Abstract
Histone deacetylases (HDACs) are chromatin-modifying enzymes that are involved in the regulation of proliferation, differentiation and development. HDAC inhibitors induce cell cycle arrest, differentiation, or apoptosis in tumor cells and are therefore promising antitumor agents. Numerous genes were found to be deregulated upon HDAC inhibitor treatment; however, the relevant target enzymes are still unidentified. HDAC1 is required for mouse development and unrestricted proliferation of embryonic stem cells. We show here that HDAC1 reversibly regulates cellular proliferation and represses the cyclin-dependent kinase inhibitor p21 in embryonic stem cells. Disruption of the p21 gene rescues the proliferation phenotype of HDAC1−/− embryonic stem cells but not the embryonic lethality of HDAC1−/− mice. In the absence of HDAC1, mouse embryonic fibroblasts scarcely undergo spontaneous immortalization and display increased p21 expression. Chromatin immunoprecipitation assays demonstrate a direct regulation of the p21 gene by HDAC1 in mouse embryonic fibroblasts. Transformation with simian virus 40 large T antigen or ablation of p21 restores normal immortalization of primary HDAC1−/− fibroblasts. Our data demonstrate that repression of the p21 gene is crucial for HDAC1-mediated control of proliferation and immortalization. HDAC1 might therefore be one of the relevant targets for HDAC inhibitors as anticancer drugs.
Acetylation of core histones is linked to the opening of chromatin and transcriptional activation. Modification of lysine residues by acetylation is thought to affect gene expression either by altering the affinity of histones to the DNA, or by creating binding sites for detector proteins that regulate chromatin accessibility. The antagonistic activities of two types of enzymes, histone acetyltransferases and histone deacetylases (HDACs), control the reversible acetylation state at the N-terminal tail of histones. HDACs catalyze the removal of the acetyl moieties from acetylated histones and other proteins and are in general associated with transcriptional repression (17). Based on their homologies with yeast deacetylases mammalian HDACs have been classified into Rpd3-like (class I), Hda1-like (class II), and Sir2-like (class III) enzymes (19). HDAC11 seems to represent a class (class IV) on its own.
HDACs have been shown to regulate many important biological processes, including cell cycle progression, differentiation, and development. In agreement with this idea, HDAC inhibitor treatment leads to cell cycle arrest, differentiation, and apoptosis in cultured tumor cells and tumors in animal models. Therefore, several HDAC inhibitors are currently tested as antitumor drugs in clinical trials. A variety of HDAC inhibitors, which target class I and class II enzymes have been identified (33), and it has been shown that they exert their antiproliferative effects via transcriptional and nontranscriptional mechanisms (32). Treatment of untransformed cells with HDAC inhibitors triggers a G2 checkpoint resulting in arrest of cells in the G2 phase (50). In contrast, HDAC inhibitor treatment often affects the cell viability of tumor cells. Loss of the G2 cell cycle checkpoint is a frequent event in cancer cells and may account for the increased sensitivity of cancer cells to the proapoptotic effects of HDAC inhibitors. Up to now, many genes have been shown to respond to HDAC inhibitor treatment; however, the relevant target deacetylases for antitumor drugs have not been identified thus far.
The first steps to answer this question are loss-of-function studies for individual HDACs in mammalian cells and organisms. Gene disruption experiments in mice have shown that class II HDACs are essential for specific differentiation processes and that their loss results in cellular hyperproliferation (11, 48, 56). In contrast, ablation of certain class I HDACs in mice or human tumor cells results in reduced proliferation or cell death (5, 16, 28, 35, 40, 46). Thus, class I deacetylases might be good candidates as targets for more specific inhibitors as anticancer drugs. This idea is also supported by observations that class I HDACs act as repressors of cyclin-dependent kinase (CDK) inhibitors, differentiation factors, and proapoptotic factors (18).
We have previously shown that HDAC1 gene disruption in mice leads to severe developmental defects and reduced proliferation both in the mouse embryo and in embryonic stem (ES) cells (28). Restricted proliferation of HDAC1−/− ES cells was accompanied by increased expression of the CDK inhibitor p21/CIP1/WAF1 (referred to here as p21 for simplicity). The p21 protein is a member of the CIP/WAF family and is involved in the regulation of cell cycle progression, senescence, and differentiation (10). The p21 gene was shown to be a target of the transcriptional corepressor HDAC1 in human tumor cells (20, 27, 40) and mouse ES cells (28). In addition, the p21 gene was consistently found to be upregulated by HDAC inhibitors in different cell types (18). However, it is not clear whether p21 is one of the crucial HDAC1 targets with respect to proliferation and development, since several hundred genes are deregulated upon loss of HDAC1 in mouse ES cells (58). Therefore, we asked in the present study whether p21 plays an important role in HDAC1-dependent regulation of proliferation in murine cells. We demonstrate here that HDAC1 acts as a positive regulator of proliferation in mouse ES cells and mouse embryonic fibroblasts (MEFs) by repressing the CDK inhibitor p21. In contrast, the developmental phenotype of HDAC1-deficient mouse embryos seems to be caused by more complex changes in gene expression.
MATERIALS AND METHODS
Generation of mice.
All mice used in the present study were kept on a mixed background (C57BL/6;129SV). To obtain HDAC1−/− p21−/− mice, HDAC1+/− mice were crossed with p21−/− mice (13).
Isolation and culture of ES cells.
ES cells were isolated from blastocysts as described previously (24a). ES cell lines were cultivated in Dulbecco modified Eagle medium-high glucose (Sigma) supplemented with antibiotics and 15% (vol/vol) fetal calf serum and either supplemented with 10 3 U of leukemia inhibitory factor/ml on gelatinized culture dishes or without leukemia inhibitory factor on feeder cell layers. All ES cell experiments were performed with cell lines obtained from littermates. To reintroduce HDAC1 into HDAC1-deficient ES cell lines, pMSCVpuro-HDAC1 (or empty pMSCVpuro as vector control) was linearized and electroporated into ES cells by using a Bio-Rad Gene Pulser II with 0.4-cm-wide sterile cuvettes (165-2088; Bio-Rad), with a single pulse at 230V and 500 μF. Electroporated ES cells were incubated for 5 min at room temperature and then plated onto puromycin-resistant feeder cells. After 24 h, puromycin (5 μg/ml) was applied to select for transfected cells. The selection procedure was continued for 14 days, and single clones were picked, expanded, and analyzed for expression of HDAC1.
Isolation and culture of MEFs.
HDAC1-deficient ES cells were injected into B6/CBA blastocysts and transferred into pseudopregnant females. Eleven days later, fibroblasts were isolated from chimeric embryos and cultivated in Dulbecco modified Eagle medium supplemented with 10% (vol/vol) fetal calf serum and antibiotics. Cells were grown for 24 h and then split at a 1:3 ratio. To select for presence of the Neo marker, cells were kept in the presence of G418 (300 μg/ml).
Immortalization of MEFs (3T3 protocol).
After we reached a critical density (subconfluent, usually day 5 to 7), fresh MEFs were split on new 60-mm dishes in 3 × 105 cell aliquots, grown for 3 days, and then split again in aliquots of 3 × 105 cells on new 60-mm dishes. This splitting was subsequently repeated until cells reached the crisis, characterized by high mortality and low proliferation rates. Then, culture medium was replaced every 3 days until the appearance of clones, which represent the immortalized fibroblasts.
Viral oncogene infection of MEFs.
MEFs were infected with pBABEpuro simian virus 40 (SV40) LT, pBABE HPV16 E6/E7 (expresses E6 and E7), pBABE HPV16 E6/E7s (expresses E6 with a stop codon in E7), or pBABE HPV16 E6s/E7 (expresses E7 with a stop codon in E6) (38). Retroviral supernatant was obtained from transiently transfected BOSC23 packaging cells as previously described (44) and subjected to selection with puromycin (1 μg/ml).
FACS analysis.
Cell cycle distribution was analyzed by fluorescence-activated cell sorter (FACS) analysis with a Partec PAS-II sorter.
SA β-Gal staining.
Senescence-associated β-galactosidase (SA β-Gal) activity was determined as described previously (14) with slight changes. Cells were washed in phosphate-buffered saline (PBS), fixed for 3 to 5 min at room temperature with 2% (vol/vol) formaldehyde-0.2% glutaraldehyde, washed with PBS, and incubated at 37°C without CO2 with freshly prepared staining solution containing 1 mg of X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside; Sigma)/ml, 30 mM citric acid, sodium phosphate (pH 5.5), 5 mM potassium ferrocyanide, 5 mM potassium ferricyanide, 150 mM NaCl, and 2 mM MgCl2 for at least 2 h.
Protein analysis and antibodies.
Whole-cell protein extraction and Western blot analysis were performed as previously described (44). The following antibodies were used for protein detection on immunoblots, indirect immunofluorescence, and chromatin immunoprecipitation assays: p21 (F5; Santa Cruz), HDAC1 polyclonal rabbit antibody and monoclonal mouse antibody from Millipore, and acetyl histone-H3 and acetyl histone-H4 (both from Millipore). The β-actin protein was visualized with a monoclonal antibody (AC74; Sigma). The proliferation marker Ki67 was detected with the monoclonal Ki67 antigen antibody (Novo Castra) by indirect immunofluorescence microscopy (Zeiss Axiovert 135TV microscope) as previously described (28). Nuclear DNA was visualized by using DAPI (4′,6′-diamidino-2-phenylindole).
