Abstract
Respiratory syncytial virus (RSV) is the major cause of lower respiratory tract infection in infants, with about half being infected in their first year of life. Yet only 2 to 3% of infants are hospitalized for RSV infection, suggesting that individual susceptibility contributes to disease severity. Previously, we determined that AKR/J (susceptible) mice developed high lung RSV titers and showed delayed weight recovery, whereas C57BL/6J (resistant) mice demonstrated low lung RSV titers and rapid weight recovery. In addition, we have reported that gene-targeted mice lacking the cystic fibrosis transmembrane conductance regulator (Cftr; ATP-binding cassette subfamily C, member 7) are susceptible to RSV infection. For this report, recombinant backcross and F2 progeny derived from C57BL/6J and AKR/J mice were infected with RSV, their lung titers were measured, and quantitative trait locus (QTL) analysis was performed. A major QTL, designated Rsvs1, was identified on proximal mouse chromosome 6 in both recombinant populations. Microarray analysis comparing lung transcripts of the parental strains during infection identified several candidate genes that mapped to the Rsvs1 interval, including Cftr. These findings add to our understanding of individual RSV susceptibility and strongly support a modifier role for CFTR in RSV infection, a significant cause of respiratory morbidity in infants with cystic fibrosis.
A major cause of lower respiratory tract infection during infancy and childhood, respiratory syncytial virus (RSV) produces annual epidemics in which >50% of infants are infected during their first RSV season, and nearly all children are infected by 2 to 3 years of age (25, 32). Approximately 2 to 3% of infected infants and an increasing number of elderly patients develop significant morbidity and mortality from RSV infection (4, 54, 59, 62). Although the mechanism is unclear, early, severe RSV infection is a risk factor for recurrent wheezing and asthma (42, 45). In infants, RSV can cause bronchiolitis, leading to severe respiratory illness requiring hospitalization (∼120,000 cases/year in the United States) (4, 54, 59). Severe RSV infections are more likely to occur in patients that are immunocompromised or in infants born prematurely. Outside of these high-risk groups, it is not clear why only a small percentage of children develop severe illness.
While almost every child becomes infected with RSV, the gene-environment interactions that predispose children to develop severe RSV infection are unknown. Previous studies of children have suggested several candidate genes associated with severe RSV infection, including polymorphisms in surfactant-associated proteins A and D (34, 36, 49) and several cytokines (21, 23, 26, 27, 50). However, these genetic differences explain only a part of the variable response to RSV infection, making it difficult to assess the specific role(s) of these genes in disease severity.
Severity of RSV infection is likely to be a complex trait and involves differences that control the initial infection, viral replication, and resolution of infection. Experimentally, the sum of these processes can be examined in mice by measurement of the lung viral titer within the first days of infection (7, 19, 47, 58). The lung viral titer varies between the AKR/J (sensitive) and C57BL/6J (resistant) mouse strains (47, 56). The transmission of resistance to F1 offspring supports the heritability of this trait (56). In this study, quantitative trait locus (QTL) and microarray analyses of mice were used to further assess the genetic determinants of RSV susceptibility.
MATERIALS AND METHODS
Viral and cell culture.
RSV strain A2 was used for all infection studies. Although mice develop pneumonia rather than bronchiolitis following infection with this virus, the pathological and immune responses to the RSV A2 strain have been studied intensively and well characterized for several mouse strains by several laboratories (11, 19, 37). To generate RSV preparations, stocks were plaque purified in HEp2 cells, and virus titer assays were performed as previously described (19). The virus and HEp2 cells were gifts from Barney Graham. Cell culture supernatant obtained from uninfected cells was used as a control medium, as described previously (19).
Infection of mice with RSV and viral titer assays.
All studies were performed with pathogen-free mice (female and male, 7 to 10 weeks of age) obtained from the Jackson Laboratory (Bar Harbor, ME) and housed in microisolator cages in an AALAC-certified animal facility. Sentinel mice were monitored for Sendai virus, pneumonia virus of mice, murine hepatitis virus, minute virus of mice, Theiler's virus of mice, reovirus 3, and mouse parvovirus. After a 1-week acclimation period, mice were sedated by ketamine (90 μg given intraperitoneally [i.p.]/mg of body weight) and exposed by intranasal inoculation to 1 × 107 PFU of RSV (100 μl) or to an uninfected cell extract that was prepared and frozen identically to the RSV preparation (19, 56).
Based on our previous results (56), mice were anesthetized (100 mg/kg pentobarbital i.p.) and killed by exsanguination on day 4 postinfection to obtain peak viral titers. The lungs were removed under sterile conditions, weighed, frozen in liquid nitrogen, and stored at −80°C until RSV titers were determined. At the same time, livers and kidneys (as a backup DNA source) were retrieved and stored at −80°C for subsequent DNA isolation. On day 4 postinfection, the lungs were homogenized at 4°C (Tissue-Tearor; BioSpec, Bartlesville, OK) (25,000 rpm for 20 s) in 2.0 ml prechilled minimum essential medium (MEM), and viral susceptibility was quantified by plaque assay, as described in detail previously (56). To assess viral recovery, lung homogenates were prepared from one set of infected C57BL/6J mice within 1 h of viral instillation, and RSV titers were determined. In these studies, a titer of 3.2 × 105 ± 0.6 × 105 PFU/gram was recovered following an instillation of 1 × 107 PFU. All procedures were performed using protocols approved by the Institutional Animal Care and Use Committee at the University of Missouri-Columbia and the University of Texas Health Science Center, Houston, TX.
Mouse breeding for genetic studies.
To determine whether susceptibility to RSV infection was a heritable trait, progeny from crosses between the sensitive AKR/J and resistant C57BL/6J strains were infected (56), and RSV titers were determined at 4 days postinfection. To generate mice for genetic analyses, the following breeding protocols were conducted by Jackson Laboratory West (Sacramento, CA); per standard nomenclature rules for mice, females are listed first in each pairing: F1, AKR/J × C57BL/6J [i.e., (AK.B6)F1] or the reciprocal, C57BL/6J × AKR/J [i.e., (B6.AK)F1]; F2, (AK.B6)F1 × (AK.B6)F1, (AK.B6)F1 × (B6.AK)F1, (B6.AK)F1 × (AK.B6)F1, and (B6.AK)F1 × (B6.AK)F1; and backcrosses, (AK.B6)F1 × AKR/J, (B6.AK)F1 × AKR/J, AKR/J × (AK.B6)F1, and AKR/J × (B6.AK)F1.
In addition, equal numbers of male and female AKR/J and C57BL/6J parental mice were obtained in every shipment and infected to serve as internal controls for the quality of the infections.
Genotyping.
Genomic DNA was isolated from liver samples by use of a commercial DNA extraction kit (Wizard genomic DNA; Promega, Madison, WI). Samples were analyzed for purity (A260/A280) and DNA content (A260), using a 96-well quartz plate and a SpectraMAX 190 spectrophotometer (Molecular Devices, Sunnyvale, CA). Microsatellite markers (i.e., primer pairs used in PCR to amplify regions of known polymorphisms between the C57BL/6J and AKR/J strains) were purchased from Research Genetics/Invitrogen (Frederick, MD) or Integrated DNA Technologies (Coralville, IA). Microsatellite markers were used to track the parental origin of each chromosomal region in the offspring. For microsatellite markers with a <5% difference in allele sizes between strains, PCR amplification was performed using fluorescent primers synthesized by Applied Biosystems, Inc. (ABI), Foster City, CA, and samples were genotyped with an ABI-3730xL sequencer. For PCR analysis, DNA was diluted to 20 ng/μl for microsatellite markers requiring agarose gel electrophoresis separation and ethidium staining or to 5 ng/μl for fluorescently labeled microsatellite markers (ABI), as described previously (48). Fluorescence genotypes were ascertained using GeneMapper software (V3.5; ABI).
QTL mapping and linkage analyses.
