Abstract
Ribosome biogenesis in eukaryotes is a major cellular activity mobilizing the products of over 200 transcriptionally coregulated genes referred to as the rRNA and ribosome biosynthesis regulon. We investigated the function of an essential, uncharacterized gene of this regulon, renamed RRP36. We show that the Rrp36p protein is nucleolar and interacts with 90S and pre-40S preribosomal particles. Its depletion affects early cleavages of the 35S pre-rRNA and results in a rapid decrease in mature 18S rRNA levels. Rrp36p is a novel component of the 90S preribosome, the assembly of which has been suggested to result from the stepwise incorporation of several modules, including the tUTP/UTP-A, PWP2/UTP-B, and UTP-C subcomplexes. We show that Rrp36p depletion does not impair the incorporation of these subcomplexes and the U3 small nucleolar RNP into preribosomes. In contrast, depletion of components of the UTP-A or UTP-B modules, but not Rrp5p, prevents Rrp36p recruitment and reduces its accumulation levels. In parallel, we studied the human orthologue of Rrp36p in HeLa cells, and we show that the function of this protein in early cleavages of the pre-rRNA has been conserved through evolution in eukaryotes.
Ribosome biogenesis begins with the transcription of the ribosomal DNA units by RNA polymerase I (RNA Pol I) in the nucleolus, generating a primary transcript cotranscriptionally assembled within nascent preribosomal particles. In Saccharomyces cerevisiae, the earliest described preribosome, defined as the 90S preribosome or small subunit (SSU) processome, contains the 35S rRNA precursor (pre-rRNA), resulting from the cotranscriptional cleavage of the primary transcript by Rnt1p (10, 15). This 35S precursor contains the sequences corresponding to three of the four RNA components of mature ribosomes, namely, the 18S, 5.8S, and 25S rRNAs, flanked by external transcribed spacers (5′ETS and 3′ETS) and separated by internal transcribed spacers (ITS1 and ITS2). The precursor to the fourth mature rRNA (5S) is synthesized independently by RNA Pol III and is incorporated within early 90S preribosomes as part of a small ribonucleoprotein particle (46). During the maturation of the preribosomal particles, the 35S pre-rRNA undergoes a complex processing pathway consisting of (i) the introduction of posttranscriptional modifications (mainly 2′-O ribose methylations and pseudouridylations) and (ii) a complex series of endonucleolytic and exonucleolytic processing events (see Fig. 3C) that remove the spacer sequences and release the mature rRNAs. Simultaneously with these processing events, the ribosomal proteins are progressively incorporated into the maturing particles. (For a recent, more detailed review on the maturation of the preribosomal particles, see reference 16.)
FIG. 3.
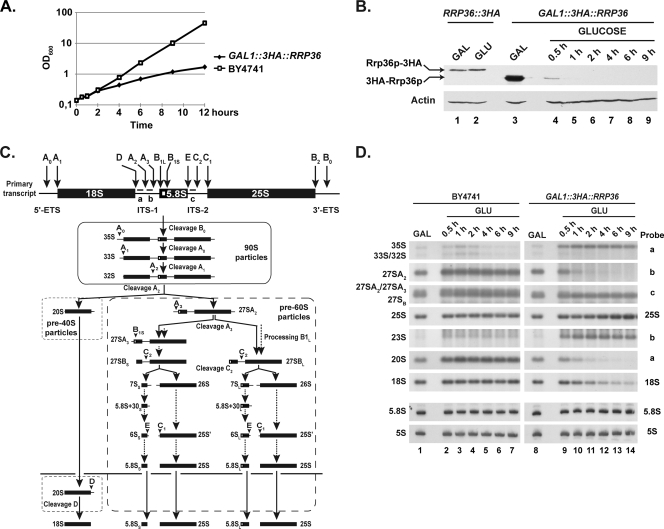
Rrp36p depletion affects early cleavages of the pre-rRNA and the production of the mature 18S rRNA in yeast cells. The GAL1::3HA::RRP36 or BY4741 (WT) strain was shifted from a galactose- to a glucose-based medium and maintained under conditions of exponential growth. (A) Growth curve of yeast cells undergoing Rrp36p depletion. Growth of the strains was followed by measuring the optical density of the cultures at 600 nm (OD600) at different times after the nutritional shift. (B) Depletion of 3HA-Rrp36p following the nutritional shift. Total proteins were extracted from the RRP36::3HA strain grown on galactose (GAL)- or glucose (GLU)-containing medium and from the GAL1::3HA::RRP36 strain before transfer to the glucose-based medium (GAL) and at different times after transfer to the glucose-based medium (GLUCOSE). The accumulation of 3HA-tagged Rrp36p proteins and actin (loading control) was assessed by Western blotting using anti-HA and actin-specific antibodies, respectively. Note that Rrp36p-3HA is slightly larger than 3HA-Rrp36p due to the insertion of a linker sequence, in addition to the 3HA tag, into the Rrp36p-3HA fusion protein. (C) Scheme of the pre-rRNA processing pathway in S. cerevisiae. Endonucleolytic cleavages and exonucleolytic trimmings are marked by solid and dotted arrows, respectively. The positions of the oligonucleotide probes used to detect the different pre-rRNAs analyzed in this study (a, b, and c) are shown on the initial precursor; their sequences are detailed in Table S1 in the supplemental material. (D) Early pre-rRNA processing defects in cells undergoing Rrp36p depletion. The BY4741 and GAL1::3HA::RRP36 strains were shifted from a galactose- to a glucose-based medium, and culture samples were collected before the nutritional shift (GAL) and at different times after the nutritional shift (GLU). Total RNAs were extracted from these cell samples, and the accumulation of the different (pre-)rRNAs was analyzed by Northern blotting using the indicated oligonucleotide probes (see Fig. 3C and Table S1 in the supplemental material).
Different versions of the early 90S preribosomal particle have been purified from yeast cells, and the protein composition has been determined by mass spectrometry (9, 12). These particles contain the 35S pre-rRNA, a specific subset of SSU ribosomal proteins, the U3 small nucleolar RNP (snoRNP), and more than 30 so-called “nonribosomal” or “trans-acting” factors, which associate transiently with the preribosomal particles and are not present in the mature ribosomes. The previously uncharacterized factors identified in these particles were named “Utp” for U three-associated protein (3, 9) and were shown to be required for early cleavages of the 35S pre-rRNA and the synthesis of the small ribosomal subunit (3, 7, 9, 12). Although many components of the 90S preribosomes have been identified and characterized, it remains unclear how they cotranscriptionally assemble with the nascent transcript to yield a processing-competent particle. Recent reports have suggested that the assembly of the 90S preribosomes results from the cotranscriptional association of several preformed modules, interacting with the nascent transcript in a stepwise and ordered manner. At least five such modules have been isolated or suggested to exist in yeast, namely, the tUTP/UTP-A (11, 18), PWP2/UTP-B (8, 12, 18), and UTP-C (18) subcomplexes, as well as the Imp3p-Imp4p-Mpp10p ternary complex (43) and the Rcl1p-Bms1p module (4, 17, 42). Components of the tUTP/UTP-A complex (11, 25, 44) as well as Mdr1p (33) associate very early with the nascent transcript, and their incorporation is a prerequisite for the subsequent association of other subcomplexes. Following association of the tUTP/UTP-A complex, the PWP2/UTP-B and UTP-C modules seem to assemble independently from one another, but the order of incorporation remains unclear (25). Integration of the U3 snoRNP and the Imp3p-Imp4p-Mpp10p module within nascent preribosomes requires prior loading of the PWP2/UTP-B subcomplex (8). Association of another component of the SSU processome, Rrp5p (37), is required for the interaction between the UTP-C module and the 35S pre-rRNA (25) and also for the recruitment of the DEAD-box protein Rok1p (38), a putative RNA helicase required for early cleavages of the pre-rRNA and the production of mature 18S rRNA (36). The basic mechanisms of this assembly process seem to be evolutionarily conserved in eukaryotes, since complexes homologous to the yeast tUTP/UTP-A (27) and Imp3p-Imp4p-Mpp10p (13) modules have been characterized in mammalian cells, as has a novel SSU processome assembly intermediate containing the U3 snoRNP, nucleolin, RRP5, and DBP4 (35). In yeast, proper assembly of the SSU processome has recently been shown to be monitored by a nucleolar surveillance mechanism responsible for degradation by the TRAMP5 complex of the RNA components of aberrant particles (44). Interestingly, the assembly and maturation of the preribosomal particles in yeast cells have recently been proposed to be coordinated with expression of the ribosomal proteins via a mechanism implicating a direct interaction between the UTP-C complex and Ifh1p, a factor controlling transcription of the ribosomal protein genes (30).
In yeast, numerous genes encoding factors required for rRNA synthesis and preribosome maturation are coregulated at the transcriptional level as cells pass through a variety of physiological transitions (23, 39, 40). These genes have been collectively referred to as the rRNA and ribosome biosynthesis (RRB) regulon. In a search for novel yeast trans-acting factors involved in ribosome biogenesis, we analyzed a recently extended list of more than 200 genes constituting the regulon (40). We noticed the presence of a noncharacterized, essential gene identified as YOR287C. We undertook the characterization of this gene in both yeast and mammalian cells, and we show that it fulfills an evolutionarily conserved function in early cleavages of the pre-rRNA and the synthesis of the 40S ribosomal subunit. We renamed this gene RRP36, since the encoded protein is a novel ribosomal RNA processing factor of 36 kDa in yeast.
MATERIALS AND METHODS
Yeast strains, mammalian cells, plasmids, and media.
