Abstract
The ETS family transcriptional repressor Yan is an important downstream target and effector of the receptor tyrosine kinase (RTK) signaling pathway in Drosophila melanogaster. Structural and biochemical studies have shown that the N-terminal sterile alpha motif (SAM) of Yan is able to self associate to form a helical polymeric structure in vitro, although the extent and functional significance of self-association of full-length Yan remain unclear. In this study, we demonstrated that full-length Yan self associates via its SAM domain to form higher-order complexes in living cells. Introduction of SAM domain missense mutations that restrict Yan to a monomeric state reduces Yan's transcriptional repression activity and impairs its function during embryonic and retinal development. Coexpression of combinations of SAM domain mutations that permit the formation of Yan dimers, but not higher-order oligomers, increases activity relative to that of monomeric Yan, but not to the level obtained with wild-type Yan. Mechanistically, self-association directly promotes transcriptional repression of target genes independent of its role in limiting mitogen-activated protein kinase (MAPK)-mediated phosphorylation and nuclear export of Yan. Thus, we propose that the formation of higher-order Yan oligomers contributes to proper repression of target gene expression and RTK signaling output in developing tissues.
Transcription factors play an important role in mediating the output of upstream signaling networks to control cell fate specification, proliferation, differentiation, and survival. In Drosophila melanogaster, two ETS (E-26-specific) family transcription factors, Pointed (Pnt) and Yan, mediate the transcriptional response downstream of the receptor tyrosine kinase (RTK) signaling pathway (3, 5, 13, 18, 22, 28, 32). Transcription from alternate promoters of the pnt locus produces two protein isoforms, PntP1 and PntP2, which function as constitutive and inducible transcription activators, respectively (12, 18, 26). yan was identified genetically as a negative regulator of photoreceptor development and was subsequently shown to antagonize RTK signaling in multiple developmental contexts (5, 13, 18). Thus, in the absence of RTK signaling, Yan represses target gene expression to prevent inappropriate developmental outcomes. When upstream signaling is activated, Yan is phosphorylated by mitogen-activated protein kinase (MAPK), transported out of the nucleus, and degraded in the cytoplasm (18, 21, 24, 30). In parallel, MAPK phosphorylates and activates PntP2, which then binds to the previously repressed target genes and induces their expression (3, 18) (Fig. 1A).
FIG. 1.
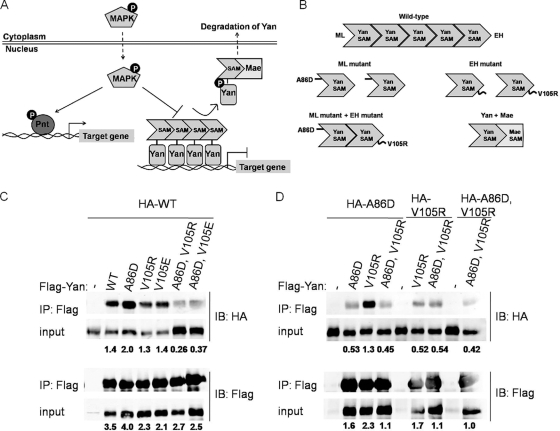
ML and EH surfaces mediate self-association of Yan. (A) Regulation of Yan and Pnt by RTK signaling. In the absence of RTK signaling, Yan represses target gene expression. Activation of RTK signaling leads to Yan's phosphorylation by MAPK and nuclear export facilitated by Mae. In parallel, MAPK phosphorylates and activates Pnt, which then induces the expression of target genes previously repressed by Yan. (B) Model of self-association between ML and EH surfaces of the Yan-SAM domain. ML (A86D) and EH (V105R) mutants exist as monomers, while they interact with each other to form dimers. Mae-SAM interacts with the EH but not the ML surface of Yan-SAM. (C and D) Different combinations of HA- and Flag-tagged wild-type (WT), ML mutant (A86D), EH mutant (V105R; V105E), and ML, EH double mutant (A86D, V105R; A86D, V105E) Yan were cotransfected into Drosophila S2 cells. Immunoprecipitation (IP) was performed using anti-Flag agarose, and the precipitated proteins were detected by immunoblotting with anti-Flag or anti-HA antibodies. (C) Co-IP between wild-type HA-Yan and different mutant forms of Flag-Yan. (D) Co-IP between mutant forms of Yan. IB, Western blotting; input, 2% of the lysate. Numbers below the blots are IP/input ratios.
Yan is evolutionarily conserved (8, 14, 17), and its mammalian homologue Tel is a transcriptional repressor frequently mutated in human leukemias (2). Yan and Tel share two conserved domains: the DNA binding ETS domain and an N-terminal protein-protein interaction sterile alpha motif (SAM) (8, 14, 17). Previous structural and biochemical analyses showed that the isolated SAM domain from Yan/Tel self associates through two interacting surfaces, the midloop (ML) and end-helix (EH) surfaces, to form a head-to-tail polymeric structure (10, 20, 31). Replacement of key hydrophobic residues mediating the ML-EH interaction with charged amino acids (for Yan, A86D on the ML surface or V105R on the EH surface) restricts the SAM domain to a monomeric state. However, combined ML mutant and EH mutant Yan/Tel-SAM domains can form dimers because each retains an intact interacting surface (Fig. 1B).
These biochemical findings have led to the proposal that self-association through the SAM domain might promote the formation of a higher-order complex required for Yan/Tel-mediated transcriptional repression (10, 20). Consistent with such a model, assays with transfected cultured cells showed that ML or EH mutation impairs Yan-mediated transcriptional repression of a synthetic reporter construct (20). In similar assays with mammalian cultured cells, deletion of the SAM domain abolished the repressor function of Tel, while replacing the Tel-SAM domain with a heterologous dimerization domain restored repression activity (16). While the latter result suggests that the formation of higher-order polymers might not be important for function, studies assaying the transforming ability of Tel oncogenic fusion proteins found that replacement of the SAM domain with a heterologous dimerization motif did not confer full activity (29). Thus, the extents to which self-association occurs and is required for full Yan/Tel function remain open questions.
A potential mechanism for regulating the extent of Yan self-association has been suggested by studies of Drosophila Mae (modulator of the activity of ETS), another SAM domain-containing protein, which binds to Yan and facilitates its MAPK-mediated phosphorylation and nuclear export (1, 30). Mae-SAM participates in a one-sided interaction with the EH surface of Yan-SAM but cannot interact with the ML surface (Fig. 1B) (20). Because the Kd (dissociation constant) of Yan-SAM binding to Mae-SAM is ∼1,000 times lower than the Kd of Yan-SAM self-association, it has been proposed that Mae-Yan interactions could efficiently disrupt and hence regulate the polymeric structure of Yan (20, 27).
In this study, we tested the hypothesis that the ML and EH surfaces of the SAM domain mediate self-interaction of the full-length Yan protein and evaluated the importance of self-association of Yan in developing Drosophila tissues. We observed self-association of full-length Yan through the ML and EH surfaces of its SAM domain and found that wild-type Yan exists in a larger complex than that formed by either individual or coexpressed ML and EH mutant variants. Disruption of Yan's self-association ability reduces its transcriptional repression of target genes and ability to complement the lethality of yan null mutants. Coexpression of the ML and EH mutants to create dimers confers significant but not wild-type levels of activity. Mechanistically, although self-association influences Yan's nuclear localization and phosphorylation by MAPK, as previously proposed by others, we showed that it also contributes directly to transcriptional repression activity. Together these data led us to propose that SAM domain-mediated self-association is important for Yan to function as a transcriptional repressor during development and that higher-order complexes beyond dimers may be required for full function.
MATERIALS AND METHODS
Molecular biology.