Chromatin immunoprecipitation assays.
Chromatin immunoprecipitation assays were carried out as described previously (23) with some modifications. Chromatin was cross-linked for 10 min with 1% formaldehyde and then sonicated. Equal amounts of sonicated chromatin were diluted 10-fold and precipitated overnight with the following antibodies: HDAC1, acetyl-histone H3, acetyl-histone H4, and preimmune serum as a control. The chromatin-antibody complexes were isolated by incubation with 30 μl of protein A-Sepharose beads (50% slurry, 100 μg of salmon sperm DNA/ml, and 500 μg of bovine serum albumin/ml), while rocking at 4°C for 2 h. The beads were harvested and washed as described previously (23). Chromatin-antibody complexes were eluted from the protein A-Sepharose beads by addition of 2% sodium dodecyl sulfate, 0.1 M NaHCO3, and 10 mM dithiothreitol. Cross-linking was reversed by addition of a 0.05 volume of 4 M NaCl and incubation of the eluted samples for 6 h at 65°C. The DNA was extracted with phenol-chloroform, precipitated with ethanol, and dissolved in water.
PCR analysis of immunoprecipitated DNA.
All PCRs were performed on a Thermocycler T3 (Biometra) by using Promega PCR Master Mix. The linear range for each primer pair was determined empirically using different amounts of genomic DNA. PCRs with 1:40 dilutions of genomic DNA (input) were carried out along with the immunoprecipitated DNA. PCR products were resolved on 2% agarose-Tris-acetate-EDTA gels. Primer sequences are available upon request.
Whole-mount in situ hybridization.
Whole-mount in situ hybridization was performed essentially as described previously (24) using BM purple (Roche) as the color substrate. The digoxigenin-labeled antisense riboprobe mouse HDAC1 cDNA (3′ fragment, bp 1041 to 1550) was used for hybridization (4).
RNA isolation and RT-PCR analysis.
Total cellular RNA was isolated with TRIzol reagent (Gibco-BRL) as recommended by the manufacturer. For cDNA, 1 μg of total RNA was reversely transcribed with the iScript cDNA synthesis kit (Bio-Rad). Real-time reverse transcription-PCRs (RT-PCRs) were performed with 0.5 μl of the RT reaction mixture by the iCycler iQ system (Bio-Rad), using SYBR green (Molecular Probes) for labeling. Primer sequences are available upon request.
Proliferation assays.
Proliferation assays were performed as previously described (40). A total of 1,000 viable cells/well were plated into a 96-well tissue culture dish. The day after (day 0), the cells were treated with the indicated concentrations of trichostatin A (TSA). Every 24 h, a 96-well tissue culture dish was stained with crystal violet solution. Crystal violet incorporated in the cellular membrane was solubilized with a solution of 10% acetic acid-phosphate-buffered saline (PBS), and the absorbance was measured at 595 nm with an MRX microplate reader (Dynatech). The increase in cell numbers at a given day is obtained as the ratio of absorbance at a given time (“time i”) relative to the zero time point (time i is thus 0, 24, 48, or 72 h after the addition of drug). Each experimental time point is the average of three independent experiments, with the respective standard deviation.
RESULTS
The proliferation phenotype of HDAC1−/− ES cells is reversible.
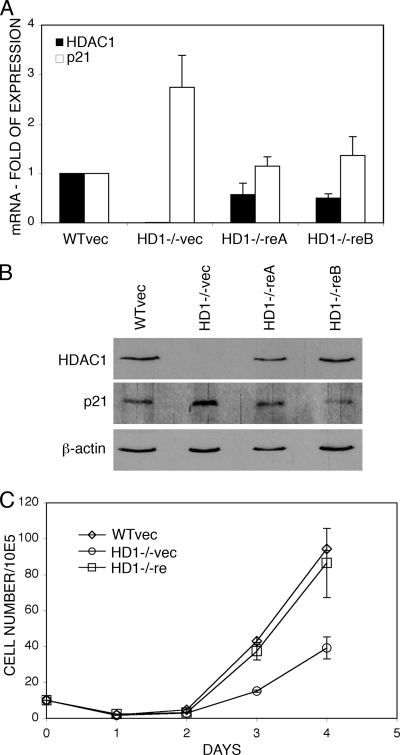
We have previously demonstrated that germ line mutation of HDAC1 is embryonic lethal (28). HDAC1−/− embryos are severely growth retarded and die before embryonic day 10.5 (E10.5). The proliferation of HDAC1-deficient ES cells was impaired, suggesting a role for HDAC1 in regulating cellular proliferation. To analyze the proliferation phenotype of HDAC1−/− ES cells in more detail, we first sought to determine whether the reduced proliferation rate could be reversed by reintroduction of HDAC1. To this end, HDAC1−/− ES cells were transfected with a pMSCV-HDAC1 expression vector. As controls, HDAC1+/+ and HDAC1−/− ES cells were transfected with empty vector. HDAC1 expression levels of reintroduced ES cell clones were analyzed for mRNA expression by real-time RT-PCR and for protein expression by immunoblots (Fig. 1A and B). Two clones (HD1−/− reA and HD1−/− reB) that displayed 70 and 50% of HDAC1+/+ mRNA levels and comparable HDAC1 protein levels were chosen for further analysis. To examine proliferation rates, equal numbers of vector transfected HDAC1+/+ cells, vector transfected HDAC1−/− cells and HDAC1 reintroduced ES cells (HD1−/− reA) were plated, and cell numbers were determined daily on a Casy Coulter Counter over a period of 4 days. As observed previously (28), HDAC1−/− ES cells showed decreased proliferation rates compared to HDAC1+/+ cells (Fig. 1C). Reintroduction of HDAC1 into HDAC1-deficient ES cells restored cellular proliferation almost to HDAC1+/+ levels.
FIG. 1.
Reintroduction of HDAC1 into HDAC1−/− ES cells restores proliferation and low p21 expression. (A) Analysis of HDAC1 and p21 mRNA expression in ES cells. RNA from vector transfected HDAC1+/+ ES cells (WTvec) and HDAC1−/− ES cells (HD1−/−vec) and two different HDAC1 reintroduced ES cell lines (HD1−/−reA and HD1−/−reB) was analyzed by real-time PCR. Expression of HDAC1 and p21 is shown relative to β-actin. The relative expression in vector-transfected HDAC1+/+ cells was arbitrarily set to 1. The data are shown as mean values of three independent experiments with standard deviation (SD). (B) Immunoblot analysis of HDAC1 and p21 protein expression. Whole-cell extracts were prepared from vector-transfected HDAC1+/+ and HDAC1−/− ES cells and HDAC1 reintroduced ES cell lines (HD1−/−reA and HD1−/−reB) and analyzed for the presence of HDAC1 and p21. β-Actin was used as loading control. (C) Growth curves of vector-transfected HDAC1+/+ and HDAC1−/− ES cells and HDAC1−/− ES cells transfected with pMSCV-HDAC1 (HD1−/−re). For each cell line, 105 cells were seeded in triplicates, and aliquots were counted daily for 4 days and presented as mean values ± the SD.
Reduced proliferation rates of HDAC1−/− ES cells are accompanied by elevated expression of the CDK inhibitor p21 (28). The p21 gene was previously shown to be a direct target of the transcriptional corepressor HDAC1 (20, 27). Therefore, we investigated whether reintroduction of HDAC1 would reduce the increased levels of p21 in HDAC1−/− ES cells. As shown in Fig. 1A and B, both mRNA and protein expression of p21 were reversed to HDAC1+/+ levels in HDAC1-reintroduced ES cells. From these results we conclude that HDAC1 reversibly regulates proliferation of murine ES cells and the expression of the CDK inhibitor p21 in ES cells.
Impaired proliferation of HDAC1−/− ES cells is rescued by additional ablation of p21.
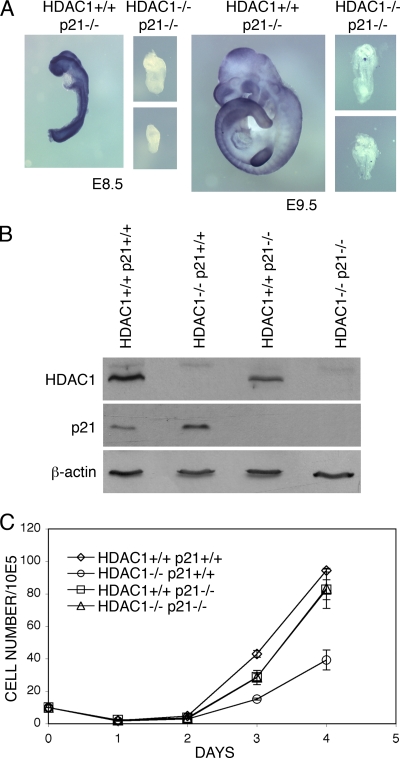
Reduced proliferation rates in HDAC1-deficient embryos and ES cells are accompanied by upregulation of p21 expression (28). To examine whether the proliferation defect of ES cells in the absence of HDAC1 might result from increased p21 levels, we ablated the expression of the CDK inhibitor in HDAC1+/+ and HDAC1−/− mice. To this purpose, we crossed HDAC1 heterozygous mice in a p21-deficient background (13). Mice lacking p21 undergo normal development. As shown in Table 1, genotyping of offspring from HDAC1+/− p21−/− breedings revealed the absence of HDAC1−/− p21−/− animals, indicating embryonic lethality. To find out whether additional loss of p21 would affect the onset of embryonic lethality, embryos from HDAC1+/− p21−/− breedings were analyzed at days E8.5 and E9.5, respectively. As shown in Fig. 2A, HDAC1−/− p21−/− embryos were severely growth retarded and resembled from their appearance HDAC1−/− p21+/+ embryos (28). Therefore, disruption of the p21 gene did not rescue or delay the onset of embryonic lethality of HDAC1-deficient mice.