Yeast strains used in this study were derivatives of either S. cerevisiae strain BY4741, originating from the S288C background (MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0), or of S. cerevisiae strain BMA64 [MATa ura3-1 trp1Δ2 ade2-1 (ochre) leu2-3,112 his3-11,15 can1-100 (ochre)] (2). UTP15::TAP, UTP17::TAP, UTP22::TAP, and PWP2::TAP strains were purchased from Open Biosystems. In these strains, insertion of the tandem affinity purification (TAP) cassette has been selected using the HIS3MX6 marker. The RRP36::3HA strain, encoding Rrp36p-3HA (HA indicates influenza virus hemagglutinin epitopes) under the control of the endogenous RRP36 promoter, was generated as follows: a PCR cassette containing the 3HA tag sequence and a kanamycin resistance (kanrMX6) selectable marker generated using OHA282 and OHA283 oligonucleotides (see Table S1 in the supplemental material) was inserted by homologous recombination downstream of the chromosomal RRP36 ORF in the BY4741, UTP15::TAP, UTP17::TAP, UTP22::TAP, and PWP2::TAP strains as described in reference 22. The GAL1::TAP::RRP36 strains, expressing TAP-Rrp36p under the control of the GAL1 promoter, was constructed as follows: a PCR cassette containing the Kluyveromyces lactis TRP1 selectable marker, the GAL1 promoter, and the TAP tag sequence was amplified by PCR from plasmid pBS1761 (28) by the use of primers OHA292 and OHA293 (see Table S1 in the supplemental material). The PCR fragment was inserted by homologous recombination upstream of the chromosomal RRP36 open reading frame (ORF) in strain BMA64, resulting in the deletion of 50 nucleotides upstream of the ORF in the RRP36 promoter and the in-frame fusion of the TAP tag sequence to the RRP36 ORF. The GAL1::3HA::RRP36 strain, allowing conditional expression of 3HA-Rrp36p, was constructed as follows: a PCR cassette containing the kanrMX6 selectable marker followed by the GAL1 promoter and the 3HA tag sequence, generated with OHA211 and OHA212 oligonucleotides (see Table S1 in the supplemental material), was inserted by homologous recombination upstream of the chromosomal RRP36 ORF in the BY4741, UTP15::TAP, UTP17::TAP, UTP22::TAP, and PWP2::TAP strains as described in reference 22. Insertion of the cassette resulted in the deletion of 50 nucleotides upstream of the ORF in the RRP36 promoter and the in-frame fusion of the 3HA tag sequence to the RRP36 ORF. When grown in a rich medium containing 2% galactose, the GAL1::3HA::RRP36 strain displays the same generation time as a wild-type (WT) strain, suggesting that the N-terminal fusion to the 3HA tag does not interfere with Rrp36p function. To generate the GAL1::UTP17, GAL1::PWP2, and GAL1::RRP5 strains expressing Rrp36p-3HA, PCR cassettes containing the HIS3MX6 selectable marker and the GAL1 promoter (generated with oligonucleotide pair OMG054 and OMG055, oligonucleotide pair OMG050 and OMG051, and oligonucleotide pair OMG058 and OMG059, respectively [described in Table S1 in the supplemental material]) were inserted by homologous recombination upstream of the chromosomal UTP17, PWP2, and RRP5 ORFs, respectively, in the previously described RRP36::3HA strain, as described in reference 22. In each case, insertion of the cassette resulted in the deletion of a region of the targeted promoter corresponding to the 50 nucleotides upstream of the ORF. The oligonucleotides used to generate the different PCR cassettes have been designed according to the method described in reference 22.
S. cerevisiae strains were grown either in YP medium (1% yeast extract, 1% peptone) (Becton-Dickinson) supplemented with 2% galactose or 2% glucose as the carbon source (rich medium) or in YNB medium [0.17% yeast nitrogen base (MP Biomedicals), 0.5% (NH4)2SO4] supplemented with 2% galactose or 2% glucose and the required amino acids (minimal medium). Selection of the Kanr transformants was achieved by addition of G418 at a final concentration of 0.2 mg/ml.
HeLa cells were grown in Dulbecco's modified Eagle medium (DMEM) supplemented with 10% fetal bovine serum (Gibco), 1 mM sodium pyruvate (Gibco), and 10 U/ml penicillin and 10 μg/ml streptomycin (Gibco) at 37°C with 5% CO2.
Plasmid pUN100-GFP-NOP1 was generated in the Hurt laboratory. Plasmid pEGFP-RRP36 was constructed as follows: the RRP36 coding sequence was amplified by PCR starting from an IMAGE clone containing the full-length C6orf153 cDNA (clone 4300313; Open Biosystems), with Phusion DNA polymerase (Finnzymes) in the optional “GC buffer.” The primers used for this reaction were designed to insert an XhoI site upstream (OCB021) and a BamHI site downstream (OCB022) of the RRP36 coding sequence in the resulting PCR product. This product was digested by XhoI and BamHI and inserted into the pEGFP-C1 vector (Clontech) digested by the same enzymes.
Sucrose gradient sedimentation experiments.
Approximately 6 × 109 cells growing exponentially were treated for 10 min with 50 μg/ml cycloheximide (Sigma) added directly to the culture medium. Cells were collected by centrifugation, rinsed with buffer K (20 mM Tris-Cl [pH 7.4], 50 mM KCl, 10 mM MgCl2) supplemented with 50 μg/ml cycloheximide, and collected again by centrifugation. Dry pellets were resuspended with approximately 1 volume of ice-cold buffer K supplemented with 1 mM dithiothreitol, 1× Complete EDTA-free protease inhibitor cocktail (Roche), 0.1 U/μl RNasin (Promega), and 50 μg/ml cycloheximide. About 250 μl of ice-cold glass beads (Sigma) was added to 500-ml aliquots of the resuspended cells, which were broken by vigorous shaking three times for 2 min each time, separated by 2-min incubations on ice. Extracts were clarified through three successive centrifugations at 13,000 rpm and 4°C for 5 min and quantified by measuring absorbance at 260 nm. About 30 A260 units were loaded on 4.5% to 45% sucrose gradients in buffer K, prepared using a Gradient Master 107ip (Biocomp) gradient maker. Gradients were centrifuged for 150 min at 39,000 rpm and 4°C in an Optima L-100XP Ultracentrifuge (Beckman-Coulter) using the SW41Ti rotor without brake. Following centrifugation, 18 fractions of 500 μl each were collected from the top of the gradients by use of a Foxy Jr. fraction collector (Teledyne ISCO). The absorbance at 254 nm was measured during collection with a UA-6 device (Teledyne ISCO).
Immunoprecipitation experiments.
The protocol used to immunoprecipitate TAP-tagged proteins was adapted from the method described in reference 29. Cell pellets corresponding to about 6 × 109 cells were resuspended with approximately 1 volume of ice-cold A200 KCl buffer (20 mM Tris-Cl [pH 8.0], 5 mM MgAc, 200 mM KCl, 0.2% Triton X-100) supplemented with 1 mM dithiothreitol, 1× Complete EDTA-free protease inhibitor cocktail (Roche), and 0.1 U/μl RNasin (Promega). About 250 ml of ice-cold glass beads (Sigma) was added to 500-μl aliquots of the resuspended cells, which were broken by vigorous shaking three times for 2 min each time, separated by 2-min incubations on ice. Extracts were clarified through three successive centrifugations at 13,000 rpm and 4°C for 5 min and quantified by measuring absorbance at 260 nm. About 60 A260 units were incubated for 2 h at 4°C with 15 μl (bed volume) of immunoglobulin G (IgG)-Sepharose beads (GE Healthcare) in a total volume of 1 ml (adjusted with A200 KCl buffer supplemented with 1 mM dithiothreitol, 1× Complete EDTA-free protease inhibitor cocktail, and 0.1 U/μl RNasin) on a rocking table. For RNA analyses, beads were washed seven times with 1 ml of ice-cold A200 KCl buffer supplemented with 1 mM dithiothreitol and 1× Complete EDTA-free protease inhibitor cocktail. RNAs were extracted from bead pellets as described in reference 34: 160 μl of 4 M guanidium isothiocyanate solution, 4 μl of glycogen (Roche), 80 ml of a mixture consisting of 10 mM Tris-Cl (pH 8.0), 1 mM EDTA (pH 8.0), and 100 mM NaAc, 120 μl of phenol, and 120 μl of chloroform were added. Tubes were shaken vigorously, incubated 5 min at 65°C, and centrifuged 5 min at 13,000 rpm (4°C). Aqueous phases (240 μl) were mixed vigorously with 120 μl of phenol and 120 μl of chloroform and centrifuged 5 min at 13,000 rpm (4°C), and the resulting aqueous phases were ethanol precipitated. For protein analyses, beads were washed five times with 1 ml of ice-cold A200 KCl buffer supplemented with 1 mM dithiothreitol and 1× Complete EDTA-free protease inhibitor cocktail and two additional times with 1 ml of A200 NaCl buffer (20 mM Tris-Cl [pH 8.0], 5 mM MgAc, 200 mM NaCl, 0.2% Triton X-100) supplemented with 1 mM dithiothreitol and 1× Complete EDTA-free protease inhibitor cocktail. Beads were resuspended directly with 2× sodium dodecyl sulfate (SDS) gel-loading buffer (100 mM Tris-Cl [pH 6.8], 4% SDS, 20% glycerol, 200 mM dithiothreitol, 0.2% bromophenol blue).
Tandem affinity purifications.
Cell pellets (corresponding to about 5 × 1010 cells) frozen in liquid nitrogen were broken with a planetary mill (Pulverisette 6; Fritsch) two times for 5 min at 500 rpm in the presence of 25 ml of ice-cold glass beads and 10 ml of A200 KCl buffer (20 mM Tris-Cl [pH 8.0], 5 mM MgAc, 200 mM KCl, 0.2% Triton X-100) supplemented with 1 mM dithiothreitol, 1× Complete EDTA-free protease inhibitor cocktail (Roche), and 0.5 U/μl RNasin (Promega). For TAP purifications under native conditions, cell extracts were clarified by centrifugation at 13,000 rpm (20,400 × g) for 15 min at 4°C in a Ti 50.2 rotor (Beckman). For TAP purifications under stringent conditions, cell extracts were clarified by centrifugation at 180,000 × g for 45 min at 4°C in a Ti 50.2 rotor (Beckman) to pellet high-molecular-weight complexes and retain only smaller modules as described in reference 18. Clarified extracts were incubated with 200 μl (bed volume) of IgG-Sepharose beads (GE Healthcare) for 2 h on a rocking table. Beads were washed with 80 ml of ice-cold A200 KCl buffer supplemented with 1 mM dithiothreitol and then with 30 ml of ice-cold TEV cleavage buffer (10 mM Tris-Cl [pH 8.0], 150 mM NaCl, 0.1% NP-40, 0.5 mM EDTA, 1 mM dithiothreitol). Beads were resuspended with 1 ml of TEV cleavage buffer and incubated for 2 h at 16°C on a rocking table in the presence of 100 units of ActTEV protease (Invitrogen). Eluted samples (about 1 ml) were mixed with 3 ml of calmodulin binding buffer (10 mM Tris-Cl [pH 8.0], 150 mM NaCl, 1 mM MgAc, 1 mM imidazole, 2 mM CaCl2, 0.1% NP-40, 10 mM ß-mercaptoethanol) and 3 μl of 1 M CaCl2 and incubated with 200 μl of calmodulin beads (Stratagene) at 4°C for 1 h on a rocking table. Beads were washed with 40 ml of calmodulin binding buffer, and proteins were eluted by the addition of 6 × 200 μl of calmodulin elution buffer (10 mM Tris-Cl [pH 8.0], 150 mM NaCl, 1 mM MgAc, 1 mM imidazole, 2 mM EGTA, 0.1% NP-40, 10 mM β-mercaptoethanol). Eluted proteins were precipitated with 20% trichloroacetic acid (TCA), rinsed with acetone, and resuspended with 15 μl of 1× SDS gel-loading buffer (40 mM Tris-Cl [pH 6.8], 2% SDS, 10% glycerol, 25 mM dithiothreitol, 0.1% bromophenol blue). The samples were reduced for 30 min at 53°C in a shaking thermomixer (Eppendorf) and then alkylated for 30 min at room temperature in a shaking thermomixer in the presence of 75 mM iodoacetamide (Sigma). Samples were loaded on 10% acrylamide-bisacrylamide (29:1)-SDS gels, run through the stacking gel, and allowed to migrate through about 0.5 cm of the resolving gel. Gels were briefly (15 min) stained with PageBlue protein staining solution (Fermentas), and pieces of gels containing the samples were excised. The proteins contained in these samples were analyzed by mass spectrometry as described in reference 19.
