.), indicating that the loss of MIST1 specifically affects levels of the two RABs. WT, wild type.
Expression of pepsinogen-RFP in MIST1-expressing stable lines causes formation of large exocrine secretory granules.
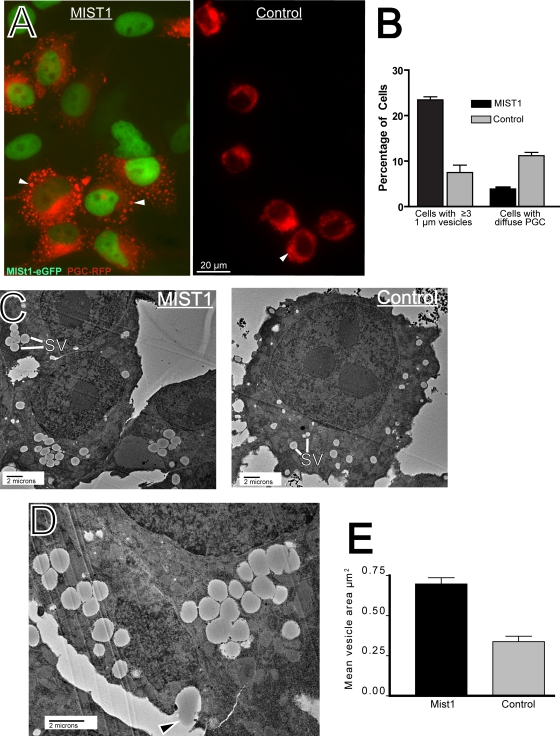
Our results so far showed that (i) loss of MIST1 leads to small secretory granules and defects in the apical compartment where those granules mature; (ii) expression of two putative regulators of exocrine granule homeostasis, RAB3D and RAB26, is directly regulated in vitro by MIST1; and (iii) RAB3D and RAB26 expression is MIST1 regulated in vivo specifically in Mist1-expressing cells. Thus, the data indicated that MIST1 regulates vesicle maturation via direct transcriptional activation of RAB3D and RAB26. To functionally test that hypothesis, we needed an in vitro system where the role of RABs in large-secretory-vesicle dynamics could be assessed mechanistically. We designed a construct comprising one of the principal constituents of ZC secretory granules, PGC, fused at the carboxyl terminus to RFP. Figure 6 shows that stable HGC-MIST1 cells transiently transfected with this construct developed multiple large PGC-containing granules. The results of multiple (n = 8) separate transfections showed, highly reproducibly, that 23.0% ± 1.2% of the cells with RFP fluorescence detectable above the background had ≥3 vesicles of ≥1 μm in size. In control HGC-27 cultures stably expressing eGFP alone (without MIST1), cells with these large secretory granules were comparatively rare (7.5% ± 1.5%; P < 0.001 by one-tailed t test comparing MIST1-expressing cells and the controls), whereas cells with bright diffuse PGC-RFP were far more common (4.3% ± 0.2% for MIST1-expressing cells versus 11.2% ± 0.6% for the controls; P < 0.001) (Fig. 6A and B).
FIG. 6.
HGC-MIST1 cells form large exocrine granules upon PGC-RFP transfection. (A) Fluorescence microscopy image of HGC-27 cells stably expressing MIST1-eGFP (green nuclear staining) showing large PGC vesicles (arrowheads) that do not form in control cells (stably expressing eGFP alone) upon transfection with PGC-RFP. (B) The fraction of cells with multiple (≥3) large (≥1-μm) vesicles and diffuse, bright vesicles is quantified across multiple experiments (note that the remaining cells showed above-background RFP but were too dim to categorize). (C) TEM image of ZCs from HGC-MIST1 and control cells. Note the reduced size of secretory vesicles in the (control) HGC-27 cells. SV, secretory vesicle. (D) TEM image showing exocytosis of vesicle contents (arrowhead) in an HGC-MIST1 cell. (E) Vesicle sizes were quantified from TEM of multiple HGC-MIST1 and control cells.
When examined on their tissue culture substrate by using transmission EM, both MIST1-expressing and control cells with PGC-RFP contained multiple secretory vesicles (Fig. 6C), whereas nontransfected cells did not (not shown). However, HGC-MIST1 cells formed vesicles that tended to cluster together and were each over twice as large as the vesicles found in control cells (0.70 ± 0.12 μm2 versus 0.34 ± 0.11 μm2; P < 1 × 106) (Fig. 6C to E). Figure 6D demonstrates how vesicles could be seen undergoing exocytosis. We observed no consistent difference in cell proliferation, apoptosis, or cell morphology between MIST1-expressing cell lines and controls (not shown). Finally, we treated cultures of MIST1-expressing, PGC-RFP-transfected cells with the secretagogue carbachol (100 μm), which greatly increased formation and trafficking of large PGC-RFP granules (not shown), indicating that these cell lines can respond to a physiologic, muscarinic agonist that induces release of zymogen-containing granules in ZCs in vivo.
Large exocrine granule formation requires RAB prenylation and RAB26 activity.
