Abstract
Heterochromatin assembly in fission yeast relies on the processing of cognate noncoding RNAs by both the RNA interference and the exosome degradation pathways. Recent evidence indicates that splicing factors facilitate the cotranscriptional processing of centromeric transcripts into small interfering RNAs (siRNAs). In contrast, how the exosome contributes to heterochromatin assembly and whether it also relies upon splicing factors were unknown. We provide here evidence that fission yeast Spf30 is a splicing factor involved in the exosome pathway of heterochromatin silencing. Spf30 and Dis3, the main exosome RNase, colocalize at centromeric heterochromatin and euchromatic genes. At the centromeres, Dis3 helps recruiting Spf30, whose deficiency phenocopies the dis3-54 mutant: heterochromatin is impaired, as evidenced by reduced silencing and the accumulation of polyadenylated centromeric transcripts, but the production of siRNAs appears to be unaffected. Consistent with a direct role, Spf30 binds centromeric transcripts and locates at the centromeres in an RNA-dependent manner. We propose that Spf30, bound to nascent centromeric transcripts, perhaps with other splicing factors, assists their processing by the exosome. Splicing factor intercession may thus be a common feature of gene silencing pathways.
In eukaryotic cells, RNA processing plays a key role in proper genetic expression, heterochromatin assembly, and mitotic chromosome segregation (9, 12, 41). Processing reactions, such as 5′ capping, splicing, and 3′ cleavage and polyadenylation, are generally coupled with RNA polymerase II (RNAPII) transcription and are thought to take place in the context of dynamic factories built on nascent transcripts (5). For instance, intron removal is catalyzed within the spliceosome, an intricate ribonucleoparticule (RNP) that assembles de novo on pre-mRNA through the temporally ordered binding of the U1, U2, and triple U4/U6.U5 small nuclear RNPs (snRNPs) (24). Spliceosome assembly and in some cases splicing itself are completed cotranscriptionally (17, 29). Splicing factors promote RNAPII elongation, physically interact with the cleavage and polyadenylation apparatus and contribute to efficient pre-mRNA 3′ end processing (28, 33). Furthermore, splicing factors have recently been shown to physically interact with the RNA interference (RNAi) machinery and to facilitate the cotranscriptional processing of double-stranded RNAs that derive from centromeric heterochromatin in fission yeast (4), providing further support to the idea of functionally integrated processing systems.
RNAi is a conserved silencing mechanism that involves the cleavage of long double-stranded RNAs into ∼25-nucleotide (nt) siRNAs by the RNase Dicer. In fission yeast, RNAi is required for the assembly of heterochromatin (18, 38). There are three main heterochromatic domains in the fission yeast genome: centromeres, telomeres, and the silent loci of the mating-type region. Heterochromatin assembly at these locations involves an ordered series of reactions leading to the methylation of histone H3 at lysine 9 (H3K9-me) by the methyltransferase Clr4/Suv39, which creates a binding site for the chromodomain protein Swi6/HP1. Thus far, most of the work on RNAi-mediated heterochromatin assembly has focused on centromeric heterochromatin. Fission yeast centromeres consist of repetitive DNA elements (outer repeats) coated with heterochromatin and flanking a nonheterochromatic central domain, which is the site of kinetochore assembly (46). A robust transcriptional silencing is exerted on class II marker genes when inserted into the centromeric heterochromatin (1). Paradoxically, both strands of outer repeats are transcribed, and a bona fide promoter drives transcription of the reverse strand by RNAPII (14, 53). Double-stranded centromeric transcripts are processed by Dicer (Dcr1 in fission yeast) (53). Centromeric siRNAs (cen-siRNAs) are loaded onto the RITS complex (named for RNA-induced initiation of transcriptional gene silencing) which associates with outer repeats, presumably through base pairing between cen-siRNAs and cognate nascent transcripts, and recruits RDRC (named for RNA-directed RNA polymerase complex) (11, 40, 52). RDRC increases the production of double-stranded centromeric precursors, thereby reinforcing the cotranscriptional production of cen-siRNAs. Through an unknown mechanism, the processing of centromeric RNA precursors into siRNAs targets Clr4 at outer repeats. Neither swi6 and clr4, nor RNAi genes are essential in fission yeast. However, their deletions strongly alleviate silencing at the centromeric outer repeats and impair proper chromosome segregation, presumably because heterochromatin is required for cohesin recruitment to centromeres (7, 45). Numerous splicing factors physically associate with Cid12, a subunit of RDRC in fission yeast, and one of them, Cwf10, has been shown to locate at centromeric outer repeats in a Dcr1-dependent manner (4, 40). Remarkably, specific mutations in several Cid12-associated splicing factors affect the production of cen-siRNAs and centromeric heterochromatin, but not splicing itself, arguing for two separable processes. Based on these observations, it has been proposed that spliceosomal complexes form a processing platform that facilitates the amplification of cen-siRNAs by RDRC (4). How the splicing machinery could promote the cotranscriptional processing of centromeric transcript precursors into siRNAs in an apparent splicing-independent manner is currently unknown.
In fission yeast, heterochromatic gene silencing relies also on an additional pathway that involves the exosome (9, 41, 54), a conserved RNase complex present in the nucleus and the cytoplasm (23). The exosome has also been linked to heterochromatin in budding yeast (which lacks the RNAi equipment) (22, 51), indicating that this chromatin-based silencing mechanism has an ancient evolutionary origin. Dis3/Rrp44, which belongs to the RNase II family, is the principal if not the only catalytic subunit of the budding yeast core exosome (15, 32). The nuclear form contains an additional RNase called Rrp6. However, unlike Dis3, Rrp6 is not essential in budding and fission yeasts. The exosome is a multitask machine, involved in various processing and degradation pathways, and little is known about the molecular mechanisms through which it chooses between the processing and degradation modes and recognizes its targets among the plethora of cellular RNAs (23, 31). Several reports have revealed the existence of cofactors that assist the RNases and/or select the substrates. Some, such as the Nrd1-Nab3 complex in budding yeast (50), are sequence specific RNA-binding proteins, whereas others, such as TRAMP, act on many different transcripts devoid of sequence similarity and are thought to recognize their targets in a structured-based mechanism. TRAMP has been identified in budding yeast (23, 30, 58), where it contains the noncanonical poly(A) polymerase Trf4, a putative RNA-binding protein (either Air1 or Air2), and the RNA helicase Mtr4. Compelling evidence indicates that Trf4-mediated polyadenylation of RNA species contributes to their targeting for degradation (23, 30, 58). However, in vitro experiments suggest that polyadenylation is not sufficient per se for exosomal degradation, stimulating activities being also required (30). These can be provided by TRAMP itself and/or additional cofactors such as the Ran GTPase Gsp1/Cnr1, which directly binds Dis3 (30, 44). Gsp1 deficiency interferes with the 3′-end processing of 5.8S rRNA precursors, leading to maturation defects that can be compensated for by Dis3 or Mtr4 overexpression (49).
RNAi components are required to establish and maintain repressive heterochromatin at centromeres but are dispensable for its maintenance at the silent domain of the mating type (20). In contrast, silencing is alleviated at centromeres, at telomeres, and at the mating type when fission yeast TRAMP poly(A) polymerase Cid14 or the exosome is impaired (9, 41, 54). The accumulation of heterochromatic transcripts in cid14, dis3, or rrp6 mutants has led to the idea that their degradation by the exosome contributes to heterochromatin-mediated gene silencing (9, 41, 54). However, the molecular mechanisms through which this processing and/or degradation takes place are still enigmatic. The centromeric transcripts which accumulate in a dis3 mutant are polyadenylated even in the absence of Cid14 (9, 41, 54), suggesting that the TRAMP poly(A) polymerase plays a minor role if any in their polyadenylation-assisted degradation. Furthermore, Cid14 and exosome RNases play apparently distinct roles with respect to centromeric RNAs and heterochromatin integrity. Dis3 deficiency lessens the amount of Swi6 at centromeres but does not affect notably the amount of cen-siRNAs (41). Similarly, H3K9-me is reduced at centromeres when Rrp6 is missing, albeit a seemingly normal quantity of cen-siRNAs is produced (9, 43). In contrast, Cid14 loss of function impairs the production of cen-siRNAs but does not impact Swi6 and H3K9-me levels at the outer repeats (9, 10). In fact, Cid14's role in centromeric siRNAs generation consists in preventing the entry of spurious euchromatic RNAs into the RNAi pathway at the expense of centromeric transcripts (10). Cid14 cooperates with the histone variant H2A.Z at euchromatic genes to promote the exosomal degradation of antisense transcripts that result from readthrough transcription (59). H2A.Z is barely present at pericentromeric heterochromatin, the mating-type region and telomeres, and its loss of function does not significantly modify H3K9-me distribution at these locations. Thus, at least partially distinct systems may operate at heterochromatin and euchromatin regions to stimulate exosome activity. These observations therefore raise the question of the cofactors that assist the exosome for the processing of heterochromatic transcripts. Furthermore, whether this reaction takes place co- or posttranscriptionally and whether it also integrates splicing factors are unknown.