Protein extractions, TCA precipitations, and Western blot analyses.
Total protein extracts used for the Western blot analyses (see Fig. 3B and Fig. 5B) were prepared as follows: cell pellets corresponding to approximately 108 cells were resuspended with 200 μl of 2× SDS gel-loading buffer (100 mM Tris-Cl [pH 6.8], 4% SDS, 20% glycerol, 200 mM dithiothreitol, 0.2% bromophenol blue), and 500 μl of glass beads was added. Cells were broken by vigorous agitation at 4°C for 5 min. Another 200 μl of 2× SDS gel-loading buffer was added, and samples were homogenized by vigorous agitation for 1 min at 4°C, incubated 10 min at 65°C, and centrifuged 15 min at 10,000 rpm and 4°C. Proteins contained in the supernatants were separated on SDS-10% polyacrylamide gels and transferred to Amersham Hybond-C Extra membranes (GE Healthcare). HA-tagged Rrp36p was detected using horseradish peroxidase (HRP)-conjugated anti-HA antibodies (clone 3F10; Roche) diluted to 1:1,000 in PBST buffer (137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 2 mM KH2PO4, 0.1% Tween 20) containing 5% (wt/vol) milk (Régilait, Pâturage, or Delisse without added vitamins). Actin was detected using mouse antiactin monoclonal antibodies (clone ACTN05; Thermo Scientific) diluted to 1:500 in PBST buffer containing 5% milk. Lcp5p was detected using specific polyclonal antibodies (generously provided by Thomas Wiederkehr, Basel, Switzerland) diluted to 1:1,000 in the same buffer. For actin and Lcp5p detection, membranes were washed three times for 10 min with PBST buffer containing 1% milk following incubation with the primary antibodies, incubated for 2 h with the relevant HRP-conjugated secondary antibodies diluted to 1:10,000 in PBST buffer containing 1% milk, and finally washed three times for 10 min each time with PBST buffer. Detections were achieved using either regular enhanced chemoluminescence (ECL) Western blotting detection reagents (GE Healthcare) or a more sensitive “ECL Advance” Western blotting detection kit (GE Healthcare), following the provided protocol.
FIG. 5.
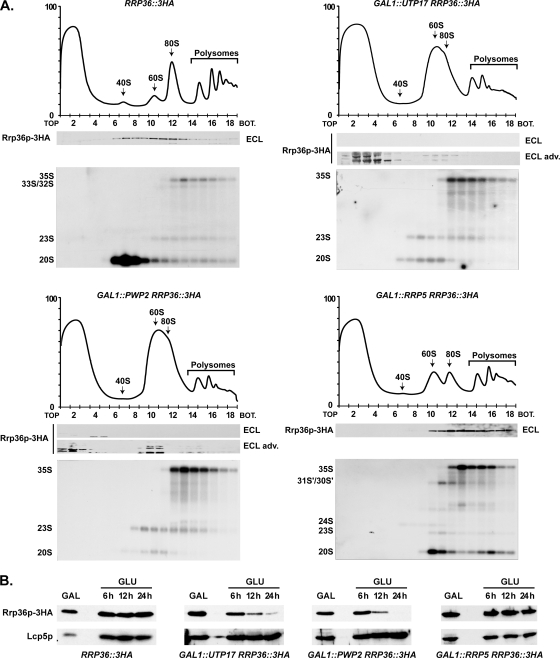
Components of the UTP-A and UTP-B complexes, but not Rrp5p, are required for the incorporation of Rrp36p into preribosomes. (A) Sedimentation profile of Rrp36p-3HA in WT cells (upper left panel) or in cells lacking Utp17p (upper right panel), Pwp2p (lower left panel), or Rrp5p (lower right panel). The GAL1::UTP17, GAL1::PWP2, and GAL1::RRP5 strains expressing Rrp36p-3HA were shifted from galactose- to glucose-containing medium and grown for 16, 18, or 14 h, respectively, to deplete the corresponding protein according to the method described in reference 25. As a control, the RRP36::3HA strain grown in the presence of glucose was used. Total extracts prepared from these cell samples were sedimented through sucrose gradients. The proteins contained in one-half of each fraction were analyzed by Western blotting using anti-HA antibodies to detect Rrp36p-3HA (note that Rrp36p-3HA was detected using regular ECL in WT and Rrp5p-depleted cells and the more sensitive “ECL Advance” [ECL adv.] kit in Utp17p- and Pwp2p-depleted cells). Total RNAs were extracted from the other half of each fraction, and the pre-rRNAs were detected by Northern blotting using probe a (Fig. 3C). BOT, bottom. (B) Accumulation levels of Rrp36p-3HA in WT or Utp17p-, Pwp2p-, or Rrp5p-depleted cells. The WT, GAL1::UTP17, GAL1::PWP2, and GAL1::RRP5 strains expressing Rrp36p-3HA were transferred from a galactose- to a glucose-containing medium, and cultures were maintained under conditions of exponential growth. Total protein extracts prepared from cell samples harvested before the nutritional shift (GAL) and 6, 12, and 24 h after the nutritional shift (GLU) were analyzed by Western blotting. Rrp36p-3HA was detected using anti-HA antibodies and regular ECL. Lcp5p was detected using specific antibodies.
Proteins contained in sucrose gradient fractions (Fig. 1B; also see Fig. 4 and 5A) were concentrated by TCA precipitation as follows: ice-cold TCA (Sigma) was added to 250 μl (about one-half) of each fraction to achieve a final concentration of 15%. Samples were mixed by inversion, incubated about 75 min on ice, and centrifuged 20 min at 13,000 rpm and 4°C. To wash the pellets, 500 ml of acetone was added and the samples were centrifuged 5 min at 13,000 rpm and 4°C. Supernatants were removed completely, and pellets were air dried and resuspended with 30 μl of NuPAGE lithium dodecyl sulfate sample buffer supplemented with NuPAGE sample-reducing agent (Invitrogen). These protein samples were heated 15 min at 65°C, loaded on NuPAGE Novex 4-12% Bis-Tris gradient gels (Invitrogen), and transferred to nitrocellulose membranes by using an iBlot dry blotting system (Invitrogen). 3HA-tagged Rrp36p was detected as described above. TAP-tagged proteins were detected using rabbit peroxidase antiperoxidase (PAP; Dako) diluted to 1:10,000 in PBST buffer containing 5% milk.
FIG. 1.
Rrp36p is a nucleolar protein cosedimenting with preribosomes in yeast cells. (A) Subcellular localization of Rrp36p. The RRP36::3HA strain transformed with a plasmid expressing GFP-Nop1p was grown exponentially, and cell samples were treated for fluorescence microscopy. The two rightmost columns show merged images. (B) Sedimentation profile of Rrp36p-3HA on a sucrose gradient. A total extract prepared from RRP36::3HA cells growing exponentially was sedimented through a sucrose gradient, and 18 fractions were collected. The corresponding A254 profile is displayed with the characteristic annotated peaks. One-half of each fraction was precipitated by TCA, and Rrp36p-3HA was detected in these samples by Western blotting using anti-HA antibodies. RNAs contained in the other half of each fraction were purified, and specific molecules were detected by Northern blotting using the indicated probes (see Fig. 3C and Table S1 in the supplemental material for details).
FIG. 4.
Depletion of Rrp36p in yeast does not affect the incorporation of components of the UTP-A, UTP-B, or UTP-C modules within preribosomes. Strains expressing TAP-tagged versions of Utp15p, Utp17p, Pwp2p, or Utp22p that were otherwise WT (lower panels) or expressing 3HA-Rrp36p under the control of the GAL1 promoter (upper panels) were transferred from galactose- to glucose-based medium and grown for 6 h. Total extracts prepared from these cells were sedimented through sucrose gradients. Western blot experiments were carried out using rabbit PAP to detect TAP-tagged proteins in the different fractions. BOT, bottom.
RNA extractions and Northern blot and primer extension analyses.
RNA extractions from yeast cells were performed as described in reference 34. RNA extractions from HeLa cells were performed using Trizol (Invitrogen). For Northern blot analyses of low-molecular-weight species (see Fig. 3D), 4 μg of total RNAs was separated on polyacrylamide gels (6% acrylamide-bisacrylamide [19:1]-8 M urea-1× Tris-borate-EDTA buffer). For Northern blot analyses of higher-molecular-weight species (Fig. 1B, 2A, and 3D; also see Fig. 5A and 7B), 4 μg of total RNAs, or half of the RNA samples purified from about 250 ml of the sucrose gradient fractions, was separated as described previously (31). In all cases, RNAs were transferred to Amersham Hybond N+ membranes (GE Healthcare), and membranes were hybridized with 32P-labeled oligodeoxynucleotides by use of Rapid-hyb buffer (GE Healthcare). The sequences of the oligodeoxyribonucleotides used as probes in these studies are described in Table S1 in the supplemental material. Primer extension analyses to detect the 35S pre-rRNA were performed using 32P-labeled 5′ 35S-3 oligodeoxyribonucleotide (see Table S1 in the supplemental material) and SuperScript II reverse transcriptase (Invitrogen), following the protocol provided with the enzyme. Following cDNA synthesis, RNAs were hydrolyzed for 1 h at 55°C in the presence of 100 mM NaOH. Samples were neutralized with 100 mM HCl, and the cDNAs were ethanol precipitated in the presence of glycogen. Pellets were resuspended with stop solution (5 mg/ml Dextran Blue 2000 in deionized formamide), and cDNAs were separated by electrophoresis on polyacrylamide gels (6% acrylamide-bisacrylamide [19:1]-8 M urea-1× Tris-borate-EDTA buffer). Gels were dried, and the signals were detected by autoradiography.
FIG. 2.
Rrp36p interacts with 90S and pre-40S preribosomal particle components in yeast cells. (A) Immunoprecipitation (IP) experiments using IgG-conjugated Sepharose and total cellular extracts prepared from strain BMA64 (devoid of TAP-tagged protein; lanes 1 and 2) and from strains expressing TAP-Rrp36p (lanes 3 and 4) or Pwp2p-TAP (lanes 5 and 6). About 1/5 of the proteins retained in each purified sample (IP) and 1/50 of the proteins contained in each input extract (T) were analyzed by Western blotting using rabbit PAP to detect the TAP-tagged proteins (WB PAP). The RNAs extracted from each purified sample (IP) and from a fraction of each input extract corresponding to 1/50 of the amount used as starting material for the immunoprecipitation (T) were analyzed by primer extension (PE) and/or Northern blot analyses. Signal intensities were quantified from PhosphorImager data using MultiGauge software, and the percentages of input RNAs precipitated in each experiment are indicated below the IP lanes (bg, background level). (B) TAP of TAP-Rrp36p under native and stringent conditions. TAP-Rrp36p was affinity purified from GAL1::TAP::RRP36 cells under both native (20,400 × g; lane 2) and stringent (180,000 × g; lane 4) conditions (see Materials and Methods). As a control, the parental BMA64 WT strain, devoid of TAP-tagged protein, was handled similarly in each case. Approximately 1/10 of the final purified samples were separated by SDS-polyacrylamide gel electrophoresis and stained with PageBlue protein staining solution. The remaining samples were subjected to mass spectrometry analyses. CBP-tagged Rrp36p is indicated with a dot in lanes 2 and 4. Lane 1, molecular weight markers (MWM). (C) Proteins specifically associated with Rrp36p. The list includes all the nonribosomal proteins detected by mass spectrometry in the TAP-Rrp36p purifications but not in the mock purifications. Ribosomal proteins have been omitted from this list.