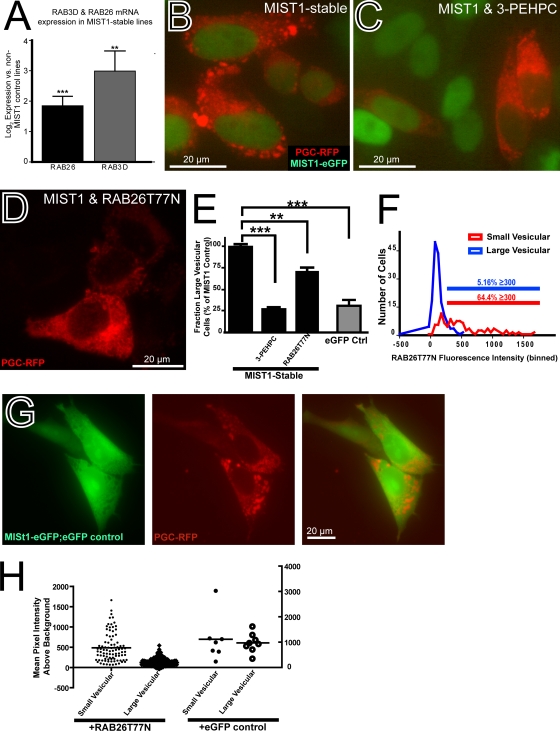
Using qRT-PCR, we confirmed that cells stably expressing MIST1, as expected, constitutively expressed higher levels of RAB transcripts. RAB26 and RAB3D were constitutively expressed in HGC-MIST1 cells at (4.1 ± 0.9)- and (15.0 ± 8.2)-fold-higher levels, respectively (6 independent experiments; respective P values of <0.001 and <0.005), than in those stably expressing a control eGFP vector alone (Fig. 7A).
FIG. 7.
Large PGC-RFP vesicles in HGC-MIST1 cells are RAB function dependent. (A) qRT-PCR from 6 separate cultures of stable HGC-MIST1 cells showing constitutively increased expression of RAB26 and RAB3D in MIST1-expressing cells relative to cells stably expressing eGFP instead of MIST1-eGFP. (B) Fluorescence microscopy image showing PGC-RFP (red) vesicles after transient transfection in HGC-MIST1 cells. (C) Dispersion of PGC-RFP vesicles in HGC-MIST1 cells treated with 5 mM RAB inhibitor 3-PEHPC. (D) Dispersion of vesicles in HGC-MIST1 cells transfected with RAB26T77N, a dominant negative RAB26 construct. (E) Effects of inhibition of RAB function on the fraction of cells with large PGC vesicles are quantified. (P values were determined by one-way ANOVA test with Dunnett's multiple-comparison correction.) eGFP Ctrl, stable eGFP-expressing control cells. (F) The effects of dominant negative RAB26 transfection on PGC-RFP vesicle formation were assessed by quantifying green (RAB26T77N) fluorescence intensity in each cell, correlated with the distribution of red (PGC-RFP) fluorescence in the same cell. “Large vesicular” cells were those with multiple (≥3) large (≥1-μm) vesicles; “small vesicular” cells were those with substantial (see Materials and Methods) red fluorescence that was not distributed in large vesicles. The large-vesicle phenotype occurred almost exclusively in cells with limited to undetectable expression of dominant negative RAB26; only 5.16% of such cells had a green MFI of ≥300 in 16-bit images following background subtraction (blue line). In contrast, 64.4% of cells effectively (MFI, ≥300) cotransfected with RAB26T77N had the diffuse or small-vesicle phenotype, indicating that those cells did not form large secretory vesicles (red line). (G) Transiently transfected eGFP has minimal effects on PGC-RFP vesicle formation, indicating that inhibition of large PGC granules is specific to effects of RAB26 and not cytoplasmic GFP. (H) Graphical representation of mean green-pixel intensity above the background in HGC-MIST1 cells with either small (diffuse)- or large-PGC-RFP-vesicle phenotype following transient transfection with meGFP-RAB26T77N or eGFP. Note that the large-vesicle cells show little to no meGFP-RAB26T77N expression. There is no correlation between expression of eGFP alone and PGC vesicle phenotype.
We next sought to ascertain whether formation of large PGC-RFP granules required RAB expression induced by MIST1. We transfected HGC-MIST1 cells with PGC-RFP in the presence of 5 mM 3-PEHPC (3, 10), an inhibitor of RAB geranylgeranyl transferase (RGGT), the enzyme that catalyzes the addition of geranylgeranyl groups to the carboxy-terminal dicysteine (CC or CXC) motifs in RAB3D and RAB26 (55). We used the minimal concentration of 3-PEHPC that completely dispersed the normal cellular localization of an eGFP-RAB26 construct. Once RABs lose the ability to localize to their defined cellular compartments, they no longer function. As hypothesized, inhibition of RAB function led to the loss of the large PGC-RFP granules (Fig. 7B and C). Cells with large granules decreased ∼4-fold in the presence of inhibitor (P < 0.001), equivalent to levels in the non-MIST1-expressing control cells (Fig. 7E).
The cellular localization and expression of RAB3D in exocrine secretion have been relatively well studied (38, 65). RAB3D is important for secretory-granule homeostasis in diverse secretory cells. Although there is some controversy about its primary function (51, 63), it is generally thought that RAB3D regulates the release of mature secretory granules, likely by tethering them to elements of the cortical actomyosin mesh (7, 37). Given that RAB3D already had an established role in exocrine granule homeostasis, we decided to further examine the role of RAB26 in this process.
To determine if more targeted disruption of RAB26 function could inhibit large-zymogenic-vesicle formation, we designed a point mutant RAB26 (eGFP-RAB26T77N) based on close conservation of the RAB26 GTP binding region with the analogous RAB3D sequence (RAB3DT36N), which acts as a dominant negative when the equivalent threonine is mutated to asparagine in the G2 box region responsible for coordinating Mg2+ binding (7); the structure of RAB26 is presented in the PDBsum database (accession no. 2g6b). As expected, eGFP-RAB26T77N did not localize to a specific membrane compartment like wild-type RAB26, which is perinuclear (Jin et al., unpublished observations). Rather, eGFP-RAB26T77N distributed throughout the cytoplasm in a pattern similar to that of wild-type RAB26 in the presence of 3-PEHPC (not shown).