TABLE 1.
Genotype of offspring from HDAC1+/− p21−/− × HDAC1+/− p21−/− matings
| Cell phenotype | No. of offspring | % |
|---|---|---|
| HDAC1+/+ p21−/− | 77 | 38.5 |
| HDAC1+/− p21−/− | 123 | 61.5 |
| HDAC1−/− p21−/− | 0 | 0 |
FIG. 2.
Loss of HDAC1 in a p21-deficient background rescues impaired proliferation of HDAC1-null ES cells. (A) Phenotype of HDAC1+/+ and HDAC1−/− embryos in a p21 null background. Embryos were obtained from HDAC1+/− p21−/− crossbreeding at E8.5 and E9.5. The genotype of the embryos was visualized by in situ hybridization with an HDAC1 riboprobe. (B) Immunoblot analysis of HDAC1 and p21 protein expression. Expression of HDAC1 and p21 was examined in protein extracts prepared from HDAC1+/+ p21+/+, HDAC1−/− p21+/+, HDAC1+/+ p21−/−, and HDAC1−/−p21−/− ES cell lines. (C) Growth curves of HDAC1+/+ p21+/+, HDAC1−/− p21+/+, HDAC1+/+ p21−/−, and HDAC1−/−p21−/− ES cell lines. For each cell line, 105 cells were seeded in triplicates, and aliquots were counted daily during a time period of 4 days and shown as mean values ± the SD.
To investigate a possible effect of p21 ablation on the proliferation phenotype of HDAC1-deficient cells, ES cells from HDAC1+/− p21−/− intercrossings were generated by blastocyst outgrowth. We obtained several cell lines for each of the expected genotypes. For further analysis, an HDAC1−/− p21−/− cell line and an HDAC1+/+ p21−/− cell line derived from littermates, was used (Fig. 2B and C). The expression of HDAC1 and p21 in the respective cell lines was controlled by immunoblot analysis (Fig. 2B). To investigate proliferation rates, equal numbers of HDAC1+/+ p21+/+, HDAC1−/− p21+/+, HDAC1+/+ p21−/−, and HDAC1−/− p21−/− ES cells were plated, and cell numbers were determined daily on a Casy Coulter Counter over a period of 4 days. In line with previous observations (28), HDAC1−/− p21+/+ ES cells showed reduced proliferation rates compared to HDAC1+/+ p21+/+ cells (Fig. 2C). HDAC1+/+ cells displayed comparable proliferation rates in the presence (HDAC1+/+ p21+/+) or absence (HDAC1+/+ p21−/−) of p21, indicating that in undifferentiated ES cells loss of p21 does not affect proliferation. Remarkably, HDAC1−/− cells regained normal proliferation rates in the absence of p21 (compare HDAC1−/− p21−/− and HDAC1+/+ p21−/−), indicating that the proliferation of HDAC1-deficient ES cells was rescued by additional ablation of p21. Thus, the CDK inhibitor p21 is a crucial target of HDAC1 as regulator of proliferation in mouse ES cells.
Primary HDAC1−/− MEFs show decreased proliferation rates and are lost during immortalization.
To further investigate a potential role of HDAC1 in cellular proliferation, we intended to generate HDAC1-deficient MEFs. The generation of MEFs derived from embryos as early as E8.5 has been previously described (see, for instance, reference 55). Therefore, we isolated HDAC1+/+ and HDAC1−/− embryos at E8.5 and established primary cell cultures. Several different fibroblast lines were obtained from HDAC1+/+ embryos. However, we did not obtain a single HDAC1 mutant cell line, possibly due to the fact that the HDAC1 mutant embryos are too retarded in development to allow the generation of MEFs. To circumvent this problem, we injected HDAC1−/− ES cells into blastocysts and generated MEFs from chimeric E13.5 embryos (Table 2). Chimeric embryos were identified by PCR with primers specific for the HDAC1-null allele, and fibroblast cultures were prepared. The ratio of HDAC1−/− MEFs to HDAC1+/+ MEFs was analyzed by indirect immunofluorescence microscopy with HDAC1 specific antibodies at passage 1. Several mixed MEF cell populations with up to 80% of HDAC1−/− cells were obtained (Table 2). These cell mixtures were subjected to the 3T3 immortalization protocol, and the fraction of HDAC1-deficient cells at different passages was monitored by indirect immunofluorescence microscopy.
TABLE 2.
Number of chimeric E13.5 embryosa and chimerism of MEFs at passage 1, as identified by immunofluorescence with an HDAC1-specific antibody
| Genotype of injected ES cells | No. of injected blastocysts | No. of chimeric embryos | Avg chimerism (%) |
|---|---|---|---|
| HDAC1−/− p21+/+ | 19 | 4 | 44 |
| HDAC1−/− p21−/− | 21 | 5 | 59 |
Identified by PCR genotyping.
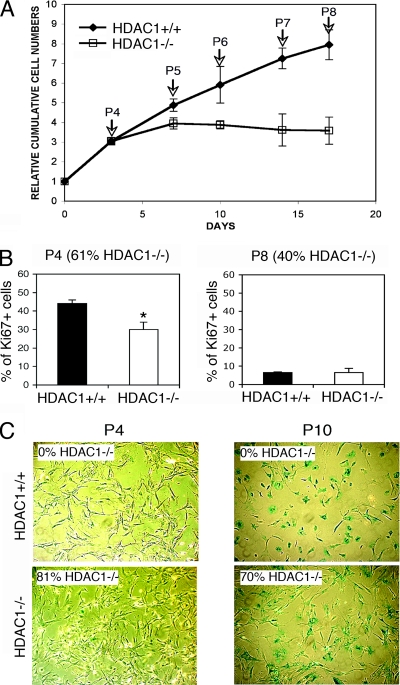
The fraction of HDAC1−/− MEFs was consistently found to decrease during culturing of mixed cell populations derived from different chimeric embryos (Fig. 3A). Therefore, we sought to determine whether the loss of HDAC1−/− MEFs during immortalization is due to reduced proliferation or premature senescence. Immunofluorescence analysis of the proliferation marker Ki67 at passage 4 revealed a significant reduction in proliferation for the HDAC1−/− MEFs compared to HDAC1+/+ MEFs (Fig. 3B). At passage 8, when cells approached the senescence state both HDAC1+/+ and HDAC1−/− MEFs contained a similar small fraction of Ki67-positive cells. When the SA β-Gal activity was measured, almost no senescent cells were observed at passage 4 in both HDAC1+/+ MEFs and MEF populations with 81% HDAC1-deficient cells (Fig. 3C). At passage 10, the fibroblasts entered senescence, as demonstrated by the appearance of SA β-Gal activity and the reduction of Ki67 positive cells (Fig. 3B and C). Importantly, MEF populations that contain ca. 70% of HDAC1−/− cells show similar numbers of SA-β-Gal positive cells at passages 7 and 10 as cell populations that consist exclusively of HDAC1+/+ cells (Fig. 3C and data not shown). Taken together, loss of HDAC1−/− in MEFs leads to reduced proliferation but not premature senescence.
FIG. 3.
Immortalization defect of HDAC1-deficient primary MEFs. (A) Growth curves of HDAC1+/+ and HDAC1−/− MEFs. A total of 3 × 105 MEFs derived from chimeric embryos (mixed HDAC1+/+ and HDAC1−/−) were passaged every 3 to 4 days in triplicates and HDAC1+/+ and HDAC1−/− MEFs were identified by indirect immunofluorescence microscopy. Cumulative cell numbers are presented relative to cell numbers of day 0 as mean values ± the SD. Arrows indicate the passage number. (B) Proliferation of HDAC1+/+ and HDAC1−/− MEFs. Primary fibroblasts isolated from chimeric embryos were analyzed at passages 4 and 8 by indirect immunofluorescence microscopy for the presence of HDAC1 and the proliferation marker Ki67. Nuclear DNA was stained with DAPI. The numbers of Ki67-positive cells were determined of triplicates and are shown as mean values ± the SD. Asterisks indicate statistically significant differences (P = 0.0056 [Student t test]). (C) Senescence of HDAC1+/+ and HDAC1−/− MEFs. Senescent fibroblasts were identified by detection of SA β-Gal activity at passages 4 and 10. The percentage of HDAC1−/− cells is indicated in the upper left corner or each subpanel.