In the present study we show that Dis3 associates with pericentric heterochromatin and helps recruiting Spf30, a conserved splicing factor that binds nascent centromeric transcripts. Spf30 deficiency alters the processing of polyadenylated centromeric RNAs but not the production of cen-siRNAs and reproduces the heterochromatic defects of a catalytically reduced Dis3 RNase. Our data argue that Spf30 is recruited along with Dis3 at the pericentric heterochromatin to facilitate the processing of centromeric transcript by the exosome. Thus, two mechanistically distinct RNA degradation systems, the RNAi and the exosome, seem to have integrated splicing factors as processing facilitators.
MATERIALS AND METHODS
Media, strains, and molecular genetics.
Media and molecular genetics methods were as described previously (39). Complete, YES+A medium was used unless otherwise stated. The synthetic medium is PMG. The spf30 gene was deleted according to the protocol available at http://mendel.imp.ac.at/pombe/. Deletion was confirmed by PCR. To tag Spf30 with green fluorescent protein (GFP), spf30 stop codon was replaced by a BamHI restriction site by PCR amplification, and a GFP cassette was inserted in the BamHI site. Plasmid DNA encoding Spf30 C terminus (amino acids 204 to 311) fused to GFP was linearized by cutting within spf30 coding sequence and inserted in the genome at the spf30 locus. The dis3 gene was appended with a 3× hemagglutinin (HA) cassette by using a PCR-based gene targeting method (3). Minichromosome loss rates were measured as described previously (1). For temperature shift experiments, cells grown at 25°C were shifted to 37°C in a water bath and incubated for 6 h, unless otherwise stated.
Coimmunoprecipitations.
Coimmunoprecipitations were carried out as described previously (6) with 5 × 108 cells per assay and analyzed by Western blotting with 1/20 of the input and flowthrough fractions and 1/5 of the immunoprecipitate. The antibodies used were polyclonal anti-GFP A-11122 and monoclonal anti-myc 9E10.
RNA extraction and Northern blots.
Total RNA was extracted from 108 exponentially growing cells by using a standard hot-phenol method. For small RNA preparations, 2.5 × 108 log-phase cells were subjected to total RNA extraction, followed by PEG 8000 precipitation. Samples were resuspended in 50% formamide. To detect centromeric transcripts, ∼20 μg of total RNA were separated on a 1% agarose-6% formaldehyde gel and blotted overnight to a Nytran SPC nylon membrane (Whatman). 32P-labeled DNA probes complementary to centromeric dg-dh repeats and act1 transcripts were generated by PCR amplification, labeled by using a RediPrime II Random-Prime DNA labeling system (GE Healthcare), and hybridized to the membrane overnight at 65°C. For cen-siRNA, ∼25-μg portions of total small RNAs were separated on a 8% urea-denaturing polyacrylamide gel and blotted onto a Hybond-NX membrane (GE Healthcare). The 32P-labeled DNA probes were a 741-nt PCR product complementary to centromeric dh repeats, labeled as described above, and an oligonucleotide complementary to snR58 labeled using T4 polynucleotide kinase. Probes were hybridized to the membrane overnight at 42°C.
RT-PCR, strand-specific analysis of cen-dh transcripts, and polyadenylation assay.
Reverse transcription (RT) was carried out using Superscript II (Invitrogen). For the splicing assay and measurements of the steady-state levels of 25S RNA, act1 and ura4+ transcripts, 500 ng of total RNA was reverse transcribed by using oligo(dT) primers. Outer repeat dh strand-specific RT was carried out on 50 ng of total RNA, using either forward or reverse strand-specific primers. cDNAs were amplified using both primers. PCR products, stained with SYBR green, were separated on an agarose gel and quantified. Polyadenylation assay was performed as described previously (54, 56), using the primers RT1-dh to amplify forward cen-dh RNA and the primer RT2-dh for the reverse.
ChIP and RNA-IP.
Swi6, H3K9-me, and Dis3-HA chromatin immunoprecipitation (ChIP) analyses were performed as described previously (6). For Spf30-GFP, cells were fixed with a 1/10 culture volume of fixative (33% formaldehyde, 100 mM NaCl, 1 mM EDTA, 0.5 mM EGTA, 50 mM Tris-Cl [pH 8.1]) for 30 min at 20°C. Chromatin was sheared to ca. 0.5- to 1-kb fragments by using a Bioruptor sonicator (Diagenode). Immunoprecipitation was carried out in IP buffer (1% Triton X-100, 2 mM EDTA, 150 mM NaCl, 20 mM Tris-Cl [pH 8.1]). Immunocomplexes were collected by using ChIP-Adembeads (Ademtech catalog no. 04240), washed, and recovered according to manufacturer instructions. For RNase ChIP, chromatin was treated with 1 μg of RNase A or 1 μg of RNase A plus 400 U of RNaseOUT (Invitrogen) for 30 min at 37°C before IP. RNA-IP was carried out as described previously (40), with the following modifications. A total of 109 cells were fixed with a 1/10 volume of above-described fixative for 30 min and lysed in ChIP lysis buffer (6) in the presence of 0.5 U of RNasin (Sigma)/μl. Extracts were sonicated by using Bioruptor (7 min of 30 s on and 30 s off at maximum power), clarified by centrifugation, and treated with DNase I (∼300 U/μl) for 1 h at 32°C. IP was performed overnight at 4°C in ChIP IP buffer. Immune complexes were collected by using ChIP-Adembeads (Ademtech catalog no. 04240), and cross-links were reversed for 2 h at 65°C. Nucleic acids were extracted with phenol-chloroform (5:1, pH 4.8) and recovered by NaCl-ethanol precipitation. RNA in the input and immunoprecipitated fractions was reverse transcribed by using random hexamers. ChIP and RNA-IP quantifications were performed by real-time PCR. The following antibodies were used: GFP (A-11122; Invitrogen), Swi6 (Abcam catalog no. 14898), H3K9-me (Abcam catalog no. 1220), and HA (Sigma catalog no. HA-7).
Quantitative PCR (qPCR) and primers.
Real-time PCR was performed in the presence of SYBR green on a Stratagene Mx3000P cycler. The primers used in the present study are listed in Table 1 .
TABLE 1.
Primers used in this study
| Analysis | Primer sequence(s) (5′-3′) |
|---|---|
| Splicing assay | |
| cut23 | GTGTATTTCTGAATGCTCTGAAAGAGG, CACCTGCCAAGTACTTACTATACAATCG |
| mis4 | AAGCCTCTAACTCTTTTGAATAGAGG, TTCAACTTGATCCTCGCCAG |
| bub3 | GACAGTTACTACCCTCGATCTTAATACG, CCTAGAACCAATATATTATCTCTCGAGC |
| rpb4 | GACAATAAACAGACTTGCCTGAGA, CTCGGTTTAAAACGGCTGAA |
| dis3 | TGATATACCCTGTCAAAGTCGGGC, TTACGGTCATTAGCAGATTCATCATCC |
| qPCR, ChIP, and RNA-IP | |
| 25S rRNA | CTGAACGCCTCTAAGCCAGA, GATTCGATTCCGCACAAGAT |
| act1 | CGAACGTGAAATTGTTCGTG, GGAGGAAGATTGAGCAGCAG |
| dh3 | CTTGGCAAACAGACCCTCAT, TATGTACCCGTGCATTGGAG |
| dg1 | ATGTGATGAGAGAACAAGTCAGGGTGGTCG, TGACGAGGCACATTCCTTATACGCAAACG |
| cnt1 | CACCACTCACTTACGCTTCACCTAGTTTCC, CCAGTATGCTGATGTAAGTTGCAACGGT |
| cut23 | TGTTGTGTGAAATCATCCTTCC, AGGATTCATTCCATTGAGCATT |
| fbp1 | CGTTCGACTTCATCGCAGTA, CATTGGTTTATCGGGAATCG |
| U2 snRNA | TCGCTGAAATCACCTCACTG, CAGTGTCGGAAAGCATAGCA |
| dh strand-specific RT-PCR | |
| RT1-dh | GAAAACACATCGTTGTCTTCAGAG |
| RT2-dh | CGTCTTGTAGCTGCATGTGAA |
| act1-1 | GAGTCATCTTCTCACGGTTGG |
| act1-2 | TCCTACGTTGGTGATGAAGC |
| Northern blot | |
| Probe for detection of siRNAs | |
| RT1-dh | GAAAACACATCGTTGTCTTCAGAG |
| dh-HsiRNA | TACTGTCATTAGGATATGCTCA |
| Probe for detection of centromeric transcripts | |
| dg-FOR | CTACTCTTCTCGATGATCCTG |
| dg-REV | GTAGTACGACGATGATGTGTTTTC |
| snoRNA58 | GATGAAATTCAGAAGTCTAGCATC |
Microscopy.
Indirect immunofluorescence was performed as described previously (6). α-Tubulin was revealed by using Tat1 monoclonal antibodies (a gift from K. Gull) (57). Spindle length measurement was done by using Metamorph software.