Transfection of the dominant negative construct inhibited large-granule formation in HGC-MIST1 cells cotransfected with PGC-RFP (Fig. 7D). Overall, the percentage of cells expressing large granules (3 independent experiments) decreased 28% ± 5% (P < 0.01) (Fig. 7E). When the effects of transfection of the dominant negative construct on the large-PGC-RFP-granule phenotype were correlated on a cell-by-cell basis, almost none of the cells with large zymogenic granules were seen to express cytoplasmic eGFP-RAB26T77N (Fig. 7F and H). Conversely, among those cells transfected with eGFP-RAB26T77N above the background, almost none showed the large-vesicle phenotype (Fig. 7F), confirming that expression of eGFP-RAB26T77N prevented formation of the large PGC-RFP vesicles. In a control experiment, we transfected a plasmid with eGFP alone in lieu of eGFP-RAB26T77N (Fig. 7G and H). Expression of eGFP alone had no effect on formation of large PGC-RFP granules, indicating that the effects of eGFP-RAB26T77N were specific to its function as a dominant negative RAB. Together, the data demonstrate that RAB26 function is required for the formation of the large PGC granules.
A previous study showed that RAB3D was not sufficient to rescue granulogenesis in Mist1−/− cells (26). We tested whether RAB26 was sufficient to induce large granules in control HGC-27 cells stably transfected with eGFP and not MIST1. Transfection of eGFP-RAB26 in these cells in three independent experiments led to a higher fraction of cells with large PGC granules (from 7.5% ± 2.3% to 10.9% ± 0.4%) (data not shown), which approached a statistically significant increase (P < 0.06 by one-tailed t test) but did not approach the level seen in HGC-MIST1 cells. Thus, although required for maturation of granules, RAB26, like RAB3D, does not seem to be sufficient to rescue the loss of MIST1 function phenotype in the absence of other MIST1 targets.
DISCUSSION
Here, we demonstrated the role of MIST1 in establishing the characteristic secretory phenotype of zymogen exocrine cells through direct transcriptional activation of RAB3D and RAB26. These two RABs were the only ones increased following MIST1 transfection and the only RABs of those thought to be associated with secretory granule homeostasis that had conserved intronic MIST1-binding E-box elements. Furthermore, ChIP confirmed that MIST1 directly associated with RAB3D and RAB26 E-boxes. We next showed that RAB3D and RAB26 expression was specific to MIST1-expressing cells in vivo and was also MIST1 dependent. Finally, we established an in vitro system to test the functional importance of RAB3D and RAB26. Cells stably expressing MIST1, as expected, had constitutively increased levels of both RAB3D and RAB26. When transfected with an RFP-tagged zymogenic granule component, PGC, these cells, but not controls, formed large zymogenic-secretory granules. Large-granule formation required increased RAB activity, because inhibition of RAB prenylation resulted in cells with smaller granules, indistinguishable from those of non-MIST1-expressing control cells. Previous studies have shown that RAB3D participates in exocrine granule homeostasis but is not itself sufficient to induce large-granule formation in MIST1-expressing cells (26, 38, 42, 51, 65). Here we show that forced expression of a RAB26 dominant negative construct completely abrogates large-vesicle formation, yet transfection of RAB26 alone does not substantially rescue the MIST1 loss-of-function granule phenotype. Thus, like RAB3D, RAB26 is required but not sufficient for exocrine granulogenesis.
MIST1 is expressed by a limited number of specialized secretory cells in diverse tissues. Loss of Mist1, in all reported cases, did not lead to a decrease in steady-state survival or formation of these cell lineages, nor did it lead to a change in their cell fate. The fundamental feature of the loss of Mist1 function is that the cells that would express Mist1 are still generated, occupy their same in vivo niches, and secrete the same substances (32, 50, 72). However, the loss of MIST1 has profound effects on cell shape/architecture and cell function. With the loss of MIST1, zymogen-secreting cells in the stomach, pancreas, and salivary gland do not achieve their full size, and their zymogenic granules become smaller as well. Pancreatic acinar cells grown in vitro show smaller zymogen stores, altered Ca2+ currents, and a loss of normal apical, perinuclear Golgi localization (34).
MIST1 protein is highly conserved across species. The Drosophila ortholog of Mist1, DIMMED, shares 78% identity with Mist1 through the bHLH region and binds the same CATATG E-box sequence in its known target gene (46, 62). Conservation between the two genes in these distantly related organisms is such that expression of MIST1 can induce a DIMMED target gene in vitro (45). The zebrafish Mist1 ortholog shares 77% identity in the bHLH domain with the mouse MIST1 gene (18). The loss of zebrafish Mist1 leads to morphological abnormalities in pancreatic enzyme-secreting cells (18). Gain-of-function experiments show that DIMMED is sufficient to orchestrate increased secretory vesicle size (19, 20, 45), analogous to the MIST1 gain of function in the current manuscript. Thus, MIST1 is a developmentally regulated TF whose expression is induced during terminal differentiation of selected secretory cells in diverse tissues with a highly evolutionarily conserved function.
Which genes might a single TF activate to effect complete remodeling of cellular architecture? Two previously reported targets of MIST1 in vitro in pancreatic cells are the gap junction gene Connexin32 (encoding GJB1) (52) and the cell cycle-regulating p21(CIP1/WAF1) gene (Cdkn1a) (24). We did not detect GJB1 expression in the gastric cell lines used in the manuscript by use of GeneChips or qRT-PCR (X. Tian and J. Mills, unpublished observations), so we do not think that this is a MIST1 target in our system. CDKN1 is expressed in ZCs in vivo and in gastric cell lines, but expression is not MIST1 dependent in cell lines. Furthermore, in vivo, expression of CDKN1 in ZCs is lower than expression in other MIST1-negative gastric epithelial cells (Tian and Mills, unpublished observations), so this also seems an unlikely target to mediate the morphological changes induced by MIST1.