Next, we attempted to obtain pure populations of HDAC1−/− MEFs. The HDAC1−/− allele contains a Neo selection marker that was previously used to identify ES cells with a recombined HDAC1 locus (28). Consequently, MEF populations from chimeras containing HDAC1-deficient cells (as analyzed by PCR of DNA from chimeras) were subjected to selection with G418 (300 μg/ml) from passage 1 onward. Surprisingly, not only HDAC1+/+ cells, but also HDAC1-deficient cells, which were expected to be resistant to selection with G418, underwent excessive cell death after 1 week of G418 selection and were completely lost within 2 weeks. This is not due to silencing of the neo transgene because SV40-LT transformed HDAC1−/− fibroblasts (described below) were still G418 resistant (data not shown). The survival of HDAC1−/− fibroblasts could be dependent on the presence of HDAC1+/+ cells as a consequence of factors secreted or presented by HDAC1 expressing cells. To test this hypothesis, we performed the G418 selection with conditioned medium obtained as supernatant from HDAC1+/+ MEFs. Again, HDAC1−/− fibroblasts failed to survive, suggesting that their proliferation does not depend entirely on factors secreted by HDAC1+/+ cells (data not shown).
Expression of viral oncogenes rescues the proliferation/immortalization defect of HDAC1−/− primary fibroblasts.
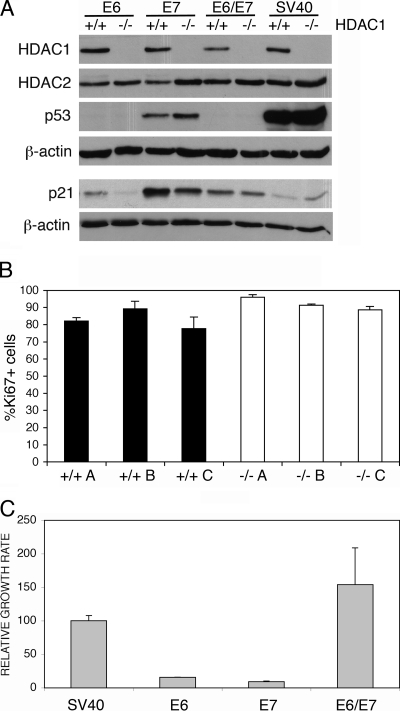
In MEFs spontaneous immortalization is usually accompanied by mutational inactivation of the tumor suppressors p53 (22) and/or p19 (26). Simian virus 40 large T antigen (SV40 LT) transforms cells by inhibiting the ability of pRb family members to repress E2F-dependent transcription (41), as well as by interfering with the ability of p53 to activate transcription of its target genes (3). To analyze whether immortalization mediated by SV40 LT is still functional in HDAC1-deficient MEFs, we transfected mixtures of primary HDAC1+/+ and HDAC1−/− MEFs isolated from chimeric embryos at passage 1 with a retroviral pBABEpuro SV40 LT expression vector. After puromycin selection, single clones were isolated and analyzed by indirect immunofluorescence for HDAC1 expression (data not shown). As previously described (45), Western blot analysis revealed high p53 protein expression upon SV40 LT expression in both HDAC1+/+ and HDAC1−/− MEFs (Fig. 4A). Three clones of each genotype were used for further analysis. As expected, SV40 LT transformed HDAC1+/+ cells showed unrestricted proliferation and did not enter senescence. Importantly, SV40 LT transformed HDAC1−/− MEFs showed similar proliferation rates and did not display any signs of senescence (data not shown). Moreover, Ki67 staining of several SV40 transformed cell lines derived from single clones did not reveal significant differences in the numbers of Ki67-positive cells between SV40 LT transformed HDAC1+/+ and HDAC1−/− fibroblasts (Fig. 4B). We conclude that SV40 LT expression rescues the proliferation/immortalization phenotype of HDAC1-deficient fibroblasts.
FIG. 4.
Transfection with viral oncogenes can rescue the proliferation/immortalization phenotype of primary HDAC1−/− MEFs. (A) Immunoblot analysis of HDAC1, HDAC2, p53, and p21 protein expression. Whole-cell extracts were prepared from HDAC1+/+ and HDAC1−/− MEF single clones obtained after transfection with HPV16 E6, HPV16 E7, HPV16 E6/E7, and SV40 LT. β-actin was used as loading control. (B) Proliferation of transformed HDAC+/+ and HDAC1−/− fibroblasts. SV40 LT-transformed HDAC+/+ and HDAC1−/− fibroblast lines were analyzed by indirect immunofluorescence microscopy for both HDAC1 and Ki67 expression. The fraction of Ki67-positive cells is depicted as the mean value ± the SD of triplicates for three independent cell lines of each genotype. (C) Relative proliferation rates of E6, E7, E6/E7, and SV40 LT-transfected MEFs. A total of 105 cells of at least three different single clones for each transfection were seeded and counted after time period of 3 days. Proliferation rates were determined and are presented as cell numbers per day relative to the value of SV40 LT cell lines in percentages as mean values ± the SD.
Next, we sought to determine whether it would be sufficient to impair either the p53 or the pRB pathway for the rescue of HDAC1−/− MEFs. To this end we transfected primary MEFs isolated from chimeric embryos of mixed WT and HDAC1−/− background with retroviral expression vectors encoding either human papillomavirus 16 (HPV16) E6 or E7 oncoproteins or both oncoproteins (E6/E7). HPV16 E6 protein is known to mediate p53 polyubiqitination and therefore cause rapid degradation of this tumor suppressor gene that directly affects levels of p53 target genes (39). As expected, p53 protein levels were low in E6 and E6/E7 transfected cells (Fig. 4A). In addition, expression of the p53 target gene p21 was reduced in the absence of functional p53 caused by enhanced degradation (E6, E6/E7) or inactivation (SV40 LT). On the other hand, HPV16 E7 protein can directly bind to the pRB tumor suppressor leading to its inactivation and fast synthesis of S phase proteins (15).
Transfection with either HPV16 E6 or E7 resulted in transformation of both HDAC1+/+ and HDAC1−/− cells, which displayed similar proliferation rates regardless of the viral protein transfected. However, the proliferation rates of E6 or E7 only transfected MEFs were more than five times lower than proliferation rates of same cells transfected with SV40 LT antigen regardless of their genotype (Fig. 4C). Transfection of primary MEFs with both E6 and E7 proteins (E6/E7) led to a dramatic increase in the proliferation rate, which was comparable to the proliferation rate of SV40 LT transformed MEFs. Thus, we conclude that inactivation of either p53 or pRb is sufficient to rescue the immortalization phenotype of primary HDAC1−/− MEFs. Inactivation of both p53 and pRB by HPV16 E6/E7 or SV40 LT led to enhanced proliferation in both transformed HDAC1+/+ and HDAC1−/− MEFs.
HDAC inhibitor treatment impairs proliferation of transformed HDAC1+/+ and HDAC1−/− fibroblasts.
Treatment of tumor cells with HDAC inhibitors leads to impaired proliferation. However, the relevant target enzymes are not yet identified. To test the importance of HDAC1 for the growth inhibitory effect of HDAC inhibitors, we treated HDAC1+/+ and HDAC1−/− SV40 LT and E6/E7 HPV16 transformed fibroblasts for 3 days with different concentrations of the general HDAC inhibitor TSA. Treatment with TSA resulted in dose- and time-dependent growth inhibition of all tested cell lines (Fig. 5 and data not shown). Interestingly, both HDAC1+/+ and HDAC1−/− cells were inhibited at similar levels. Similar results were obtained with the HDAC inhibitor suberoylanilide hydroxamic acid also known as Vorinostat or Zolinza (data not shown). Thus, in agreement with several reports on proliferation-associated functions of other HDACs (16, 29, 51, 57), our data suggest that HDAC1 is not the only deacetylase that is relevant for the growth-inhibitory effect of HDAC inhibitors.
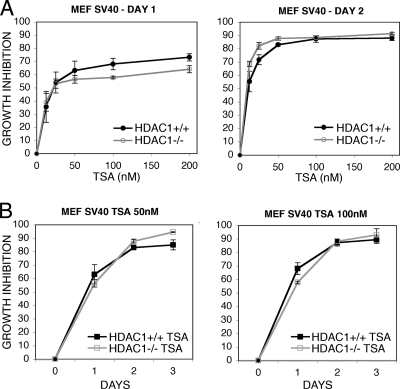
FIG. 5.
Long-term treatment with HDAC inhibitors TSA impairs the proliferation of transformed HDAC1+/+ and HDAC1−/− fibroblasts. (A) Concentration-dependent inhibition of proliferation. The same number of SV40 LT transformed HDAC1+/+ and HDAC1−/− MEFs was plated and treated with the indicated concentrations of TSA. Proliferation in the absence or presence of TSA was measured at days 1 and 2 as described in Materials and Methods, and growth inhibition was calculated relative to untreated control. (B) Time-dependent inhibition of proliferation. Growth curves of SV40-transformed HDAC1+/+ and HDAC1−/− fibroblasts treated with 50 or 100 nM TSA. Growth inhibition was calculated as described for panel A.
Impaired proliferation in HDAC1−/− immortalized MEFs is accompanied by increased levels of p21 and hyperacetylation at the p21 promoter.
In our numerous attempts to isolate fibroblast lines by spontaneous immortalization without viral oncogene transfection we obtained only two HDAC1-deficient MEF lines (HDAC1−/− 1 and HDAC1−/− 2) (Fig. 6A). Compared to HDAC1+/+ cells, the HDAC1−/− fibroblast lines showed reduced proliferation rates (Fig. 6B) and never reached the confluence of HDAC1+/+ fibroblasts (data not shown). To determine whether the cell cycle distribution of HDAC1-deficient fibroblasts was affected, we performed FACS analysis of logarithmically growing HDAC1+/+ and HDAC1−/− cell lines (Fig. 6C). Importantly, HDAC1−/− cells displayed reduced numbers of cycling cells in S phase, while the amount of cells in G1 was increased by ca. 20%. Furthermore, no apoptotic sub-G1 peak was observed, suggesting that impaired proliferation of HDAC1-deficient fibroblasts was not caused by increased apoptosis.