Wild-type Yan was subcloned into the pENTR3C vector of the Drosophila Gateway system (Invitrogen), using the PCR primers 5′-TAGGTACCTCCAAAATGAAAATGCTCCC-3′ and 5′-TACTCGAGCTACTGCTGCATGTGTCGG-3′. Gateway reactions were carried out as described by the manufacturer to recombine the Yan open reading frame into the pAHW and pAFW vectors, which contain N-terminal 3× hemagglutinin (HA) and 3× Flag tags, respectively. The ML and EH Yan mutants and the ML+EH Yan double mutant were generated by site-directed mutagenesis, using the following PCR primer sets: (i) A86D, 5′-ATGAACGGCAAGGATCTGTGCTTGCTGAC-3′ and 5′-GTCAGCAAGCACAGATCCTTGCCGTTCAT-3′; (ii) V105R, 5′-CTGGGGCAGGAGATCGGCTCCACAATGTGC-3′ and 5′-GCACATTGTGGAGCCGATCTCCTGCCCCAG-3′; and (iii) V105E, 5′-CTGGGGCAGGAGATGAGCTCCACAATGTGCT-3′ and 5′-AGCACATTGTGGAGCTCATCTCCTGCCCCAG-3′.
A Yan-EGFP fusion construct had previously been generated (A. Ghosh and R. DeSouza, unpublished data) in the custom pPL17 expression vector containing the armadillo promoter, which is constitutively active in Drosophila Schneider 2 (S2) cells. The mutation A206K was added to enhanced green fluorescent protein (EGFP) to convert it into its monomeric mutant form (mEGFP) (35). For generation of Yan-mEGFP transgenic flies, this construct was subcloned as a NotI fragment into the pUAST vector. An internal nuclear localization sequence (IntNLS) identical to that previously described (30) was added to the Yan-mEGFP, Yan/pRmHa3, and YanAct/pRmHa3 (YanAct is the constitutively active form of the Yan repressor) constructs by “piecewise” site-directed mutagenesis, using the primers 5′-GTGACCCCTGCTGGAGGATCTACCCCGCCAAAGAAGAAGCGCAAGGT-3′ and 5′-GGTAGGTGCGGAGATGGATCCGTCCTCCACCTTGCGCTTCTTCTTTGG-3′. Briefly, partially overlapping primers were annealed and extended by PCR to generate full-length primers for site-directed mutagenesis. Detailed methods are available upon request. To generate Yan2xNLS constructs, an additional N-terminal NLS was added to YanIntNLS constructs (Yan with an IntNLS added) by piecewise site-directed mutagenesis, using the primers 5′-GTCCCCCAGCGCCGTCCAGCCAGACCCCACCTAAGAAGAAGCGCAAGGTG-3′ and 5′- GCGTCTTAAGCTCCGCCAGCTGGGAGTCCTCCACCTTGCGCTTCTTCTTAG-3′. MaeA141D (a Mae mutant that cannot interact in vitro with Yan [20]) was generated by regular site-directed mutagenesis, using the primers 5′-CCCATGAACGGCAAGGATTTGTGCCTGATGAGT-3′ and 5′-ACTCATCAGGCACAAATCCTTGCCGTTCATGGG-3′.
Constructs used in transcription assays and/or subcellular localization experiments include argos-luciferase, mae-luciferase, and eve-luciferase sequences in the pBluescript vector and the following sequences in the inducible metallothionein promoter vector pRmHa3: lacZ, Flag-PntP1, YanWT (wild-type Yan), YanA86D, YanV105R, YanV105E, YanAct, WT, YanAct, A86D, YanAct, V105R, YanIntNLS, WT, YanIntNLS, A86D, YanIntNLS, V105R, YanAct, IntNLS, WT, YanAct, IntNLS, A86D, and YanAct, IntNLS, V105R.
Cell culture and transfections.
Drosophila S2 cells (25a) were grown in either Gibco Schneider's Drosophila medium supplemented with 10% (vol/vol) Gibco insect medium supplement (IMS) or Gibco Sf-900 serum-free medium (Invitrogen). Comparable results were obtained in both media. S2 cells were transfected by lipofection with dimethyldioctadecyl ammonium bromide (DDAB). For fluorescence recovery after photobleaching (FRAP) and coimmunoprecipitation (co-IP) experiments, transfections were performed in 6-well plates with cells plated at 2.7 × 106 cells per well. Fifty microliters of 0.25 mg/ml DDAB solution in double-distilled water (ddH2O) and 100 μl of medium (per well of a 6-well plate) were mixed and allowed to incubate at room temperature for 5 min. Following incubation, the lipid-medium mix was added to DNA in Eppendorf tubes and incubated for an additional 15 min at room temperature, after which the transfection mixes were added to S2 cells.
For localization experiments, the subcellular localization of Yan in S2 cells was scored qualitatively as cytoplasmic, nuclear, or both. DAPI (4′,6-diamidino-2-phenylindole) staining was used to visualize the location of the nucleus. To prevent bias, scoring was performed without knowledge of the identity of each sample.
Coimmunoprecipitation.
Flag- and HA-tagged Yan were cotransfected into S2 cells. Forty-eight hours after being transfected, cells were lysed in interactor buffer (50 mM HEPES, 150 mM NaCl, 1 mM EDTA, 0.5 mM EGTA, 0.9 M glycerol, 0.5 mM dithiothreitol [DTT], 0.1% Triton X-100 [pH 7.4]) supplemented with a protease inhibitor cocktail (Roche). The clarified lysates were incubated with anti-Flag M2 agarose (Sigma) at 4°C for 2 h, followed by three washes with the interactor buffer. Samples were run on SDS-PAGE gels and transferred for immunoblotting. The blots were probed with rabbit anti-Flag (1:1,000; Sigma) or rabbit anti-HA (1:1,000; Rockland) and IRDye secondary antibodies (1:5,000; Li-Cor Biosciences), and images were captured and quantified with an Odyssey infrared imaging system (Li-Cor Biosciences).
FRAP experiments.
FRAP experiments were performed on a Zeiss 510 confocal microscope, using the accompanying software package. A 63× oil immersion objective was used at 20× zoom, and 128 by 128 pixel images were acquired at a rate of one frame every 98.30 ms. The 488-nm line of an Ar laser (at 50% of its maximum output) was used at 5% of its total intensity to excite EGFP for imaging. Rectangular stripes of 32 by 150 pixels (extending slightly outside of the field of view) were bleached for two frames in the center of the field. All available lines of the Ar laser were used at maximum power for bleaching. Recovery of fluorescence was measured by integrating pixel intensities in the bleached region and dividing by the sum of pixel intensities in the two flanking unbleached rectangular regions. Time constants were obtained by fitting recovery curves to single exponentials. Image analysis and curve fitting were performed with a homemade Python script that is available upon request. Statistical significance of the data was assessed with Origin 7 software, using one-way analysis of variance (ANOVA) followed by a Tukey post hoc test.
Transcription assays.
Transfections were performed in 12-well plates with cells plated at a density of 1.1 × 106 cells per well, and 30 μl of 0.25 mg/ml DDAB solution in ddH2O and 60 μl of medium (per well of a 12-well plate) were used. Twenty-four hours after being transfected, cells were treated with 0.7 mM CuSO4 to induce expression from the metallothionein promoter. Forty-eight hours after being transfected, cells were collected, and transcription assays were performed as previously described, using a luciferase assay kit (Applied Biosystems) (30). Relative luciferase activity was defined as a ratio of luciferase to β-galactosidase activity. Each transfection was performed in duplicate, and the average and standard deviation were calculated. The average relative luciferase activity for the reporter alone was set to 1, and the other conditions were normalized to this value. Microsoft Excel was used to process and graph the data.
Fly strains and genetics.
lozenge-Gal4, GMR-Gal4 (Freeman), rhomboid-Gal4, twist-Gal4 (gla/CyO, twist-Gal4, UAS-GFP), and dpp-Gal4 were used to drive the expression of the following transgenes in eye discs, wing discs, and embryos: (i) UAS-YanAct; (ii) UAS-YanAct, V105R, UAS-YanAct, V105R; and (iii) UAS-YanAct, A86D, UAS-YanAct, V105R. For examining the expression of D-Pax2 genes, the following fly strains carrying the SME-lacZ transgene were crossed with the GMR-Gal4 strain: (i) the UAS-YanAct/CyO, actin-GFP; SME-lacZ/TM6,Tb strain; (ii) the UAS-YanAct, V105R, UAS-YanAct, V105R/CyO, actin-GFP; SME-lacZ/TM6,Tb strain; and (iii) the UAS-YanAct, A86D, UAS-YanAct, V105R/CyO, actin-GFP; SME-lacZ/TM6,Tb strain.