FIG. 7.
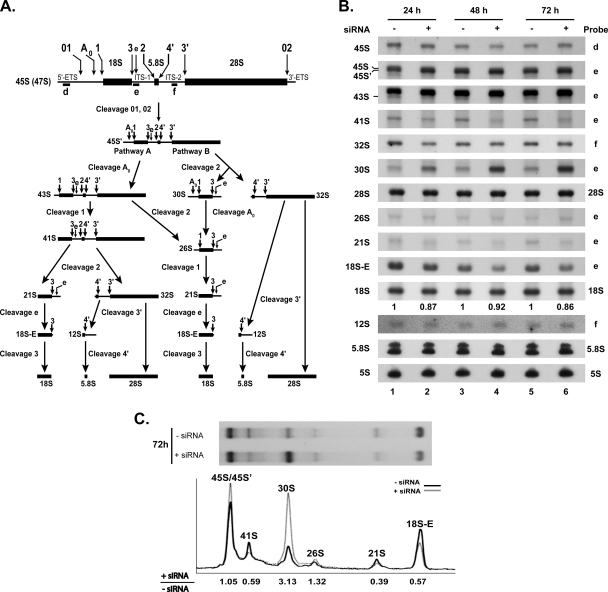
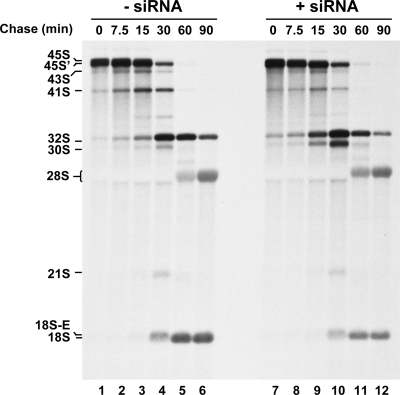
Depletion of RRP36 mRNA in HeLa cells impairs early cleavages of the pre-rRNA. (A) Scheme of the pre-rRNA processing pathway in HeLa cells. The positions of the oligonucleotide probes used to detect the pre-rRNAs analyzed in this study (d, e, and f) are shown on the initial precursor; their sequences are described in Table S1 in the supplemental material. (B) Pre-rRNA processing defects in siRNA-treated HeLa cells. Northern blot analysis of total RNAs extracted from HeLa cells treated (+) with siRNAs against RRP36 mRNA for 24, 48, or 72 h or left untreated (−). The different (pre-)rRNA species were detected using the specified oligonucleotide probes (Fig. 7A; also see Table S1 in the supplemental material). The second and third hybridization panels from the top correspond to different exposures of the same hybridized RNAs. The values shown below the 18S rRNA panel correspond to the 18S/28S ratios calculated from PhosphorImager quantifications of the signals corresponding to each species in four experiments, with the value for control cells arbitrarily set to 1 (−siRNA). (C) Quantification of the accumulation levels of precursors to 18S rRNA in siRNA-treated and control cells. The accumulation of precursors to 18S rRNA in siRNA-treated cells and control cells after 72 h of treatment (Fig. 7B [72 h, probe e]) was quantified from PhosphorImager data. The intensity profiles of the different maturation intermediates in siRNA-treated cells (gray line) and control cells (black line) are depicted. The value given below each species designation corresponds to the ratio of its accumulation levels in siRNA-treated cells (+siRNA) to those in control cells (−siRNA), after normalization to the amount of 28S rRNA.
Immunofluorescence microscopy using yeast cells.
About 2 × 108 cells growing exponentially were fixed with 4% paraformaldehyde (Electron Microscopy Science) during 30 min under conditions of gentle shaking, collected by centrifugation, rinsed with buffer B (8.6 mM K2HPO4, 1.3 mM KH2PO4, 1.2 M sorbitol), and collected again by centrifugation. Cell pellets were resuspended with 500 μl of digestion buffer obtained by mixing 1 ml buffer B with 10 ml of 200 μM vanadium ribonucleoside complex, 2 μl of 0.1 M phenylmethylsulfonyl fluoride, 2 μl of 14.3 M β-mercaptoethanol, and 10 μl of 5 mg/ml Zymolyase 100T (Seikagaku Corporation). After 45 min at 30°C, spheroplasts were collected by gentle centrifugation, resuspended with 500 μl of buffer B, spotted on eight-well multitest slides previously coated with poly-l-lysine (Sigma), and incubated for 1 h at room temperature to allow cell adhesion. Excess cells were removed, and slides were incubated for 5 min in phosphate-buffered saline (PBS) containing 0.1% bovine serum albumin (BSA) and then two times for 5 min each time in PBS containing 0.1% BSA and 0.1% NP-40. To detect HA-tagged proteins, slides were incubated for 2 h at room temperature with anti-HA monoclonal antibodies (clone 12CA5; Roche) diluted to 1:200 in PBS containing 0.1% BSA. Slides were rinsed once for 5 min with PBS containing 0.1% BSA, once for 5 min with PBS containing 0.1% BSA and 0.1% NP-40, and once again for 5 min with PBS containing 0.1% BSA. Slides were then incubated for 2 h at room temperature with Alexa Fluor 555-conjugated goat anti-mouse IgGs (Invitrogen) diluted to 1:1,000 in PBS containing 0.1% BSA. Slides were then rinsed three times as described above. DNA was stained with 0.1 μg/ml DAPI (4′,6-diamidino-2-phenylindole) in Moviol. Images were captured with an Olympus IX-81 microscope using Metamorph acquisition software.
Transfection of HeLa cells with pEGFP-RRP36 and immunofluorescence microscopy.
Exponentially growing HeLa cells were electrotransformed with pEGFP-RRP36 plasmid by the use of 10 μg of DNA for 10 × 106 HeLa cells in 200 μl Opti-MEM (Gibco). The mix was transferred into a 4-mm cuvette (Eurogentec) and incubated 5 min on ice, and cells were electrotransformed with a Bio-Rad Gene Pulser apparatus (250 V, 950 μF). Cells were then grown on microscope cover glass in six-well plates for 48 h and fixed for 20 min with 4% paraformaldehyde. Fixed cells were washed twice with PBS, permeabilized with 0.4% Triton X-100 in PBS for 5 min, and washed twice with PBS. The cells were incubated for 1 h with 1% BSA in PBS. Cells were then incubated 2 h in the same solution containing antifibrillarin monoclonal antibodies (clone 72B9, diluted to 1:200). Cells were washed twice with 1% BSA in PBS and subsequently incubated 1 h with tetramethyl rhodamine isothiocyanate-conjugated anti-mouse antibodies (Dako) diluted to 1:200 in PBS containing 1% BSA. Cells were washed three times with 1% BSA in PBS and one time with PBS only, and the nuclei were stained with Hoescht solution. The coverslips were mounted in Moviol. Images were captured on a DMRB microscope (Leika) using Metavue software.
Knockdown of RRP36 expression by RNA interference (RNAi).
The small interfering RNA (siRNA) duplex matching a region corresponding to nucleotides +692 to +710 from the ATG of C6orf153 cDNA (5′-GCAGCAAGAAAUUGGAGAA-3′) was purchased from Eurogentec. Exponentially growing HeLa cells were trypsinized, washed in Opti-MEM (Gibco), and resuspended in Opti-MEM at a density of 5 × 107 cells/ml. Forty microliters of siRNA solution (100 μM) was added to 200 ml of the cell suspension, the mix was transferred into a 4-mm cuvette (Eurogentec) and incubated 5 min on ice, and cells were electrotransformed with a Bio-Rad Gene Pulser apparatus (250 V, 950 μF). Cells were immediately resuspended and divided in three 10-cm-diameter plates. Cells were harvested 24, 48, and 72 h after transfection.
Pulse-chase analysis.
HeLa cells were electrotransformed simultaneously with two siRNA duplexes targeting RRP36 mRNA, the one described previously (see “Knockdown of RRP36 expression by RNAi” above) and a second one matching a region corresponding to nucleotides +131 to +150 from the ATG of C6orf153 cDNA (5′-GCACAUCUAACAUGUCAUU-3′). Forty microliters of each siRNA duplex (100 μM) was added to 10 × 106 HeLa cells in a total volume of 240 μl of Opti-MEM. Electrotransformation was performed as described previously (see “Knockdown of RRP36 expression by RNAi” above). Cells were then grown on 12-well plates for 48 h. The pulse-chase protocol was adapted from reference 26. Cells were incubated in methionine-free DMEM (Invitrogen) for 30 min at 37°C, labeled for 15 min with l-methyl 3H methionine (50 μCi/ml), rinsed with regular DMEM, and incubated for 0 min (i.e., immediately rinsed with PBS; see below) or 7.5, 15, 30, 60, and 90 min in regular DMEM. Cells were rinsed with cold PBS and immediately resuspended with 500 μl of Trizol (Invitrogen). RNAs were extracted, separated on a 1.2% agarose gel as described previously (see “RNA extractions and Northern blot and primer extension analyses” above), transferred to Amersham Hybond N+ membranes (GE Healthcare), and exposed for several days using Biomax KODAK MS films and a KODAK BioMax TranScreen LE apparatus.
RESULTS
Rrp36p displays a predominant nucleolar localization and interacts with 90S and pre-40S preribosomes in vivo.
To investigate the subcellular localization of endogenous Rrp36p, we constructed a strain expressing under the control of the endogenous RRP36 promoter a modified version of Rrp36p bearing the 3HA epitope tag at its C terminus (the RRP36::3HA strain). This strain was grown exponentially, and cell samples were analyzed by fluorescence microscopy. The anti-HA immunofluorescent signal observed in these cells is restricted to the nucleus (Fig. 1A), which is consistent with the presence of a putative nuclear localization signal (NLS) at the C terminus of the protein (see Fig. 6A). Within the nucleus, Rrp36p-3HA is present at low levels in the nucleoplasmic area and is highly enriched within a crescent-shaped region also containing a green fluorescent protein (GFP)-fused version of Nop1p (GFP-Nop1p), a core component of box C/D snoRNPs involved in early, nucleolar stages of the pre-rRNA maturation pathway. We conclude that Rrp36p accumulates predominantly in yeast cell nucleoli.