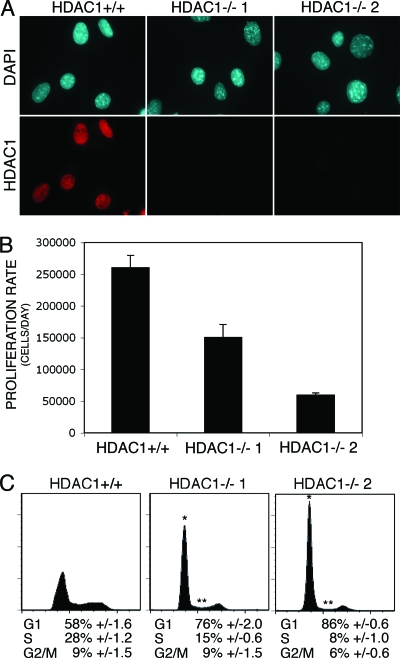
FIG. 6.
Characterization of immortalized HDAC1-deficient fibroblasts. (A) One immortalized HDAC1+/+ (WT) and two immortalized HDAC1−/− fibroblast lines were analyzed for the presence of HDAC1 by indirect immunofluorescence microscopy. Nuclei were visualized by using DAPI. (B) Proliferation rates of HDAC1+/+ and HDAC1−/− immortalized fibroblasts. A total of 105 cells were seeded and counted after time period of 3 days. The data are averages of triplicate determinations and are expressed as number of grown cells per day ± the SD. (C) FACS analysis of HDAC1+/+ and HDAC1−/− fibroblast lines. The experiment shown is representative for three independent experiments for each cell line. Asterisks indicate statistically significant differences in G1 (*) and S (**) phases between HDAC1+/+ and HDAC1−/− cells (for HDAC1−/− 1, PG1 = 0.00028 and PS = 0.00001 and for HDAC1−/− 2, PG1 = 0.0000004 and PS = 0.0000084 [Student t test]).
Since the reduced proliferation rate of HDAC1−/− ES cells was linked with increased levels of p21 (Fig. 2), we analyzed the expression of this CDK inhibitor in the immortalized MEF lines. Intriguingly, the two HDAC1−/− MEF lines showed elevated p21 levels both on the mRNA and the protein level compared to HDAC1+/+ cells (Fig. 7A and B).
FIG. 7.
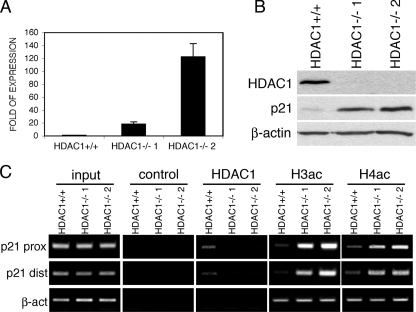
Increased levels of p21 in HDAC1-deficient fibroblasts. (A) Analysis of HDAC1 and p21 mRNA expression by real-time RT-PCR. Expression of p21 is shown relative to β-actin as mean values ± the SD. (B) Analysis of HDAC1 and p21 protein expression. Whole-cell extracts from HDAC1+/+ and HDAC1−/− fibroblast lines were analyzed on an immunoblot by sequential incubation with antibodies specific for HDAC1, p21, and β-actin (as a loading control). (C) Formaldehyde cross-linked chromatin from HDAC1+/+ and HDAC1−/− MEF lines was immunoprecipitated with control antibody or antibodies specific for HDAC1, acetylated H3 (AcH3), and acetylated H4 (AcH4). DNA isolated from immunoprecipitated fractions, as well as total input chromatin, was analyzed by semiquantitative PCR specific for the distal and proximal p21 promoter.
To examine whether HDAC1 directly controls the expression of the p21 gene in MEFs, we performed chromatin immunoprecipitation experiments. To this end, chromatin from HDAC1+/+, HDAC1−/− 1, and HDAC1−/− 2 cell lines was prepared and immunoprecipitated with antibodies specific for HDAC1, acetylated histone H3, acetylated histone H4, and an unspecific control antibody. As shown in Fig. 7C, HDAC1 was present at the proximal and distal promoter region of the p21 gene in HDAC1+/+ cells but not at the control gene β-actin. Loss of HDAC1 led to increased acetylation levels of histones H3 and H4 at both promoter regions of the p21 gene in HDAC1−/− 1 and HDAC1−/− 2 cells, whereas no significant differences in the histone acetylation levels were detected at the β-actin promoter. These data strongly suggest that the increased levels of p21 in HDAC1−/− MEF lines are directly caused by loss of HDAC1.
The immortalization phenotype of HDAC1−/− fibroblasts is rescued by additional ablation of p21.
In ES cells, additional deletion of p21 rescues the proliferation defect of HDAC1−/− ES cells (Fig. 2). Therefore, we wanted to investigate whether the observed immortalization phenotype of HDAC1-deficient MEFs would be rescued in a p21 null background. To this end, HDAC1−/− p21−/− ES cells were injected into blastocysts and MEFs were established from chimeric embryos as described above. Five different MEF populations with up to 73% HDAC1−/− p21−/− primary MEFs were obtained (Table 2). HDAC1+/+ and HDAC1−/− cells were identified by immunofluorescence analysis, and cell numbers were measured over eight passages. In contrast to HDAC1−/− p21+/+ MEFs, the proportion of HDAC1−/− p21−/− MEFs did increase at higher passages in this competitive proliferation assay (Fig. 8A). In line with this observation, HDAC1−/−p21−/− MEFs showed significantly higher numbers of Ki67-positive cells compared to HDAC1+/+ cells (Fig. 8B, compare to Fig. 3B). Moreover, we detected only low numbers of SA β-Gal-positive cells in HDAC1−/− p21−/− cells at passage 10, indicating that these primary fibroblasts enter senescence with delay (Fig. 8C, compare to Fig. 3C). However, from passage 13 onward, the number of Ki67-positive HDAC1−/− p21−/− MEFs was close to that of HDAC1+/+ cells, and the HDAC1−/− p21−/− MEFs displayed a senescence phenotype (data not shown). Nevertheless, the period until spontaneous immortalization occurred seemed to be shortened in HDAC1−/− p21−/− MEFs compared to HDAC1+/+ MEFs. Finally, in contrast to HDAC1−/− p21+/+ ES cells, injection of HDAC1−/− p21−/− ES cells gave rise to immortalized HDAC1 deficient fibroblast lines with high efficiency. In conclusion, our data indicate that impaired spontaneous immortalization of HDAC1-deficient MEFs is rescued by ablation of p21.
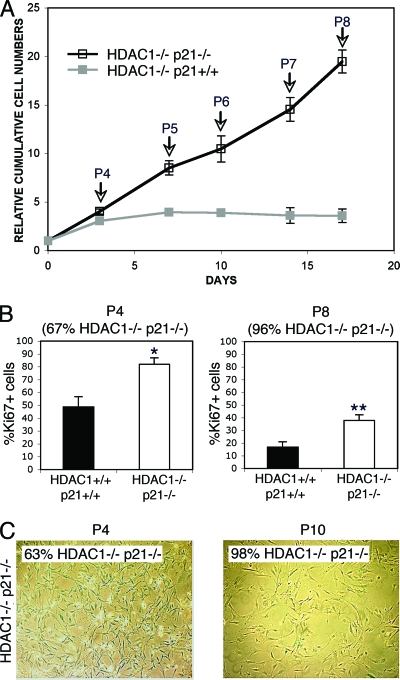
FIG. 8.
Ablation of p21 rescues the immortalization phenotype of HDAC1-deficient fibroblasts. (A) Competitive proliferation assays of HDAC1−/− and HDAC1−/− p21−/− primary fibroblasts. MEFs derived from chimeric mice (mixed HDAC1+/+/HDAC1−/− p21+/+ and mixed HDAC1+/+/HDAC1−/− p21−/−) were passaged every 3 to 4 days in triplicates, and HDAC1-deficient cells were identified by indirect immunofluorescence microscopy. Cumulative cell numbers are presented relative to cell numbers at day 0 as mean values ± the SD (arrows indicate the passage number). (B) Primary fibroblasts isolated from chimeric embryos obtained after injection of HDAC1−/− p21−/− ES cells were analyzed by indirect immunofluorescence microscopy using antibodies specific for HDAC1 and Ki67. Fractions of Ki67 positive HDAC1−/− p21−/− primary fibroblasts are shown for passages 4 and 8. The fraction of HDAC1−/− p21−/− fibroblasts is indicated. Asterisks (* and **) indicate statistically significant differences (*, P = 0.0037; **, P = 0.0041 [Student t test]). (C) Delayed senescence of HDAC1−/− p21−/− fibroblasts. Senescent fibroblasts were detected by SA β-Gal activity at passages 4 and 10. The percentage of HDAC1−/− p21−/− fibroblasts is indicated.
DISCUSSION
Reversibility of HDAC1-mediated effects.