For the rescue assays, flies carrying yanER443, armadillo-Gal4/CyO, twist-Gal4, UAS-GFP were crossed with the following strains to generate yan null mutant embryos and to ubiquitously express Yan in this genetic background: (i) the yanE833/CyO, twist-Gal4, UAS-GFP strain; (ii) the yanE833, UAS-YanWT/CyO, twist-Gal4, UAS-GFP strain; (iii) the yanE833, UAS-YanA86D/CyO, twist-Gal4, UAS-GFP strain; (iv) the yanE833, UAS-YanV105R/CyO, twist-Gal4, UAS-GFP strain; (v) the yanE833, UAS-YanV105E/CyO, twist-Gal4, UAS-GFP strain; (vi) the yanE833, UAS-YanA86D, UAS-YanV105R/CyO, twist-Gal4, UAS-GFP strain; and (vii) the yanE833, UAS-YanA86D, UAS-YanV105E/CyO, twist-Gal4, UAS-GFP strain. About 300 to 500 GFP-negative embryos for each genotype were selected, and the numbers of hatched embryos were counted. From 60 to 160 surviving first instar larvae were picked for each genotype (except for the A86D line), and pupal survival rates were scored.
Immunostaining and antibodies.
S2 cells were fixed in 4% formaldehyde solution in phosphate-buffered saline (PBS) for 10 min at room temperature, washed five times in PBS, and stained with anti-Yan monoclonal antibody (MAb) 8B12 (1:500) followed by Cy3-conjugated goat anti-mouse secondary antibody (1:1,000; Jackson ImmunoResearch) and DAPI staining.
Third instar eye-antenna and wing imaginal discs were dissected in Schneider's Drosophila medium (Invitrogen) and fixed at room temperature for 10 min in 4% formaldehyde solution in PBT (PBS with 0.1% Triton X-100). After three 10-min washes in PBT, the discs were blocked in PNT (PBS, 1% normal goat serum, 0.1% Triton X-100) for 30 min and stained with anti-Yan MAb 8B12 (1:750), mouse anti-Prospero (1:10; Developmental Studies Hybridoma Bank), or mouse anti-β-galactosidase (1:1,000; Promega), followed by secondary antibody staining as above.
Stage 11 embryos were collected on apple juice plates, dechorionated in 50% bleach, and fixed at room temperature for 10 min in a 1:1 mixture of n-heptane and 4% formaldehyde solution in PBS containing 0.3% Triton X-100, with vigorous shaking. The vitellin membrane was removed by replacing the formaldehyde solution with methanol. After three washes with methanol and three with PBS, the embryos were blocked in PNT (PBS, 1% normal goat serum, 0.3% Triton X-100) for 30 min and stained with anti-Eve MAb 3C10 (1:10), followed by secondary antibody staining as mentioned above.
Immunostained eye discs, wing discs, and embryos were examined under a Zeiss 510 confocal microscope, and pictures of 0.3-μm sections were taken and stacked across the z axis, using LSM Image Browser software. For quantification of Yan protein levels in eye discs, z projection was carried out using ImageJ by averaging the signals across sections. The Yan-expressing region was circled by hand, and the average fluorescence intensity within the region was measured by ImageJ.
In situ hybridization.
Embryos were collected and fixed as described above for immunostaining. A digoxigenin (DIG)-labeled argos probe was generated according to the manufacturer's protocol (Roche) from a 1.0-kb template amplified by PCR, using 5′-TGCAGTTTGTTCCAAGATCG-3′ and 5′-GCCATGCAGTAACCGGAATA-3′. Embryos were incubated with the DIG-labeled probe overnight at 48°C, washed at 48°C in hybridization buffer (HB), 50% HB and 50% PBT, 2× PBT, and 3× PBT at room temperature. The embryos were subsequently incubated for 2 h at room temperature with AP-conjugated anti-DIG antibody (1:2,000 Roche). The signal was developed using Nitro Blue Tetrazolium (NBT) and BCIP (5-bromo-4-chloro-3-indolylphosphate).
RESULTS
Yan self associates via its N-terminal SAM domain.
While self-association of the isolated Yan-SAM domain has been demonstrated in vitro (20), self-association of the full-length protein in living cells has not previously been studied. To test if full-length Yan protein interacts with itself through the ML and EH surfaces of its N-terminal SAM domain, we performed coimmunoprecipitation (co-IP) experiments from lysates of transfected S2 cells. Interactions between different combinations of Flag or HA-tagged wild-type, ML mutant (A86D), EH mutant (V105R; V105E), or ML, EH double mutant (A86D, V105R; A86D, V105E) Yan constructs were analyzed (Fig. 1C and D). Efficient co-IP was observed between Flag and HA-tagged wild-type Yan. Wild-type Yan also coprecipitated with the ML or EH mutant proteins, consistent with each retaining one intact interaction surface to bind to the complementary surface on wild-type Yan. In contrast, mutations of both ML and EH surfaces in the same Yan molecule dramatically reduced association with wild-type Yan (Fig. 1C). Examination of interactions between different mutant Yan forms revealed the expected strong association of ML mutant Yan with EH mutant Yan but weaker interactions between either mutant with itself or with the ML, EH double mutant (Fig. 1D). Thus, interactions between the ML and EH surfaces of the SAM domain mediate self-association of full-length Yan in living cells.
Self-association is required for Yan's function during development.
In order to investigate the functional relevance of Yan self-association, we tested the ability of wild-type, ML mutant, EH mutant, or coexpressed ML and EH mutant Yan transgenes to complement the lethality of yan null mutants. As shown in Fig. 2A, whereas loss of yan leads to fully penetrant late embryonic lethality, ubiquitous expression of the wild-type Yan transgene under the armadillo-Gal4 driver rescued about 85% of the yan null mutant embryos to the larval stage; the incomplete rescue presumably reflects the fact that the Gal4 driver does not adequately recapitulate the endogenous Yan expression pattern. In contrast, three independent ML or EH mutant Yan transgenic lines exhibited reduced rescue rates, with the variation in rescue activity correlating with Yan expression levels as assayed by Western blotting (Fig. 2B). Importantly, the wild-type Yan transgenic line expressed protein levels lower than those of the ML and EH mutants, indicating that the reduced rescue activity observed for the ML and EH mutants is not due to a lower level of transgene expression. One limitation of the embryonic rescue experiment is the potential for maternally provided Yan to form a complex with Yan monomers and augment their activity, which would lead us to overestimate function. Therefore, to obtain a more accurate assessment of the ability of monomeric Yan to complement the null mutation, we extended the rescue assay to postembryonic stages, when all maternal product has been depleted.
FIG. 2.
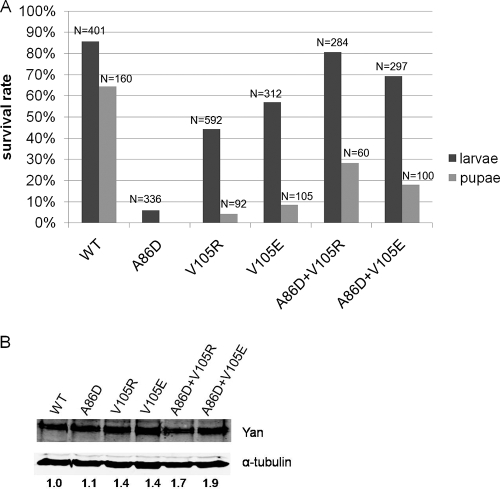
Rescue of yan null mutants. (A) Wild-type Yan, ML mutant (A86D) Yan, EH mutant (V105R or V105E) Yan, or the combination of both (A86D+V105R; A86D+V105E) was expressed under the control of the armadillo-Gal4 driver in yanER443/yanE833 null mutant embryos, and the percentages of animals that developed to the larval and pupal stages were calculated (see Materials and Methods). Pupal survival for A86D was not quantified because of the low rate of larval hatching. N, number of animals scored in survival rate calculation. (B) Quantification of Yan protein levels by Western blotting. Numbers below the blots are relative Yan/α-tubulin ratios. The ratio for WT is set to 1.
The functional differences between wild-type and SAM domain mutant Yan transgenes became even more apparent when rescue was scored at a later developmental stage. Approximately 64% of yan mutants expressing the wild-type Yan transgene survived to the pupal stage, compared to less than 10% of those expressing the EH mutants. Pupal survival rates were not measured for the ML A86D transgene, owing to the extremely low numbers of recovered larvae. These genetic rescue results thus demonstrate that Yan mutants with disrupted self-association ability cannot effectively complement the loss of endogenous yan during Drosophila development, although they do retain partial function.