To assess linkage (i.e., a significant association between phenotype and genotype) and possible gene-gene interactions (epistasis), all backcross (n = 246) and F2 (n = 235) mice were initially genotyped for the same set of 73 polymorphic microsatellite markers, spaced at 35 to 40 million base pairs (Mbp) throughout the genome. Because the distribution of RSV titers at 4 days postinoculation was skewed for the backcross and F2 populations (ranging from near zero to more than 10 million PFU/gram of lung), both data sets were log transformed to normalize distributions for QTL analyses. Phenotype (i.e., mouse RSV titer at 4 days postinfection) and microsatellite marker genotype data were evaluated for linkage and main-effect QTLs in the separate populations by using the “scanone” function in R/QTL (5), a freely available QTL analysis computer software package. R/QTL was also used to determine the corresponding experiment-wise threshold values for significant linkage. Specifically, significance threshold values for the separate backcross and F2 populations and the combined population were established using 10,000 permutations of each data set (9). An estimate of linkage, presented as the LOD score, is the log10 ratio of the likelihood of linkage to the likelihood of no linkage. Accordingly, theoretical threshold LOD scores of 3.3 for a backcross and 4.3 for an F2 mouse indicate a likelihood of linkage of 2,000/1 (corresponding to a P value of 5 ×10−5) and were historically considered “significant” (35). In our analysis, regions on chromosomes 1, 2, 6, and 16 suggested possible linkage, as indicated by LOD scores of ≥2.0 for one or both populations. Additional microsatellite markers located near these intervals were genotyped, with a focus on proximal chromosome 6, because it had the highest LOD score. A total of 85 microsatellite markers were typed for all recombinant (backcross and F2) progeny.
To examine potential gene-gene interactions, separate scans of the entire genome were performed on backcross and F2 populations, using the “scantwo” function of R/QTL, which searches through all pairs of QTL locations for an association of genotypes with the phenotype. In particular, LOD scores were obtained for joint interactions (which tested a two-locus model) and epistatic interactions (which tested for interactions between loci). Empirical threshold significance values for interactions were determined using 10,000 permutations of each data set, which required writing specific programming code in the computer language R to run batches of the data simultaneously in a cluster of microprocessors. Batch results were then compiled from all nodes to establish significance threshold values at 0.01, 0.05, and 0.1, for highly significant, significant, and suggestive linkages for interactions, respectively.
Microarray analysis, data normalization, and statistical analysis.
Female C57BL/6J and AKR/J mice at 6 to 8 weeks of age were treated with 1 × 107 PFU RSV (100 μl) or with control cell extract. Lungs were harvested at 0, 6, 12, 24, 48, 72, 96, or 192 h, frozen in liquid nitrogen, and stored at −80°C until processed by homogenization in TRIzol (Invitrogen, Frederick, MD) (n = 9 mice/time point, pooled to generate 3 mouse lungs/microarray and 3 microarrays/sample period; the 12-h time point had two replicates due to the loss of one sample). The extracted RNA was subjected to repeated ethanol precipitation, resuspended in water, and reprecipitated, and the A260 and A260/A280 values were measured to quantify the RNA and to assess purity, respectively. RNA (12 μg) was resuspended in sterile, RNase-free water at a concentration of 1 μg/μl for microarray analysis.
Microarrays were generated with a mouse library consisting of 35,302 70-mer oligonucleotides (MEEBO library, version 1.05; Invitrogen, Carlsbad, CA), which were resuspended in 3× SSC (30 μM; 1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) and printed on aminosilane-coated slides (Cel Associates, Inc., Pearland, TX) by use of a high-speed robot (Omnigrid; GeneMachines, San Carlos, CA). The complete list of genes spotted on the microarrays can be viewed at http://microarray.uc.edu. Microarray analysis was performed using previously described methods (40, 51, 63). In short, lung RNA samples from control and RSV-infected AKR/J and C57BL/6J mice at different times postinfection were compared using 20 μg total RNA/array. Each sample of mRNA was reverse transcribed and randomly tagged in reciprocal fashion with fluorescent cyanine 3 (Cy3) or cyanine 5 (Cy5) (e.g., Cy3 for control and Cy5 for RSV-infected mice, and then vice versa). Cy3- and Cy5-labeled samples were cohybridized with the printed 70-mer oligonucleotides. Following hybridization, slides were washed and scanned at 635 (Cy5) and 532 (Cy3) nm, using a GenePix 4000B microarray scanner (Axon Instruments, Inc., Union City, CA), and the associated images were captured as JPEG and TIFF files. The intensities of the Cy3 and Cy5 fluorescence signals were normalized, and the results are presented as differentially expressed genes (GenePix Pro, version 5.0, software). Using R statistical software and the limma Bioconductor package (55), the raw data generated by GenePix Pro, version 5.0, software were analyzed to identify differentially expressed genes. Data normalization was performed in two steps for each microarray separately, as described previously (51, 52). After normalization, fluorescence intensity ratios were adjusted according to the estimated dye bias, and separate analysis of variance (ANOVA) models were calculated for each strain-time combination. Estimated fold changes were calculated with ANOVA models, and the resulting t statistics from each comparison were modified using an intensity-based empirical Bayes method (IBMT) (52). Genes with a false discovery rate (FDR; the expected proportion of false-positive results among the declared significant results) of <0.20 (3) were initially considered significantly differentially expressed. To estimate differences in response to infection between strains, additional t tests were performed between strains, using estimated log fold changes. Differences were also estimated between strains at baseline (no infection) by the following method. First, Cy3 and Cy5 values were separately normalized among arrays by quantile normalization. Then, an ANOVA was performed using dye and strain as cofactors. Fold changes were calculated as described above for each time point. Consistent strain differences were then determined by performing t tests on the estimated log fold changes for AKR/J and C57BL/6J controls and adjusting the P values using the FDR (3).
Evaluation of biological function using GO analyses.
The discovery of gene categories enriched with differentially expressed genes was performed using DAVID 2008 (database for annotation, visualization and integrated discovery) (12). The DAVID 2008 analysis uses a Bonferroni multiplicity correction to test each subset list against the population of all genes detected, and a “group enrichment score” is determined as the geometric mean (on a log scale) of member's P values in a corresponding annotation cluster. To help determine the biological functions and locations of these gene lists, each group score was assessed for its likelihood of overrepresentation in the Gene Ontology (GO) Consortium categories, i.e., GO biological process, GO cell component, and GO molecular function, as well as KEGG pathways (Kyoto Encyclopedia of Genes and Genomes; a collection of online databases dealing with genomes, enzymatic pathways, and biological chemicals). FDRs were then calculated (48), and GO categories with FDRs of <0.05 were considered for further analysis.
To visualize the enriched category, the resulting elements contained in each significant gene category were placed in GO Tree Machine (GOTM) (64) and displayed as the most parsimonious directed acyclic graph (DAG). Following assessment of significance, the identified transcripts were further annotated using Ensembl accession numbers and/or reference sequence (RefSeq) format, gene symbol, and name. Each transcript was then matched to its current GeneID at the NCBI Entrez Gene website (http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?CMD=search&DB=gene). Retired records were renamed with the current replacement GeneID. Genes labeled as unknown RIKEN cDNA clones (http://fantom.gsc.riken.jp/4/), DNA segments, hypothetical proteins, or unresolved NCBI cDNAs were eliminated from the visualization but may be obtained with the complete data set. All LocusLink IDs, symbols, and annotated genes were displayed using the TreeView program (http://rana.lbl.gov/EisenSoftware.htm) (13). The complete data set from all analyzed microarrays is available via MIAME.
Assessment of candidate genes from linkage and microarray analyses.
The region of interest identified by genomewide linkage analysis was evaluated for candidate genes by two methods. First, all candidates mapping within 0 to 30.1 Mbp on chromosome 6, as identified by NCBI Mapview and Mouse Genome Informatics (MGI; http://www.informatics.jax.org), were evaluated for biological relevance and the existence of nonsynonymous single nucleotide polymorphisms (SNPs), which can result in a different polypeptide sequence (i.e., a change in an amino acid or a premature stop signal that terminates translation). SNPs in coding regions (i.e., exons), together with SNPs in regulatory regions, are believed to have the most impact on phenotype. Previously, we determined the relative RSV infection susceptibilities of eight inbred mouse strains (listed in the order of most resistant to most susceptible, they are C57BL/6J, A/J, DBA/2J, C3H/HeJ, CBA/J, BALB/cJ, 129P3/J, and AKR/J) (56), and all but strain 129P3/J were interrogated using the Celera2, Perlegen2, and Broad1 SNP databases. Second, all candidate genes in this region were evaluated for differential expression between the C57BL/6J and AKR/J strains at baseline and at all the sampling periods. Using an FDR of <0.10 and the Mouse Phenome Database (http://phenome.jax.org/pub-cgi/phenome/mpdcgi?rtn=docs/home) (20), the resulting candidate genes were evaluated for SNPs contained in the 5′-untranslated region (5′-UTR), splice sites (i.e., intron-exon boundaries), and the first two exons.
RESULTS
RSV titers.