It is possible that MIST1 activates both tissue-specific and universal sets of genes. In this work, we identified 16 genes whose expression is activated by MIST1 in two different gastric cell lines. The majority of these genes are thought to function in secretion or membrane trafficking, reinforcing our understanding of MIST1 as a TF that enables terminally differentiating cells to upregulate cellular effectors that establish a high-capacity secretory architecture. In addition to the RAB3D and RAB26 genes, the subjects of the current manuscript, four other genes, including the CCPG1 gene, were also direct targets of MIST1, as indicated by our chromatin immunoprecipitation studies (Fig. 4). CCPG1 did not appear to be regulated by MIST1 in gastric ZCs (Fig. 5C and data not shown); however, the CCPG1 gene is a large gene with multiple splice variants, and we have assayed only one variant so far. We are currently analyzing the 10 genes without conserved CATATG sequences to determine whether any of them binds MIST1 in a noncanonical fashion.
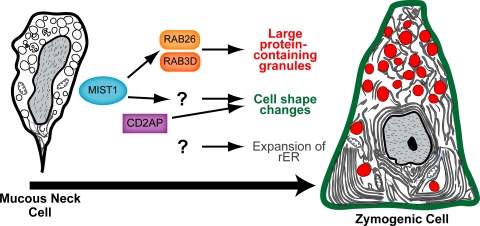
Are the increases in granule size and trafficking sufficient in themselves to effect the apical-basal cell shape reorganization also induced by MIST1, or are other MIST1 targets involved? We already know from previous studies that the cytoskeletal adaptor CD2AP is critical in the absence of MIST1 for maintaining cell shape, specifically cell height and ordered cell-cell adhesion (Fig. 8), though CD2AP does not appear to be a target of MIST1(5). The experiments do suggest, in any case, a role for other genes besides just RAB26 and RAB3D in the cell shape changes MIST1 induces.
FIG. 8.
Schematic illustrating how MIST1 expression is induced as mucous neck cells differentiate into zymogenic cells. MIST1 is modeled as acting primarily through RAB3D and RAB26 to regulate large zymogenic granules, though a role for other MIST1 targets in this process has not necessarily been ruled out. Other targets of MIST1 are probably involved in the apical-basal cell shape reorganization induced by MIST1. Based on an earlier study, the cytoskeletal adaptor CD2AP plays a role in this process but is not a target of MIST1 (5). Expansion of rER is not affected by loss of MIST1 expression.
RAB26 clusters by sequence homology with the RAB37, RAB27, and RAB3 subfamilies, which are critical for regulated secretion. However, whereas RAB27 and RAB3 family members (such as RAB3D) are relatively ubiquitously expressed in specialized secretory cells in numerous tissues, expression of RAB26 appears to be more limited, with the highest previously reported expression in pancreatic and submandibular enzyme-secreting acinar cells, both of which express MIST1 (40, 64, 67). A functional genomic screen also identified the RAB26 gene as a normal gastric gene whose expression is downregulated in human cancer (66). The only previous functional study of RAB26 showed that in salivary gland zymogen-secreting cells, RAB26 was associated with secretory granules (by cell fractionation and Western blotting) and injection of anti-RAB26 antibodies inhibited exocytosis, consistent with our findings (40).
It is currently unclear what mechanisms RAB3D and RAB26 use to induce formation of larger secretory granules. Our current results and those of previous studies indicate that both RAB26 and RAB3D are required but not individually sufficient for the formation of large secretory granules. The simplest model would be that MIST1-mediated vesicle maturation and trafficking are dependent on a balance between RAB26 and RAB3D activities (Fig. 8), though we cannot currently rule out a role for other direct targets. In a series of ongoing experiments (R. Jin and J. Mills, unpublished observations), we have learned that RAB26 helps organize the apical cellular compartment where immature secretory granules form and mature. RAB3D has been implicated downstream in tethering vesicles to the apical cytoskeleton. Tethering vesicles would encourage vesicle enlargement, because it would delay vesicle secretion and facilitate fusion of smaller, immature vesicles. Thus, RAB3D and RAB26 might coordinate sequential membrane compartments, RAB26 encouraging homotypic fusion of immature granules and/or larger budding from the trans-Golgi and RAB3D tethering maturing granules to the cortical cytoskeleton to await a signal for granule fusion with the plasma membrane. Together, these two RABs may represent the key cellular effectors that carry out the developmental signal encoded by the MIST1 gene.
Prospectus.
The process of cellular differentiation is typically understood as a sequence of transcription factor inductions, each of which specifies the next step in the developmental program of a given cell lineage. Any given terminally differentiated cell is thus ultimately specified by its unique history of sequential transcription factor induction steps. That view explains well how each differentiated cell in an organism ultimately comes to produce the unique set of proteins that distinguish it from all other cells, but what induces the genes that high-capacity secretory cells all share, no matter what they actually secrete, the genes that lay down the secretory architecture? Given the parsimony of evolution, it would make sense if these secretory cassettes were induced by a limited number of tissue-independent but still developmentally regulated transcription factors. Indeed, our recent survey of gene expression across multiple adult tissues indicates that this is so (13). In the present paper, we show how MIST1, a transcription factor critical for enacting cellular architecture, activates RAB26 and RAB3D, which in turn organize vesicle trafficking to maximize apical production and maintenance of large secretory granules. The mechanism we describe here will undoubtedly be only part of a much larger story as we continue to dissect the mechanisms of transcriptional regulation of cell architecture change.