To determine whether the requirement for Yan self-association can be fulfilled by putative Yan dimers, we coexpressed the ML (A86D) mutant transgene with each of the two EH mutant transgenes. The rescue activity of coexpressed ML and EH transgenes was increased compared to that of the ML or EH mutants alone and appeared comparable to that seen with wild-type Yan with respect to larval survival. However, when we followed the development of the surviving first instar larvae to pupal stages, we found that while the putative Yan dimers showed enhanced rescue ability relative to that of the individual EH mutant (18 to 28% versus <10%, respectively), they were markedly less active than wild-type Yan (64%), despite expressing higher Yan protein levels. In conclusion, ML and EH mutant Yan, alone or coexpressed, are less able than wild-type Yan to complement the lethality of yan null mutants, suggesting that formation of a higher-order oligomeric complex may be important for the biological function of Yan.
Self-association is required for Yan to repress gene transcription.
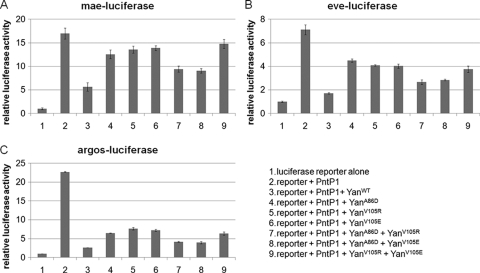
Is the formation of higher-order complexes required for Yan's transcriptional repressor function? To answer this question, we compared the repression activity of wild-type Yan, the ML mutant, the EH mutant, and the coexpressed ML and EH mutants in transcription assays in cultured S2 cells, using luciferase reporter constructs containing previously characterized Yan binding sites from its known target genes argos (5, 6), mae (33), and even skipped (eve) (7). As shown in Fig. 3, wild-type Yan effectively repressed the transcription of all reporters, the ML or EH mutants alone had reduced repression activity, and the coexpressed ML and EH mutants showed intermediate levels of activity, indicating that self-association and formation of oligomeric complexes may be important for Yan to repress the transcription of target genes. These findings agree with the results of analogous previous experiments that used an artificial luciferase reporter construct carrying tandem multimerized ETS binding consensus sequences (20). In our study, we found that the repression activity of ML or EH mutant Yan at different reporter genes varied, with a relatively high level of activity at the argos reporter, an intermediate level of activity at the eve reporter, and a minimal level of activity at the mae reporter. This could reflect the different extents to which the repression of distinct target genes requires self-association of Yan. Alternatively, the differences may reflect how well the cloned genomic fragments in our reporter constructs encompass sequences needed for regulation of the endogenous genes.
FIG. 3.
Monomeric Yan has reduced activity in repressing reporter gene expression in S2 cells. Transcription assays were performed with wild-type Yan, ML mutant (A86D) Yan, EH mutant (V105R or V105E) Yan, or the combination of both (A86D+V105R; A86D+V105E) (V105R+V105E served as a control). Error bars indicate standard deviations between the results of two transfections. Results are shown for mae-luciferase (A), eve-luciferase (B), and argos-luciferase (C).
We next asked if self-association of Yan is important for regulating transcriptional targets during eye development by examining the effects of ectopically expressed wild-type, ML mutant, EH mutant, or coexpressed ML and EH mutant Yan. Expressing the constitutively active form of the Yan repressor (YanAct) that is refractory to MAPK-mediated phosphorylation (21) with the lozenge-Gal4 driver in cone cells and R1, R6, and R7 photoreceptor cells leads to significant disruption of adult eye morphology, whereas expression of two copies of the EH mutant YanAct (2× YanAct, V105R) has no observable effect (Fig. 4A to C). Coexpression of ML and EH mutant YanAct transgenes (YanAct, A86D + YanAct, V105R) also disrupts the adult eye structure, but to a lesser extent than YanAct, as evidenced by the partial restoration of pigmentation and the slightly less “glassy” appearance of the eye (Fig. 4D).
FIG. 4.
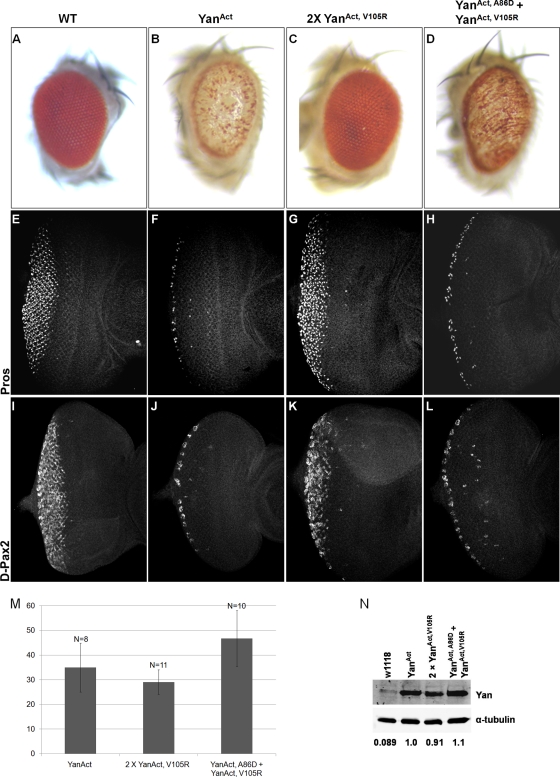
Monomeric Yan fails to inhibit eye development and repress target genes prospero and D-Pax2 in third instar eye imaginal discs. (A to D) Pictures of eyes of adult wild-type flies (A) and flies expressing YanAct (B), two copies of YanAct, V105R (C), or YanAct, A86D + YanAct, V105R (D) under the lozenge-Gal4 driver. (E to L) Third instar wild-type eye discs (E and I) and discs expressing YanAct (F and J), two copies of YanAct, V105R (G and K), or YanAct, A86D + YanAct, V105R (H and L) under the GMR-Gal4 driver. (E to H) Prospero expression. (I to L) β-Galactosidase staining revealing the expression of the D-Pax2 gene. (M) Quantification of Yan protein levels by immunostaining (see Materials and Methods). Error bars represent standard deviations. N, number of discs measured for each genotype. (N) Quantification of Yan protein levels by Western blotting. Numbers below the blots are relative Yan/α-tubulin ratios. The ratio for YanAct is set to 1.
To investigate whether the inability of the EH mutant YanAct to disrupt eye development reflected loss of transcriptional repression activity, we tested the ability of different forms of YanAct to regulate the expression of two direct target genes, prospero (pros) and D-Pax2 (4, 34). In wild-type third instar eye imaginal discs, pros is expressed in the R7 photoreceptors and cone cells while D-Pax2 is expressed only in the cone cells (Fig. 4E and I). Expressing YanAct with the GMR-Gal4 driver strongly inhibited Pros and D-Pax2 expression, while expression of EH mutant YanAct had no observable effect (Fig. 4F and G and J and K). Coexpression of ML and EH mutant YanAct transgenes inhibited Pros and D-Pax2 expression to an extent comparable to that of YanAct (Fig. 4H and L). To rule out the possibility that the different effects of YanAct and its SAM domain mutant variants were due to differences in their expression levels, we quantified the levels of Yan protein by immunostaining and Western blotting. As shown in Fig. 4M and N, comparable Yan protein levels were detected in eye discs expressing the various YanAct transgenes.