RESULTS
Human splicing factor Spf30 is conserved in fission yeast.
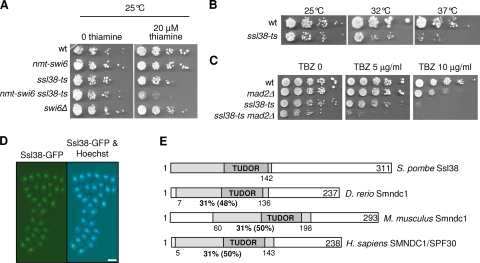
The ssl38-ts mutation has been isolated through a genetic screen for mutants synthetically lethal with swi6 (Fig. 1A) (6). The ssl38-ts mutant also exhibited thermosensitivity for growth at 37°C (Fig. 1B) and hypersensitivity to the microtubule poison thiabendazole at 25°C (Fig. 1C), indicating that the ssl38-ts allele is at least partly defective even at 25°C. Complementation and sequencing revealed that ssl38 corresponds to the SPCC1281.02c open reading frame. To determine whether ssl38 was an essential gene, it was deleted in a diploid strain, and the growth of isolated haploid spores was examined at 32°C. Spores bearing the ssl38Δ::ura4+ null allele germinated but rapidly ceased proliferating (results not shown), demonstrating that ssl38 is essential for growth at 32°C.
FIG. 1.
The ssl38 gene, synthetically lethal with swi6 when mutated, encodes a protein structurally similar to the human splicing factor SPF30. (A to C) The ssl38-ts mutation is synthetically lethal with the swi6Δ mutation, thermosensitive for growth, and hypersensitive to thiabendazole (TBZ). (A) Cells cultured in the absence of thiamine were serially diluted and spotted on indicated synthetic media. Thiamine represses nmt-swi6 transcription. (B and C) Cells were serially diluted, spotted onto YES+A (B) or YES+A plus TBZ (C). (D) Ssl38 is a nuclear protein. Exponentially growing ssl38-GFP cells were treated with Hoechst 33342 to stain DNA and then directly observed under a fluorescence microscope. Bar, 5 μm. (E) Ssl38 is similar to the Smndc1/Spf30 splicing factor. Region of sequence identity between Ssl38 and Spf30 orthologs, determined by using BLASTP, are indicated as shaded areas with the percentages underneath.
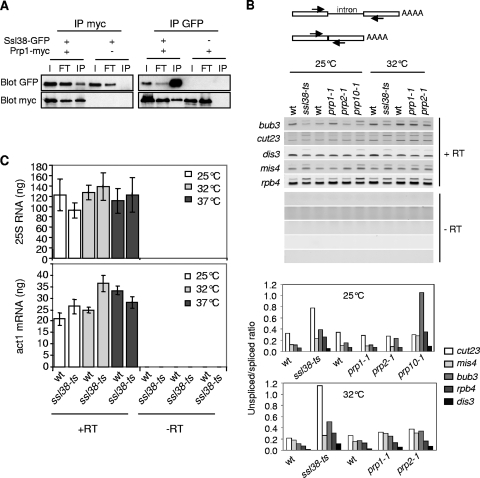
Ssl38 is an abundant nuclear protein (Fig. 1D) that resembles the splicing factor Smndc1/Spf30 (Fig. 1E) (37, 48), harboring a Tudor domain that is frequently found in RNA-binding proteins and related to the chromodomain (36). Corroborating the structural similarity between Ssl38 and Spf30 proteins, Carnahan et al. identified peptide fragments corresponding to Ssl38 that copurified with spliceosome proteins (13). Indeed, Ssl38 and Prp1, a component of the U4/U6.U5 tri-snRNP (13), coimmunoprecipitated (Fig. 2A), confirming that Ssl38 and Prp1 belong to a same complex, most likely the spliceosome.
FIG. 2.
Ssl38 is required for efficient splicing. (A) Ssl38 coimmunoprecipitates with Prp1. Soluble protein extracts were subjected to immunoprecipitation with anti-GFP or anti-myc antibodies. Inputs (lanes I), immunoprecipitated (lanes IP) fractions, and proteins washed away (lanes FT) were analyzed by Western blotting with anti-myc or anti-GFP antibodies. (B) Unspliced transcripts accumulate in ssl38-ts. The scheme of the RT-PCR assay used to assess splicing efficiency is shown. Total RNA extracted from indicated cells was reverse transcribed in the presence (+RT) or absence (−RT) of reverse transcriptase. Complementary DNAs (cDNAs) were PCR amplified and in-gel quantified. Unspliced, upper band; spliced, lower band. (C) The steady-state level of act1 transcripts or 25S RNA is not overtly affected by ssl38-ts. Same procedure as in panel B, except that cDNAs were quantified by real-time PCR. The indicated values correspond to averages and mean deviations (m.d.) calculated from three (25S RNA) or two (act1) independent samples.
This prompted us to investigate whether Ssl38 was required for splicing. Mutants prp1-1, prp2-1, and prp10-1 were used as controls for defective splicing (19, 47). Five genes were examined, and, for all of them, unspliced transcripts accumulated when Ssl38 was impaired (Fig. 2B). Pre-mRNAs showed variable sensitivities toward Ssl38 deficiency; however, those corresponding to the cut23 gene were particularly affected. A similar heterogeneity was observed in the prp10-1 mutant, with a strong increase in bub3 pre-mRNAs. Surprisingly, the differential between the ssl38-ts mutant and the wild type was not significantly deepened at 37°C compared to that seen at 32°C (data not shown). To ascertain the splicing origin of these defects, we examined ribosomal 25S RNA and act1 transcripts, both being intronless. No significant accumulation was observed in ssl38-ts mutant cells (Fig. 2C). Altogether, these data indicate that Ssl38 is a splicing factor that is structurally related to human Spf30. Therefore, ssl38 was renamed spf30, and the thermosensitive allele was named spf30-38.
Spf30 disrupts proper centromere functioning when impaired.
The characterization of Spf30 as a splicing factor raised the question of the biological significance of the synthetically lethal interaction between swi6Δ and spf30-38 at the permissive temperature. Most ssl mutations reside in genes involved in sister chromatid cohesion (6). However, spf30 involvement in cohesion appeared improbable since spf30-38 cells exhibited seemingly normal chromosomal association of cohesin and cohesion (data not shown). Defective splicing of pre-mRNA molecules encoding factors whose loss of function becomes lethal in a swi6Δ background was another possibility. However, among thermosensitive mutations dim1-35, prp1-1, prp2-1, and prp10-1, all characterized as splicing deficient (13, 19, 47), only dim1-35 showed colethality with swi6Δ at 25°C (data not shown). Furthermore, prp10-1 was not colethal with swi6Δ despite a splicing defect similar to if not slightly stronger than the one caused by spf30-38 (Fig. 2B). Therefore, being deficient for splicing is not sufficient for being synthetically lethal with swi6. The corollary is that spf30-38 and dim1-35 might affect another function related to swi6.
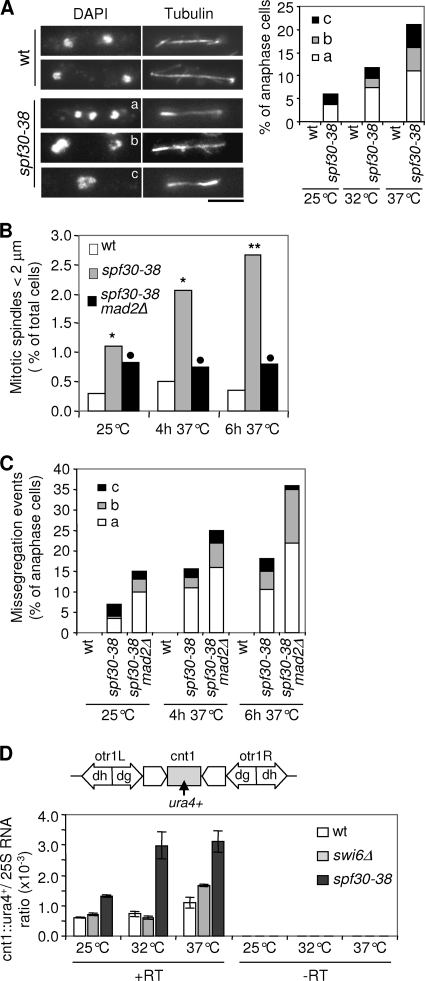
The dim1-35 mutation disrupts accurate chromosome segregation (8). Interestingly, spf30-38 cells also exhibited defective chromosome segregation during anaphase (Fig. 3A) and were delayed in prophase/prometaphase at 37°C (Fig. 3B). The spindle assembly checkpoint is a mitotic surveillance mechanism that detects faulty kinetochore to microtubule attachments and, in response, inhibits anaphase onset, thereby providing time for their correction (42). Mad2 is an essential component of this checkpoint. The mitotic delay was greatly attenuated when Mad2 was missing in spf30-38 mutant cells (Fig. 3B), and defective anaphases occurred more frequently (Fig. 3C). Note that chromosome segregation during anaphase is barely affected by the mad2Δ mutation alone, with ∼1% of lagging chromosomes (n = 300). In addition, the absence of mad2 enhanced spf30-38 hypersensitivity to thiabendazole (Fig. 1C). These data indicate that Spf30 deficiency activates the spindle assembly checkpoint, probably because of disrupted chromosome-to-microtubule attachment.