We analyzed the function of HDAC1 in proliferation of murine ES cells and immortalization of MEFs. Loss of HDAC1 was previously shown to result in reduced proliferation rates of ES cells accompanied by increased expression of the CDK inhibitors p21 (28). Reintroduction of HDAC1 led to reversion of the proliferation phenotype and the downregulation of p21 (Fig. 1). These results are in line with previous observations on changes in gene expression in HDAC1-deficient ES cells, which were mostly reversible upon reintroduction of HDAC1 (58). Given that acetylation of core histones is a highly dynamic and reversible process, the reversibility of HDAC1-mediated effects in ES cells is not unexpected.
Additional ablation of p21 rescues the proliferation defect of HDAC1−/− ES cells.
Treatment of tumor cells with HDAC inhibitors results in growth inhibition and/or cell death. Several studies have identified the CDK inhibitor p21 as one of the most reliable target genes of HDAC inhibitors in different cell systems (reviewed in reference 36). HDAC1 was identified as one of the members of the HDAC family that negatively regulate p21 expression in human tumor cells (27). We show here that additional loss of p21 rescues the proliferation phenotype of HDAC1-null ES cells (Fig. 2). Reduced proliferation of HDAC1-deficient ES cells is completely abolished upon deletion of p21. In contrast, ablation of p21 alone does not lead to increased proliferation of HDAC1+/+ ES cells. The latter finding is in accordance with previous results published for p21-deficient MEFs (8).
Cell cycle regulation of ES cells is different from other cell types, such as fibroblasts and tumor cells. ES cells display short gap phases, which are achieved by the constitutive, cell cycle, and mitogen-independent activity of CycE/Cdk2 and CycA/Cdk2 complexes, whereas CycD/Cdk4/6 activity is almost not detected in ES cells (9, 43). Increased expression of p21 (and p27) was found to correlate with severely decreased cyclin A- and cyclin E-associated kinase activity in HDAC1-deficient ES cells (28). Therefore, the reduced cyclin A- and cyclin E-associated kinase activity in HDAC1−/− ES cells might be less efficient in phosphorylating pRb, leading to partial sequestration of E2F by hypophosphorylated pRb and incomplete activation of E2F-dependent S-phase genes. On the other hand, pRb was shown to recruit HDAC1 to target promoters and loss of HDAC1 should therefore result in the activation of E2F target genes (6, 31). However, comparison of the gene expression profiles of HDAC1+/+ and HDAC1−/− ES cells did not reveal significant changes in the expression of E2F target genes (58). We conclude that p21 mediates the antiproliferative effect in murine ES cells by an alternative pathway. One possible explanation is the repressive function of p21 on the essential replication factor proliferating-cell nuclear antigen (PCNA) (49).
Ablation of p21 fails to rescue the developmental phenotype of HDAC1-null embryos.
Disruption of the HDAC1 gene led to severe developmental abnormalities during mouse embryogenesis (28). In contrast to the effect on proliferation, loss of p21 was not sufficient to rescue the developmental phenotype of HDAC1−/− embryos (Fig. 2). The HDAC1−/− p21−/− embryos still died before E10.5 and showed the same developmental defects, including malformed allantois and defects in head formation, as previously described for HDAC1−/− p21+/+ embryos (28). Since p27, another member of the CIP/KIP family of CDK inhibitors, is also upregulated in HDAC1−/− ES cells (28), we ablated HDAC1 in a p27−/− background. However, the embryonic lethality of HDAC1−/− embryos was not rescued in either a p27−/− or a p21−/−p27−/− background (D. Meunier and C. Seiser, unpublished data). Given the role of the tumor suppressor p53 as important regulator of p21 expression, we examined in addition the phenotype of HDAC1 deficient mice in a p53 null background. Again, HDAC1−/− p53−/− embryos were lost before E10.5 (Meunier and Seiser, unpublished). These data suggest that the reduced proliferation rates are not the sole reason for the aberrant development of HDAC1-null embryos but that additional developmental programs are affected by the absence of HDAC1. This idea is supported by the fact that several genes with functions during development are deregulated in HDAC1−/− ES cells (58).
HDAC1-deficient primary MEFs show impaired spontaneous immortalization.
According to our previous findings HDAC1 is crucial for undisturbed cell proliferation in ES cells. However, as mentioned above, ES cells show fundamental differences in cell cycle regulation from somatic cells. Therefore, we aimed to determine the growth properties of HDAC1-deficient fibroblasts. Chimeric embryos containing HDAC1−/− fibroblasts were obtained (see Table 1), indicating that fibroblast development is not abrogated in the absence of HDAC1. This is in line with the finding that embryoid bodies derived from HDAC1−/− ES cells also contained fibroblasts (28). However, HDAC1−/− fibroblasts showed reduced proliferation and were mostly lost during the 3T3 immortalization process. This was not due to premature senescence as previously shown for fibroblasts lacking the Polycomb protein Bmi (25), since no differences in SA β-Gal staining of HDAC1+/+ and MEFs enriched for HDAC1−/− cells were observed (Fig. 3).
In addition to reduced proliferation, HDAC1-deficient primary MEFs also were severely impaired in their ability to spontaneously immortalize. One reason for this effect might be an increased sensitivity of HDAC1-deficient MEFs to the oxidative stress generated by the culture conditions. It is conceivable that HDAC1 is important for the adaptation to stress during immortalization but is dispensable when cells have escaped senescence. In accordance with this idea ablation of HDAC1 in immortalized fibroblasts does not affect the viability and proliferation rate (Patrick Matthias, FMI, unpublished data; J. Jurkin and C. Seiser, unpublished observations).
p21 gene as target for HDACs and antitumor drugs.
The expression of p21 is transiently induced during the course of senescence and might therefore contribute to the cell cycle exit of aging fibroblasts. However, data obtained in studies with human and mouse fibroblasts showed in part divergent results with respect to the role of p21 during immortalization (7, 37). The two HDAC1−/− fibroblast lines that survived the immortalization process displayed reduced proliferation rates accompanied by increased p21 protein expression (Fig. 6 and 7). Strikingly, the severely reduced spontaneous immortalization observed for HDAC1-deficient MEFs is abrogated in a p21-deficient background, suggesting that p21 also plays an important role during immortalization. In accordance with this finding SV40 LT and HPV E6 and E7 rescued the immortalization phenotype. All three viral oncoproteins negatively affect either p21 induction via inhibiting p53 and pRB or p21 function (12, 42). Interestingly, HDAC1-deficient fibroblasts transformed with either SV40 LT or HPV16 E6/E7 show no signs of apoptosis as described for HDAC1-deficient human tumor cells (40). However, in the absence of both HDAC1 and HDAC2, transformed mouse fibroblasts show a severe inhibition of proliferation, suggesting partially redundant functions of the two class I HDACs (21).
The HDAC1 protein was found to be present at the p21 gene in HDAC1+/+ fibroblasts, indicating that HDAC1 is necessary for transcriptional repression of p21 by establishing a hypoacetylated p21 promoter region (Fig. 7). In human tumor cells the HDAC1 repressor protein was shown to be recruited to the p21 promoter by SP1 (27). In addition, other HDACs, such as HDAC2, HDAC3, and HDAC4, were identified as regulator of the p21 gene (30, 53, 54). Correspondingly, the p21 gene is activated by most of the tested HDAC inhibitors, suggesting that p21 might in part mediate the antiproliferation effect of these drugs (18). Indeed, a tumor suppressor function of p21 was proposed in several reports. For instance, the frequency of tumor formation is increased in aged p21−/− mice (34). Moreover, loss of p21 accelerates ras oncogenesis in a mammary cancer model (1). However, there are contradictory reports on whether or not p21 mediates the growth-inhibitory effect of diverse HDAC inhibitors in tumor cells and other cell types (2, 47, 52). Recently, the role of individual HDACs for proliferation of human tumor cells was analyzed. Based on knockdown experiments, the class I deacetylases HDAC1 and HDAC3 were proposed to be relevant targets for anticancer drugs (16, 40). In this setting, siRNA-mediated knockdown of HDAC1 in human tumor cells led to apoptosis independent of p21 presence (40; Susanna Chiocca, unpublished data). The fact that, in addition to HDAC1, other enzymes are also important for the maintenance of unrestricted proliferation of tumor cells.
Redundancy and cell type-specific function of HDAC1.
Enzymatic activity assays performed with ES cells and fibroblasts indicate that HDAC1, as 1 of 18 members of the mammalian HDAC family, significantly contributes to the total cellular deacetylase activity (28; Jurkin and Seiser, unpublished). The actual contribution of HDAC1 might be even underestimated, since HDAC2 as the closest HDAC1 homologue can in part compensate for the loss of HDAC1 (35, 58). The present study, together with previous findings in ES cells (28), demonstrates a positive function of HDAC1 in proliferation. In contrast, loss of HDAC1 in T cells surprisingly leads to increased proliferation, indicating a negative regulatory role of HDAC1 for proliferation in these cell types (R. Grausenburger et al., unpublished data). Obviously, HDAC1 regulates different sets of target genes mediating distinct proliferation signals in particular cell types. These findings suggest a cell-type-specific regulatory role of HDAC1 for proliferation. In order to reveal individual and synergistic activities of HDAC1 and HDAC2, it will be necessary to perform conditional knockout studies for both class I deacetylases in different cell types and tissues.