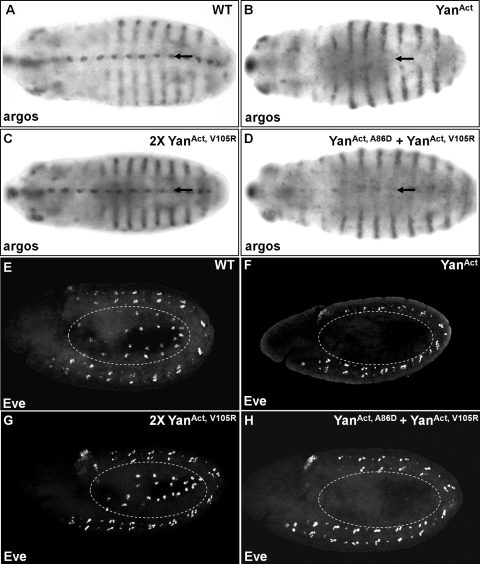
To compare the effects of different forms of Yan on target gene expression in another developmental context, we analyzed the expression in embryos of two additional Yan target genes, argos and eve (5-7). In wild-type embryos, argos is expressed in the ventral epidermal cells, in the head region, and in lateral stripes, while eve is expressed in the mesodermal cardiac progenitor cells and in the central nervous system (CNS) (Fig. 5A and E). Consistent with our observations with pros and D-Pax2 in eye discs, overexpression of the YanAct transgene in the ventral epidermis with the rhomboid-Gal4 driver essentially abolished argos expression in this region, as revealed by in situ hybridization (Fig. 5B). Similarly, expressing YanAct under the twist-Gal4 driver completely inhibited Eve expression in the mesoderm (Fig. 5F). When the ML and EH mutant YanAct transgenes were coexpressed, Eve expression was inhibited to an extent comparable to that when YanAct was expressed, while argos expression was also significantly reduced (Fig. 5D and H). In contrast, the expression of two copies of the EH mutant YanAct had no observable effect on argos or eve expression (Fig. 5C and G). Taken together, these results show that self-association is important for Yan to regulate its target genes in multiple developmental contexts and that at least under conditions in which Yan is rendered insensitive to MAPK-mediated phosphorylation and downregulation, Yan dimers can confer significant repression activity.
FIG. 5.
Monomeric Yan fails to repress the expression of target genes argos and even skipped in embryos. (A to D) In situ hybridization of argos in wild-type embryos (A) and embryos expressing YanAct (B), two copies of YanAct, V105R (C), or YanAct, A86D + YanAct, V105R (D) under the control of the rhomboid-Gal4 driver. (E to H) Eve staining in wild-type embryos (E) and embryos expressing YanAct (F), two copies of YanAct, V105R (G), or YanAct, A86D + YanAct, V105R (H) under the control of the twist-Gal4 driver. YanAct and YanAct, A86D + YanAct, V105R strongly repress the expression of argos in the ventral epidermis (arrows) and Eve in the mesoderm (dashed circles), whereas YanAct, V105R has minimal effects.
Yan self-association functions downstream of nuclear localization and phosphorylation.
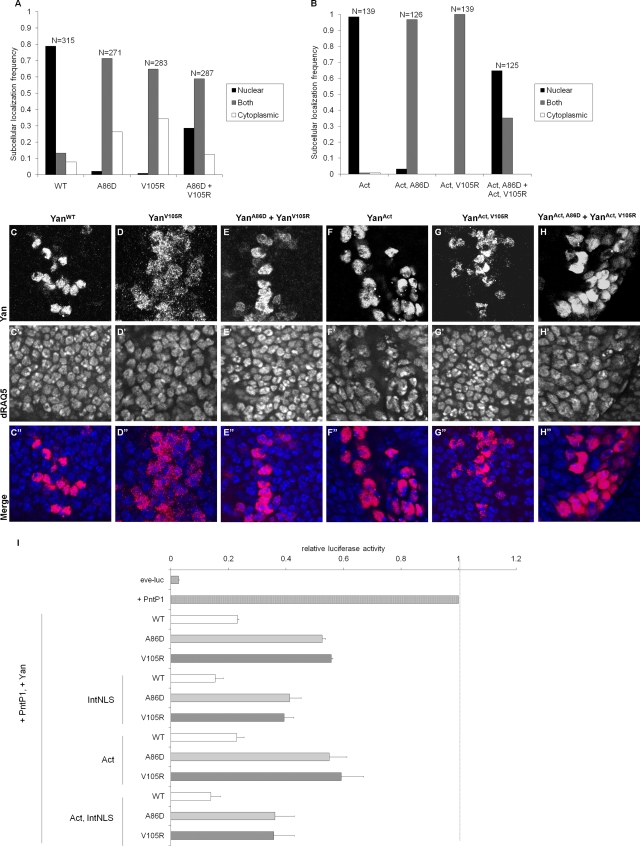
It was previously shown that MAPK-mediated phosphorylation leads to nuclear export and downregulation of Yan and that ML or EH mutants localize less exclusively to the nucleus than wild-type Yan, although they have equivalent DNA binding affinity (20, 27, 30). Indeed, cotransfection of the complementary ML (YanA86D) and EH (YanV105R) mutants into cultured S2 cells or coexpression of the ML and EH Yan transgenes in wing discs led to increased nuclear localization compared to that for the ML or EH mutants alone (Fig. 6A and C to E). The increased level of nuclear localization of coexpressed ML and EH mutants relative to that for individually expressed ML or EH mutants appears to be partially independent of MAPK-mediated phosphorylation, as it was still observed in the background of the nonphosphorylatable YanAct mutant (Fig. 6B and F to H). However, all YanAct-based mutants localize more tightly to the nucleus than their phosphorylatable Yan counterparts (Fig. 6B and F to H versus 6A and C to E). Thus, subcellular localization of ML or EH Yan appears to depend partially, but not entirely, on levels of Yan phosphorylation.
FIG. 6.
Yan self-association functions downstream of nuclear localization and phosphorylation. (A) Localization of the YanWT, YanA86D, YanV105R, and Yan “dimer” made by cotransfecting YanA86D and YanV105R. (B) Localization of YanAct, YanAct, A86D, YanAct, V105R, and YanAct, A86D + YanAct, V105R. Subcellular localization of Yan was scored by eye with reference to nuclear staining by DAPI. Localization was judged nuclear if anti-Yan staining was clearly brighter in the region of the nucleus than in the surrounding cytoplasm. Conversely, localization was judged cytoplasmic if the staining intensity in the nuclear region appeared to be less than that in the surrounding cytoplasm. Cells were classified as having both nuclear and cytoplasmic staining if the relative intensities of the signals in the two regions appeared comparable. Scoring was performed without knowledge of the identity of the samples. Yan is expressed endogenously in S2 cells but at low levels that are essentially undetectable relative to the proteins expressed from our transfected constructs. N, number of cells scored. (C to H) Immunostaining of Yan (red) and dRAQ5 (blue), which stains DNA, in third instar wing discs expressing YanWT (C), YanV105R (D), YanA86D + YanV105R (E), YanAct (F), YanAct, V105R (G), and YanAct, A86D + YanAct, V105R (H) under the dpp-Gal4 driver. (I) Repression of the PntP1-activated eve-luciferase reporter in Drosophila S2 cells by various Yan constructs. The data have been normalized such that the fully activated reporter (with PntP1) is set to 1. All Yan constructs with a wild-type SAM domain (white bars) repress the reporter approximately fivefold, while all monomeric constructs (gray bars) repress the reporter approximately twofold. Even when the protein is rendered nonphosphorylatable (Act), restricted to the nucleus (IntNLS), or both (Act, IntNLS), the monomeric mutants are still less active than wild-type Yan.
Given that Yan functions in the nucleus as a transcriptional repressor, a reasonable hypothesis is that the reduced function of the Yan ML and EH mutants is due primarily to their disrupted nuclear localization. To test this, we introduced the ML and EH mutants into a Yan construct carrying a strong exogenous nuclear localization sequence from the simian virus 40 (SV40) large T antigen (30) and assessed the ability of these constructs to repress expression of the eve-luciferase reporter in cultured cells. While YanIntNLS, ML and YanIntNLS, EH mutants were strongly nuclear in virtually all cells (data not shown), they still repressed eve-luciferase less efficiently than YanIntNLS or wild-type Yan (Fig. 6I). This suggests that the reduced repression activity of the Yan ML and EH mutants cannot be explained by their disrupted localization alone.
Self-association is thought to sterically occlude phosphorylation of Yan by MAPK (20). To examine whether increased susceptibility to phosphorylation accounts for the reduced activity of ML or EH mutant Yan, we tested YanAct, A86D and YanAct, V105R mutants in transcriptional reporter assays. YanAct, A86D and YanAct, V105R mutants still exhibited reduced repression activity of eve-luciferase compared with YanAct (Fig. 6I), consistent with our results with Drosophila tissues. These results suggest that while ML or EH mutant Yan may be more accessible to MAPK, this alone is not sufficient to explain their reduced repression activity compared with that of wild-type Yan.