Acknowledgments
We thank the Washington University Multiplexed Gene Analysis Core of the Siteman Cancer Center (supported in part by National Cancer Institute Grant P30-CA-91842). Further support came from NIH R01 DK-079798-01 and ACS DDC-115769 (to J.C.M.), T32-AI007172 (to X.T.), and a grant from Procter & Gamble (to C.E.M.).
Thanks also go to Karen Green for EM, to Liwei Zhang for ideas on gene alignments, and to Indira Mysorekar for thoughtful review of the manuscript.
Footnotes
Published ahead of print on 30 December 2009.
The authors have paid a fee to allow immediate free access to this article.
REFERENCES
- 1.Barnes, W. M. 2003. Ribocloning, chapter 29. In C. W. Dieffenbach and G. S. Dveksler (ed.), PCR primer: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- 2.Barnes, W. M. 1994. PCR amplification of up to 35-kb DNA with high fidelity and high yield from lambda bacteriophage templates. Proc. Natl. Acad. Sci. U. S. A. 91:2216-2220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baron, R. A., R. Tavare, A. C. Figueiredo, K. M. Blazewska, B. A. Kashemirov, C. E. McKenna, F. H. Ebetino, A. Taylor, M. J. Rogers, F. P. Coxon, and M. C. Seabra. 2009. Phosphonocarboxylates inhibit the second geranylgeranyl addition by Rab geranylgeranyl transferase. J. Biol. Chem. 284:6861-6868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bhattacharya, D., M. T. Cheah, C. B. Franco, N. Hosen, C. L. Pin, W. C. Sha, and I. L. Weissman. 2007. Transcriptional profiling of antigen-dependent murine B cell differentiation and memory formation. J. Immunol. 179:6808-6819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bredemeyer, A. J., J. H. Geahlen, V. G. Weis, W. J. Huh, B. H. Zinselmeyer, S. Srivatsan, M. J. Miller, A. S. Shaw, and J. C. Mills. 2009. The gastric epithelial progenitor cell niche and differentiation of the zymogenic (chief) cell lineage. Dev. Biol. 325:211-224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Capoccia, B. J., W. J. Huh, and J. C. Mills. 2009. How form follows functional genomics: gene expression profiling gastric epithelial cells with a particular discourse on the parietal cell. Physiol. Genomics 37:67-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen, X., J. A. Edwards, C. D. Logsdon, S. A. Ernst, and J. A. Williams. 2002. Dominant negative Rab3D inhibits amylase release from mouse pancreatic acini. J. Biol. Chem. 277:18002-18009. [DOI] [PubMed] [Google Scholar]
- 8.Cohen, M. M., Jr. 2003. The hedgehog signaling network. Am. J. Med. Genet. A 123A:5-28. [DOI] [PubMed] [Google Scholar]
- 9.Cox, G. A., C. L. Mahaffey, A. Nystuen, V. A. Letts, and W. N. Frankel. 2000. The mouse fidgetin gene defines a new role for AAA family proteins in mammalian development. Nat. Genet. 26:198-202. [DOI] [PubMed] [Google Scholar]
- 10.Coxon, F. P., F. H. Ebetino, E. H. Mules, M. C. Seabra, C. E. McKenna, and M. J. Rogers. 2005. Phosphonocarboxylate inhibitors of Rab geranylgeranyl transferase disrupt the prenylation and membrane localization of Rab proteins in osteoclasts in vitro and in vivo. Bone 37:349-358. [DOI] [PubMed] [Google Scholar]
- 11.Davis, R. L., A. E. Shrimpton, R. W. Carrell, D. A. Lomas, L. Gerhard, B. Baumann, D. A. Lawrence, M. Yepes, T. S. Kim, B. Ghetti, P. Piccardo, M. Takao, F. Lacbawan, M. Muenke, R. N. Sifers, C. B. Bradshaw, P. F. Kent, G. H. Collins, D. Larocca, and P. D. Holohan. 2002. Association between conformational mutations in neuroserpin and onset and severity of dementia. Lancet 359:2242-2247. [DOI] [PubMed] [Google Scholar]
- 12.Davis, R. L., A. E. Shrimpton, P. D. Holohan, C. Bradshaw, D. Feiglin, G. H. Collins, P. Sonderegger, J. Kinter, L. M. Becker, F. Lacbawan, D. Krasnewich, M. Muenke, D. A. Lawrence, M. S. Yerby, C. M. Shaw, B. Gooptu, P. R. Elliott, J. T. Finch, R. W. Carrell, and D. A. Lomas. 1999. Familial dementia caused by polymerization of mutant neuroserpin. Nature 401:376-379. [DOI] [PubMed] [Google Scholar]
- 13.Doherty, J. M., M. J. Geske, T. S. Stappenbeck, and J. C. Mills. 2008. Diverse adult stem cells share specific higher-order patterns of gene expression. Stem Cells 26:2124-2130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ea, C. K., L. Sun, J. Inoue, and Z. J. Chen. 2004. TIFA activates IkappaB kinase (IKK) by promoting oligomerization and ubiquitination of TRAF6. Proc. Natl. Acad. Sci. U. S. A. 101:15318-15323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fukuda, M. 2008. Regulation of secretory vesicle traffic by Rab small GTPases. Cell Mol. Life Sci. 65:2801-2813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gerson, K. D., C. D. Foster, P. Zhang, Z. Zhang, M. M. Rosenblatt, and S. H. Guttentag. 2008. Pepsinogen C proteolytic processing of surfactant protein B. J. Biol. Chem. 283:10330-10338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Grosshans, B. L., D. Ortiz, and P. Novick. 2006. Rabs and their effectors: achieving specificity in membrane traffic. Proc. Natl. Acad. Sci. U. S. A. 103:11821-11827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Guo, X., L. Cheng, Y. Liu, W. Fan, and D. Lu. 2007. Cloning, expression, and functional characterization of zebrafish Mist1. Biochem. Biophys. Res. Commun. 359:20-26. [DOI] [PubMed] [Google Scholar]