Acknowledgments
We thank E. Pineda for help with the proliferation assays and N. Foeger for helpful discussions. We are grateful to E. F. Wagner for kindly providing the p21-deficient mice, D. J. McCance for the HPV16 E6/E7 constructs, and I. Mudrak for the pBABE SV40 LT vector.
C.S. was supported by the Austrian Science Fund (FWF P16443 and P18746) and the Herzfelder Family Foundation and the GEN-AU project Epigenetic Plasticity of the Mammalian Genome (Federal Ministry for Education, Science, and Culture), and S.C. was supported by the Associazione Italiana Ricerca sul Cancro. R.G., G.Z., and S.L. were fellows of the Vienna Biocenter PhD program (Austrian Science Fund), and R.B. was a fellow of the Ernst Schering Foundation.
Footnotes
Published ahead of print on 22 December 2009.
REFERENCES
- 1.Adnane, J., R. J. Jackson, S. V. Nicosia, A. B. Cantor, W. J. Pledger, and S. M. Sebti. 2000. Loss of p21WAF1/CIP1 accelerates Ras oncogenesis in a transgenic/knockout mammary cancer model. Oncogene 19:5338-5347. [DOI] [PubMed] [Google Scholar]
- 2.Archer, S. Y., S. Meng, A. Shei, and R. A. Hodin. 1998. p21Waf1 is required for butyrate-mediated growth inhibition of human colon cancer cells. Proc. Natl. Acad. Sci. U. S. A. 95:6791-6796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bargonetti, J., I. Reynisdottir, P. N. Friedman, and C. Prives. 1992. Site-specific binding of wild-type p53 to cellular DNA is inhibited by SV40 T antigen and mutant p53. Genes Dev. 6:1886-1898. [DOI] [PubMed] [Google Scholar]
- 4.Bartl, S., J. Taplick, G. Lagger, H. Khier, K. Kuchler, and C. Seiser. 1997. Identification of mouse histone deacetylase 1 as a growth factor-inducible gene. Mol. Cell. Biol. 17:5033-5043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bhaskara, S., B. J. Chyla, J. M. Amann, S. K. Knutson, D. Cortez, Z. W. Sun, and S. W. Hiebert. 2008. Deletion of histone deacetylase 3 reveals critical roles in S phase progression and DNA damage control. Mol. Cell 30:61-72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brehm, A., E. A. Miska, D. J. McCance, J. L. Reid, A. J. Bannister, and T. Kouzarides. 1998. Retinoblastoma protein recruits histone deacetylase to repress transcription. Nature 391:597-601. [DOI] [PubMed] [Google Scholar]
- 7.Brown, J. P., W. Wei, and J. M. Sedivy. 1997. Bypass of senescence after disruption of p21CIP1/WAF1 gene in normal diploid human fibroblasts. Science 277:831-834. [DOI] [PubMed] [Google Scholar]
- 8.Brugarolas, J., C. Chandrasekaran, J. I. Gordon, D. Beach, T. Jacks, and G. J. Hannon. 1995. Radiation-induced cell cycle arrest compromised by p21 deficiency. Nature 377:552-557. [DOI] [PubMed] [Google Scholar]
- 9.Burdon, T., A. Smith, and P. Savatier. 2002. Signalling, cell cycle and pluripotency in embryonic stem cells. Trends Cell Biol. 12:432-438. [DOI] [PubMed] [Google Scholar]
- 10.Campisi, J. 2000. Cancer, aging and cellular senescence. In Vivo 14:183-188. [PubMed] [Google Scholar]
- 11.Chang, S., T. A. McKinsey, C. L. Zhang, J. A. Richardson, J. A. Hill, and E. N. Olson. 2004. Histone deacetylases 5 and 9 govern responsiveness of the heart to a subset of stress signals and play redundant roles in heart development. Mol. Cell. Biol. 24:8467-8476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Decesse, J. T., S. Medjkane, M. B. Datto, and C. E. Cremisi. 2001. RB regulates transcription of the p21/WAF1/CIP1 gene. Oncogene 20:962-971. [DOI] [PubMed] [Google Scholar]
- 13.Deng, C., P. Zhang, J. W. Harper, S. J. Elledge, and P. Leder. 1995. Mice lacking p21CIP1/WAF1 undergo normal development, but are defective in G1 checkpoint control. Cell 82:675-684. [DOI] [PubMed] [Google Scholar]
- 14.Dimri, G. P., X. Lee, G. Basile, M. Acosta, G. Scott, C. Roskelley, E. E. Medrano, M. Linskens, I. Rubelj, O. Pereira-Smith, et al. 1995. A biomarker that identifies senescent human cells in culture and in aging skin in vivo. Proc. Natl. Acad. Sci. U. S. A. 92:9363-9367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dyson, N., P. M. Howley, K. Munger, and E. Harlow. 1989. The human papillomavirus-16 E7 oncoprotein is able to bind to the retinoblastoma gene product. Science 243:934-937. [DOI] [PubMed] [Google Scholar]
- 16.Glaser, K. B., J. Li, M. J. Staver, R. Q. Wei, D. H. Albert, and S. K. Davidsen. 2003. Role of class I and class II histone deacetylases in carcinoma cells using siRNA. Biochem. Biophys. Res. Commun. 310:529-536. [DOI] [PubMed] [Google Scholar]
- 17.Glozak, M. A., N. Sengupta, X. Zhang, and E. Seto. 2005. Acetylation and deacetylation of non-histone proteins. Gene 363:15-23. [DOI] [PubMed] [Google Scholar]
- 18.Glozak, M. A., and E. Seto. 2007. Histone deacetylases and cancer. Oncogene 26:5420-5432. [DOI] [PubMed] [Google Scholar]
- 19.Gregoretti, I. V., Y. M. Lee, and H. V. Goodson. 2004. Molecular evolution of the histone deacetylase family: functional implications of phylogenetic analysis. J. Mol. Biol. 338:17-31. [DOI] [PubMed] [Google Scholar]
- 20.Gui, C. Y., L. Ngo, W. S. Xu, V. M. Richon, and P. A. Marks. 2004. Histone deacetylase (HDAC) inhibitor activation of p21WAF1 involves changes in promoter-associated proteins, including HDAC1. Proc. Natl. Acad. Sci. U. S. A. 101:1241-1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Haberland, M., A. Johnson, M. H. Mokalled, R. L. Montgomery, and E. N. Olson. 2009. Genetic dissection of histone deacetylase requirement in tumor cells. Proc. Natl. Acad. Sci. U. S. A. 106:7751-7755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Harvey, M., A. T. Sands, R. S. Weiss, M. E. Hegi, R. W. Wiseman, P. Pantazis, B. C. Giovanella, M. A. Tainsky, A. Bradley, and L. A. Donehower. 1993. In vitro growth characteristics of embryo fibroblasts isolated from p53-deficient mice. Oncogene 8:2457-2467. [PubMed] [Google Scholar]
- 23.Hauser, C., B. Schuettengruber, S. Bartl, G. Lagger, and C. Seiser. 2002. Activation of the mouse histone deacetylase 1 gene by cooperative histone phosphorylation and acetylation. Mol. Cell. Biol. 22:7820-7830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Henrique, D., J. Adam, A. Myat, A. Chitnis, J. Lewis, and D. Ish Horowicz. 1995. Expression of a Delta homologue in prospective neurons in the chick. Nature 375:787-790. [DOI] [PubMed] [Google Scholar]
- 24a.Hogan, B., R. Beddington, F. Constantini, and E. Lacy. 1994. Manipulating the mouse embryo: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 25.Jacobs, J. J., K. Kieboom, S. Marino, R. A. DePinho, and M. van Lohuizen. 1999. The oncogene and Polycomb-group gene bmi-1 regulates cell proliferation and senescence through the ink4a locus. Nature 397:164-168. [DOI] [PubMed] [Google Scholar]
- 26.Kamijo, T., F. Zindy, M. F. Roussel, D. E. Quelle, J. R. Downing, R. A. Ashmun, G. Grosveld, and C. J. Sherr. 1997. Tumor suppression at the mouse INK4a locus mediated by the alternative reading frame product p19ARF. Cell 91:649-659. [DOI] [PubMed] [Google Scholar]
- 27.Lagger, G., A. Doetzlhofer, B. Schuettengruber, E. Haidweger, E. Simboeck, J. Tischler, S. Chiocca, G. Suske, H. Rotheneder, E. Wintersberger, and C. Seiser. 2003. The tumor suppressor p53 and histone deacetylase 1 are antagonistic regulators of the cyclin-dependent kinase inhibitor p21/WAF1/CIP1 gene. Mol. Cell. Biol. 23:2669-2679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lagger, G., D. O'Carroll, M. Rembold, H. Khier, J. Tischler, G. Weitzer, B. Schuettengruber, C. Hauser, R. Brunmeir, T. Jenuwein, and C. Seiser. 2002. Essential function of histone deacetylase 1 in proliferation control and CDK inhibitor repression. EMBO J. 21:2672-2681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lee, Y. S., K. H. Lim, X. Guo, Y. Kawaguchi, Y. Gao, T. Barrientos, P. Ordentlich, X. F. Wang, C. M. Counter, and T. P. Yao. 2008. The cytoplasmic deacetylase HDAC6 is required for efficient oncogenic tumorigenesis. Cancer Res. 68:7561-7569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lin, Y. C., J. H. Lin, C. W. Chou, Y. F. Chang, S. H. Yeh, and C. C. Chen. 2008. Statins increase p21 through inhibition of histone deacetylase activity and release of promoter-associated HDAC1/2. Cancer Res. 68:2375-2383. [DOI] [PubMed] [Google Scholar]