FIG. 3.
Chromosome segregation is defective in spf30-38 cells. (A) Spf30 deficiency affects chromosome segregation during mitosis. Cells fixed at 25, 32, or 37°C were processed for immunofluorescence against α-tubulin (Tubulin). DNA was stained with DAPI (4′,6′-diamidino-2-phenylindole), and chromosome segregation was examined in late-anaphase cells (mitotic spindle length, >5 μm; n > 100). Mutant cells exhibited three distinct phenotypes. (a) Lagging chromosomes: DAPI signals away from one spindle pole. (b) Uneven chromosome distribution: unbalanced DAPI signals at the poles. (c) Absence of or sustained delay in chromosome movement: DAPI signals stuck around the spindle center. Bar, 5 μm. (B) spf30-38 cells exhibit a Mad2-dependent delay in early mitosis. Fixed cells were processed as in panel A and prophase/prometaphase figures (mitotic spindle length, <2 μm) scored (n = 400 to 1,000). **, the difference with the wild-type (wt) control is statistically highly significant (P < 0.01, as determined by the chi-square test); *, borderline of statistical significance (P < 0.05); •, statistically not significant. (C) The absence of Mad2 increases the rate of missegregation events in spf30-38 cells. The same cells as in panel B were examined for chromosome segregation defects in late anaphase (n > 100). (D) spf30-38 alleviates silencing at the central domain of centromere 1. Centromere 1 with the central domain (cnt1) surrounded by outer repeats left and right (otr1), made of dg and dh elements, is represented, and the location of the ura4+ marker gene is indicated. Strains whose sole ura4+ gene was inserted within cnt1 (cnt1::ura4+), were grown at 25 or 32°C or shifted to 37°C for 6 h. A total of 500 ng of total RNA was reverse transcribed by using oligo(dT) primers, in the presence (+RT) or absence (−RT) of reverse transcriptase. ura4+ and 25S cDNAs were quantified by real-time PCR, and their ratios were determined. The indicated values correspond to averages and m.d. calculated based on two PCRs.
To investigate this further, the transmission fidelity through mitoses of the 36-kb centric circular minichromosome CM3112 was assessed. CM3112 bears a functional albeit truncated centromere (35). In agreement with previous reports, the loss rate per division was estimated at between 1 and 2% in the wild type (Table 2). In contrast, in a spf30-38 background CM3112 was lost in 12 and 20% of cell divisions at 25 and 32°C, respectively. Acentric minichromosomes or ars plasmids are lost in 20 to 50% of mitotic divisions in fission yeast (21, 35). Therefore, this strongly suggests that proper centromere functioning is disrupted by spf30-38.
TABLE 2.
The spf30-38 mutation lessens the transmission fidelity of the CM3112 circular minichromosome through mitosis
| Temp (°C) | Strain | Sectoring | Total | Loss per division (%)a | Fold increaseb |
|---|---|---|---|---|---|
| 25 | Wild type | 13 | 805 | 2 | |
| bub1Δ mutant | 82 | 712 | 12 | 6 | |
| spf30-38 mutant | 84 | 685 | 12 | 6 | |
| 32 | Wild type | 5 | 526 | 1 | |
| bub1Δ mutant | 115 | 830 | 14 | 14 | |
| spf30-38 mutant | 179 | 886 | 20 | 20 |
That is, the number of colonies with a red sector covering at least half of the colony divided by the total number of colonies (white plus half-sectored).
Versus the wild type.
Spf30 preserves the structural integrity of heterochromatin.
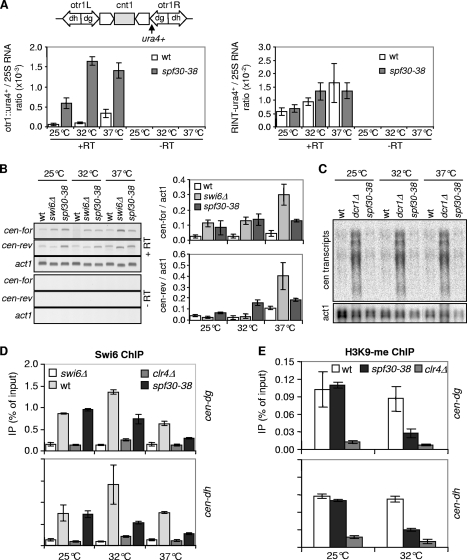
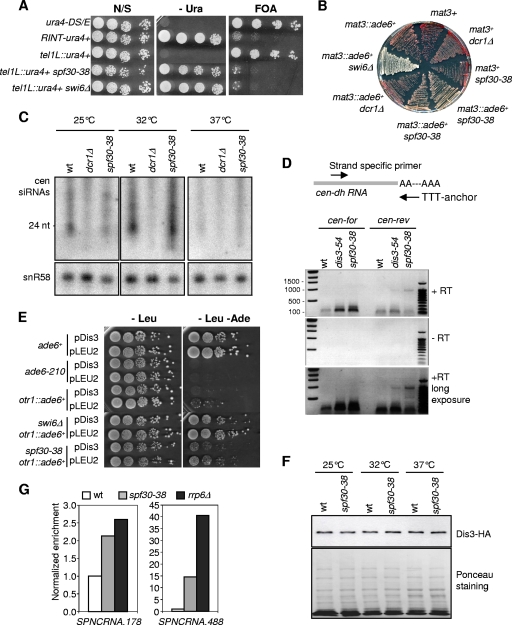
To get some insight into how Spf30 might contribute to centromere functioning, silencing of an ura4+ reporter gene inserted within the outer repeats (otr1::ura4+) of centromere 1 was assessed in spf30-38 cells. As shown in Fig. 4A, otr1::ura4+ transcripts clearly accumulated in spf30-38 cells. Silencing at the central domain was also alleviated by Spf30 deficiency (Fig. 3D). In contrast, the steady-state level of ura4 transcripts appeared not modified when ura4+ was inserted at a control, random location in the genome (RINT-ura4+) (Fig. 4A, right panel). Furthermore, spf30-38 cells also exhibited an increased expression of a distinct, otr1::ade6+ reporter gene, ruling out any ura4-biased effect (not shown). Thus, spf30-38 causes a centromeric silencing defect.
FIG. 4.
Spf30 preserves the integrity centromeric heterochromatin. (A) spf30-38 alleviates silencing of the otr1::ura4+ marker gene. The location of the ura4+ marker gene within centromere 1 is indicated. Total RNA was reverse transcribed in the presence (+RT) or absence (−RT) of reverse transcriptase, and ura4+ and 25S cDNAs were quantified by real-time PCR. Indicated values correspond to averages and m.d. calculated from duplicated samples. (B and C) Endogenous centromeric transcripts accumulate in spf30-38 cells. (B) Total RNA was reverse transcribed using a primer complementary to the forward (cen-for) or the reverse (cen-rev) strand of dh repeat, in the presence (+RT) or absence (−RT) of reverse transcriptase. act1 transcripts were used as control. The indicated values are averages and m.d. calculated from two independent experiments. (C) Centromeric transcripts were detected by Northern blotting, and blots were reprobed with act1 for a loading control. (D and E) Decreased amounts of Swi6 and H3K9-me at the centromeric outer repeats in spf30-38 cells. The Swi6 and H3K9-me levels at the outer repeats (cen-dg and cen-dh) were assessed by ChIP. The indicated values are averages and m.d. calculated from two qPCRs.
Noncoding centromeric dh transcripts were also assessed, using swi6Δ cells as a control. At 25 and 32°C, the forward transcript level was fourfold increased in spf30-38 and swi6Δ mutants (Fig. 4B). Reverse transcripts were also affected by Spf30 deficiency, being, respectively, two- and fivefold increased at 25 and 32°C, whereas swi6Δ displayed no accumulation, as expected (53). Deeper inactivation of spf30-38 at 37°C did not further increase forward and reverse transcript amounts. Unexpectedly, reverse transcripts accumulated in swi6Δ cells at this temperature. As a complementary approach, the relative amount of centromeric transcripts over act1 was determined by Northern blotting. We observed a twofold increase in centromeric RNA levels in spf30-38 compared to wild-type cells (Fig. 4C [note that less material was loaded in the spf30-38 lanes]), confirming that Spf30 contributes to the silencing of natural centromeric outer repeats.
Next, we sought to determine whether Spf30 might play a role in the assembly of heterochromatin and examined the association of Swi6 with centromeric outer repeats by using ChIP. At 25°C, spf30-38 cells exhibited normal Swi6 levels at the dg and dh repeats (Fig. 4D). In contrast, Swi6 binding was clearly reduced at 32 and 37°C but not abolished (compare to the swi6Δ and clr4Δ controls). In good agreement, H3K9-me was also clearly reduced in the spf30-38 context, at 32°C (Fig. 4E). A high background in clr4Δ control precluded any analysis at 37°C (data not shown). Together, these results indicate that Swi6 is severely, but not entirely, delocalized from centromeric outer repeats when Spf30 is deficient and strongly suggest that this stems from a reduced amount of H3K9-me.