RSV lung titers were significantly lower in the resistant C57BL/6J mice than in the sensitive AKR/J strain (0.79 × 105 ± 0.08 × 105 PFU/mouse versus 9.07 × 105 ± 2.5 × 105 PFU/mouse [mean ± standard deviation {SD}]; P < 0.01), as previously described (56). F1 mice demonstrated RSV titers intermediate to those of the two parent strains (4.16 × 105 ± 0.78 × 105 PFU/mouse). The F2 and backcross populations had RSV titers that were more variable, with mean RSV titers also falling between those demonstrated by the parent strains (2.4 × 105 ± 7.6 × 105 and 1.3 × 105 ± 2.7 × 105 PFU/mouse, respectively). As expected for a complex trait, individual F2 and backcross mice had RSV titers that fell anywhere along the entire susceptibility range identified by the sensitive and resistant parent strains.
Mouse QTL analysis.
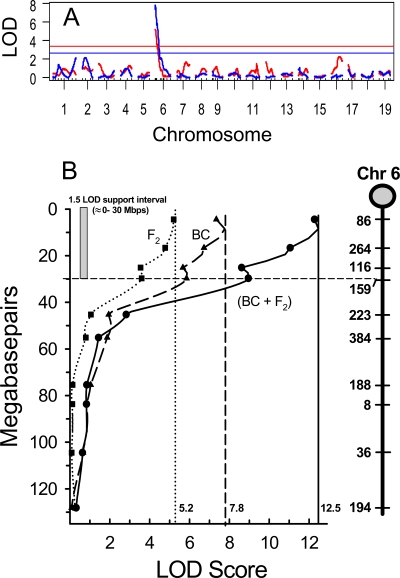
Experiment-wise levels of statistical significance were established by R/QTL, using 10,000 permutations of the respective data sets (9). For the backcross data set, LOD scores of ≥3.4, ≥2.6, and ≥2.3 were determined to represent highly significant, significant, and suggestive linkages, respectively (Fig. 1A). A QTL at the proximal end of chromosome 6 was highly significant, reaching a peak LOD score of 7.8 for the microsatellite marker D6Mit86, positioned at 4.41 Mbp (Fig. 1B). This QTL was designated Rsvs1, for RSV susceptibility QTL 1. The 1.5-LOD support interval for Rsvs1 was ∼0 to 30 Mbp for the backcross population and explained 15% of the variance in RSV titer levels between the parental C57BL/6J and AKR/J strains. No other QTL reached the level of suggestive linkage in the backcross population. Similar results were obtained for the F2 progeny, for which statistical threshold levels were established at ≥4.2, ≥3.4, and ≥3.1, for highly significant, significant, and suggestive linkages, respectively. Rsvs1 was highly significant in the F2 population, reaching an LOD score of 5.2 (Fig. 1A and B) and explaining 16% of the variation. No other marker reached the level of suggestive linkage for the F2 population.
FIG. 1.
(A) Genomewide LOD plot of total RSV lung titers at 4 days postinoculation. Linkage analyses were performed on the separate backcross (blue; n = 246) and F2 (red; n = 235) populations derived from sensitive (AKR/J) and resistant (C57BL/6J) strains. Analyses were performed with the R/QTL computer package (5). Significance thresholds were established by performing 10,000 permutations of each data set at 1-Mb steps and were plotted as F2 LOD scores of ≥3.4 and backcross LOD scores of ≥2.5. (B) Chromosome 6 LOD plots of the combined (•) and separate backcross (BC) (▴) and F2 (▪) populations for total RSV lung titers at 4 days postinoculation. Symbols represent corresponding mitochondrial microsatellite markers at Mbp positions (MGI map locations) along the chromosome (far right). The support interval is displayed along the y axis and represents the approximate Mbp span encompassing a bidirectional 1.5-LOD score drop from the QTL peak.
QTL analysis of the combined data set was accomplished using R/QTL, and results were verified using QTL Express (53). Statistical threshold levels for the combined data set were established by 10,000 permutations and were set at ≥4.1, ≥3.4, and ≥3.0 for highly significant, significant, and suggestive linkages, respectively. In agreement with the analyses of individual backcross and F2 progeny, the combined data set identified a highly significant LOD score of 12.5 for D6Mit86 (the peak microsatellite marker representing Rsvs1), demonstrating a nearly additive value of the separate crosses and suggesting a single gene or small set of closely linked genes. In addition, a suggestive linkage on chromosome 2 (at D2Mit237; LOD score = 3.0) was revealed in the combined data set. To test whether chromosome 6 included a second peak around 30 Mbp, D6Mit86 was fixed to remove its variance from the analysis and the chromosome was rescanned with MapManager QTX (38) to identify any additional QTLs that could explain further variance. No evidence was found for a second QTL peak on chromosome 6.
Pairwise interactions.
To investigate whether gene-gene interactions could contribute to the phenotype, the backcross and F2 data sets were tested for all pairwise (i.e., marker-marker) comparisons, using the “twoscan” function of R/QTL. This analysis yields outputs for the epistatic LOD score (likelihood of a gene-gene interaction for each marker pair) and the joint LOD score (likelihood of a two-locus additive effect) and plots them in a single figure, with epistatic interactions in the top left half and joint effects in the bottom right half (Fig. 2A). For the backcross progeny, no significant or suggestive epistatic interactions were noted (Fig. 2A). A suggestive joint LOD score was detected between Rsvs1 on chromosome 6 and the suggestive QTL on chromosome 2 (joint LOD score = 10.6). For the F2 population, the highest epistatic LOD score was 5.6, between Rsvs1 and D10Mit106. This pairwise LOD score fell short of the 5.9 threshold value for significance but was suggestive of an epistatic interaction. Combining this epistatic LOD score with the additive LOD score yielded a highly significant joint LOD score of 9.5 (Fig. 2B), suggesting a positive interaction between the two markers/genes on chromosomes 6 and 10.
FIG. 2.
Pairwise analyses of gene interactions contributing to RSV resistance in the backcross and F2 progeny. Backcross (A) and F2 (B) populations were analyzed separately for all pairwise interactions. An analysis was performed for epistasis (top left half) and joint (additive plus epistatic) effects (bottom right half), using R/QTL. Significance thresholds are given on the colored bar to the right of each figure, with the scale on the left corresponding to epistasis LOD scores and the scale on the right showing joint LOD scores. (A) For the backcross progeny, no significant or suggestive epistatic (gene-gene) interactions were noted. A suggestive joint LOD score (additive effects) was detected between Rsvs1 on chromosome 6 and the suggestive QTL on chromosome 2 (joint LOD score = 10.6). (B) For the F2 population, the highest epistatic LOD score was 5.6, between Rsvs1 and D10Mit106. This pairwise LOD score fell short of the 5.9 threshold value for significance but was suggestive of an epistatic interaction. Combining this epistatic LOD score with the additive LOD score yielded a highly significant joint LOD score of 9.5, suggesting a positive interaction between the two markers/genes on chromosomes 6 and 10.
Microarray analysis.
To identify transcripts that may be related to the difference in phenotype with the induction of infection, the transcripts that differed between C57BL/6J and AKR/J mouse lungs at any time point (6, 12, 24, 48, 96, or 192 h) were identified using a statistical threshold of an FDR of <0.05 (equivalent to an error probability of ∼0.01), a ≥3.5-fold difference, and an average intensity of ≥80 fluorescence units. These criteria yielded a total of 1,389 elements consisting of 1,350 unique transcripts (i.e., 39 elements were duplicates or triplicates) and included 1,121 annotated genes, 190 nonannotated RIKEN cDNAs (DNA sequences deposited in the RIKEN Bioresource Center Data Bank), and 39 unnamed or pseudogenes. For inclusion, a gene had to be differentially expressed between the two strains at any time point (see Table S1 in the supplemental material).
To characterize transcriptional changes that were similar or different between strains, a total of 1,744 elements (1,626 unique genes) were identified as differentially expressed at least once in the lungs of AKR/J or C57BL/6J mice compared to strain-matched controls, using a statistical threshold of an FDR of <0.05 (equivalent to an error probability of ∼0.01). The stringency of the threshold of significance was further increased to a ≥2-fold difference and an average intensity of ≥80 fluorescence units. This filtering yielded a total of 571 elements consisting of 535 unique transcripts (i.e., 36 elements were duplicates or triplicates) and included 479 annotated genes, 49 nonannotated RIKEN cDNAs, and 7 unnamed or pseudogenes (see Table S2 in the supplemental material).
Discovery of GO categories enriched with differentially expressed transcripts.
To better characterize the temporal transcript changes that occurred during RSV infection, we analyzed the 571 elements that were changed in either strain by using DAVID 2008. The group enrichment score was determined, and categories with an FDR of <0.05 were considered for further analysis. Of the 571 elements, 199 transcripts were identified throughout the enriched categories and were then placed into a self-organizing map. Four groups or clusters of transcripts that were differentially expressed by the resistant and sensitive mouse strains at different times following RSV infection were identified in the data sets: group 1 mRNAs increased at 12 to 48 h, group 2 mRNAs decreased at 6 h to 96 h, group 3 mRNAs increased in the C57BL/6J mice at 48 h, and group 4 mRNAs increased in both mouse strains 192 h following RSV inoculation.