Results from the YanAct and YanIntNLS mutant constructs made it clear that neither altered phosphorylation nor altered nuclear localization alone could explain the difference in activity between monomeric and wild-type Yan. However, it remained possible that both enhanced phosphorylation and disrupted nuclear localization could together explain the reduced activity of ML or EH mutant Yan compared to that of wild-type Yan. To rule this out, we inserted the NLS sequence into the YanAct construct to generate nonphosphorylatable Yan restricted to the nucleus. Even in the YanAct, IntNLS background, constructs containing ML and EH mutations repressed eve-luciferase less efficiently than either wild-type Yan or a YanAct, IntNLS construct with a wild-type SAM domain (Fig. 6I). These results imply that Yan self-association promotes transcriptional repression independent of its contribution to the regulation of MAPK-mediated phosphorylation and nuclear localization.
Yan forms a higher-order complex in living cells.
The observation that full-length Yan self associated via its SAM domain and that ML or EH mutants, either alone as monomers or coexpressed to generate dimers, had reduced transcriptional repression activity led us to hypothesize that Yan functions in higher-order complexes beyond dimers in vivo. We reasoned that measuring the relative diffusion rates of fluorescently tagged Yan proteins in living cells using FRAP (15) might provide an informative first test of this model. In this method, the fluorescence label on a protein is irreversibly photobleached in a defined region, and the recovery of fluorescence due to the influx of fluorescent protein from other regions of the cell is measured over time. The prediction was that if wild-type Yan oligomerizes, it would exhibit a slower recovery rate than monomeric or dimeric Yan.
We generated a Yan-mEGFP construct by fusing the monomeric EGFP mutant (A206K) (35) in-frame to the Yan C terminus. To counteract the partial mislocalization to the cytoplasm caused by the EGFP fusion and to study signaling conditions in which Yan would normally be subject to nuclear export and rapid degradation, an internal nuclear localization sequence (IntNLS) was included (30) to achieve nuclear localization comparable to that of wild-type Yan. The Yan-mEGFP construct produced a functional protein of the expected size, as demonstrated by normal transcriptional repression activity in cultured cell assays, and expected mobility, as judged by immunoblotting with anti-Yan and anti-GFP antibodies (data not shown).
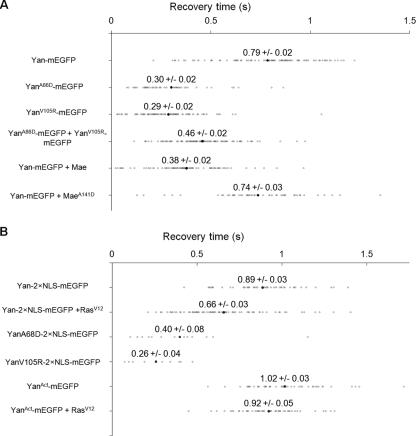
Diffusion rate measurements by FRAP showed that Yan-mEGFP diffused into a 1.66-μm-wide rectangular bleach region with an average recovery time of 0.79 ± 0.02 s, compared with average recovery times of 0.30 ± 0.02 s and 0.29 ± 0.02 s for YanA86D-mEGFP and YanV105R-mEGFP, respectively (Fig. 7A). The increase in the diffusion rate of ML or EH mutant Yan-mEGFP was statistically significant (P < 0.001). This suggests that wild-type Yan exists as part of a larger, more slowly diffusing complex than ML or EH Yan monomers. Coexpressed YanA86D-mEGFP and YanV105R-mEGFP had a recovery time (0.46 ± 0.02 s) intermediate between and significantly different (P < 0.001) from both wild-type and the individual ML or EH mutants (Fig. 7A), consistent with ML-EH dimers forming an intermediate-sized complex. It should be noted that while these results are consistent with Yan oligomerization, we cannot exclude the possibility that heterotypic interactions specific to wild-type Yan, rather than homotypic polymerization, might be responsible for the slower diffusion rate.
FIG. 7.
Yan forms a large complex in living cells. Recovery time measurements by FRAP of Yan-mEGFP in Drosophila S2 cells. Data points in gray are for individual cells. Black points are averages of between 49 and 101 cells per condition. The mean ± standard error of the mean are reported above each black point. (A) Recovery time of wild-type, A86D, V105R, and coexpressed A86D and V105R Yan-mEGFP and the effect of Mae on Yan's diffusion rate. (B) The effect of RasV12 on the diffusion rates of Yan and YanAct.
The Yan antagonist protein Mae has been proposed to disrupt Yan polymers through one-sided interactions with the Yan SAM domain (20). Consistent with this hypothesis, the diffusion rate of Yan-mEGFP increased significantly (P < 0.001) upon coexpression of wild-type Mae (recovery time, 0.37 ± 0.02 s) (Fig. 7A). However, the diffusion rate did not significantly change upon cotransfection of MaeA141D (20), a mutant that cannot interact in vitro with Yan (recovery time, 0.74 ± 0.02 s). Both Mae and MaeA141D were comparably expressed, as judged by immunofluorescence staining of the same population of cells used for the FRAP experiments (data not shown).
Finally, we asked whether MAPK-mediated phosphorylation, which is required for nuclear export and downregulation of Yan (21, 30), alters the diffusion rate of Yan-mEGFP. Two separate nuclear localization sequences (Yan2×NLS-mEGFP) were inserted into Yan to block phosphorylation-induced nuclear export. The diffusion rates of single and double NLS-tagged Yan constructs were comparable (Fig. 7A and B). Stimulation of MAPK by coexpression of an activated Ras construct (RasV12) significantly increased the diffusion rate of Yan2×NLS-mEGFP (recovery time, 0.66 ± 0.03 s versus 0.86 ± 0.03 s in the absence of RasV12; P < 0.001). The increased diffusion rate is due to phosphorylation, since RasV12 did not increase the diffusion rate of the nonphosphorylatable YanAct-mEGFP (Fig. 7B). These results suggest that MAPK-mediated phosphorylation partially disrupts the higher-order structure of the Yan complex.
DISCUSSION
This study supports the hypothesis that full-length Yan self associates to form an oligomeric complex in living cells via the ML and EH surfaces of its N-terminal SAM domain. Self-association appears critical for in vivo function, as evidenced by the reduced ability of monomeric mutant Yan to complement the lethality of yan mull mutants and to repress target gene expression. Mechanistically, we show that self-association directly promotes Yan-mediated transcriptional repression independent of its effects on subcellular localization and MAPK-mediated phosphorylation.
Previous structural and biochemical analyses of Yan and Tel revealed that the ML and EH surfaces mediate self-association of isolated SAM domains. Despite forming very similar polymeric structures, the Kd measured for Yan-SAM interactions was significantly higher than that for Tel-SAM (∼10 μM versus 2 nM, respectively) (10, 20, 31). This raises the possibility that the interaction might not be of sufficient affinity to promote stable association of full-length Yan. Arguing against this, our coimmunoprecipitation studies demonstrate that full-length Yan interacts with itself via the two complementary ML and EH surfaces to form stable SAM-mediated complexes. However, we also detected residual self-association with ML or EH mutant Yan proteins that would not be expected to interact based on in vitro studies of the corresponding SAM domains. This could reflect other regions of Yan contributing to self-association, either directly or via interactions with other proteins or DNA. Given the higher affinity of Tel-SAM self-association, such extra interactions might not be needed for oligomerization of full-length Tel. In fact, in co-IP experiments analogous to ours, SAM domain mutants of full-length Tel completely lacked self-association ability (25). Thus, while SAM-SAM interactions promote and are required for self-association, in the case of Yan additional interactions may stabilize binding.
Based on crystal structure and electron microscope images showing that the isolated Yan/Tel-SAM domains can form large filaments, it has been proposed that Yan/Tel forms a head-to-tail helical polymer that might wrap the DNA around itself to mediate transcriptional repression (10, 20). Unfortunately, difficulties in purifying sufficient soluble, full-length proteins have to date hindered further biochemical investigation of these models. As an alternate approach, we assessed the extent of self-association in living cells, using FRAP to compare the relative diffusion rates of the putative monomeric, dimeric, and oligomeric Yan complexes. While our results are consistent with wild-type Yan forming higher-order oligomers, the larger, slower-diffusing complexes could also reflect differential heterotypic interactions. Thus, it is possible that the ML-EH mutant combination used to generate Yan dimers does not permit interactions with the same set of corepressors or other factors that dimers made of wild-type Yan proteins normally bind and thus forms a faster-diffusing complex. Biochemical purification and characterization of complexes isolated from living cells expressing monomeric versus dimeric versus wild-type Yan will be needed to determine the composition and stoichiometry of the different mobility complexes detected in our FRAP studies.