Mutations that affect centromeric silencing do not necessarily have a similar impact at the mating-type locus or telomeres (4, 16, 20). However, spf30-38 clearly alleviated the silencing at telomeres, as revealed by the increased expression of a subtelomeric ura4+ marker gene (Fig. 5A). The mating-type region contains three mat cassettes; mat1 is expressed, whereas silent, heterochromatic mat2 and mat3 serve as donors for switching the genetic information expressed at mat1. In contrast to the dcr1Δ mutation, spf30-38 alleviated silencing of a mat3::ade6+ reporter and impaired the efficient switching of the mating-type throughout cell divisions (Fig. 5B and data not shown). Collectively, these data indicate that Spf30 is required for the structural integrity of heterochromatin at centromeres, telomeres, and the mating-type region.
FIG. 5.
Spf30 functions in the exosome-mediated silencing pathway. (A) Spf30 deficiency alleviates silencing of a subtelomeric tel1L::ura4+ marker gene. Cells of indicated genotypes were serially diluted and spotted on nonselective (N/S), selective (−Ura), and counterselective (FOA) synthetic medium. 5-Fluoroorotic acid (5-FOA) is toxic to Ura+ cells. Plates were incubated at 32°C. (B) spf30-38 affects silencing at the mating-type locus. A colony color assay for ade6+ silencing when it is inserted within the mating-type locus (mat3::ade6+) was performed. The indicated strains were grown at 32°C on YES containing a low amount of adenine. (C) Spf30 deficiency does not overtly affect the production of siRNAs. Centromeric siRNAs were detected by Northern blotting, and blots were reprobed with snR58 for a loading control. (D) Polyadenylated cen-dh transcripts accumulate when Spf30 is deficient. The scheme of the polyadenylation assay is shown. Total RNA extracted from indicated strains grown at 30.5°C was reverse transcribed with the poly(dT) anchor primer and PCR amplified by using cen-dh strand-specific and anchor primers. (E) Dis3 overexpression partly suppresses spf30-38 silencing defect at 32°C. Strains transformed with the pCD174 episomal multicopy plasmid (pDis3) (26) or with an empty vector (pLEU2) were serially diluted and spotted onto selective media. (F) The steady-state amount of Dis3 protein is not altered in the spf30-38 mutant. Soluble protein extracts prepared from wild-type (wt) and spf30-38 cells expressing Dis3-HA and grown at 25 or 32°C or shifted at 37°C for 6 h were subjected to Western blotting with monoclonal 12CA5 anti-HA antibodies. Equal loading was verified by Ponceau staining. (G) Cryptic unstable transcripts accumulate in spf30-38 cells. Total RNA from indicated strains was reverse transcribed by using random hexamers, and cDNAs were quantified by real-time PCR.
The exosome pathway of heterochromatin assembly seems defective in spf30-38.
Spf30 may take part in a heterochromatin assembly pathway unrelated to Dcr1, and by extension the RNAi pathway, since silencing is alleviated at the mating type in spf30-38 cells but not in dcr1Δ cells. To directly test this, we looked at cen-siRNAs by Northern blotting. As shown in Fig. 5C, the spf30-38 mutant displayed no striking modification in the abundance or length of cen-siRNAs compared to the wild type and was clearly distinct from dcr1Δ. The production of cen-siRNAs is suspected to be inhibited at elevated temperature in fission yeast (27). Indeed, wild-type and spf30-38 cells exhibited a sharp decline in their respective amounts of cen-siRNAs at 37°C. These data indicate that the processing of centromeric transcripts into siRNAs is not affected in spf30-38 cells.
Based on these results, we envisaged that centromeric RNA precursors might escape destruction by the exosome when Spf30 is defective. To explore this possibility, their polyadenylation state in spf30-38 cells was determined by carrying out a polyadenylation assay (54). As a control, we used the dis3-54 exosome mutant, in which Dis3 RNase is catalytically reduced (41). In agreement with previous studies (54, 56), 0.1- to 0.2-kb and 0.9-kp products corresponding, respectively, to polyadenylated forward and reverse cen-dh transcripts were abundant in dis3-54 compared to the wild type (Fig. 5D). These two bands were even more intense in the spf30-38 mutant. Thus, the forward and reverse cen-dh transcripts that accumulate in spf30-38 cells are polyadenylated.
At this point it became manifest that the spf30-38 and dis3-54 mutations led to the same heterochromatic phenotypes (41), suggesting that Spf30 and Dis3 might act together with respect to the processing of polyadenylated heterochromatic transcripts. Accordingly, dis3 overexpression in spf30-38 cells partially restored the silencing of the otr1::ade6+ marker gene (Fig. 5E). Even in wild-type or swi6Δ backgrounds, silencing appeared to be increased by dis3 overexpression. These results suggest that Dis3 is rate limiting for heterochromatin-mediated silencing in wild-type cells and are consistent with Dis3 being even less effective in the spf30-38 mutant, despite a normal steady-state amount (Fig. 5F). In good agreement, we found that two cryptic noncoding transcripts, which are normally maintained at a low level in wild-type cells through the action of the exosome (55), were more abundant in the absence of a fully functional Spf30 protein (Fig. 5G).
Dis3 binds pericentric heterochromatin and helps recruiting Spf30.
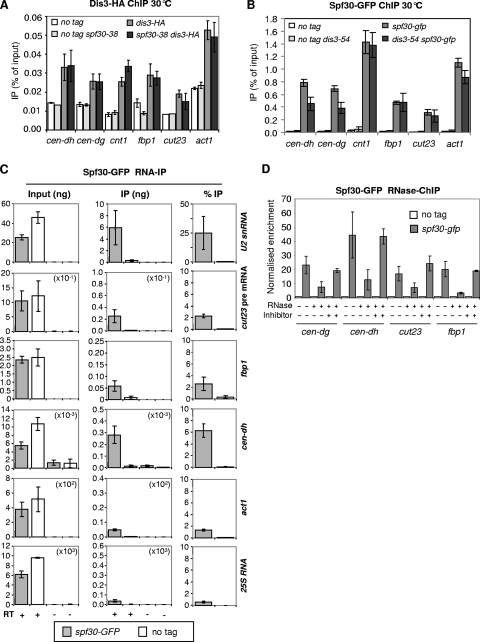
To investigate further the functional link between Dis3 and Spf30, we first sought to determine whether they interacted with chromatin, and notably with centromeres. By ChIP, both were found to be associated with the heterochromatic outer repeats of centromeres, the central domain, and euchromatin genes such as cut23, fbp1, and act1 (Fig. 6A and B). Thus, consistent with a functional overlap, Spf30 and Dis3 colocalize at centromeres, as well as at multiple sites along the chromosome arms.
FIG. 6.
Spf30, recruited to centromeric heterochromatin in a Dis3-dependent manner, binds nascent centromeric transcripts. (A) The chromosomal association of Dis3 is unaffected in spf30-38 cells. Dis3 levels at indicated chromosomal sites were assessed by ChIP. The indicated values correspond to averages and m.d. calculated from four (wild type) and six (spf30-38) experiments. (B) Spf30 colocalizes with Dis3 at centromeres and euchromatic genes and is reduced at the pericentric heterochromatin in dis3-54 cells. Spf30 binding was assessed by ChIP. The indicated values correspond to averages and m.d. calculated from four experiments. (C) Spf30 binds RNA species, among which were centromeric transcripts. The indicated strains were processed for RNA-IP by using polyclonal anti-GFP antibodies. RNA recovered in the input and immunoprecipitated fractions was reverse transcribed in the presence (+) or absence (−) of reverse transcriptase. cDNAs were quantified by real-time PCR. The indicated values correspond to averages and m.d. calculated from two independent experiments with two reverse transcriptions per experiment. (D) Spf30 associates with chromatin in an RNA-dependent manner. Chromatin extracts prepared for ChIP were treated with RNase or RNase plus inhibitor before immunoprecipitation. Enrichments in Spf30-GFP immunoprecipitated fractions were calculated as a ratio, i.e., the quantity in the immunoprecipitate/the quantity in the input, normalized to the ratio of the corresponding untagged control. The indicated values correspond to averages and m.d. calculated from two experiments.
Next, their mutual dependencies for chromosomal association were assessed. As shown in Fig. 6A, ChIP revealed no significant modification of Dis3 binding in a spf30-38 genetic background, implying that spf30-38 does not alter the chromosomal association of Dis3. In contrast, we reproducibly observed a 40% reduction in the amount of Spf30 bound to centromeric outer repeats in dis3-54 cells, whereas chromosome arm association remained unaffected (Fig. 6B). This shows that Dis3 helps recruiting Spf30 at centromeric heterochromatin and raises the intriguing possibility of a causal relationship between the reduced association of Spf30 and the accumulation of polyadenylated centromeric transcripts in dis3-54.