- 31.MagnaghiJaulin, L., R. Groisman, I. Naguibneva, P. Robin, S. Lorain, J. P. LeVillain, F. Troalen, D. Trouche, and A. HarelBellan. 1998. Retinoblastoma protein represses transcription by recruiting a histone deacetylase. Nature 391:601-605. [DOI] [PubMed] [Google Scholar]
- 32.Marks, P. A., V. M. Richon, T. Miller, and W. K. Kelly. 2004. Histone deacetylase inhibitors. Adv. Cancer Res. 91:137-168. [DOI] [PubMed] [Google Scholar]
- 33.Marks, P. A., V. M. Richon, and R. A. Rifkind. 2000. Histone deacetylase inhibitors: inducers of differentiation or apoptosis of transformed cells. J. Natl. Cancer Inst. 92:1210-1216. [DOI] [PubMed] [Google Scholar]
- 34.Martin-Caballero, J., J. M. Flores, P. Garcia-Palencia, and M. Serrano. 2001. Tumor susceptibility of p21Waf1/Cip1-deficient mice. Cancer Res. 61:6234-6238. [PubMed] [Google Scholar]
- 35.Montgomery, R. L., C. A. Davis, M. J. Potthoff, M. Haberland, J. Fielitz, X. Qi, J. A. Hill, J. A. Richardson, and E. N. Olson. 2007. Histone deacetylases 1 and 2 redundantly regulate cardiac morphogenesis, growth, and contractility. Genes Dev. 21:1790-1802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ocker, M., and R. Schneider-Stock. 2007. Histone deacetylase inhibitors: signalling toward p21cip1/waf1. Int. J. Biochem. Cell Biol. 39:1367-1374. [DOI] [PubMed] [Google Scholar]
- 37.Pantoja, C., and M. Serrano. 1999. Murine fibroblasts lacking p21 undergo senescence and are resistant to transformation by oncogenic Ras. Oncogene 18:4974-4982. [DOI] [PubMed] [Google Scholar]
- 38.Patel, D., A. Incassati, N. Wang, and D. J. McCance. 2004. Human papillomavirus type 16 E6 and E7 cause polyploidy in human keratinocytes and up-regulation of G2-M-phase proteins. Cancer Res. 64:1299-1306. [DOI] [PubMed] [Google Scholar]
- 39.Scheffner, M., B. A. Werness, J. M. Huibregtse, A. J. Levine, and P. M. Howley. 1990. The E6 oncoprotein encoded by human papillomavirus types 16 and 18 promotes the degradation of p53. Cell 63:1129-1136. [DOI] [PubMed] [Google Scholar]
- 40.Senese, S., K. Zaragoza, S. Minardi, I. Muradore, S. Ronzoni, A. Passafaro, L. Bernard, G. F. Draetta, M. Alcalay, C. Seiser, and S. Chiocca. 2007. Role for histone deacetylase 1 in human tumor cell proliferation. Mol. Cell. Biol. 27:4784-4795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sheng, Q., D. Denis, M. Ratnofsky, T. M. Roberts, J. A. DeCaprio, and B. Schaffhausen. 1997. The DnaJ domain of polyomavirus large T antigen is required to regulate Rb family tumor suppressor function. J. Virol. 71:9410-9416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shin, M. K., S. Balsitis, T. Brake, and P. F. Lambert. 2009. Human papillomavirus E7 oncoprotein overrides the tumor suppressor activity of p21Cip1 in cervical carcinogenesis. Cancer Res. 69:5656-5663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stead, E., J. White, R. Faast, S. Conn, S. Goldstone, J. Rathjen, U. Dhingra, P. Rathjen, D. Walker, and S. Dalton. 2002. Pluripotent cell division cycles are driven by ectopic Cdk2, cyclin A/E and E2F activities. Oncogene 21:8320-8333. [DOI] [PubMed] [Google Scholar]
- 44.Taplick, J., V. Kurtev, K. Kroboth, M. Posch, T. Lechner, and C. Seiser. 2001. Homo-oligomerisation and nuclear localisation of mouse histone deacetylase 1. J. Mol. Biol. 308:27-38. [DOI] [PubMed] [Google Scholar]
- 45.Tiemann, F., and W. Deppert. 1994. Stabilization of the tumor suppressor p53 during cellular transformation by simian virus 40: influence of viral and cellular factors and biological consequences. J. Virol. 68:2869-2878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Trivedi, C. M., Y. Luo, Z. Yin, M. Zhang, W. Zhu, T. Wang, T. Floss, M. Goettlicher, P. R. Noppinger, W. Wurst, V. A. Ferrari, C. S. Abrams, P. J. Gruber, and J. A. Epstein. 2007. Hdac2 regulates the cardiac hypertrophic response by modulating Gsk3 beta activity. Nat. Med. 13:324-331. [DOI] [PubMed] [Google Scholar]
- 47.Vaziri, C., L. Stice, and D. V. Faller. 1998. Butyrate-induced G1 arrest results from p21-independent disruption of retinoblastoma protein-mediated signals. Cell Growth Differ. 9:465-474. [PubMed] [Google Scholar]
- 48.Vega, R. B., K. Matsuda, J. Oh, A. C. Barbosa, X. Yang, E. Meadows, J. McAnally, C. Pomajzl, J. M. Shelton, J. A. Richardson, G. Karsenty, and E. N. Olson. 2004. Histone deacetylase 4 controls chondrocyte hypertrophy during skeletogenesis. Cell 119:555-566. [DOI] [PubMed] [Google Scholar]
- 49.Waga, S., G. J. Hannon, D. Beach, and B. Stillman. 1994. The p21 inhibitor of cyclin-dependent kinases controls DNA replication by interaction with PCNA. Nature 369:574-578. [DOI] [PubMed] [Google Scholar]
- 50.Warrener, R., H. Beamish, A. Burgess, N. J. Waterhouse, N. Giles, D. Fairlie, and B. Gabrielli. 2003. Tumor cell-selective cytotoxicity by targeting cell cycle checkpoints. FASEB J. 17:1550-1552. [DOI] [PubMed] [Google Scholar]
- 51.Weichert, W., A. Roske, S. Niesporek, A. Noske, A. C. Buckendahl, M. Dietel, V. Gekeler, M. Boehm, T. Beckers, and C. Denkert. 2008. Class I histone deacetylase expression has independent prognostic impact in human colorectal cancer: specific role of class I histone deacetylases in vitro and in vivo. Clin. Cancer Res. 14:1669-1677. [DOI] [PubMed] [Google Scholar]
- 52.Wharton, W., J. Savell, W. D. Cress, E. Seto, and W. J. Pledger. 2000. Inhibition of mitogenesis in BALB/c-3T3 cells by Trichostatin A. Multiple alterations in the induction and activation of cyclin-cyclin-dependent kinase complexes. J. Biol. Chem. 275:33981-33987. [DOI] [PubMed] [Google Scholar]
- 53.Wilson, A. J., D. S. Byun, S. Nasser, L. Murray, K. Ayyanar, D. Arango, M. Figueroa, A. Melnick, G. D. Kao, L. H. Augenlicht, and J. M. Mariadason. 2008. HDAC4 promotes growth of colon cancer cells via repression of p21. Mol. Biol. Cell. 19:4062-4075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wilson, A. J., D. S. Byun, N. Popova, L. B. Murray, K. L'Italien, Y. Sowa, D. Arango, A. Velcich, L. H. Augenlicht, and J. M. Mariadason. 2006. Histone deacetylase 3 (HDAC3) and other class I HDACs regulate colon cell maturation and p21 expression and are deregulated in human colon cancer. J. Biol. Chem. 281:13548-13558. [DOI] [PubMed] [Google Scholar]
- 55.Ylikorkala, A., D. J. Rossi, N. Korsisaari, K. Luukko, K. Alitalo, M. Henkemeyer, and T. P. Makela. 2001. Vascular abnormalities and deregulation of VEGF in Lkb1-deficient mice. Science 293:1323-1326. [DOI] [PubMed] [Google Scholar]
- 56.Zhang, C. L., T. A. McKinsey, S. Chang, C. L. Antos, J. A. Hill, and E. N. Olson. 2002. Class II histone deacetylases act as signal-responsive repressors of cardiac hypertrophy. Cell 110:479-488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zimmermann, S., F. Kiefer, M. Prudenziati, C. Spiller, J. Hansen, T. Floss, W. Wurst, S. Minucci, and M. Gottlicher. 2007. Reduced body size and decreased intestinal tumor rates in HDAC2-mutant mice. Cancer Res. 67:9047-9054. [DOI] [PubMed] [Google Scholar]
- 58.Zupkovitz, G., J. Tischler, M. Posch, I. Sadzak, K. Ramsauer, G. Egger, R. Grausenburger, N. Schweifer, S. Chiocca, T. Decker, and C. Seiser. 2006. Negative and positive regulation of gene expression by mouse histone deacetylase 1. Mol. Cell. Biol. 26:7913-7928. [DOI] [PMC free article] [PubMed] [Google Scholar]