- 19.Hamanaka, Y., D. Park, Y. Ping, T. Edwards, J. W. Sweedler, I. A. Meinertzhagen, and P. H. Taghert. 2010. Transcriptional orchestration of the regulated secretory pathway in neurons by the bHLH protein DIMM. Curr. Biol. 20:9-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hewes, R. S., D. Park, S. A. Gauthier, A. M. Schaefer, and P. H. Taghert. 2003. The bHLH protein Dimmed controls neuroendocrine cell differentiation in Drosophila. Development 130:1771-1781. [DOI] [PubMed] [Google Scholar]
- 21.Hulsen, T., J. de Vlieg, and W. Alkema. 2008. BioVenn—a web application for the comparison and visualization of biological lists using area-proportional Venn diagrams. BMC Genomics 9:488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Im, H., J. A. Grass, K. D. Johnson, M. E. Boyer, J. Wu, and E. H. Bresnick. 2004. Measurement of protein-DNA interactions in vivo by chromatin immunoprecipitation. Methods Mol. Biol. 284:129-146. [DOI] [PubMed] [Google Scholar]
- 23.Inase, N., K. Fushimi, K. Ishibashi, S. Uchida, M. Ichioka, S. Sasaki, and F. Marumo. 1995. Isolation of human aquaporin 3 gene. J. Biol. Chem. 270:17913-17916. [DOI] [PubMed] [Google Scholar]
- 24.Jia, D., Y. Sun, and S. F. Konieczny. 2008. Mist1 regulates pancreatic acinar cell proliferation through p21 CIP1/WAF1. Gastroenterology 135:1687-1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jiang, M., X. Gu, X. Feng, Z. Fan, F. Ding, and Y. Liu. 2009. The molecular characterization of the brain protein 44-like (Brp44l) gene of Gekko japonicus and its expression changes in spinal cord after tail amputation. Mol. Biol. Rep. 36:215-220. [DOI] [PubMed] [Google Scholar]
- 26.Johnson, C. L., A. S. Kowalik, N. Rajakumar, and C. L. Pin. 2004. Mist1 is necessary for the establishment of granule organization in serous exocrine cells of the gastrointestinal tract. Mech. Dev. 121:261-272. [DOI] [PubMed] [Google Scholar]
- 27.Kanamori, M., H. Suzuki, R. Saito, M. Muramatsu, and Y. Hayashizaki. 2002. T2BP, a novel TRAF2 binding protein, can activate NF-kappaB and AP-1 without TNF stimulation. Biochem. Biophys. Res. Commun. 290:1108-1113. [DOI] [PubMed] [Google Scholar]
- 28.Karam, S. M., and C. P. Leblond. 1993. Dynamics of epithelial cells in the corpus of the mouse stomach. I. Identification of proliferative cell types and pinpointing of the stem cell. Anat. Rec. 236:259-279. [DOI] [PubMed] [Google Scholar]
- 29.Karam, S. M., and C. P. Leblond. 1993. Dynamics of epithelial cells in the corpus of the mouse stomach. III. Inward migration of neck cells followed by progressive transformation into zymogenic cells. Anat. Rec. 236:297-313. [DOI] [PubMed] [Google Scholar]
- 30.Kaser, A., A. H. Lee, A. Franke, J. N. Glickman, S. Zeissig, H. Tilg, E. E. Nieuwenhuis, D. E. Higgins, S. Schreiber, L. H. Glimcher, and R. S. Blumberg. 2008. XBP1 links ER stress to intestinal inflammation and confers genetic risk for human inflammatory bowel disease. Cell 134:743-756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kostenko, E. V., O. O. Olabisi, S. Sahay, P. L. Rodriguez, and I. P. Whitehead. 2006. Ccpg1, a novel scaffold protein that regulates the activity of the Rho guanine nucleotide exchange factor Dbs. Mol. Cell. Biol. 26:8964-8975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kowalik, A. S., C. L. Johnson, S. A. Chadi, J. Y. Weston, E. N. Fazio, and C. L. Pin. 2007. Mice lacking the transcription factor Mist1 exhibit an altered stress response and increased sensitivity to caerulein-induced pancreatitis. Am. J. Physiol. Gastrointest. Liver Physiol. 292:G1123-G1132. [DOI] [PubMed] [Google Scholar]
- 33.Lee, A. H., G. C. Chu, N. N. Iwakoshi, and L. H. Glimcher. 2005. XBP-1 is required for biogenesis of cellular secretory machinery of exocrine glands. EMBO J. 24:4368-4380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Luo, X., D. M. Shin, X. Wang, S. F. Konieczny, and S. Muallem. 2005. Aberrant localization of intracellular organelles, Ca2+ signaling, and exocytosis in Mist1 null mice. J. Biol. Chem. 280:12668-12675. [DOI] [PubMed] [Google Scholar]
- 35.Ma, Y., A. Erkner, R. Gong, S. Yao, J. Taipale, K. Basler, and P. A. Beachy. 2002. Hedgehog-mediated patterning of the mammalian embryo requires transporter-like function of dispatched. Cell 111:63-75. [DOI] [PubMed] [Google Scholar]