Spf30 binds nascent centromeric transcripts.
Dis3/exosome takes part in the processing of centromeric transcripts, and Spf30 may be able to bind RNA. Thus, we sought to determine whether Spf30 might bind centromeric RNAs. For this, an RNA-IP experiment was performed. Whole lysates from fixed cells were treated with DNase, Spf30 was immunoprecipitated, and associated (coimmunoprecipitated) RNA molecules were analyzed by RT-PCR. Human Spf30 associates with the U2 snRNP (37). As shown in Fig. 6C, U2 snRNA clearly coimmunoprecipitated with Spf30, as expected. In contrast, neither ribosomal 25S RNA nor act1 transcripts were efficiently coimmunoprecipitated despite their abundance in the inputs. Therefore, the experiment seemed to reveal specific interactions between Spf30 and target RNA molecules. We found that unspliced cut23 pre-mRNA and fbp1 transcripts were also slightly enriched in the immunoprecipitated fraction compared to 25S RNA and act1. Remarkably, centromeric dh transcripts were efficiently coimmunoprecipitated with Spf30. Furthermore, they still coimmunoprecipitated in a dcr1Δ background (data not shown). These data indicate that Spf30 associates with specific RNA species, including centromeric transcripts, and this even when the RNAi pathway of heterochromatin assembly is defective.
Given that RNA processing can be cotranscriptional, we were curious to see whether Spf30 might associate with nascent transcripts. To test this idea, we sought to determine whether Spf30 binding to chromatin was dependent on RNA species. Spf30-ChIP was carried out on chromatin extracts treated or not treated with active RNase before IP. RNase treatment somehow increased the background (not shown). To circumvent this problem, the percentage of IP measured in each Spf30-GFP sample was normalized to its respective untagged control, allowing direct comparison between samples treated or not treated with active RNase. As shown in Fig. 6D, the centromeric dg and dh repeats, cut23 and fbp1, showed a sharp decline in their normalized enrichment when chromatin was treated with active RNase prior to Spf30-GFP IP. Similar observations were made at the central domain of centromere 1 (data not shown). We conclude that Spf30 binding to chromatin is RNA dependent. Altogether, these data are consistent with the idea that Spf30 associates with nascent transcripts.
DISCUSSION
Transcript processing by the RNAi and the exosome contributes to transcriptional silencing and heterochromatin assembly. Recently, splicing factors have been proposed to improve the production of centromeric siRNA in fission yeast. Whether splicing factors also played a role in the exosome branch of the silencing pathway was unknown. We characterized here the fission yeast counterpart of the human splicing factor Spf30. We show that Spf30 contributes to the structural integrity of heterochromatin and efficient silencing and provide circumstantial evidence that Spf30, recruited in a Dis3-dependent manner at centromeric heterochromatin, binds cognate nascent transcripts, and assists their degradation by the exosome.
Spf30 as a splicing factor.
Spf30 has been identified from purified human spliceosome complexes. It bridges an interaction between the U2 and U4/U6.U5 snRNPs and takes part in spliceosome assembly in vitro (34, 37, 48). Spf30 is apparently missing in budding yeast, and little is known about its involvement in vivo. Here we show that unspliced RNA species accumulate in spf30-38 mutant cells and that Spf30 interacts with two spliceosomal components in vivo: Prp1, which belongs to the U4/U6.U5 tri-snRNP (13), and the U2 snRNA (Fig. 2 and 6C). These data are consistent with the idea that Spf30 is a splicing factor in fission yeast and suggest that its bridging function has been conserved. Thus, defective splicing in spf30-38 may stem from an impaired assembly of the spliceosome.
Spf30 deficiency leads to chromosome segregation defects.
Spf30 loss of function disrupts accurate chromosome segregation during anaphase and alleviates the silencing imposed on a marker gene inserted within the central domain of fission yeast centromeres (Fig. 3). Chromatin at the central domain possesses specific attributes that dictate kinetochore assembly, and silencing at the central domain is thought to reflect this fact (46). Thus, proper kinetochore assembly may be disrupted in spf30-38 cells, and this in a way that would become lethal in the absence of pericentric heterochromatin. One trivial explanation would be a splicing defect of a critical kinetochore component. On the other hand, both Spf30 and Dis3, which are functionally related (see below), bind the central domain (Fig. 6A and B), and dis3-54 alleviates silencing at the central domain and is also colethal with swi6Δ (41), raising the possibility of a direct role at the kinetochore. Swi6 is reduced but not absent from centromeric outer repeats in dis3-54 or spf30-38 mutant cells (Fig. 4D) (41). Residual centromeric heterochromatin may become vital for proper chromosome segregation when Dis3 or Spf30 is defective. Clearly, further investigations are required to precise the nature of the link between Spf30, Dis3 and the kinetochore.
Spf30 is required for exosome-mediated silencing and heterochromatin assembly.
Polyadenylated centromeric transcripts accumulate in spf30-38 cells, silencing is alleviated, and pericentric heterochromatin integrity is altered, as revealed by reduced H3K9-me and Swi6 levels at outer repeats (Fig. 4 and 5D). Several arguments indicate that Spf30 may contribute to the exosome rather than the RNAi pathway. First, the steady-state amount of centromeric siRNA appears unaffected in spf30-38 (Fig. 5C), implying that polyadenylated centromeric transcripts accumulate not because of an impaired conversion into siRNAs, though it remains formally possible that siRNAs are not functional and/or improperly loaded onto RITS. Second, Spf30 is required for the maintenance of robust silencing at the mating-type locus (Fig. 5B), in sharp contrast with Dcr1 and other factors of the RNAi pathway (4, 20), implying that an RNAi-independent silencing process is disrupted by Spf30 deficiency. The striking similarities between spf30-38 phenotypes and those exhibited by rrp6Δ and dis3-54 mutants with respect to heterochromatic silencing place Spf30 in the exosome branch of the silencing pathway (Fig. 4 and 5) (9, 41). Assuming that a fraction of centromeric transcripts is degraded by the exosome, their accumulation in spf30-38 cells may therefore stem from an impaired degradation.
What could be the molecular basis of Spf30 contribution? Since the accumulating centromeric transcripts are polyadenylated in spf30-38 cells, Spf30 deficiency must interfere with a reaction posterior or concomitant with 3′-end cleavage and polyadenylation, possibly with substrate targeting and/or exosome stimulation. A possibility, inferred merely from Spf30 involvement in splicing, would be an indirect effect through the defective splicing of an as-yet-unidentified cofactor of the exosome. Although we cannot formally rule out this possibility, we found that Spf30 depends on Dis3 for its association with pericentric heterochromatin, a fact that would not be expected if the link between Spf30 and Dis3/exosome was indirect. Rather, the data presented are most consistent with a direct, facilitating role for Spf30. We show that Spf30 associates with nascent centromeric transcripts and Spf30 binding to centromeric outer repeats (transcription sites) is altered in a dis3 mutant, implying that this association is relevant to exosome functioning and providing a straightforward explanation for the strikingly similar phenotypes of spf30-38 and dis3-54 mutants with respect to heterochromatin silencing. Furthermore, reminiscent of Ran GTPase Gsp1 which regulates rRNA processing by the exosome (49), overexpressing Dis3 partially restores the centromeric silencing defect in spf30-38 cells (Fig. 5E). Spf30 is an abundant nuclear protein and increasing Dis3 level could possibly compensate for Spf30-38 deficiency by allowing more mutant protein to be recruited at centromeric outer repeats. We therefore propose that Spf30, recruited at pericentric heterochromatin through Dis3, facilitates the degradation of polyadenylated centromeric transcripts by the exosome. Furthermore, Spf30 intercession may be not limited to centromeric RNAs, as suggested by the accumulation of euchromatic, cryptic noncoding transcripts in spf30-38 cells (Fig. 5G).
The molecular relationships between Dis3, Spf30, and RNA targets remain unclear. We failed to coimmunoprecipitate Dis3 and Spf30, and our data do not allow determining whether Spf30 binds RNA directly or indirectly. Spf30 does not copurify with Cid12 (4), and Spf30 remained bound to centromeric transcripts in the absence of Dcr1 (data not shown). Therefore, Spf30 association with outer repeats is probably not directly mediated by contacts with Dis3, RDRC, and/or RITS. Spf30 may fulfill its task as part of an RNP, perhaps a dedicated spliceosomal snRNP.