FIG. 6.
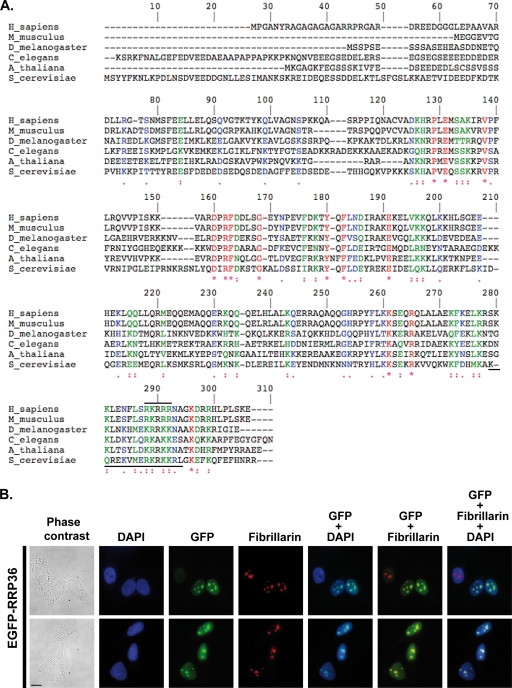
RRP36 accumulates in the nucleoli of HeLa cells. (A) Alignment of S. cerevisiae Rrp36p with homologous proteins in Arabidopsis thaliana (NP_172725.2), Caenorhabditis elegans (NP_503179.1), Drosophila melanogaster (NP_650367.1), Mus musculus (NP_659106.1), and Homo sapiens (NP_149103.1), identified using the NCBI BLAST program and aligned using ClustalW. Identical residues are indicated by red characters and stars, strongly similar residues by green characters and double dots, and weakly similar residues by blue characters and single dots. The horizontal solid black lines mark potential nuclear localization signals in the yeast and human sequences. (B) Nucleolar localization of RRP36 in HeLa cells. An immunofluorescence experiment was performed using antifibrillarin antibodies and fixed HeLa cells electroporated with plasmid pEGFP-RRP36. From left to right, images represent cells visualized by phase-contrast microscopy, DAPI staining (blue), EGFP-RRP36 signal (green), and antifibrillarin signal (red); the rightmost three columns represent merged images. Scale bar, 10 μm.
To obtain insights into the nature of the macromolecular complexes containing Rrp36p, we compared the sedimentation profile of Rrp36p with that of (pre)ribosomal particles on density gradients (Fig. 1B). Rrp36p-3HA concentrates in two sets of fractions of the gradient. The first set (fractions 6 to 8) contains the pre-40S particles associated with the 20S pre-rRNA, and the second set (fractions 10 to 13) includes the 23S pre-rRNA-containing preribosomes (fractions 10 to 12), the pre-60S particles (fractions 10 to 12), and the early preribosomes associated with the 35S and 33S/32S precursors (fractions 12 to 16). Most of the fractions in which Rrp36p is detected also contain different levels of U3 snoRNA. Thus, most Rrp36p-3HA is engaged within large complexes, the density of which is consistent with that of various preribosomal particles. We next carried out immunoprecipitation experiments to assess whether Rrp36p interacts physically with preribosomal particles (Fig. 2A). We constructed a strain expressing Rrp36p fused at the N terminus with the TAP tag under the control of the GAL1 promoter (the GAL1::TAP::RRP36 strain). This strain was grown in the presence of galactose, and TAP-Rrp36p was precipitated from a total cellular extract by the use of IgG-conjugated Sepharose. A fraction of the precipitated material was analyzed by Western blotting to assess the precipitation efficiency of TAP-Rrp36p. The RNAs contained in the remaining sample were purified, and specific molecules were identified by primer extension and/or Northern blot analyzes. The parental BMA64 WT strain was used as a negative control in this experiment, and, for reasons that become obvious later in this report, we chose a strain expressing a TAP-tagged component of the SSU processome, Pwp2p, as a positive control. The RNA components of 90S preribosomal particles, the 35S and 33S/32S pre-rRNAs, are coprecipitated with TAP-Rrp36p, as well as the 23S pre-rRNA and the 20S precursor to the mature 18S rRNA to a lesser extent (Fig. 2A). PhosphorImager quantifications indicate that about 2% of the 35S pre-rRNA and approximately 1% of the 23S and 20S precursors are coprecipitated with TAP-Rrp36p. In contrast, the 7S pre-rRNA, a component of late pre-60S particles, is detected at background levels in the precipitate. These interactions suggest that Rrp36p is a component of the 90S preribosomal particles, a conclusion further supported by the presence in the TAP-Rrp36p immunoprecipitate of the U3 snoRNA but not of other abundant noncoding RNAs such as the RNA component of the signal recognition particle (SCR1) or the U2 spliceosomal small nuclear RNA. Consistent with the observed cosedimentation of Rrp36p and pre-40S particles on sucrose gradients (Fig. 1B), Rrp36p seems also to interact to a lesser extent with pre-40S particles containing the 23S and 20S intermediates, suggesting that the protein remains associated with the pre-40S preribosomal particles following early cleavages of the 35S pre-rRNA. Interestingly, the profiles of the RNA species associated with TAP-Rrp36p and Pwp2p-TAP are qualitatively very similar, strengthening the conclusion that Rrp36p is a component of the 90S preribosomes. However, these RNAs are precipitated much more efficiently with Pwp2p-TAP than with TAP-Rrp36p, suggesting that the latter protein interacts with preribosomal particles through more labile interactions.
To confirm that Rrp36p is associated with preribosomal particles, we performed TAPs (Fig. 2B and C). TAP-Rrp36p was purified under native (Fig. 2B, lane 2) or stringent (Fig. 2B, lane 4) conditions (see Materials and Methods), and the copurified proteins were identified by mass spectrometry (Fig. 2C). The BMA64 strain devoid of TAP-tagged protein was used as a control in each experiment (Fig. 2B, lanes 3 and 5). Consistent with our previous observations, several factors shown or suggested to be components of the 90S preribosomal particles are specifically detected in the TAP-Rrp36p purification under native conditions. In addition, two protein components of pre-60S particles also copurify with Rrp36p. Interestingly, the 90S preribosome factors associated with Rrp36p include all but one of the components of the previously described UTP-C complex (18), namely, Utp22p and the four subunits of casein kinase II (Cka1p, Cka2p, Ckb1p, and Ckb2p). Two of these, Cka2p and Ckb1p, remain associated with TAP-Rrp36p under stringent conditions, suggesting that Rrp36p may be preferentially associated with the UTP-C module.
To provide further evidence that Rrp36p is a novel component of 90S preribosomes, we showed that Rrp36p-3HA partially cosediments with TAP-tagged versions of known components of these particles such as Utp15p, Utp17p, Pwp2p, and Utp22p on sucrose gradients (see Fig. S1A in the supplemental material) and interacts physically with these factors, as assessed by immunoprecipitation experiments (see Fig. S1B in the supplemental material).
Depletion of Rrp36p affects early cleavages of the pre-rRNA and inhibits the production of the 40S ribosomal subunit in yeast.
To investigate the function of Rrp36p in ribosome biogenesis, we produced a strain that conditionally expresses 3HA-tagged Rrp36p in the presence of galactose and undergoes Rrp36p depletion when glucose is added (the GAL1::3HA::RRP36 strain). Upon transfer to a glucose-containing medium, this strain displays a rapid growth defect compared to the BY4741 WT strain (Fig. 3A). To monitor 3HA-Rrp36p depletion, the accumulation levels of the protein were monitored by Western blot analysis at different time points. These levels were compared to those of the Rrp36p-3HA protein expressed from the endogenous RRP36 promoter in the RRP36::3HA strain grown on galactose- and glucose-containing medium (Fig. 3B, lanes 1 and 2, respectively), which likely reflect the physiological levels. As soon as 30 min after transcriptional repression, 3HA-Rrp36p accumulation dropped below the levels observed with the RRP36::3HA strain grown in the presence of glucose (Fig. 3B; compare lanes 4 and 2) and became barely detectable after 1 h (lane 5). To determine whether Rrp36p depletion affects the production of ribosomes, we compared the ribosome profiles in the presence and absence of Rrp36p on sucrose gradients (see Fig. S2 in the supplemental material). Depletion of 3HA-Rrp36p resulted in a clear reduction of the sedimentation peak corresponding to the free 40S subunits and a correlated strong increase of the 60S subunit sedimentation peak. To delineate the origin of this phenotype, we compared pre-rRNA processing in WT and Rrp36p-depleted cells by Northern blot analysis (Fig. 3D). As early as 30 min after the nutritional shift, the GAL1::3HA::RRP36 strain accumulated the 35S pre-rRNA (Fig. 3D, lanes 9 to 14) in comparison to the WT control experiment results (lanes 2 to 7), suggesting that the maturation of this precursor was impaired. Concomitantly, four intermediates of the processing pathway were very rapidly depleted: the 33S and 32S pre-rRNAs, resulting from cleavage of the 35S transcript at sites A0 and A1 respectively, as well as the 20S and the 27SA2 intermediates produced from the 32S pre-rRNA following A2 cleavage. The accumulation of the 35S precursor and the disappearance of the 33S, 32S, 20S, and 27SA2 intermediates strongly suggest that early cleavages of the 35S pre-rRNA at sites A0, A1, and A2 were rapidly and strongly inhibited in cells undergoing Rrp36p depletion. As a consequence, the 23S species, resulting from cleavage of the 35S precursor at site A3 without prior cleavage at sites A0, A1, and A2, clearly accumulated. The strong depletion of the 20S pre-rRNA reduced the synthesis of the mature 18S rRNA and therefore the production of the 40S ribosomal subunit (see Fig. S2 in the supplemental material). In contrast, production of the RNA components of the 60S ribosomal subunit, the 5S, 5.8S, and 25S rRNAs, was not affected by the absence of Rrp36p. We conclude that Rrp36p is specifically required for early cleavages of the 35S pre-rRNA at sites A0, A1, and A2 in the processing pathway leading to the mature 18S rRNA and the production of the 40S ribosomal subunit. This conclusion is fully consistent with the results of a large-scale microarray-based study reporting that transcriptional repression of the YOR287C ORF induces a U3 processome-like mutant phenotype (24) and with the partial characterization of the function of YOR287C in ribosome biogenesis published while this work was in progress (21). Probably as a consequence of the rapid depletion of the 18S rRNA compromising translation and growth, Rrp36p inactivation resulted in the accumulation of cells in the G1 phase of the cell cycle (see Fig. S3 in the supplemental material).
Incorporation of Rrp36p into nascent preribosomes in yeast cells requires prior loading of the tUTP/UTP-A and PWP2/UTP-B complexes but not Rrp5p.