Group 1 consists of 96 elements that clustered with a temporal pattern of increased expression levels 12 to 48 h following RSV exposure (Fig. 3A). The mean tendency for this cluster demonstrated that increased transcripts were not significantly different between the two strains (Fig. 3B). Organizational relationships of group 1 transcripts are presented as a directed acyclic graph (DAG), which displays the GO categories generated by GO Tree for transcripts identified by DAVID 2008 (see Fig. S1 in the supplemental material). Statistically overrepresented categories included increases in early defense responses (including T-cell differentiation and activation, innate immune responses, antigen presentation, early response to virus, inflammatory response and chemotaxis [P < 0.001], and interferon [IFN] type I biosynthesis [P < 0.005]). Representative individual transcripts included several essential transcription factors responsible for virus-induced transcription of the type I IFNs (Stat1, Stat2, and Irf7) (Fig. 3C and D). Increased chemokine transcripts included chemokine (CC motif) ligand 2 (Ccl2; also called Mcp-1), Ccl4 (also called Mip-1β), and chemokine (CXC motif) ligand 10 (Cxcl10; also called IP10). In addition, numerous transcripts previously associated with antiviral properties increased, and these included transcripts for RNA-specific adenosine deaminase (Adar), eukaryotic translation initiation factor 2α kinase 2 (Eif2ak2; also called PKR), myxovirus resistance 1 (Mx1) and Mx2, 2′-5′-oligoadenylate synthase 1A (Oas1a), 2′-5′-oligoadenylate synthase-like 1 (Oasl1), ISG15 ubiquitin-like modifier (Isg15), IFN-induced protein with tetratricopeptide repeats 1 (Ifit1), and thymidylate kinase family lipopolysaccharide (LPS)-inducible member (Tyki). These transcripts can be induced by the type I and type III IFNs. Taken together, the group I transcripts function in early host cellular antiviral responses and were expressed similarly in both resistant (C57BL/6J) and sensitive (AKR/J) mouse strains.
FIG. 3.
Group 1 genes cluster with a temporal pattern of increased levels at 12 to 48 h during RSV infection. (A) Group 1 consists of 96 elements clustering together on a self-organizing map of transcripts significantly (FDR < 0.05) altered in microarray analysis. (B) Comparison of mean tendencies of transcripts that increased in C57BL6/J and AKR/J mouse lungs. The responses of the mouse strains were comparable and increased nearly equally during infection. Values are presented as means ± standard errors of the means (SEM) for the 96 elements in group 1. (C) Representative transcripts increased in C57BL/6J and AKR/J mouse lungs during RSV infection. Activating kinases (Adar, Eif2aka), transcription factors (Stat1, Stat2, Irf7), and cytokines (Ccl2, Ccl4, Cxcl10) increased in both strains. (C) Representative transcripts increased in C57BL/6J and AKR/J mouse lungs during RSV infection and indicative of an interferon response. Antiviral proteins (Mx1, Mx2), oligoadenylate synthetases induced by IFN (Oas1a, Oasl1), and IFN-induced transcripts (Isg15, Ifit1, Tyki) increased in both strains. The results in panels C and D are mean ± SEM values obtained at 12 h postinoculation. Adar, adenosine deaminase, RNA-specific; Eif2ak2, eukaryotic translation initiation factor 2-alpha kinase 2; Stat1, signal transducer and activator of transcription 1; Stat2, signal transducer and activator of transcription 2; Irf7, interferon regulatory factor 7; Ccl2, chemokine (CC motif) ligand 2 (or MCP-1); Ccl4, chemokine (CC motif) ligand 4 (or MIP-1 beta); Cxcl10, chemokine (CXC motif) ligand 10 (or IP-10); Mx1, myxovirus (influenza virus) resistance 1; Mx2, myxovirus (influenza virus) resistance 2; Oas1a, 2′-5′ oligoadenylate synthetase 1A; Oasl1, 2′-5′ oligoadenylate synthetase-like 1; Isg15, ISG15 ubiquitin-like modifier; Ifit1, interferon-induced protein with tetratricopeptide repeats 1; Tyki, thymidylate kinase family LPS-inducible member.
Group 2 consists of 18 transcripts which decreased in both the AKR/J and C57BL/6J lungs following RSV exposure (Fig. 4A). The expression of group 2 genes was lowest 6 h following RSV exposure in the AKR/J mice and returned to baseline by 12 to 24 h (Fig. 4B). In contrast, the group 2 transcripts in C57BL/6J mice decreased to a lesser extent by 6 h, recovered to baseline, and then demonstrated a second decrease at 96 h before again recovering to baseline (Fig. 4B). Overrepresented GO categories contained transcripts involved in muscle contraction, organelle organization (P < 0.001), epidermis development, and amino acid metabolism (P < 0.005) (see Fig. S2 in the supplemental material). The members of group 2 functioning in muscle contraction and regulation of contraction included components of the troponin-actin complex (troponin C2 [Tnnc2], Tnnt3, and actin alpha1 [Acta1]) and the troponin-tropomyosin complex (Tnni1 and tropomyosin 2, beta [Tpm2]) (Fig. 4C). Other decreased GO categories include epithelial development or differentiation (myosin light [Myl1] and heavy [Myh2] chains, creatine kinase, muscle [Ckm], loricrin [Lor], and chloride channel calcium-activated 3 [Clca3; also called Gob5]) and cytoskeletal remodeling (keratins 5 and 13 [Krt5 and Krt13] and eukaryotic translation elongation factor alpha 2 [Eef1a2]). These transcripts suggest a greater degree of initial epithelial injury in the sensitive AKR/J strain than in the resistant C57BL/6J strain (Fig. 4D).
FIG. 4.
Group 2 genes cluster with a temporal pattern of decreased levels at 6 h, more in AKR/J than in C57BL/6J mice, during RSV infection. (A) Group 2 consists of 18 elements that cluster together in the self-organizing map of transcripts significantly (FDR < 0.05) altered in microarray analysis. (B) Comparison of mean tendencies of transcripts that increased in C57BL6/J and AKR/J mouse lungs. The responses of the mouse strains were comparable, but AKR/J mice had a greater response at 6 h of infection. Values are presented as means ± SEM for the 18 elements in group 2. (C) Representative transcripts decreased more in AKR/J mouse lungs during RSV infection. Components of the troponin-actin complex (Tnnc2, Acta1, Tnnt3) and troponin-tropomyosin (Tpm2) and cytokeratin filaments (Krt1-13) decreased markedly in AKR/J mouse lungs. (D) Representative transcripts decreased in AKR/J lungs more than in C57BL/6J lungs during RSV infection and indicative of epithelial cytoskeletal involvement. Accompanying altered troponin-actin, myosin light and heavy chains (Myl1, Myh2) and muscle creatine kinase (Ckm) levels decreased in the mouse lung. In addition, components of the cornified epithelial cell envelope in terminally differentiated (Lor) and developing (Krt2-5) epidermal cells were decreased. Several gene products of this cluster have functional interactions with calcium (Clca3) and regulate cytoskeletal remodeling (Eef1a2). The results in panels C and D are mean ± SEM values obtained at 6 h postinoculation. Tnnc2, troponin C2, fast; Acta1, actin, alpha 1, skeletal muscle; Tnnt3, troponin T3, skeletal, fast; Tpm2, tropomyosin 2, beta; Tnni2, troponin I, skeletal, fast 2; Krt13, keratin 13; Myl1, myosin, light polypeptide 1; Myh2, myosin, heavy polypeptide 2, skeletal muscle, adult; Clca3, chloride channel calcium-activated 3 (or Gob5); Ckm, creatine kinase, muscle; Lor, loricrin; Krt5, keratin 5; Eef1a, eukaryotic translation elongation factor 1 alpha 2.