Consistent with the previously proposed hypothesis that Yan-Mae interactions provide a mechanism for limiting polymer length (20, 27), coexpression of Mae significantly increases the diffusion rate of Yan-mEGFP. Activation of the MAPK-signaling cascade, which normally results in phosphorylation, nuclear export, and degradation of Yan (18, 21, 24), also increases the diffusion rate of Yan-mEGFP restricted to the nucleus. However, the diffusion rate is slower than that of the ML or EH mutants. One possible explanation is that the Yan-mEGFP fusion protein restricted to the nucleus might be less efficiently targeted by MAPK than wild-type Yan. Alternatively, MAPK-mediated phosphorylation may on its own only partially disrupt Yan oligomers. Full disruption of Yan complexes may require interaction with Mae and nuclear export.
Despite a general consensus that Yan/Tel self-association is important for transcriptional repression (16, 20), controversy remains as to whether higher-order complexes beyond dimers are necessary. As a framework for discussing the relationship between Yan/Tel polymerization, dimerization, and transcriptional repression functions, it is helpful to consider two not-necessarily-mutually-exclusive models. An assumption common to both models and supported by our FRAP and genetic rescue results as well as by previous in vitro studies and analyses of leukemic Tel fusion proteins (10, 20, 29) is that Yan/Tel complexes extend beyond a dimeric form. Therefore, in the first model, as originally suggested by Bowie and colleagues (10, 20), higher-order oligomers participate directly in transcriptional repression complexes. Alternatively, the active unit directly mediating transcriptional repression could be a dimer, with polymers providing a dynamic nucleoplasmic pool of Yan/Tel from which repression complexes can be rapidly assembled and targeted to appropriate regulatory elements.
Supporting this second scenario, our analysis of Yan's ability to repress the expression of four different target genes in developing Drosophila tissues revealed essentially identical activity for wild-type and dimeric Yan, while rescue assays demonstrated a functional requirement for higher-order Yan complexes. One caveat to this interpretation is that the gene expression analyses were performed in the constitutively active YanAct background; this was necessary because MAPK-signaling levels are sufficiently robust to downregulate even high levels of overexpressed wild-type Yan (21). Thus, it is possible that only when Yan protein levels are unusually high, as they are in the YanAct backgrounds, are dimers sufficient for repression, but that under more physiological protein concentrations, a polymeric complex might be required for full repression. Arguing that this could be the case and favoring the first model, dimeric Yan is consistently less active than polymerization-competent Yan in cultured cell transcription assays. Because only a limited number of Yan target genes are currently known, identification and analysis of additional direct targets will be needed to assess more comprehensively in what cases polymers are required for full activity and in what cases dimers may be sufficient.
Through what mechanisms might self-association contribute to repression activity? Both our experiments and previously published reports indicate that self-association enhances nuclear localization of Yan (27), which logically should increase repression activity. In addition, it has been suggested that self-association may prevent downregulation of Yan by occluding critical MAPK phosphorylation sites on the protein (20, 27). Either or both mechanisms could in principle entirely explain the differences in repression activity among monomeric, dimeric, and wild-type Yan. Arguing against this, nuclearly restricted and/or nonphosphorylatable monomeric Yan mutants have reduced repression activity compared to their counterparts with a wild-type SAM domain, indicating that self-association contributes to repression activity independently of nuclear localization or MAPK-mediated phosphorylation. An alternate explanation of the differences in repression activity among monomeric, dimeric, and wild-type Yan is that they might have different abilities to recruit essential cofactors. For example, Yan polymers might form complexes more effectively with critical corepressors than Yan monomers, resulting in stronger transcriptional repression. Varying Yan polymer lengths and repression complex composition could provide a dynamic mechanism for fine tuning transcriptional output.
Intriguingly, a number of corepressor proteins have themselves been shown to form polymers. Among the most interesting to consider in the context of SAM domain-mediated polymerization mechanisms are two Polycomb group (PcG) proteins, Sex comb on midleg (Scm) and Polyhomeotic (Ph) (9, 11, 19). PcG proteins form repression complexes that occupy large chromatin domains and recruit histone-modifying proteins to maintain the repressed status of genes throughout development. Both Scm and Ph can form homo- or heteropolymers via their SAM domains, and it is thought that such interactions are important for the formation of large PcG complexes and maintenance of repression of their target genes (9, 11, 19, 23). Whether and how interactions with PcG proteins or other corepressor proteins influence Yan polymerization and repression activity will be an important and interesting area for further exploration.
Unlike Scm and Ph, which do not have a DNA binding domain and depend on specific protein-protein interactions to be recruited to targeted chromatin loci, Yan can interact specifically and with high affinity to DNA via its ETS DNA binding domain (13, 20). It has been suggested that Yan/Tel binds to the ETS binding sites (EBS) of a functional cis-regulatory module and nucleates the formation of polymers on chromatin (10, 20). Such polymers could either passively block access to the transcriptional machinery or actively recruit a chromatin-modifying repression complex. Whether stability and function of the postulated Yan/Tel polymers depend on multiple interactions with DNA elements remains an open question. An alternate possibility is that Yan/Tel participates in two entirely different repression complexes, one in which they operate as sequence-specific DNA binding proteins and another in which they function as corepressors independently of DNA binding. While our data suggest that dimerization, at least, is required in the first context, higher-order polymers might be more crucial in the second context. Discriminating between the various mechanistic possibilities linking Yan/Tel polymerization and repression will require a high-resolution system level analysis of chromatin occupancy, binding partners, repressed target genes, and DNA regulatory elements.
Acknowledgments
We thank S. Morillo and W. Xiong for comments on the manuscript, Rebay and Fehon lab members for helpful discussions, M. DiMarco for confocal assistance, and the Developmental Studies Hybridoma Bank for antibodies.
This research was supported by a Women's Board Fellowship of the University of Chicago to J.Z., by NIH Roadmap Physical and Chemical Biology Training Program grant T90-DK070076 to T.G.W.G., and by NIH grants R01 GM80372 and P50 GM081892 to I.R.
Footnotes
Published ahead of print on 4 January 2010.