As previously shown, Rrp36p is a novel component of the SSU processome and is required for early cleavages of the 35S pre-rRNA. The assembly of this initial preribosomal particle has been suggested to result from the sequential loading of several preformed subcomplexes. We determined whether Rrp36p is required for the incorporation of the UTP-A, UTP-B, and UTP-C modules and several snoRNPs within the SSU processome and conversely whether components of the UTP-A and UTP-B subcomplexes as well as Rrp5p are necessary for Rrp36p recruitment. To study the importance of Rrp36p in the assembly of the 90S preribosome, we compared the sedimentation profiles of components of the UTP-A, UTP-B, or UTP-C subcomplexes and different snoRNAs in WT and Rrp36p-depleted cells (Fig. 4; also see Fig. S2 in the supplemental material). For this analysis, we placed the RRP36 ORF under the control of the conditional GAL1 promoter in strains expressing TAP-tagged versions of Utp15p or Utp17p (UTP-A complex), Pwp2p (UTP-B complex), or Utp22p (UTP-C complex). The A254 absorbance profiles presented in the upper panels in Fig. 4 confirm that depletion of Rrp36p induced a severe defect in the production of the mature 40S ribosomal subunit. Yet the Utp15p-TAP, Utp17p-TAP, Pwp2p-TAP, or Utp22p-TAP proteins were still detected in fractions of the gradient containing high-molecular-weight complexes, and none of them was detected in the top fractions containing small particles and free proteins. These data suggest that the stalled preribosomal particles lacking Rrp36p contain the tested components of the different UTP modules and that Rrp36p is therefore probably not required for their incorporation. Likewise, the sedimentation profiles of U3 snoRNA and of two other essential snoRNAs also crucial for early processing of the pre-rRNAs (U14 and snR30) were not modified in Rrp36p-depleted cells (Fig. S2 in the supplemental material and data not shown). We conclude that Rrp36p is likely not necessary for the insertion of U3, U14, and snR30 snoRNPs into preribosomes.
We next studied whether Utp17p (UTP-A complex), Pwp2p (UTP-B complex), or Rrp5p is required for Rrp36p recruitment to the nascent preribosomal particles. We replaced the endogenous promoter of the UTP17, PWP2, or RRP5 gene with the conditional GAL1 promoter in the previously described RRP36::3HA strain. These strains were shifted from a galactose- to a glucose-containing medium and grown for different times to deplete Utp17p, Pwp2p, or Rrp5p. Extracts prepared from the depleted cells were analyzed by sedimentation on sucrose gradients. The absorbance profiles observed in Utp17p-depleted cells (Fig. 5A, upper right panel) and Pwp2p-depleted cells (Fig. 5A, bottom left panel) are consistent with those reported previously by others (8, 25) and very similar to those observed in the absence of Rrp36p (Fig. 4; also see Fig. S2 in the supplemental material). Interestingly, Rrp36p-3HA could not be detected using conventional methods in the fractions of the gradient containing the preribosomal particles in Utp17p- and Pwp2p-depleted extracts, in contrast with the WT control experiment results (Fig. 5A, upper left panel). Thus, Rrp36p does not accumulate within preribosomal particles in the absence of Utp17p or Pwp2p. Using a highly sensitive detection kit, we could nevertheless detect trace amounts of partially degraded Rrp36p-3HA protein in the top fractions of the gradient containing free proteins and low-molecular-weight complexes (see the legend of Fig. 5A for more details). This result prompted us to assess whether the accumulation of Rrp36p-3HA is affected in cells undergoing Utp17p or Pwp2p depletion (Fig. 5B). The BY4741, GAL1::UTP17, and GAL1::PWP2 strains expressing Rrp36p-3HA were subjected to a galactose-to-glucose nutritional shift, and total protein extracts were prepared from cell samples harvested before the shift (galactose) and 6, 12, and 24 h after the shift (glucose). Western blot analyses show that depletion of Utp17p and of Pwp2p clearly affected Rrp36p-3HA protein levels (Fig. 5B, second and third panels from the left, respectively) compared to control cell results (left panel). In contrast, the stability of Lcp5p, another nucleolar factor required for early cleavages of the 35S pre-rRNA at sites A0, A1, and A2 (45), was not affected under the same conditions. Together, these data suggest that Rrp36p recruitment to nascent preribosomes requires prior loading of the UTP-A and UTP-B subcomplexes. Defective integration of either of these modules affects the loading of Rrp36p and results in a pool of free proteins that rapidly turn over. Alternatively, the UTP-A and UTP-B complexes may be directly required for the stability of Rrp36p.
Recently, Rrp5p has been shown to be required for the association of the UTP-C subcomplex with the 35S pre-rRNA (25). We studied the effects of Rrp5p depletion on the stability and sedimentation profile of Rrp36p. In contrast to what has been observed upon depletion of Utp17p and Pwp2p, the absence of Rrp5p did not affect the steady-state accumulation of Rrp36p-3HA (Fig. 5B, right panel) and the protein was still detected in the fractions of the gradient containing preribosomal particles (Fig. 5A, bottom right panel). Surprisingly, however, the sedimentation profile of Rrp36p-3HA in Rrp5p-depleted cells appears significantly different from that observed with WT cells. The protein was not detected in fractions 6 to 9 of the gradient and appeared more concentrated in the bottom fractions containing polysomes and other high-molecular-weight complexes. These data indicate that the incorporation of Rrp36p into preribosomes is not prevented in the absence of Rrp5p. The observed shift in sedimentation may suggest that the incorporation of Rrp36p is, on the contrary, facilitated under these conditions or, alternatively, that the protein remains associated for a longer period of time with preribosomal particles and/or is trapped within aberrant particles cosedimenting with polysomes.
A human protein homologous to yeast Rrp36p accumulates in the nucleoli of HeLa cells and is required for early cleavages of the pre-rRNAs.
By sequence comparison, we could identify proteins homologous to yeast Rrp36p in all available eukaryotic genomes. Alignment of these homologous proteins shows that they display several blocks of identical or similar residues separated by more variable regions (Fig. 6A). These conserved blocks do not correspond to any recognizable protein motif, except for the lysine- and arginine-rich domains systematically found at the very end of all sequences, predicted to correspond to nuclear localization signals (6). The human genome contains a single, uncharacterized ORF (named C6orf153) encoding a hypothetical protein predicted to display 21% identity and 44% similarity with yeast Rrp36p. We initiated the characterization of this human protein in HeLa cells and renamed it RRP36, since our results strongly suggest that this protein is the orthologue of yeast Rrp36p in mammalian cells.
To investigate the subcellular localization of RRP36 in HeLa cells, we constructed a plasmid expressing, under the control of the cytomegalovirus promoter, RRP36 fused at the N terminus to a variant of the GFP (EGFP). This plasmid was transfected into HeLa cells, and the localization of EGFP-RRP36 was analyzed by fluorescence microscopy. Figure 6B shows that EGFP-RRP36 was exclusively detected within the nucleus of HeLa cells, where it was found homogeneously distributed throughout the nucleoplasm and concentrated, in addition, in the nucleoli identified by the presence of fibrillarin, a core component of box C/D snoRNPs. In agreement with our observations, the protein product of the C6orf153 gene has been detected by mass spectrometry in highly purified preparations of human nucleoli (1, 20). The presence of RRP36 in the nucleoli of human cells suggests that the protein functions in early steps of ribosome synthesis. To investigate this hypothesis further, we reduced the expression of RRP36 in HeLa cells by RNAi and assessed the effects on the pre-rRNA processing pathway (Fig. 7). The pre-rRNA maturation process in higher eukaryotes (5, 14, 32) is depicted in Fig. 7A. HeLa cells were transfected with or without siRNAs targeting RRP36 mRNA and were harvested after 24, 48, or 72 h of treatment. Total RNAs were extracted and analyzed by Northern blotting to detect the various pre-rRNAs (Fig. 7B). The accumulation levels of some of these precursors were quantified from PhosphorImager data (Fig. 7C). We could not evaluate the impact of the siRNA treatment on RRP36 protein levels, since we lacked specific antibodies. However, we showed by reverse transcription combined with semiquantitative PCR that the accumulation of RRP36 mRNA is specifically reduced by approximately 70% in siRNA-treated cells compared to the accumulation of the irrelevant GAPDH (glyceraldehyde 3-phosphate dehydrogenase) mRNA (see Fig. S4A in the supplemental material). Partial depletion of RRP36 mRNA did not affect the steady-state accumulation of the initial 45S pre-rRNA, which remained roughly constant at the different time points (Fig. 7B). This result suggests that inactivation of RRP36 does not affect transcription by RNA Pol I. In that respect, RRP36 differs from early-associating t-UTP proteins shown to be required for this process in HeLa cells (27). In contrast, the 45S′ precursor, resulting from cleavages of the 45S transcript at site 01 in the 5′ETS and at site 02 in the 3′ETS, accumulated to some extent at 24 h and afterwards. This result suggests that cleavages of the 45S pre-rRNA at sites 01 and 02 are not drastically affected in siRNA-treated cells but that the downstream maturation of the resulting 45S′ species is delayed. In WT cells, the 45S′ transcript is processed following two alternative pathways. One pathway consists of cleavages at sites A0 and A1, generating the 43S and 41S precursors. The accumulation of these species was clearly reduced in siRNA-treated cells (by approximately 40% for the 41S pre-rRNA [Fig. 7C]; the accumulation levels of the 43S pre-rRNA could not be quantified accurately because of the close proximity of the 45S′) despite the increased levels of the 45S′ pre-rRNA, suggesting that RRP36 is required for cleavages at sites A0 and A1. The alternative 45S′ pre-rRNA maturation pathway involves cleavage at site 2 within ITS1, which splits the transcript into the 30S intermediate, containing the mature 18S rRNA, and the 32S precursor to the mature 5.8S and 28S rRNAs. Noticeably, the 30S species rapidly accumulated in siRNA-treated cells and became enriched threefold compared to control cells after 72 h of treatment (Fig. 7B and C), suggesting that this intermediate is produced efficiently from the 45S′ precursor but that its downstream maturation is impaired. We propose from these observations that RRP36 is not required for cleavage at site 2 within ITS1 but is important for the processing of the 30S intermediate. This processing consists of cleavage at site A0, which generates the 26S species, followed by cleavage at site 1, producing the 21S pre-rRNA and 18S-E intermediates, precursors to the 18S rRNA. The accumulation levels of the 21S and 18S-E pre-rRNAs were reduced by 60% and 40%, respectively, in siRNA-treated cells, suggesting again that cleavages at sites A0 and A1 are impaired in cells partially deprived of RRP36. In contrast, the processing pathway generating the mature RNA components of the 60S ribosomal subunit was not affected by siRNA treatment, since the accumulation of the 5.8S and 28S rRNAs and their immediate precursors (the 32S and 12S pre-rRNAs) remained unchanged (Fig. 7B). These results strongly suggest that RRP36 is specifically required for processing at sites A0 and A1 of the pre-rRNAs in the maturation pathway leading to the 18S rRNA. Depletion of a core component of the U3 snoRNP (hU3-55K) or UTP proteins in HeLa cells has been shown to induce the accumulation of a processing intermediate comprising the 5′ETS, 18S, ITS1, and 5.8S sequences (27). Consistent with these results, such an intermediate accumulated to some extent in siRNA-treated cells compared to control cells (see Fig. S4B in the supplemental material). However, unlike Prieto and McStay, we could also detect this precursor in untreated cells. This discrepancy may be due to differences in growth conditions or HeLa cells.