Group 3 consists of 68 elements that increased in both the AKR/J and C57BL/6J mice 48 h following RSV exposure, more in C57BL/6J mice than in AKR/J mice (Fig. 5A and B). Significant increases were noted in biologic processes contributing to T-cell differentiation and activation (P < 0.001) (see Fig. S3 in the supplemental material). Representative transcripts involved enhanced T-cell activity, including increases in surface CD antigens that characterized T cells (CD3 antigen, delta peptide [Cd3d], CD8 antigen, alpha chain [Cd8a], CD4 antigen [Cd4], and CD247 antigen [Cd247; also called CD3]), T-cell receptors (T-cell receptor beta, variable 8.2 [Tcrb-V8.2] and T-cell receptor alpha chain [Tcra]), and cytokine signal transduction molecules (interleukin-2 [IL-2]-inducible T-cell kinase [Itk] and IKAROS family zinc finger 1 [Ikzf1]) (Fig. 5C). In addition, T-cell and nucleosome involvements were increased more in C57BL/6J mice and included antigen receptor-activated T-cell signaling molecules (protein tyrosine phosphatase, receptor type C [Ptprc; also called CD45]), transcription factors (vav1 oncogene), motility molecules (dedicator of cytokinesis 2 [Dock2]), ancillary molecule transcripts (corin, actin binding protein 1a [Coro1a], and Rho, GDP dissociation inhibitor [GDI] beta [Arhgdib]), and histones (18 transcripts found in this group, including histone cluster 1 H3b [Hist1h3b]) (Fig. 5D). These changes supported T-cell activation in the lungs of C57BL/6J mice that exceeded that in the AKR/J mice at 48 h, which was coincident with the lower RSV titer in the C57BL/6J mice at 96 h postinfection.
FIG. 5.
Group 3 genes cluster with a temporal pattern of increased levels at 48 h, more in C57BL/6J than in AKR/J mouse lungs, during RSV infection. (A) Group 3 consists of 68 elements that cluster together in the self-organizing map of transcripts significantly (FDR < 0.05) altered in microarray analysis. (B) Comparison of mean tendencies of transcripts that increased in C57BL/6J and AKR/J mouse lungs. The responses of the C57BL/6J mice were greater than those of the AKR/J mouse strain at 48 h of infection. Values are presented as means ± SEM for the 68 elements in group 3. (C) Representative transcripts increased more in C57BL/6J mouse lungs during RSV infection and indicative of enhanced T-cell involvement. Increases in surface CD antigens that characterize T cells (Cd3d, Cd4, Cd8a, Cd247), T-cell receptors (Tcrb-V8.2, Tcra), and cytokine signal transduction molecules (Itk, Ikzf1) are supportive of a marked influx of T cells into the lungs of C57BL/6J mice, which exceeded that of AKR/J mice. (D) Representative transcripts increased more in C57BL/6J than in AKR/J mouse lungs during RSV infection and indicative of T-cell and nucleosome involvement. Supportive of the aforementioned T-cell signaling molecules, antigen receptor-activated (Ptprc), transcription factor (Vav1), cytokinesis (Dock2), and ancillary (Coro1a; Arhgdib) transcripts increased more in C57BL/6J mouse lungs. Transcripts for a number of histones (18 transcripts were found in this group, including Hist1h3b) contribute to altered nucleosome assembly. The results in panels C and D are mean ± SEM values obtained at 48 h postinoculation. Tcrb-V8.2, T-cell receptor beta, variable 8.2; Cd8a, CD8 antigen, alpha chain; Cd3d, CD3 antigen, delta polypeptide; Cd247, CD247 antigen; Tcra, T-cell receptor alpha chain; Itk, IL-2-inducible T-cell kinase; Cd4, CD4 antigen; Ikzf1, IKAROS family zinc finger 1; Hist1h3b, histone cluster 1, H3b; Pvrl1, poliovirus receptor-related 1; Ptprc, protein tyrosine phosphatase, receptor type, C; Coro1a, coronin, actin binding protein 1A; Vav1, vav 1 oncogene; Arhgdib, Rho, GDP dissociation inhibitor (GDI) beta; Dock2, dedicator of cytokinesis 2.
Group 4 consists of 17 elements that increased (Fig. 6A and B) more in C57BL/6J lungs than in AKR/J lungs 196 h after RSV exposure. These increases were overrepresented in the GO categories of antigen presentation and major histocompatibility complex (MHC) class I antigen processing (P < 0.001) (see Fig. S4 in the supplemental material). Representative increased transcripts that were indicative of enhanced T-cell involvement included cytokines (resistin-like alpha [Retnla; also called Fizz1], Cxcl9, Ccl5 [also called RANTES], Ccl8 [also called Mcp-2], and CXC receptor 6 [Cxcr6]) and complement components (e.g., complement component 1 q subcomponent, C chain [C1qg]). These increases are consistent with the development of determinants of immunogenicity and tolerance in C57BL/6J mouse lungs exceeding that in AKR/J mouse lungs (Fig. 6C). In addition, increased airway and alveolar remodeling was suggested by increased transcripts for epithelial products (Clca3, regenerating islet-derived 3 gamma [Reg3g], mucin 5 subtype B, tracheobronchial [Muc5b], trefoil factor 2, spasmolytic protein 1 [Tff2]) and macrophage chitinases, including Chi3l4 (Fig. 6D).
FIG. 6.
Group 4 genes cluster with a temporal pattern of increased levels at 192 h, more in C57BL/6J than in AKR/J mouse lungs, during RSV infection. (A) Group 4 consists of 17 elements that clustered in 3 regions in the self-organizing map of transcripts significantly (FDR < 0.05) altered in microarray analysis. (B) Comparison of mean tendencies of transcripts that increased in C57BL6/J and AKR/J mouse lungs. The responses of the C57BL/6J mice were greater than those of the AKR/J mouse strain 192 h after inoculation. Values are presented as means ± SEM for the 17 elements in group 4. (C) Representative transcripts increased more in C57BL/6J mouse lungs during RSV infection and indicative of enhanced T-cell involvement. Increases in cytokines (Retnla, Cxcl9, Ccl8, Cxcr6, Ccl5) and complement components (C1qg) are consistent with the development of determinants of immunogenicity and tolerance in the lungs of C57BL/6J mice, which exceeded that in AKR/J mice. (D) Representative transcripts increased more in C57BL/6J than in AKR/J mouse lungs following RSV infection and indicative of epithelial cell involvement. Increased epithelial (Clca3, Reg3g, Muc5b, Tff2) and macrophage chitinase (Chi3l4) transcripts are consistent with airway and alveolar remodeling as a sequela to infection. The results in panels C and D are mean ± SEM values obtained at 192 h postinoculation. Retnla, resistin-like alpha (or Fizz1 [found in inflammatory zone 1] or HIMF [hypoxia-induced mitogenic factor]); Cxcl9, chemokine (CXC motif) ligand 9 (or Mig [monokine induced by gamma interferon]); Ccl8, chemokine (CC motif) ligand 8 (or Mcp-2 [monocyte chemoattractant protein 2]); Cxcr6, chemokine (CXC motif) receptor 6; Ccl5, chemokine (CC motif) ligand 5 (or RANTES [regulated upon activation, normally T-expressed, and presumably secreted]); C1qg, complement component 1, q subcomponent, gamma polypeptide; Clca3, chloride channel calcium-activated 3 (or Gob5); Reg3g, regenerating islet-derived 3 gamma; Chi3l4, chitinase 3-like 4; Muc5b, mucin 5, subtype B, tracheobronchial; Tff2, trefoil factor 2 (spasmolytic protein 1).
Combining QTL and microarray analyses.
Assessment of the Rsvs1 interval on mouse chromosome 6 (build 37; NCBI) recognized 113 annotated genes and RIKEN cDNAs. Of the 113 identified sequences, 107 (95%) elements were interrogated by microarray analysis. Transcripts mapping to the Rsvs1 interval demonstrated between-strain (n = 12) and within-strain expression differences compared to controls (n = 7). One transcript, encoding RNA binding motif protein 28 (Rbm28), was contained in both groups, and therefore 18 genes were identified. Nine of these 18 genes also had SNPs that differed between AKR/J and C57BL/6J mice (Table 1). Because these candidate genes were selected based on variable expression between strains, we further filtered these candidates based on the locations of SNPs within the genes (including only transcripts that had SNPs within the 5′-UTR and intronic, nonsynonymous, and 3′-UTR variants). Five of the nine candidates with SNPs that differed between AKR/J and C57BL/6J mice met this criterion, including sterile alpha motif domain containing 9-like (Samd9l), with 1 nonsynonymous SNP (I168V; the SNP resulted in valine replacing isoleucine at amino acid position 168) and 25 intronic SNPs; procollagen, type I, alpha 2 (Col1a2), with 2 intronic and 1 3′-UTR SNP; transcription factor EC (Tcfec), with 1 intronic SNP; cystic fibrosis transmembrane regulator homolog (Cftr; ATP-binding cassette subfamily C, member 7), with 35 intronic SNPs; and RNA binding motif protein 28 (Rbm28), with 2 nonsynonymous (T387S, serine replacing threonine at amino acid 387; and A362V, valine replacing alanine at amino acid 362) and 10 3′-UTR SNPs.
TABLE 1.