The cotranscriptional recruitment of the exosome may allow very rapid degradation of RNA targets (2, 22, 50). We show that Dis3 binds pericentric heterochromatin and euchromatic genes in both wild-type and spf30-38 cells (Fig. 6A), suggesting that Spf30 does not significantly contribute to this chromosomal association, which may rather rely on interactions between the exosome and the RNAPII machinery, as observed in Drosophila and budding yeast (2, 50). Dis3 may thus process centromeric transcripts cotranscriptionally. However, despite Dis3 presence at outer repeats, centromeric polyadenylated transcripts accumulate and heterochromatin integrity is altered in spf30-38 cells (Fig. 4D and E and Fig. 5D). As such, spf30-38 mimics the dis3-54 mutation that reduces Dis3 RNase activity (41). Thus, the mere presence of Dis3 at outer repeats does not allow efficient degradation of polyadenylated centromeric transcripts when Spf30 is impaired. The most straightforward interpretation is that Dis3 activity toward centromeric transcripts requires Spf30. This is consistent with Dis3 helping recruiting Spf30 at pericentric heterochromatin (Fig. 6B). Spf30's Tudor domain might contact a methyl-lysine containing histone (25), and RNAPII-associated Dis3 could promote and/or stabilize this association. In turn, Spf30 and its associated factors, bound to nascent centromeric transcripts, would assist Dis3/exosome in substrate recognition and/or facilitate its exonucleolytic activity, hence playing the role of an exosome cofactor.
Recently, splicing factors have been involved in the pathway leading to the production of centromeric siRNA in fission yeast. Similarly, the data presented here strongly suggest that Spf30, perhaps in combination with other splicing factors, cotranscriptionally binds centromeric transcripts to facilitate their degradation by Dis3/exosome. Versatile splicing factors may take part in the formation of RNPs to escort nascent, noncoding RNAs toward the exosome or the RNAi branch of the silencing pathway, thereby facilitating the fine tuning of their level.
Acknowledgments
We thank K. Gould, D. Moazed, M. Yanadiga, the Yeast Genetic Resource Center (YGRC), and K. Gull for strains and reagents. We are grateful to L. Bayne, K. Ekwall, A. Pidoux, and M. Motamedi for technical advice.
This study was supported by the Centre National de la Recherche Scientifique, l'Université Victor Segalen Bordeaux 2, the Région Aquitaine, and a grant from l'Association pour la Recherche sur le Cancer. J.D. was supported by a fellowship from the Ministère de la Recherche et de l'Enseignement Supérieur.
Footnotes
Published ahead of print on 22 December 2009.
REFERENCES
- 1.Allshire, R. C., E. R. Nimmo, K. Ekwall, J. P. Javerzat, and G. Cranston. 1995. Mutations derepressing silent centromeric domains in fission yeast disrupt chromosome segregation. Genes Dev. 9:218-233. [DOI] [PubMed] [Google Scholar]
- 2.Andrulis, E. D., J. Werner, A. Nazarian, H. Erdjument-Bromage, P. Tempst, and J. T. Lis. 2002. The RNA processing exosome is linked to elongating RNA polymerase II in Drosophila. Nature 420:837-841. [DOI] [PubMed] [Google Scholar]
- 3.Bahler, J., J. Q. Wu, M. S. Longtine, N. G. Shah, A. McKenzie III, A. B. Steever, A. Wach, P. Philippsen, and J. R. Pringle. 1998. Heterologous modules for efficient and versatile PCR-based gene targeting in Schizosaccharomyces pombe. Yeast 14:943-951. [DOI] [PubMed] [Google Scholar]
- 4.Bayne, E. H., M. Portoso, A. Kagansky, I. C. Kos-Braun, T. Urano, K. Ekwall, F. Alves, J. Rappsilber, and R. C. Allshire. 2008. Splicing factors facilitate RNAi-directed silencing in fission yeast. Science 322:602-606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bentley, D. L. 2005. Rules of engagement: co-transcriptional recruitment of pre-mRNA processing factors. Curr. Opin. Cell Biol. 17:251-256. [DOI] [PubMed] [Google Scholar]
- 6.Bernard, P., J. Drogat, J. F. Maure, S. Dheur, S. Vaur, S. Genier, and J. P. Javerzat. 2006. A screen for cohesion mutants uncovers Ssl3, the fission yeast counterpart of the cohesin loading factor Scc4. Curr. Biol. 16:875-881. [DOI] [PubMed] [Google Scholar]
- 7.Bernard, P., J. F. Maure, J. F. Partridge, S. Genier, J. P. Javerzat, and R. C. Allshire. 2001. Requirement of heterochromatin for cohesion at centromeres. Science 294:2539-2542. [DOI] [PubMed] [Google Scholar]
- 8.Berry, L. D., and K. L. Gould. 1997. Fission yeast dim1(+) encodes a functionally conserved polypeptide essential for mitosis. J. Cell Biol. 137:1337-1354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Buhler, M., W. Haas, S. P. Gygi, and D. Moazed. 2007. RNAi-dependent and -independent RNA turnover mechanisms contribute to heterochromatic gene silencing. Cell 129:707-721. [DOI] [PubMed] [Google Scholar]
- 10.Buhler, M., N. Spies, D. P. Bartel, and D. Moazed. 2008. TRAMP-mediated RNA surveillance prevents spurious entry of RNAs into the Schizosaccharomyces pombe siRNA pathway. Nat. Struct. Mol. Biol. 15:1015-1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Buhler, M., A. Verdel, and D. Moazed. 2006. Tethering RITS to a nascent transcript initiates RNAi- and heterochromatin-dependent gene silencing. Cell 125:873-886. [DOI] [PubMed] [Google Scholar]
- 12.Camblong, J., N. Iglesias, C. Fickentscher, G. Dieppois, and F. Stutz. 2007. Antisense RNA stabilization induces transcriptional gene silencing via histone deacetylation in Saccharomyces cerevisiae. Cell 131:706-717. [DOI] [PubMed] [Google Scholar]
- 13.Carnahan, R. H., A. Feoktistova, L. Ren, S. Niessen, J. R. Yates III, and K. L. Gould. 2005. Dim1p is required for efficient splicing and export of mRNA encoding lid1p, a component of the fission yeast anaphase-promoting complex. Eukaryot. Cell 4:577-587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Djupedal, I., M. Portoso, H. Spahr, C. Bonilla, C. M. Gustafsson, R. C. Allshire, and K. Ekwall. 2005. RNA Pol II subunit Rpb7 promotes centromeric transcription and RNAi-directed chromatin silencing. Genes Dev. 19:2301-2306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dziembowski, A., E. Lorentzen, E. Conti, and B. Seraphin. 2007. A single subunit, Dis3, is essentially responsible for yeast exosome core activity. Nat. Struct. Mol. Biol. 14:15-22. [DOI] [PubMed] [Google Scholar]
- 16.Ekwall, K., G. Cranston, and R. C. Allshire. 1999. Fission yeast mutants that alleviate transcriptional silencing in centromeric flanking repeats and disrupt chromosome segregation. Genetics 153:1153-1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gornemann, J., K. M. Kotovic, K. Hujer, and K. M. Neugebauer. 2005. Cotranscriptional spliceosome assembly occurs in a stepwise fashion and requires the cap binding complex. Mol. Cell 19:53-63. [DOI] [PubMed] [Google Scholar]
- 18.Grewal, S. I., and S. C. Elgin. 2007. Transcription and RNA interference in the formation of heterochromatin. Nature 447:399-406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Habara, Y., S. Urushiyama, T. Shibuya, Y. Ohshima, and T. Tani. 2001. Mutation in the prp12+ gene encoding a homolog of SAP130/SF3b130 causes differential inhibition of pre-mRNA splicing and arrest of cell-cycle progression in Schizosaccharomyces pombe. RNA 7:671-681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hall, I. M., G. D. Shankaranarayana, K. Noma, N. Ayoub, A. Cohen, and S. I. Grewal. 2002. Establishment and maintenance of a heterochromatin domain. Science 297:2232-2237. [DOI] [PubMed] [Google Scholar]
- 21.Heyer, W. D., M. Sipiczki, and J. Kohli. 1986. Replicating plasmids in Schizosaccharomyces pombe: improvement of symmetric segregation by a new genetic element. Mol. Cell. Biol. 6:80-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Houseley, J., K. Kotovic, A. El Hage, and D. Tollervey. 2007. Trf4 targets ncRNAs from telomeric and rDNA spacer regions and functions in rDNA copy number control. EMBO J. 26:4996-5006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Houseley, J., J. LaCava, and D. Tollervey. 2006. RNA-quality control by the exosome. Nat. Rev. Mol. Cell. Biol. 7:529-539. [DOI] [PubMed] [Google Scholar]
- 24.Jurica, M. S., and M. J. Moore. 2003. Pre-mRNA splicing: awash in a sea of proteins. Mol. Cell 12:5-14. [DOI] [PubMed] [Google Scholar]