Our Northern blot analyses showed an only marginal decrease of the 18S-to-28S rRNA ratio in siRNA-treated cells (see quantifications in Fig. 7B), probably because RRP36 depletion is not severe enough and because the half-life of this mature species is much longer than that of the various processing intermediates. To get additional evidence that RRP36 is required for production of the 18S rRNA in HeLa cells, we performed a pulse-chase analysis of neosynthesized RNAs in siRNA-treated cells (Fig. 8). This experiment allowed us to confirm the phenotypes observed by Northern blot analysis and to show convincingly that the accumulation of the mature 18S rRNA was significantly reduced after 60 and 90 min of chase in siRNA-treated cells (lanes 11 and 12) compared to control cells (lanes 5 and 6), whereas the production of the 28S rRNA was not affected. Altogether, our results show that the function of RRP36 in the production of the mature 18S rRNA has been conserved from yeast to human cells.
FIG. 8.
Depletion of RRP36 mRNA in HeLa cells affects the production of the mature 18S rRNA. Neosynthesized RNAs from HeLa cells transfected with siRNAs targeting RRP36 mRNA (lanes 7 to 12) or mock transfected (lanes 1 to 6) were pulse-labeled with l-methyl 3H methionine 48 h posttransfection. Cells were subsequently rinsed to remove radioactive methionine, incubated in regular DMEM, and harvested at the indicated time points. The different (pre-)rRNA species are indicated on the left.
DISCUSSION
The assembly and maturation of preribosomes mobilize a plethora of trans-acting factors and snoRNPs associating transiently with the preribosomal particles to promote their maturation and the formation of translation-competent ribosomal subunits. Although many components of early preribosomes have been identified and partially characterized, the current challenges in the field are (i) to establish an exhaustive list of these components, (ii) to understand their precise function in the maturation process and identify the substrate(s) of the various enzyme activities, (iii) to understand how the preribosomal particles assemble and evolve, and (iv) to determine to what extent the mechanisms elucidated in yeast are transposable to human cells. We report here the characterization of the RRP36 gene, encoding a novel nucleolar protein required for early cleavages of the pre-rRNAs in the 40S ribosomal subunit maturation pathway, in both yeast and mammalian cells. In yeast cells, Rrp36p interacts physically with 90S preribosomal particles and to a lesser extent with pre-40S particles. These interactions appear weaker than those observed with other components of the SSU processome such as Pwp2p (Fig. 2A) and Utp6p (data not shown). However, the sedimentation profile of Rrp36p on sucrose gradients shows that the protein was not detected in the top fractions containing free proteins (Fig. 1B), suggesting that the interactions between Rrp36p and preribosomes are not transient, but rather labile, and are partially lost during the immunoprecipitation procedure.
Upon RRP36 inactivation in yeast cells, we observed a surprisingly rapid depletion of the mature 18S rRNA, the accumulation of which began to decline as early as 1 h after the nutritional shift (Fig. 3D). This rapid decrease in the accumulation of the mature 18S rRNA is surprising given the relatively long half-life of mature ribosomes, generally assumed to largely exceed the generation time. Our results suggest that depletion of Rrp36p induced a prompt and potent block in early cleavages of the pre-rRNAs, rapidly abolishing the synthesis of the mature 18S rRNA. Since, as shown in Fig. 3A, cells undergoing Rrp36p depletion divide as WT cells up to 2 h after the nutritional shift, mature ribosomes produced before the block are very likely diluted during cell division, which may explain the rapid depletion of the mature 18S rRNA.
Our results show that Rrp36p depletion does not prevent the association of components of the tUTP/UTP-A, PWP2/UTP-B, and UTP-C complexes and the U3, U14, and snR30 snoRNPs with the 90S preribosomes (Fig. 4, Fig. S2 in the supplemental material, and data not shown), indicating that these modules assemble independently from Rrp36p. On this basis, we propose that Rrp36p is likely not a component of the tUTP/UTP-A subcomplex, the incorporation of which is a prerequisite for the recruitment of most subsequent modules. Our TAP data suggest that Rrp36p is preferentially associated with the UTP-C subcomplex (Fig. 2C). However, the Pwp2p-containing UTP-B module is necessary for the inclusion of Rrp36p whereas the UTP-B and UTP-C modules have been suggested to be recruited independently from each other (25). We propose that Rrp36p, although it may be preferentially associated with the UTP-C complex within 90S preribosomes, is recruited by a different mechanism than the one governing incorporation of the UTP-C module. This hypothesis is further supported by the observation that depletion of Rrp5p, required for the recruitment of the UTP-C module (25), does not seem to prevent Rrp36p recruitment, although it significantly modifies its sedimentation profile (Fig. 5A). Remarkably, the recruitment of the U3 snoRNP displays the same requirements as that of Rrp36p (it necessitates prior loading of the UTP-A and UTP-B complexes but not Rrp5p), raising the possibility that Rrp36p and the U3 snoRNP are recruited at a concomitant stage of the assembly process.
Interestingly, the steady-state accumulation of Rrp36p was strongly reduced in Utp17p- or Pwp2p-depleted cells (Fig. 5B), suggesting either that the UTP-A and UTP-B complexes are directly required for Rrp36p stability or that Rrp36p is rapidly degraded under conditions preventing its recruitment into preribosomes. Our conclusion that the tUTP/UTP-A or PWP2/UTP-B complexes are required for the assembly of Rrp36p with preribosomes originates from the shift in sedimentation and partial degradation of Rrp36p observed in the absence of Utp17p or Pwp2p. Formally, we cannot exclude the possibility that Rrp36p is properly incorporated into preribosomes in the absence of Utp17p or Pwp2p but that the stalled, improperly assembled particles are partially degraded and some of their components released as free proteins and turned over. However, the sedimentation profiles of the 35S, 23S, and 20S pre-rRNAs in cells lacking Utp17p or Pwp2p are similar to the profiles observed in WT cells (Fig. 5A), and the stability of Lcp5p is not affected (Fig. 5B), suggesting that the accumulation of free Rrp36p proteins is not due to a global degradation of the particles. In addition, the shift in Rrp36p sedimentation induced upon Utp17p or Pwp2p depletion is not a general consequence of defective assembly or maturation of the preribosomal particles, since it was not observed upon depletion of Rrp5p.
In an effort to investigate the evolutionary conservation of RRP36 function, we searched for homologous proteins in the different kingdoms. We could not identify homologues in bacteria and archeae, but in contrast, all the eukaryotic genomes we investigated contain at least one gene encoding a protein homologous to yeast Rrp36p. The product of the human RRP36 gene displays 21% identity and 44% similarity to the yeast Rrp36p protein. This degree of homology is in the range of what has been previously observed between several yeast Utp proteins and their characterized human counterparts (27). Our results strongly suggest that RRP36 is the orthologue of yeast Rrp36p in human cells, since EGFP-RRP36 is a nucleolar protein and partial depletion of RRP36 mRNA affects early processing steps of the pre-rRNA in the maturation pathway leading to the 18S rRNA. Potential orthologues of several components of the yeast 90S preribosomes in HeLa cells have been identified and studied (13, 27, 35, 41). This suggests that the fundamental mechanisms of preribosome assembly and processing are globally conserved from yeast to mammals.
Supplementary Material
Acknowledgments
We are very grateful to Christophe Dez, Isabelle Léger-Sylvestre, Jorge Pérez-Fernández, and Olivier Gadal for reagents, protocols, advice, and helpful discussions. We acknowledge Coralie Caron, Valérie Cadamuro, Marie-Françoise O'Donohue, and Pierre-Emmanuel Gleizes for reagents and help with microscopy, cell culture, and pre-rRNA processing in human cells. We also thank all the members of the group of M.C-.F. and Y.H. for advice and helpful discussions. We thank Philippe Pasero (IGH/CNRS, Montpellier, France) for fluorescence-activated cell sorting (FACS) protocols and helpful discussions. We acknowledge Carine Froment and Bernard Monsarrat from the proteomic facility of the Institut de Pharmacologie et de Biologie Structurale, CNRS UMR 5089, Toulouse, France, for mass spectrometry analyses as well as Aurélie Leru and the use of the FACS facility of CBD-IFR109. Anti-Lcp5p antibodies and pUN100-GFP-NOP1 plasmid were generously provided by Thomas Wiederkehr (Basel, Switzerland) and Ed Hurt, respectively. We thank Annie Mougin, Christophe Dez, Jorge Pérez-Fernández, Pierre-Emmanuel Gleizes, and Julien Soudet for critical reading of the manuscript and David Villa for help with figures.
This work was supported by the CNRS, Université Paul Sabatier, and grants from the Agence Nationale de la Recherche and the Ligue Contre le Cancer. M.G. is supported by a Ph.D. fellowship from the Ligue Contre le Cancer. A.K.H. is supported by a Carrier Development Award from the Human Frontier Science Program Organization (HFSPO).
Footnotes
Published ahead of print on 28 December 2009.