Candidate genes in QTL region on chromosome 6 with altered transcript levels in the lung and SNPs that vary between AKR/J and C57BL/6J strains during RSV infection
| Gene | Gene product | Start site (Mbp) | No. of SNPs | Fluorescence intensity | Fold change in transcript level | SE of change | FDR | Timea |
|---|---|---|---|---|---|---|---|---|
| Samd9l | Sterile alpha motif domain containing 9-like | 3.32 | 29 | 2,831 | 2.28 | 0.09 | 0.00805 | 1 |
| Calcr | Calcitonin receptor | 3.63 | 2 | 130 | 5.35 | 0.58 | 0.00513 | 2 |
| Col1a2 | Procollagen, type I, alpha 2 | 4.46 | 21 | 11,364 | −20.31 | 0.72 | 0.00013 | 2 |
| Phf14 | PHD finger protein 14 | 11.88 | 49 | 1,199 | −4.04 | 0.61 | 0.01958 | 2 |
| Tcfec | Transcription factor EC | 16.78 | 3 | 254 | 7.57 | 0.80 | 0.00997 | 2 |
| Cav2 | Caveolin 2 | 17.23 | 1 | 2,629 | 1.71 | 0.10 | 0.04586 | 1 |
| Cftr | Cystic fibrosis transmembrane conductance regulator homolog | 18.12 | 81 | 595 | 3.72 | 0.43 | 0.02555 | 3 |
| Rbm28 | RNA binding motif protein 28 | 29.07 | 137 | 469 | −4.91 | 0.42 | 0.00040 | 2 |
| Impdh1 | Inosine 5′-phosphate dehydrogenase 1 | 29.16 | 2 | 326 | −3.84 | 0.48 | 0.00610 | 2 |
1, AKR/J mice at 24 h; 2, AKR/J and C57BL/6J mice at 6 h; 3, AKR/J and C57BL/6J mice at 192 h.
DISCUSSION
Although half of all children become infected with RSV during the first year of life, only a small percentage go on to develop severe disease requiring hospitalization or long-term respiratory sequelae of infection (recurrent cough or wheeze). Several developmental, environmental, and genetic differences can contribute to the susceptibility of these children to RSV-induced illness.
Our previous studies with mice demonstrated that RSV titers peaked on day 4 following RSV infection and that there were quantitative differences in the titers obtained from AKR/J and C57BL/6J mouse strains (56). RSV titers are the result of complicated interactions between the virus and host, including attachment, entry, replication, release from the respiratory epithelium, and the early host inflammatory responses to this infection. Although titer is not definitively related to disease, RSV titer has been associated with disease severity in animals or humans. RSV infection does induce clinical illness in mice (e.g., predictable and reproducible ruffling and weight loss and inflammatory responses that are more difficult to quantify). In our initial report, we described a difference in weight loss between AKR/J and C57BL/6J mice, with AKR/J mice demonstrating higher titers and greater weight loss (56). One of the most convincing demonstrations of the association between RSV titers and clinical indicators of disease in mice was described by Ramilo and associates (41). Their study demonstrated that reduced RSV replication (lower titer) was associated with significant modulation of inflammatory and clinical markers of acute disease.
Clinical reports also provide evidence for the association between viral titer and disease in humans. Studies measuring passive antibody against RSV and disease in children demonstrate decreased symptoms associate with decreased titers (24). Children with immune deficiency (primary or secondary) do not limit RSV replication and have increased and prolonged viral shedding and increased clinical illness and mortality from the infection (22). Premature infants treated with palivizumab have decreased clinical illness (presumably due to limitation of virus spread in the lungs) compared to control infants (43). Although it is important to recognize that these are associations and not proven causal relationships between RSV titer and disease severity, differences in titer levels can be a useful biologic marker and provide insights into the underlying disease pathogenesis.
In this study, we used RSV titer levels at 96 h postinfection as a quantitative trait for RSV susceptibility and identified a genetic locus (QTL) containing one or more genes that control the differential susceptibility of AKR/J and C57BL/6J mice. This significant QTL, named Rsvs1 (LOD = 7.8 for backcross studies, LOD = 5.2 for F2 crosses, and LOD = 12.5 for the combined backcross and F2 data), maps to the proximal end of mouse chromosome 6. One additional putative QTL on chromosome 2 was suggestive of a linkage, but only when the total backcross and F2 populations were combined in the analysis. Further analysis suggested joint interactions between genes on chromosomes 2 and 6 (backcrosses) and between genes on chromosomes 6 and 10 (F2 crosses). Microarray analysis identified genes differentially expressed in a time-dependent and strain-dependent fashion following RSV infection. Integrating the data from the microarray and QTL studies identified 18 candidate genes for RSV susceptibility within the defined Rsvs1 interval on mouse chromosome 6.
RSV assembly requires interaction with cellular actin and microtubules (15, 30). Differences in the expression or function of genes involved in actin-cytoskeleton interactions in the C57BL/6J and AKR/J mice could result in the differences in viral titers. RSV filament and syncytium (cell-cell fusion) formation following infection is dependent on RhoA signaling activity (44). For example, RSV F associates with RhoA and caveolin-1 in plasma membrane microdomains (lipid rafts) (39). Eight genes within the chromosome 6 interval encompassing Rsvs1 have involvement in cytoskeleton elements and actin-G-protein interactions.
The QTL and microarray data suggest that early biochemical and cellular events following RSV infection determine the level of RSV titer and the ongoing cellular and immune responses to the acute infection. Moreover, strain-specific differences in these early events were consistent with the differential RSV susceptibility demonstrated by C57BL/6J and AKR/J mice. These differences could be determined, in part, by the genes that map within Rsvs1. The QTL region on mouse chromosome 6 is large (30 Mbp) and contains over 200 genes. Further studies are needed to fine map Rsvs1. Nonetheless, to begin to assess how genes in this region might respond during infection, transcriptional changes were used to identify genes that are responsive to infection and to delineate the role of mechanistic pathways and processes related to RSV pathogenesis.
The pathogenesis of RSV infection has been studied in mouse models for over 20 years. After internalization of the RSV virion, lungs of infected mice manifest an eclipse period with significantly decreased virus titers. This is followed by increasing titers that peak by 4 to 5 days postinoculation (19, 57). Despite initial viral clearing by day 7, visible illness peaked on day 7 or 8, as evidenced by ruffled fur, reduced activity, and weight loss (19). Mice (7 to 8 weeks old) infected with RSV typically lose a small amount of weight initially (56) but can lose up to 10% of body weight by day 7, with a slow recovery by 12 to 14 days postinfection (19). Lung inflammation is observable at days 2 to 4 after infection, manifested as lymphocyte margination within the pulmonary vasculature and mononuclear cell infiltrates in the perivascular spaces (19), and includes NK cell immigration to the lung (28). The pathology becomes most severe between days 5 and 8, with progression of perivascular, peribronchial, and diffuse alveolar macrophage and lymphocyte infiltrates (18, 19, 28). During this time, both CD4- and CD8-positive T lymphocytes play major roles in RSV clearance, but they also contribute to lung pathology and clinical illness in the mice (7, 16, 18). By day 10, pneumonia resolves and progression of the perivascular and peribronchiolar infiltrates ceases, and by day 15, the alveolar spaces become relatively free of lymphocytes and macrophages, but the perivascular lymphoid aggregates remain. Serum neutralizing antibody to RSV can first be detected 7 to 10 days following RSV infection and peaks at 4 to 6 weeks (17, 19).
Previous studies have demonstrated that RSV infection results in the activation of numerous transcription factors, including NF-κB and c/EBP, which in turn coordinate the expression of a cascade of proinflammatory cytokines and adhesion proteins (8). Thus, it was reasonable to examine the lung transcriptome during infection. Alterations in transcript levels noted in the microarray analysis are consistent with the pathological time course outlined above. A limitation of this approach is that the cellular source of the mRNA is uncertain. RSV infection can result in alterations in gene expression in resident cells and can cause the immigration of lymphocytes and other inflammatory cells into the lungs, resulting in changes in the RNA levels observed. Despite this limitation, several informative patterns of gene expression were identified by this method.
We used two approaches to contrast transcriptomes of the mouse strains. The first involved identifying the transcripts that differed between C57BL/6J and AKR/J mouse lungs at each time during infection; this yielded 1,350 unique transcripts, including 1,121 annotated genes and 229 nonannotated cDNAs. Interestingly, the transcripts encoded by Cftr differed between the AKR/J and C57BL/6J mice. Mice deficient in Cftr (Cftr−/−) have a decreased ability to clear RSV infection, increased inflammation, and decreased production of NO in the airway (10). The effect of Cftr alteration on RSV susceptibility could be the result of decreased production of NO species such as peroxynitrite, which inhibits viral replication. Therefore, Cftr deficiency or alteration in CFTR protein function could lead to the titer differences observed following RSV infection in the AKR/J and C57BL/6J mice.