- 25.Kim, J., J. Daniel, A. Espejo, A. Lake, M. Krishna, L. Xia, Y. Zhang, and M. T. Bedford. 2006. Tudor, MBT and chromo domains gauge the degree of lysine methylation. EMBO Rep. 7:397-403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kinoshita, N., M. Goebl, and M. Yanagida. 1991. The fission yeast dis3+ gene encodes a 110-kDa essential protein implicated in mitotic control. Mol. Cell. Biol. 11:5839-5847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kloc, A., M. Zaratiegui, E. Nora, and R. Martienssen. 2008. RNA interference guides histone modification during the S phase of chromosomal replication. Curr. Biol. 18:490-495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kyburz, A., A. Friedlein, H. Langen, and W. Keller. 2006. Direct interactions between subunits of CPSF and the U2 snRNP contribute to the coupling of pre-mRNA 3′ end processing and splicing. Mol. Cell 23:195-205. [DOI] [PubMed] [Google Scholar]
- 29.Lacadie, S. A., D. F. Tardiff, S. Kadener, and M. Rosbash. 2006. In vivo commitment to yeast cotranscriptional splicing is sensitive to transcription elongation mutants. Genes Dev. 20:2055-2066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.LaCava, J., J. Houseley, C. Saveanu, E. Petfalski, E. Thompson, A. Jacquier, and D. Tollervey. 2005. RNA degradation by the exosome is promoted by a nuclear polyadenylation complex. Cell 121:713-724. [DOI] [PubMed] [Google Scholar]
- 31.Lebreton, A., and B. Seraphin. 2008. Exosome-mediated quality control: substrate recruitment and molecular activity. Biochim. Biophys. Acta 1779:558-565. [DOI] [PubMed] [Google Scholar]
- 32.Lebreton, A., R. Tomecki, A. Dziembowski, and B. Seraphin. 2008. Endonucleolytic RNA cleavage by a eukaryotic exosome. Nature 456:993-996. [DOI] [PubMed] [Google Scholar]
- 33.Lin, S., G. Coutinho-Mansfield, D. Wang, S. Pandit, and X. D. Fu. 2008. The splicing factor SC35 has an active role in transcriptional elongation. Nat. Struct. Mol. Biol. 15:819-826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Little, J. T., and M. S. Jurica. 2008. Splicing factor SPF30 bridges an interaction between the prespliceosome protein U2AF35 and tri-small nuclear ribonucleoprotein protein hPrp3. J. Biol. Chem. 283:8145-8152. [DOI] [PubMed] [Google Scholar]
- 35.Matsumoto, T., S. Murakami, O. Niwa, and M. Yanagida. 1990. Construction and characterization of centric and acentric linear minichromosomes in fission yeast. Curr. Genet. 18:320-330. [Google Scholar]
- 36.Maurer-Stroh, S., N. J. Dickens, L. Hughes-Davies, T. Kouzarides, F. Eisenhaber, and C. P. Ponting. 2003. The Tudor domain ‘Royal Family’: Tudor, plant Agenet, Chromo, PWWP and MBT domains. Trends Biochem. Sci. 28:69-74. [DOI] [PubMed] [Google Scholar]
- 37.Meister, G., S. Hannus, O. Plottner, T. Baars, E. Hartmann, S. Fakan, B. Laggerbauer, and U. Fischer. 2001. SMNrp is an essential pre-mRNA splicing factor required for the formation of the mature spliceosome. EMBO J. 20:2304-2314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Moazed, D. 2009. Small RNAs in transcriptional gene silencing and genome defence. Nature 457:413-420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Moreno, S., A. Klar, and P. Nurse. 1991. Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Methods Enzymol. 194:795-823. [DOI] [PubMed] [Google Scholar]
- 40.Motamedi, M. R., A. Verdel, S. U. Colmenares, S. A. Gerber, S. P. Gygi, and D. Moazed. 2004. Two RNAi complexes, RITS and RDRC, physically interact and localize to noncoding centromeric RNAs. Cell 119:789-802. [DOI] [PubMed] [Google Scholar]
- 41.Murakami, H., D. B. Goto, T. Toda, E. S. Chen, S. I. Grewal, R. A. Martienssen, and M. Yanagida. 2007. Ribonuclease activity of Dis3 is required for mitotic progression and provides a possible link between heterochromatin and kinetochore function. PLoS One 2:e317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Musacchio, A., and E. D. Salmon. 2007. The spindle-assembly checkpoint in space and time. Nat. Rev. Mol. Cell. Biol. 8:379-393. [DOI] [PubMed] [Google Scholar]
- 43.Nicolas, E., T. Yamada, H. P. Cam, P. C. Fitzgerald, R. Kobayashi, and S. I. Grewal. 2007. Distinct roles of HDAC complexes in promoter silencing, antisense suppression and DNA damage protection. Nat. Struct. Mol. Biol. 14:372-380. [DOI] [PubMed] [Google Scholar]
- 44.Noguchi, E., N. Hayashi, Y. Azuma, T. Seki, M. Nakamura, N. Nakashima, M. Yanagida, X. He, U. Mueller, S. Sazer, and T. Nishimoto. 1996. Dis3, implicated in mitotic control, binds directly to Ran and enhances the GEF activity of RCC1. EMBO J. 15:5595-5605. [PMC free article] [PubMed] [Google Scholar]
- 45.Nonaka, N., T. Kitajima, S. Yokobayashi, G. Xiao, M. Yamamoto, S. I. Grewal, and Y. Watanabe. 2002. Recruitment of cohesin to heterochromatic regions by Swi6/HP1 in fission yeast. Nat. Cell Biol. 4:89-93. [DOI] [PubMed] [Google Scholar]
- 46.Pidoux, A. L., and R. C. Allshire. 2005. The role of heterochromatin in centromere function. Philos. Trans. R. Soc. Lond. B Biol. Sci. 360:569-579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Potashkin, J., R. Li, and D. Frendewey. 1989. Pre-mRNA splicing mutants of Schizosaccharomyces pombe. EMBO J. 8:551-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rappsilber, J., P. Ajuh, A. I. Lamond, and M. Mann. 2001. SPF30 is an essential human splicing factor required for assembly of the U4/U5/U6 tri-small nuclear ribonucleoprotein into the spliceosome. J. Biol. Chem. 276:31142-31150. [DOI] [PubMed] [Google Scholar]
- 49.Suzuki, N., E. Noguchi, N. Nakashima, M. Oki, T. Ohba, A. Tartakoff, M. Ohishi, and T. Nishimoto. 2001. The Saccharomyces cerevisiae small GTPase, Gsp1p/Ran, is involved in 3′ processing of 7S-to-5.8S rRNA and in degradation of the excised 5′-A0 fragment of 35S pre-rRNA, both of which are carried out by the exosome. Genetics 158:613-625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Vasiljeva, L., and S. Buratowski. 2006. Nrd1 interacts with the nuclear exosome for 3′ processing of RNA polymerase II transcripts. Mol. Cell 21:239-248. [DOI] [PubMed] [Google Scholar]
- 51.Vasiljeva, L., M. Kim, N. Terzi, L. M. Soares, and S. Buratowski. 2008. Transcription termination and RNA degradation contribute to silencing of RNA polymerase II transcription within heterochromatin. Mol. Cell 29:313-323. [DOI] [PubMed] [Google Scholar]
- 52.Verdel, A., S. Jia, S. Gerber, T. Sugiyama, S. Gygi, S. I. Grewal, and D. Moazed. 2004. RNAi-mediated targeting of heterochromatin by the RITS complex. Science 303:672-676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Volpe, T. A., C. Kidner, I. M. Hall, G. Teng, S. I. Grewal, and R. A. Martienssen. 2002. Regulation of heterochromatic silencing and histone H3 lysine-9 methylation by RNAi. Science 297:1833-1837. [DOI] [PubMed] [Google Scholar]
- 54.Wang, S. W., A. L. Stevenson, S. E. Kearsey, S. Watt, and J. Bahler. 2008. Global role for polyadenylation-assisted nuclear RNA degradation in posttranscriptional gene silencing. Mol. Cell. Biol. 28:656-665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wilhelm, B. T., S. Marguerat, S. Watt, F. Schubert, V. Wood, I. Goodhead, C. J. Penkett, J. Rogers, and J. Bahler. 2008. Dynamic repertoire of a eukaryotic transcriptome surveyed at single-nucleotide resolution. Nature 453:1239-1243. [DOI] [PubMed] [Google Scholar]
- 56.Win, T. Z., S. Draper, R. L. Read, J. Pearce, C. J. Norbury, and S. W. Wang. 2006. Requirement of fission yeast Cid14 in polyadenylation of rRNAs. Mol. Cell. Biol. 26:1710-1721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Woods, A., T. Sherwin, R. Sasse, T. H. MacRae, A. J. Baines, and K. Gull. 1989. Definition of individual components within the cytoskeleton of Trypanosoma brucei by a library of monoclonal antibodies. J. Cell Sci. 93(Pt. 3):491-500. [DOI] [PubMed] [Google Scholar]
- 58.Wyers, F., M. Rougemaille, G. Badis, J. C. Rousselle, M. E. Dufour, J. Boulay, B. Regnault, F. Devaux, A. Namane, B. Seraphin, D. Libri, and A. Jacquier. 2005. Cryptic pol II transcripts are degraded by a nuclear quality control pathway involving a new poly(A) polymerase. Cell 121:725-737. [DOI] [PubMed] [Google Scholar]
- 59.Zofall, M., T. Fischer, K. Zhang, M. Zhou, B. Cui, T. D. Veenstra, and S. I. Grewal. 2009. Histone H2A.Z cooperates with RNAi and heterochromatin factors to suppress antisense RNAs. Nature 461:419-422. [DOI] [PMC free article] [PubMed] [Google Scholar]