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Andersen, J. S., C. E. Lyon, A. H. Fox, A. K. Leung, Y. W. Lam, H. Steen, M. Mann, and A. I. Lamond. 2002. Directed proteomic analysis of the human nucleolus. Curr. Biol. 12:1-11. [DOI] [PubMed] [Google Scholar]
- 2.Baudin, A., O. Ozier-Kalogeropoulos, A. Denouel, F. Lacroute, and C. Cullin. 1993. A simple and efficient method for direct gene deletion in Saccharomyces cerevisiae. Nucleic Acids Res. 21:3329-3330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bernstein, K. A., J. E. Gallagher, B. M. Mitchell, S. Granneman, and S. J. Baserga. 2004. The small-subunit processome is a ribosome assembly intermediate. Eukaryot. Cell 3:1619-1626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Billy, E., T. Wegierski, F. Nasr, and W. Filipowicz. 2000. Rcl1p, the yeast protein similar to the RNA 3′-phosphate cyclase, associates with U3 snoRNP and is required for 18S rRNA biogenesis. EMBO J. 19:2115-2126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bowman, L. H., B. Rabin, and D. Schlessinger. 1981. Multiple ribosomal RNA cleavage pathways in mammalian cells. Nucleic Acids Res. 9:4951-4966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cokol, M., R. Nair, and B. Rost. 2000. Finding nuclear localization signals. EMBO Rep. 1:411-415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dez, C., M. Dlakic, and D. Tollervey. 2007. Roles of the HEAT repeat proteins Utp10 and Utp20 in 40S ribosome maturation. RNA 13:1516-1527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dosil, M., and X. R. Bustelo. 2004. Functional characterization of Pwp2, a WD family protein essential for the assembly of the 90 S pre-ribosomal particle. J. Biol. Chem. 279:37385-37397. [DOI] [PubMed] [Google Scholar]
- 9.Dragon, F., J. E. Gallagher, P. A. Compagnone-Post, B. M. Mitchell, K. A. Porwancher, K. A. Wehner, S. Wormsley, R. E. Settlage, J. Shabanowitz, Y. Osheim, A. L. Beyer, D. F. Hunt, and S. J. Baserga. 2002. A large nucleolar U3 ribonucleoprotein required for 18S ribosomal RNA biogenesis. Nature 417:967-970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Elela, S. A., H. Igel, and M. Ares, Jr. 1996. RNase III cleaves eukaryotic preribosomal RNA at a U3 snoRNP-dependent site. Cell 85:115-124. [DOI] [PubMed] [Google Scholar]
- 11.Gallagher, J. E., D. A. Dunbar, S. Granneman, B. M. Mitchell, Y. Osheim, A. L. Beyer, and S. J. Baserga. 2004. RNA polymerase I transcription and pre-rRNA processing are linked by specific SSU processome components. Genes Dev. 18:2506-2517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Grandi, P., V. Rybin, J. Bassler, E. Petfalski, D. Strauss, M. Marzioch, T. Schafer, B. Kuster, H. Tschochner, D. Tollervey, A. C. Gavin, and E. Hurt. 2002. 90S pre-ribosomes include the 35S pre-rRNA, the U3 snoRNP, and 40S subunit processing factors but predominantly lack 60S synthesis factors. Mol. Cell 10:105-115. [DOI] [PubMed] [Google Scholar]
- 13.Granneman, S., J. E. Gallagher, J. Vogelzangs, W. Horstman, W. J. van Venrooij, S. J. Baserga, and G. J. Pruijn. 2003. The human Imp3 and Imp4 proteins form a ternary complex with hMpp10, which only interacts with the U3 snoRNA in 60-80S ribonucleoprotein complexes. Nucleic Acids Res. 31:1877-1887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hadjiolova, K. V., M. Nicoloso, S. Mazan, A. A. Hadjiolov, and J. P. Bachellerie. 1993. Alternative pre-rRNA processing pathways in human cells and their alteration by cycloheximide inhibition of protein synthesis. Eur. J. Biochem. 212:211-215. [DOI] [PubMed] [Google Scholar]
- 15.Henras, A. K., E. Bertrand, and G. Chanfreau. 2004. A cotranscriptional model for 3′-end processing of the Saccharomyces cerevisiae pre-ribosomal RNA precursor. RNA 10:1572-1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Henras, A. K., J. Soudet, M. Gerus, S. Lebaron, M. Caizergues-Ferrer, A. Mougin, and Y. Henry. 2008. The post-transcriptional steps of eukaryotic ribosome biogenesis. Cell. Mol. Life Sci. 65:2334-2359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Karbstein, K., S. Jonas, and J. A. Doudna. 2005. An essential GTPase promotes assembly of preribosomal RNA processing complexes. Mol. Cell 20:633-643. [DOI] [PubMed] [Google Scholar]
- 18.Krogan, N. J., W. T. Peng, G. Cagney, M. D. Robinson, R. Haw, G. Zhong, X. Guo, X. Zhang, V. Canadien, D. P. Richards, B. K. Beattie, A. Lalev, W. Zhang, A. P. Davierwala, S. Mnaimneh, A. Starostine, A. P. Tikuisis, J. Grigull, N. Datta, J. E. Bray, T. R. Hughes, A. Emili, and J. F. Greenblatt. 2004. High-definition macromolecular composition of yeast RNA-processing complexes. Mol. Cell 13:225-239. [DOI] [PubMed] [Google Scholar]
- 19.Lebaron, S., C. Froment, M. Fromont-Racine, J. C. Rain, B. Monsarrat, M. Caizergues-Ferrer, and Y. Henry. 2005. The splicing ATPase prp43p is a component of multiple preribosomal particles. Mol. Cell. Biol. 25:9269-9282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Leung, A. K., L. Trinkle-Mulcahy, Y. W. Lam, J. S. Andersen, M. Mann, and A. I. Lamond. 2006. NOPdb: nucleolar proteome database. Nucleic Acids Res. 34:D218-D220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li, Z., I. Lee, E. Moradi, N. J. Hung, A. W. Johnson, and E. M. Marcotte. 2009. Rational extension of the ribosome biogenesis pathway using network-guided genetics. PLoS Biol. 7:e1000213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Longtine, M. S., A. McKenzie III, D. J. Demarini, N. G. Shah, A. Wach, A. Brachat, P. Philippsen, and J. R. Pringle. 1998. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14:953-961. [DOI] [PubMed] [Google Scholar]
- 23.Miyoshi, K., C. Shirai, and K. Mizuta. 2003. Transcription of genes encoding trans-acting factors required for rRNA maturation/ribosomal subunit assembly is coordinately regulated with ribosomal protein genes and involves Rap1 in Saccharomyces cerevisiae. Nucleic Acids Res. 31:1969-1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Peng, W. T., M. D. Robinson, S. Mnaimneh, N. J. Krogan, G. Cagney, Q. Morris, A. P. Davierwala, J. Grigull, X. Yang, W. Zhang, N. Mitsakakis, O. W. Ryan, N. Datta, V. Jojic, C. Pal, V. Canadien, D. Richards, B. Beattie, L. F. Wu, S. J. Altschuler, S. Roweis, B. J. Frey, A. Emili, J. F. Greenblatt, and T. R. Hughes. 2003. A panoramic view of yeast noncoding RNA processing. Cell 113:919-933. [DOI] [PubMed] [Google Scholar]
- 25.Pérez-Fernández, J., A. Roman, J. De Las Rivas, X. R. Bustelo, and M. Dosil. 2007. The 90S preribosome is a multimodular structure that is assembled through a hierarchical mechanism. Mol. Cell. Biol. 27:5414-5429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pestov, D. G., Y. R. Lapik, and L. F. Lau. 2008. Assays for ribosomal RNA processing and ribosome assembly. Curr. Protocols Cell Biol. Chapter 22, unit 22-11. [DOI] [PubMed]
- 27.Prieto, J. L., and B. McStay. 2007. Recruitment of factors linking transcription and processing of pre-rRNA to NOR chromatin is UBF-dependent and occurs independent of transcription in human cells. Genes Dev. 21:2041-2054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Puig, O., F. Caspary, G. Rigaut, B. Rutz, E. Bouveret, E. Bragado-Nilsson, M. Wilm, and B. Seraphin. 2001. The tandem affinity purification (TAP) method: a general procedure of protein complex purification. Methods 24:218-229. [DOI] [PubMed] [Google Scholar]
- 29.Rigaut, G., A. Shevchenko, B. Rutz, M. Wilm, M. Mann, and B. Seraphin. 1999. A generic protein purification method for protein complex characterization and proteome exploration. Nat. Biotechnol. 17:1030-1032. [DOI] [PubMed] [Google Scholar]
- 30.Rudra, D., J. Mallick, Y. Zhao, and J. R. Warner. 2007. Potential interface between ribosomal protein production and pre-rRNA processing. Mol. Cell. Biol. 27:4815-4824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 32.Savino, R., and S. A. Gerbi. 1990. In vivo disruption of Xenopus U3 snRNA affects ribosomal RNA processing. EMBO J. 9:2299-2308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Segerstolpe, A., P. Lundkvist, Y. N. Osheim, A. L. Beyer, and L. Wieslander. 2008. Mrd1p binds to pre-rRNA early during transcription independent of U3 snoRNA and is required for compaction of the pre-rRNA into small subunit processomes. Nucleic Acids Res. 36:4364-4380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tollervey, D., and I. W. Mattaj. 1987. Fungal small nuclear ribonucleoproteins share properties with plant and vertebrate U-snRNPs. EMBO J. 6:469-476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Turner, A. J., A. A. Knox, J. L. Prieto, B. McStay, and N. J. Watkins. 2009. A novel small-subunit processome assembly intermediate that contains the U3 snoRNP, nucleolin, RRP5, and DBP4. Mol. Cell. Biol. 29:3007-3017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Venema, J., C. Bousquet-Antonelli, J. P. Gelugne, M. Caizergues-Ferrer, and D. Tollervey. 1997. Rok1p is a putative RNA helicase required for rRNA processing. Mol. Cell. Biol. 17:3398-3407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Venema, J., and D. Tollervey. 1996. RRP5 is required for formation of both 18S and 5.8S rRNA in yeast. EMBO J. 15:5701-5714. [PMC free article] [PubMed] [Google Scholar]
- 38.Vos, H. R., R. Bax, A. W. Faber, J. C. Vos, and H. A. Raue. 2004. U3 snoRNP and Rrp5p associate independently with Saccharomyces cerevisiae 35S pre-rRNA, but Rrp5p is essential for association of Rok1p. Nucleic Acids Res. 32:5827-5833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wade, C., K. A. Shea, R. V. Jensen, and M. A. McAlear. 2001. EBP2 is a member of the yeast RRB regulon, a transcriptionally coregulated set of genes that are required for ribosome and rRNA biosynthesis. Mol. Cell. Biol. 21:8638-8650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wade, C. H., M. A. Umbarger, and M. A. McAlear. 2006. The budding yeast rRNA and ribosome biosynthesis (RRB) regulon contains over 200 genes. Yeast 23:293-306. [DOI] [PubMed] [Google Scholar]
- 41.Wang, Y., J. Liu, H. Zhao, W. Lu, J. Zhao, L. Yang, N. Li, X. Du, and Y. Ke. 2007. Human 1A6/DRIM, the homolog of yeast Utp20, functions in the 18S rRNA processing. Biochim. Biophys. Acta 1773:863-868. [DOI] [PubMed] [Google Scholar]
- 42.Wegierski, T., E. Billy, F. Nasr, and W. Filipowicz. 2001. Bms1p, a G-domain-containing protein, associates with Rcl1p and is required for 18S rRNA biogenesis in yeast. RNA 7:1254-1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wehner, K. A., J. E. Gallagher, and S. J. Baserga. 2002. Components of an interdependent unit within the SSU processome regulate and mediate its activity. Mol. Cell. Biol. 22:7258-7267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wery, M., S. Ruidant, S. Schillewaert, N. Lepore, and D. L. Lafontaine. 2009. The nuclear poly(A) polymerase and Exosome cofactor Trf5 is recruited cotranscriptionally to nucleolar surveillance. RNA 15:406-419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wiederkehr, T., R. F. Pretot, and L. Minvielle-Sebastia. 1998. Synthetic lethal interactions with conditional poly(A) polymerase alleles identify LCP5, a gene involved in 18S rRNA maturation. RNA 4:1357-1372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhang, J., P. Harnpicharnchai, J. Jakovljevic, L. Tang, Y. Guo, M. Oeffinger, M. P. Rout, S. L. Hiley, T. Hughes, and J. L. Woolford, Jr. 2007. Assembly factors Rpf2 and Rrs1 recruit 5S rRNA and ribosomal proteins rpL5 and rpL11 into nascent ribosomes. Genes Dev. 21:2580-2592. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.