The second approach was to evaluate the within-strain lung transcriptome, comparing untreated controls to infected mice. This yielded 535 unique transcripts (including 479 annotated genes and 49 nonannotated cDNAs). To better characterize the temporal transcriptome response to RSV, these transcripts were analyzed further using self-organizing map and GO enrichment tools. The initial response involved early antiviral defense changes, including transcripts associated with T-cell differentiation and activation, innate immune responses, antigen presentation, early response to virus, inflammatory response and chemotaxis, and IFN type I biosynthesis. For example, the expression of several essential transcription factors responsible for the virus-induced transcription of the type-I IFNs (Stat1, Stat2, and Irf7) and of chemokine genes (Ccl2 [MCP-1], Ccl4 [MIP1-β], and Cxcl10 [IP10]) increased during the initial 12 to 24 h following RSV exposure. This group also contained transcripts of genes with antiviral properties, including Adar, Eif2ak2, Mx1, Mx2, Oas1a, Oasl1 Isg15, Ifit1, and Tyki. Taken together, these changes are consistent with augmented early host cellular antiviral responses and were similar in both resistant (C57BL/6J) and sensitive (AKR/J) mouse strains. This was unexpected, because it would have been predicted that the C57BL/6J mouse would be more efficient in these early responses, resulting in the lower RSV titers observed subsequently. Although the resistant and sensitive mouse strains had many similar transcript levels during the initial response, by 6 h the AKR/J mice had more decreased transcripts involved in cytoskeletal organization and epidermis development, suggestive of a greater degree of initial epithelial injury (possibly due to preceding syncytium formation) in the sensitive AKR/J strain than the resistant C57BL/6J strain.
These initial transcriptional changes were followed by a cluster of transcripts that increased more in C57BL/6J mice than in AKR/J mice at 48 h. Significant increases were noted in biologic processes contributing to T-cell differentiation and activation, suggesting a more robust immune response in the resistant strain. Representative transcripts that increased more in the C57BL/6J mouse included CD antigens (Cd3d, Cd4, Cd8a, and Cd247 [CD3h]), T-cell receptors (Tcra and Tcrb-V8.2), cytokine signal transduction molecules (Itk and Ikzf1), and T antigen receptor-activated T-cell signaling molecules (Ptprc [CD45]). Other increased transcripts occurred for transcription factors (Vav1), motility factors (Dock2), ancillary molecules (Coro1a and Arhgdib), and a larger group of histones (18 transcripts were found in this group, including Hist1h3b). The differences in these transcripts at 48 h suggested activation of resident T cells or rapid T-cell immigration, which may account for the significant reduction in RSV titer seen in the C57BL/6J mice at 96 h postinfection.
Similarly, at 192 h, a cluster of transcripts representative of antigen presentation and MHC class I antigen processing were increased more in the resistant strain. Increased transcripts, including those for cytokines (Ccl5 [RANTES], Ccl8 [MCP-2], Cxcl9, Cxcr6, and Retnla) and complement components (C1qg), are consistent with the development of determinants of immunogenicity and tolerance in the lungs of C57BL/6J mice exceeding that in AKR/J mice. The resistant strain also had greater increases in transcripts reflecting improved epithelial cell function and lung remodeling, including transcripts for epithelial products (Clca3 [gob5], Reg3g, Muc5b, and Tff2) and macrophage chitinases (Chi3l4). These increases are consistent with the development of determinants of tolerance and epithelial cell repair in C57BL/6J mouse lungs exceeding that in AKR/J mouse lungs.
Having characterized the transcriptional difference, we then integrated the results obtained by the QTL and microarray approaches. While this could aid in uncovering transcripts altered during infection, a limitation is that variants of genes in this region may not lead to differences in expression or message stabilization, or other events reflected by measurement of steady-state mRNA levels. Nonetheless, changes in transcript levels suggest involvement in RSV pathogenesis and strengthen possible associations.
The role of CFTR in RSV infection has not been investigated fully. As noted above, we previously reported that Cftr−/− mice have a decreased ability to clear RSV infection, increased inflammation, and decreased production of NO in the airway (10). Several previous clinical studies have demonstrated that acute exacerbations in patients with cystic fibrosis (CF) can result from nonbacterial agents. Studies of infants have demonstrated that RSV infection is at least temporally associated with the initial identification (and perhaps the initial colonization) by Pseudomonas aeruginosa in the respiratory tract for a significant portion of these patients (1, 2, 29, 46). Thus, acute viral infections, particularly those caused by RSV, are likely to contribute to a significant portion of acute pulmonary exacerbations and may be a key event in bacterial colonization in the CF lung.
In addition, CFTR dysfunction predisposes the CF airway to increased inflammation by altering intracellular signaling pathways, including NF-κB, which results in the activation of inflammatory genes (61). Venkatakrishnan et al. demonstrated increased nuclear translocation of NF-κB in uninfected CF cells and a corresponding decrease in cytosolic IκB (which inhibits NF-κB nuclear translocation and subsequent gene activation) (60). The signal transducer and activation of transcription-1 (STAT-1) signaling pathway is also defective in CFTR-deficient cells, due to the increased expression of the protein inhibitor of activated STAT1 (PIAS1) and defective activation of the GTPase RhoA (31, 33). Functional CFTR was demonstrated to be necessary for normal NF-κB regulation; blocking CFTR function in normal respiratory epithelial cells or restoration of CFTR function in CF epithelial cells restores normal NF-κB regulation. RSV infection causes NF-κB activation in respiratory epithelial cell cultures (14). RSV infection in patients with CF therefore likely results in activation of NF-κB that cannot be inhibited adequately in the absence or malfunction of CFTR. These events could result in enhanced airway obstruction (pulmonary exacerbations), as seen in patients with CF. Another proposed mechanism whereby CFTR deficiency could lead to susceptibility to infection is that lymphocytes, which also express CFTR, have a diminished capacity to secrete antibodies and cytokines in response to antigens in vitro (6). Our mouse microarray data noted key differences in T-lymphocyte function and antigen presentation and are certainly consistent with this viewpoint. Although immunosuppressive therapy may contribute to the chronic illness, the reduced ability to regulate lymphocyte secretion could reduce the response to antigen presentation and may even explain why lung-transplanted patients with CF remain chronically ill.
In summary, having previously reported that gene-targeted mice lacking Cftr were susceptible to RSV infection, we have now performed a QTL analysis using AKR/J and C57BL/6J backcross and F2 progeny. A major QTL (Rsvs1) was identified on proximal mouse chromosome 6. Integrating these QTL results with those of a microarray analysis that compared the polar-responding strains during RSV infection identified several candidate genes mapping to the Rsvs1 interval, including Cftr. These findings add to our understanding of individual RSV susceptibility and implicate a modifying role for CFTR in RSV infection, which may help to explain why viral infection is a significant cause of respiratory morbidity in patients with CF.
Supplementary Material
Acknowledgments
We give special thanks to Karl Broman (Department of Biostatistics and Medical Informatics, University of Wisconsin, Madison, WI) for support and guidance through R/QTL analysis and programming. We also thank Michelle Horner at Cincinnati Children's Hospital, who performed much of the initial genotyping of backcross and F2 recombinants, and Prakash Velayutham and Michael Wagner, of Cincinnati Children's Hospital Division of Bioinformatics, for writing R scripts to run pairwise permutation tests on parallel microprocessors. Hannah Coleman at the University of Missouri-Columbia provided technical support, performing the RSV infections and tissue procurement for the QTL studies.
This research was supported by NIH grants AI046556 (J.M.S.), HG003749 and LM009662 (M.M.), HL077763, HL085655, ES015675 (G.D.L.), and HL075562 (D.R.P.).
Footnotes
Published ahead of print on 16 December 2009.