- 36.Mannan, A. U., E. Roussa, C. Kraus, M. Rickmann, J. Maenner, K. Nayernia, K. Krieglstein, A. Reis, and W. Engel. 2004. Mutation in the gene encoding lysosomal acid phosphatase (Acp2) causes cerebellum and skin malformation in mouse. Neurogenetics 5:229-238. [DOI] [PubMed] [Google Scholar]
- 37.Marchelletta, R. R., D. T. Jacobs, J. E. Schechter, R. E. Cheney, and S. F. Hamm-Alvarez. 2008. The class V myosin motor, myosin 5c, localizes to mature secretory vesicles and facilitates exocytosis in lacrimal acini. Am. J. Physiol. Cell Physiol. 295:C13-C28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Millar, A. L., N. J. Pavios, J. Xu, and M. H. Zheng. 2002. Rab3D: a regulator of exocytosis in non-neuronal cells. Histol. Histopathol. 17:929-936. [DOI] [PubMed] [Google Scholar]
- 39.Mills, J. C., N. Andersson, C. V. Hong, T. S. Stappenbeck, and J. I. Gordon. 2002. Molecular characterization of mouse gastric epithelial progenitor cells. Proc. Natl. Acad. Sci. U. S. A. 99:14819-14824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nashida, T., A. Imai, and H. Shimomura. 2006. Relation of Rab26 to the amylase release from rat parotid acinar cells. Arch. Oral Biol. 51:89-95. [DOI] [PubMed] [Google Scholar]
- 41.Obholz, K. L., A. Akopyan, K. G. Waymire, and G. R. MacGregor. 2006. FNDC3A is required for adhesion between spermatids and Sertoli cells. Dev. Biol. 298:498-513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ohnishi, H., S. A. Ernst, N. Wys, M. McNiven, and J. A. Williams. 1996. Rab3D localizes to zymogen granules in rat pancreatic acini and other exocrine glands. Am. J. Physiol. 271:G531-G538. [DOI] [PubMed] [Google Scholar]
- 43.Oka, S., H. Masutani, W. Liu, H. Horita, D. Wang, S. Kizaka-Kondoh, and J. Yodoi. 2006. Thioredoxin-binding protein-2-like inducible membrane protein is a novel vitamin D3 and peroxisome proliferator-activated receptor (PPAR)gamma ligand target protein that regulates PPARgamma signaling. Endocrinology 147:733-743. [DOI] [PubMed] [Google Scholar]
- 44.Ovcharenko, I., M. A. Nobrega, G. G. Loots, and L. Stubbs. 2004. ECR Browser: a tool for visualizing and accessing data from comparisons of multiple vertebrate genomes. Nucleic Acids Res. 32:W280-W286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Park, D., O. T. Shafer, S. P. Shepherd, H. Suh, J. S. Trigg, and P. H. Taghert. 2008. The Drosophila basic helix-loop-helix protein DIMMED directly activates PHM, a gene encoding a neuropeptide-amidating enzyme. Mol. Cell. Biol. 28:410-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Park, D., and P. H. Taghert. 2009. Peptidergic neurosecretory cells in insects: organization and control by the bHLH protein DIMMED. Gen. Comp. Endocrinol. 162:2-7. [DOI] [PubMed] [Google Scholar]
- 47.Patel, S., and M. Latterich. 1998. The AAA team: related ATPases with diverse functions. Trends Cell Biol. 8:65-71. [PubMed] [Google Scholar]
- 48.Pin, C. L., A. C. Bonvissuto, and S. F. Konieczny. 2000. Mist1 expression is a common link among serous exocrine cells exhibiting regulated exocytosis. Anat. Rec. 259:157-167. [DOI] [PubMed] [Google Scholar]
- 49.Pin, C. L., J. M. Rukstalis, C. Johnson, and S. F. Konieczny. 2001. The bHLH transcription factor Mist1 is required to maintain exocrine pancreas cell organization and acinar cell identity. J. Cell Biol. 155:519-530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ramsey, V. G., J. M. Doherty, C. C. Chen, T. S. Stappenbeck, S. F. Konieczny, and J. C. Mills. 2007. The maturation of mucus-secreting gastric epithelial progenitors into digestive-enzyme secreting zymogenic cells requires Mist1. Development 134:211-222. [DOI] [PubMed] [Google Scholar]
- 51.Riedel, D., W. Antonin, R. Fernandez-Chacon, G. Alvarez de Toledo, T. Jo, M. Geppert, J. A. Valentijn, K. Valentijn, J. D. Jamieson, T. C. Sudhof, and R. Jahn. 2002. Rab3D is not required for exocrine exocytosis but for maintenance of normally sized secretory granules. Mol. Cell. Biol. 22:6487-6497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rukstalis, J. M., A. Kowalik, L. Zhu, D. Lidington, C. L. Pin, and S. F. Konieczny. 2003. Exocrine specific expression of Connexin32 is dependent on the basic helix-loop-helix transcription factor Mist1. J. Cell Sci. 116:3315-3325. [DOI] [PubMed] [Google Scholar]
- 53.Saftig, P., D. Hartmann, R. Lullmann-Rauch, J. Wolff, M. Evers, A. Koster, M. Hetman, K. von Figura, and C. Peters. 1997. Mice deficient in lysosomal acid phosphatase develop lysosomal storage in the kidney and central nervous system. J. Biol. Chem. 272:18628-18635. [DOI] [PubMed] [Google Scholar]
- 54.Sardiello, M., M. Palmieri, A. di Ronza, D. L. Medina, M. Valenza, V. A. Gennarino, C. Di Malta, F. Donaudy, V. Embrione, R. S. Polishchuk, S. Banfi, G. Parenti, E. Cattaneo, and A. Ballabio. 2009. A gene network regulating lysosomal biogenesis and function. Science 325:473-477. [DOI] [PubMed] [Google Scholar]