REFERENCES
- 1.Baker, D. A., B. Mille-Baker, S. M. Wainwright, D. Ish-Horowicz, and N. J. Dibb. 2001. Mae mediates MAP kinase phosphorylation of Ets transcription factors in Drosophila. Nature 411:330-334. [DOI] [PubMed] [Google Scholar]
- 2.Bohlander, S. K. 2005. ETV6: a versatile player in leukemogenesis. Semin. Cancer Biol. 15:162-174. [DOI] [PubMed] [Google Scholar]
- 3.Brunner, D., K. Ducker, N. Oellers, E. Hafen, H. Scholz, and C. Klambt. 1994. The ETS domain protein pointed-P2 is a target of MAP kinase in the sevenless signal transduction pathway. Nature 370:386-389. [DOI] [PubMed] [Google Scholar]
- 4.Flores, G. V., H. Duan, H. Yan, R. Nagaraj, W. Fu, Y. Zou, M. Noll, and U. Banerjee. 2000. Combinatorial signaling in the specification of unique cell fates. Cell 103:75-85. [DOI] [PubMed] [Google Scholar]
- 5.Gabay, L., H. Scholz, M. Golembo, A. Klaes, B. Z. Shilo, and C. Klambt. 1996. EGF receptor signaling induces pointed P1 transcription and inactivates Yan protein in the Drosophila embryonic ventral ectoderm. Development 122:3355-3362. [DOI] [PubMed] [Google Scholar]
- 6.Golembo, M., R. Schweitzer, M. Freeman, and B. Z. Shilo. 1996. Argos transcription is induced by the Drosophila EGF receptor pathway to form an inhibitory feedback loop. Development 122:223-230. [DOI] [PubMed] [Google Scholar]
- 7.Halfon, M. S., A. Carmena, S. Gisselbrecht, C. M. Sackerson, F. Jimenez, M. K. Baylies, and A. M. Michelson. 2000. Ras pathway specificity is determined by the integration of multiple signal-activated and tissue-restricted transcription factors. Cell 103:63-74. [DOI] [PubMed] [Google Scholar]
- 8.Hsu, T., and R. A. Schulz. 2000. Sequence and functional properties of Ets genes in the model organism Drosophila. Oncogene 19:6409-6416. [DOI] [PubMed] [Google Scholar]
- 9.Kim, C. A., M. Gingery, R. M. Pilpa, and J. U. Bowie. 2002. The SAM domain of polyhomeotic forms a helical polymer. Nat. Struct. Biol. 9:453-457. [DOI] [PubMed] [Google Scholar]
- 10.Kim, C. A., M. L. Phillips, W. Kim, M. Gingery, H. H. Tran, M. A. Robinson, S. Faham, and J. U. Bowie. 2001. Polymerization of the SAM domain of TEL in leukemogenesis and transcriptional repression. EMBO J. 20:4173-4182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kim, C. A., M. R. Sawaya, D. Cascio, W. Kim, and J. U. Bowie. 2005. Structural organization of a Sex-comb-on-midleg/polyhomeotic copolymer. J. Biol. Chem. 280:27769-27775. [DOI] [PubMed] [Google Scholar]
- 12.Klämbt, C. 1993. The Drosophila gene pointed encodes two ETS-like proteins which are involved in the development of the midline glial cells. Development 117:163-176. [DOI] [PubMed] [Google Scholar]
- 13.Lai, Z. C., and G. M. Rubin. 1992. Negative control of photoreceptor development in Drosophila by the product of the yan gene, an ETS domain protein. Cell 70:609-620. [DOI] [PubMed] [Google Scholar]
- 14.Laudet, V., C. Hanni, D. Stehelin, and M. Duterque-Coquillaud. 1999. Molecular phylogeny of the ETS gene family. Oncogene 18:1351-1359. [DOI] [PubMed] [Google Scholar]
- 15.Lippincott-Schwartz, J., E. Snapp, and A. Kenworthy. 2001. Studying protein dynamics in living cells. Nat. Rev. Mol. Cell Biol. 2:444-456. [DOI] [PubMed] [Google Scholar]
- 16.Lopez, R. G., C. Carron, C. Oury, P. Gardellin, O. Bernard, and J. Ghysdael. 1999. TEL is a sequence-specific transcriptional repressor. J. Biol. Chem. 274:30132-30138. [DOI] [PubMed] [Google Scholar]
- 17.Mavrothalassitis, G., and J. Ghysdael. 2000. Proteins of the ETS family with transcriptional repressor activity. Oncogene 19:6524-6532. [DOI] [PubMed] [Google Scholar]
- 18.O'Neill, E. M., I. Rebay, R. Tjian, and G. M. Rubin. 1994. The activities of two Ets-related transcription factors required for Drosophila eye development are modulated by the Ras/MAPK pathway. Cell 78:137-147. [DOI] [PubMed] [Google Scholar]
- 19.Peterson, A. J., M. Kyba, D. Bornemann, K. Morgan, H. W. Brock, and J. Simon. 1997. A domain shared by the Polycomb group proteins Scm and ph mediates heterotypic and homotypic interactions. Mol. Cell. Biol. 17:6683-6692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Qiao, F., H. Song, C. A. Kim, M. R. Sawaya, J. B. Hunter, M. Gingery, I. Rebay, A. J. Courey, and J. U. Bowie. 2004. Derepression by depolymerization: structural insights into the regulation of Yan by Mae. Cell 118:163-173. [DOI] [PubMed] [Google Scholar]
- 21.Rebay, I., and G. M. Rubin. 1995. Yan functions as a general inhibitor of differentiation and is negatively regulated by activation of the Ras1/MAPK pathway. Cell 81:857-866. [DOI] [PubMed] [Google Scholar]
- 22.Rogge, R., P. J. Green, J. Urano, S. Horn-Saban, M. Mlodzik, B. Z. Shilo, V. Hartenstein, and U. Banerjee. 1995. The role of yan in mediating the choice between cell division and differentiation. Development 121:3947-3958. [DOI] [PubMed] [Google Scholar]
- 23.Roseman, R. R., K. Morgan, D. R. Mallin, R. Roberson, T. J. Parnell, D. J. Bornemann, J. A. Simon, and P. K. Geyer. 2001. Long-range repression by multiple polycomb group (PcG) proteins targeted by fusion to a defined DNA-binding domain in Drosophila. Genetics 158:291-307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Roukens, M. G., M. Alloul-Ramdhani, S. Moghadasi, M. Op den Brouw, and D. A. Baker. 2008. Downregulation of vertebrate Tel (ETV6) and Drosophila Yan is facilitated by an evolutionarily conserved mechanism of F-box-mediated ubiquitination. Mol. Cell. Biol. 28:4394-4406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Roukens, M. G., M. Alloul-Ramdhani, A. C. Vertegaal, Z. Anvarian, C. I. Balog, A. M. Deelder, P. J. Hensbergen, and D. A. Baker. 2008. Identification of a new site of sumoylation on Tel (ETV6) uncovers a PIAS-dependent mode of regulating Tel function. Mol. Cell. Biol. 28:2342-2357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25a.Schneider, I. 1972. Cell lines derived from late embryonic stages of Drosophila melanogaster. J. Embryol. Exp. Morphol. 27:353-365. [PubMed] [Google Scholar]
- 26.Scholz, H., J. Deatrick, A. Klaes, and C. Klambt. 1993. Genetic dissection of pointed, a Drosophila gene encoding two ETS-related proteins. Genetics 135:455-468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Song, H., M. Nie, F. Qiao, J. U. Bowie, and A. J. Courey. 2005. Antagonistic regulation of Yan nuclear export by Mae and Crm1 may increase the stringency of the Ras response. Genes Dev. 19:1767-1772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tei, H., I. Nihonmatsu, T. Yokokura, R. Ueda, Y. Sano, T. Okuda, K. Sato, K. Hirata, S. C. Fujita, and D. Yamamoto. 1992. pokkuri, a Drosophila gene encoding an E-26-specific (Ets) domain protein, prevents overproduction of the R7 photoreceptor. Proc. Natl. Acad. Sci. U. S. A. 89:6856-6860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tognon, C. E., C. D. Mackereth, A. M. Somasiri, L. P. McIntosh, and P. H. Sorensen. 2004. Mutations in the SAM domain of the ETV6-NTRK3 chimeric tyrosine kinase block polymerization and transformation activity. Mol. Cell. Biol. 24:4636-4650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tootle, T. L., P. S. Lee, and I. Rebay. 2003. CRM1-mediated nuclear export and regulated activity of the receptor tyrosine kinase antagonist YAN require specific interactions with MAE. Development 130:845-857. [DOI] [PubMed] [Google Scholar]
- 31.Tran, H. H., C. A. Kim, S. Faham, M. C. Siddall, and J. U. Bowie. 2002. Native interface of the SAM domain polymer of TEL. BMC Struct. Biol. 2:5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vivekanand, P., and I. Rebay. 2006. Intersection of signal transduction pathways and development. Annu. Rev. Genet. 40:139-157. [DOI] [PubMed] [Google Scholar]
- 33.Vivekanand, P., T. L. Tootle, and I. Rebay. 2004. MAE, a dual regulator of the EGFR signaling pathway, is a target of the Ets transcription factors PNT and YAN. Mech. Dev. 121:1469-1479. [DOI] [PubMed] [Google Scholar]
- 34.Xu, C., R. C. Kauffmann, J. Zhang, S. Kladny, and R. W. Carthew. 2000. Overlapping activators and repressors delimit transcriptional response to receptor tyrosine kinase signals in the Drosophila eye. Cell 103:87-97. [DOI] [PubMed] [Google Scholar]
- 35.Zacharias, D. A., J. D. Violin, A. C. Newton, and R. Y. Tsien. 2002. Partitioning of lipid-modified monomeric GFPs into membrane microdomains of live cells. Science 296:913-916. [DOI] [PubMed] [Google Scholar]