- 55.Seabra, M. C., J. L. Goldstein, T. C. Sudhof, and M. S. Brown. 1992. Rab geranylgeranyl transferase. A multisubunit enzyme that prenylates GTP-binding proteins terminating in Cys-X-Cys or Cys-Cys. J. Biol. Chem. 267:14497-14503. [PubMed] [Google Scholar]
- 56.Shaffer, A. L., M. Shapiro-Shelef, N. N. Iwakoshi, A. H. Lee, S. B. Qian, H. Zhao, X. Yu, L. Yang, B. K. Tan, A. Rosenwald, E. M. Hurt, E. Petroulakis, N. Sonenberg, J. W. Yewdell, K. Calame, L. H. Glimcher, and L. M. Staudt. 2004. XBP1, downstream of Blimp-1, expands the secretory apparatus and other organelles, and increases protein synthesis in plasma cell differentiation. Immunity 21:81-93. [DOI] [PubMed] [Google Scholar]
- 57.Shan, S. W., D. Y. Lee, Z. Deng, T. Shatseva, Z. Jeyapalan, W. W. Du, Y. Zhang, J. W. Xuan, S. P. Yee, V. Siragam, and B. B. Yang. 2009. MicroRNA MiR-17 retards tissue growth and represses fibronectin expression. Nat. Cell Biol. 11:1031-1038. [DOI] [PubMed] [Google Scholar]
- 58.Snapp, E. L., R. S. Hegde, M. Francolini, F. Lombardo, S. Colombo, E. Pedrazzini, N. Borgese, and J. Lippincott-Schwartz. 2003. Formation of stacked ER cisternae by low affinity protein interactions. J. Cell Biol. 163:257-269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sriburi, R., H. Bommiasamy, G. L. Buldak, G. R. Robbins, M. Frank, S. Jackowski, and J. W. Brewer. 2007. Coordinate regulation of phospholipid biosynthesis and secretory pathway gene expression in XBP-1(S)-induced endoplasmic reticulum biogenesis. J. Biol. Chem. 282:7024-7034. [DOI] [PubMed] [Google Scholar]
- 60.Stoeckli, E. T., P. F. Lemkin, T. B. Kuhn, M. A. Ruegg, M. Heller, and P. Sonderegger. 1989. Identification of proteins secreted from axons of embryonic dorsal-root-ganglia neurons. Eur. J. Biochem. 180:249-258. [DOI] [PubMed] [Google Scholar]
- 61.Takatsuna, H., H. Kato, J. Gohda, T. Akiyama, A. Moriya, Y. Okamoto, Y. Yamagata, M. Otsuka, K. Umezawa, K. Semba, and J. Inoue. 2003. Identification of TIFA as an adapter protein that links tumor necrosis factor receptor-associated factor 6 (TRAF6) to interleukin-1 (IL-1) receptor-associated kinase-1 (IRAK-1) in IL-1 receptor signaling. J. Biol. Chem. 278:12144-12150. [DOI] [PubMed] [Google Scholar]
- 62.Tran, T., D. Jia, Y. Sun, and S. F. Konieczny. 2007. The bHLH domain of Mistl is sufficient to activate gene transcription. Gene Expr. 13:241-253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Valentijn, J. A., L. van Weeren, A. Ultee, and A. J. Koster. 2007. Novel localization of Rab3D in rat intestinal goblet cells and Brunner's gland acinar cells suggests a role in early Golgi trafficking. Am. J. Physiol. Gastrointest. Liver Physiol. 293:G165-G177. [DOI] [PubMed] [Google Scholar]
- 64.Wagner, A. C., M. Z. Strowski, B. Goke, and J. A. Williams. 1995. Molecular cloning of a new member of the Rab protein family, Rab 26, from rat pancreas. Biochem. Biophys. Res. Commun. 207:950-956. [DOI] [PubMed] [Google Scholar]
- 65.Williams, J. A., X. Chen, and M. E. Sabbatini. 2009. Small G proteins as key regulators of pancreatic digestive enzyme secretion. Am. J. Physiol. Endocrinol. Metab. 296:E405-E414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wu, M. S., Y. S. Lin, Y. T. Chang, C. T. Shun, M. T. Lin, and J. T. Lin. 2005. Gene expression profiling of gastric cancer by microarray combined with laser capture microdissection. World J. Gastroenterol. 11:7405-7412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yoshie, S., A. Imai, T. Nashida, and H. Shimomura. 2000. Expression, characterization, and localization of Rab26, a low molecular weight GTP-binding protein, in the rat parotid gland. Histochem. Cell Biol. 113:259-263. [DOI] [PubMed] [Google Scholar]
- 68.Zhao, Y., C. Johansson, T. Tran, R. Bettencourt, Y. Itahana, P. Y. Desprez, and S. F. Konieczny. 2006. Identification of a basic helix-loop-helix transcription factor expressed in mammary gland alveolar cells and required for maintenance of the differentiated state. Mol. Endocrinol. 20:2187-2198. [DOI] [PubMed] [Google Scholar]
- 69.Zhong, S., C. Li, and W. H. Wong. 2003. ChipInfo: software for extracting gene annotation and gene ontology information for microarray analysis. Nucleic Acids Res. 31:3483-3486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zhou, B., and J. Gitschier. 1997. hCTR1: a human gene for copper uptake identified by complementation in yeast. Proc. Natl. Acad. Sci. U. S. A. 94:7481-7486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zhou, G., Z. Q. Bao, and J. E. Dixon. 1995. Components of a new human protein kinase signal transduction pathway. J. Biol. Chem. 270:12665-12669. [DOI] [PubMed] [Google Scholar]
- 72.Zhu, L., T. Tran, J. M. Rukstalis, P. Sun, B. Damsz, and S. F. Konieczny. 2004. Inhibition of Mist1 homodimer formation induces pancreatic acinar-to-ductal metaplasia. Mol. Cell. Biol. 24:2673-2681. [DOI] [PMC free article] [PubMed] [Google Scholar]