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1.Abman, S. H., J. W. Ogle, N. Butler-Simon, C. M. Rumack, and F. J. Accurso. 1988. Role of respiratory syncytial virus in early hospitalizations for respiratory distress of young infants with cystic fibrosis. J. Pediatr. 113:826-830. [DOI] [PubMed] [Google Scholar]
- 2.Armstrong, D., K. Grimwood, J. B. Carlin, R. Carzino, J. Hull, A. Olinsky, and P. D. Phelan. 1998. Severe viral respiratory infections in infants with cystic fibrosis. Pediatr. Pulmonol. 26:371-379. [DOI] [PubMed] [Google Scholar]
- 3.Benjamini, Y., and Y. Hochberg. 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. B 57:289-300. [Google Scholar]
- 4.Boyce, T. G., B. G. Mellen, E. F. Mitchel, Jr., P. F. Wright, and M. R. Griffin. 2000. Rates of hospitalization for respiratory syncytial virus infection among children in Medicaid. J. Pediatr. 137:865-870. [DOI] [PubMed] [Google Scholar]
- 5.Broman, K. W., H. Wu, S. Sen, and G. A. Churchill. 2003. R/qtl: QTL mapping in experimental crosses. Bioinformatics 19:889-890. [DOI] [PubMed] [Google Scholar]
- 6.Bubien, J. K. 2001. CFTR may play a role in regulated secretion by lymphocytes: a new hypothesis for the pathophysiology of cystic fibrosis. Pflugers Arch. 443(Suppl. 1):S36-S39. [DOI] [PubMed] [Google Scholar]
- 7.Cannon, M. J., P. J. Openshaw, and B. A. Askonas. 1988. Cytotoxic T cells clear virus but augment lung pathology in mice infected with respiratory syncytial virus. J. Exp. Med. 168:1163-1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chini, B. A., M. A. Fiedler, L. Milligan, T. Hopkins, and J. M. Stark. 1998. Essential roles of NF-kappaB and C/EBP in the regulation of intercellular adhesion molecule-1 after respiratory syncytial virus infection of human respiratory epithelial cell cultures. J. Virol. 72:1623-1626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Churchill, G. A., and R. W. Doerge. 1994. Empirical threshold values for quantitative trait mapping. Genetics 138:963-971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Colasurdo, G. N., J. J. Fullmer, O. Elidemir, C. Atkins, A. M. Khan, and J. M. Stark. 2006. Respiratory syncytial virus infection in a murine model of cystic fibrosis. J. Med. Virol. 78:651-658. [DOI] [PubMed] [Google Scholar]
- 11.Culley, F. J., J. Pollott, and P. J. Openshaw. 2002. Age at first viral infection determines the pattern of T cell-mediated disease during reinfection in adulthood. J. Exp. Med. 196:1381-1386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dennis, G., Jr., B. T. Sherman, D. A. Hosack, J. Yang, W. Gao, H. C. Lane, and R. A. Lempicki. 2003. DAVID: database for annotation, visualization, and integrated discovery. Genome Biol. 4:P3. [PubMed] [Google Scholar]
- 13.Eisen, M. B., P. T. Spellman, P. O. Brown, and D. Botstein. 1998. Cluster analysis and display of genome-wide expression patterns. Proc. Natl. Acad. Sci. USA 95:14863-14868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fiedler, M. A., K. Wernke-Dollries, and J. M. Stark. 1996. Mechanism of RSV-induced IL-8 gene expression in A549 cells before viral replication. Am. J. Physiol. 271:L963-L971. [DOI] [PubMed] [Google Scholar]
- 15.Ghildyal, R., A. Ho, and D. A. Jans. 2006. Central role of the respiratory syncytial virus matrix protein in infection. FEMS Microbiol. Rev. 30:692-705. [DOI] [PubMed] [Google Scholar]
- 16.Graham, B. S. 1995. Pathogenesis of respiratory syncytial virus vaccine-augmented pathology. Am. J. Respir. Crit. Care Med. 152:S63-S66. [DOI] [PubMed] [Google Scholar]
- 17.Graham, B. S., L. A. Bunton, P. F. Wright, and D. T. Karzon. 1991. Reinfection of mice with respiratory syncytial virus. J. Med. Virol. 34:7-13. [DOI] [PubMed] [Google Scholar]
- 18.Graham, B. S., L. A. Bunton, P. F. Wright, and D. T. Karzon. 1991. Role of T lymphocyte subsets in the pathogenesis of primary infection and rechallenge with respiratory syncytial virus in mice. J. Clin. Invest. 88:1026-1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Graham, B. S., M. D. Perkins, P. F. Wright, and D. T. Karzon. 1988. Primary respiratory syncytial virus infection in mice. J. Med. Virol. 26:153-162. [DOI] [PubMed] [Google Scholar]
- 20.Grubb, S. C., G. A. Churchill, and M. A. Bogue. 2004. A collaborative database of inbred mouse strain characteristics. Bioinformatics 20:2857-2859. [DOI] [PubMed] [Google Scholar]
- 21.Hacking, D., J. C. Knight, K. Rockett, H. Brown, J. Frampton, D. P. Kwiatkowski, J. Hull, and I. A. Udalova. 2004. Increased in vivo transcription of an IL-8 haplotype associated with respiratory syncytial virus disease-susceptibility. Genes Immun. 5:274-282. [DOI] [PubMed] [Google Scholar]
- 22.Hall, C. B., K. R. Powell, N. E. MacDonald, C. L. Gala, M. E. Menegus, S. C. Suffin, and H. J. Cohen. 1986. Respiratory syncytial viral infection in children with compromised immune function. N. Engl. J. Med. 315:77-81. [DOI] [PubMed] [Google Scholar]
- 23.Heinzmann, A., I. Ahlert, T. Kurz, R. Berner, and K. A. Deichmann. 2004. Association study suggests opposite effects of polymorphisms within IL8 on bronchial asthma and respiratory syncytial virus bronchiolitis. J. Allergy Clin. Immunol. 114:671-676. [DOI] [PubMed] [Google Scholar]
- 24.Hemming, V. G., W. Rodriguez, H. W. Kim, C. D. Brandt, R. H. Parrott, B. Burch, G. A. Prince, P. A. Baron, R. J. Fink, and G. Reaman. 1987. Intravenous immunoglobulin treatment of respiratory syncytial virus infections in infants and young children. Antimicrob. Agents Chemother. 31:1882-1886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Henderson, F. W., W. A. Clyde, Jr., A. M. Collier, F. W. Denny, R. J. Senior, C. I. Sheaffer, W. G. Conley III, and R. M. Christian. 1979. The etiologic and epidemiologic spectrum of bronchiolitis in pediatric practice. J. Pediatr. 95:183-190. [DOI] [PubMed] [Google Scholar]
- 26.Hoebee, B., E. Rietveld, L. Bont, M. Oosten, H. M. Hodemaekers, N. J. Nagelkerke, H. J. Neijens, J. L. Kimpen, and T. G. Kimman. 2003. Association of severe respiratory syncytial virus bronchiolitis with interleukin-4 and interleukin-4 receptor alpha polymorphisms. J. Infect. Dis. 187:2-11. [DOI] [PubMed] [Google Scholar]
- 27.Hull, J., A. Thomson, and D. Kwiatkowski. 2000. Association of respiratory syncytial virus bronchiolitis with the interleukin 8 gene region in UK families. Thorax 55:1023-1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hussell, T., and P. J. Openshaw. 1998. Intracellular IFN-gamma expression in natural killer cells precedes lung CD8+ T cell recruitment during respiratory syncytial virus infection. J. Gen. Virol. 79:2593-2601. [DOI] [PubMed] [Google Scholar]
- 29.Johansen, H. K., and N. Hoiby. 1992. Seasonal onset of initial colonisation and chronic infection with Pseudomonas aeruginosa in patients with cystic fibrosis in Denmark. Thorax 47:109-111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kallewaard, N. L., A. L. Bowen, and J. E. Crowe, Jr. 2005. Cooperativity of actin and microtubule elements during replication of respiratory syncytial virus. Virology 331:73-81. [DOI] [PubMed] [Google Scholar]
- 31.Kelley, T. J., and H. L. Elmer. 2000. In vivo alterations of IFN regulatory factor-1 and PIAS1 protein levels in cystic fibrosis epithelium. J. Clin. Invest. 106:403-410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kim, H. W., J. O. Arrobio, C. D. Brandt, B. C. Jeffries, G. Pyles, J. L. Reid, R. M. Chanock, and R. H. Parrott. 1973. Epidemiology of respiratory syncytial virus infection in Washington, D.C. I. Importance of the virus in different respiratory tract disease syndromes and temporal distribution of infection. Am. J. Epidemiol. 98:216-225. [DOI] [PubMed] [Google Scholar]
- 33.Kreiselmeier, N. E., N. C. Kraynack, D. A. Corey, and T. J. Kelley. 2003. Statin-mediated correction of STAT1 signaling and inducible nitric oxide synthase expression in cystic fibrosis epithelial cells. Am. J. Physiol. Lung Cell. Mol. Physiol. 285:L1286-L1295. [DOI] [PubMed] [Google Scholar]
- 34.Lahti, M., J. Lofgren, R. Marttila, M. Renko, T. Klaavuniemi, R. Haataja, M. Ramet, and M. Hallman. 2002. Surfactant protein D gene polymorphism associated with severe respiratory syncytial virus infection. Pediatr. Res. 51:696-699. [DOI] [PubMed] [Google Scholar]
- 35.Lander, E., and L. Kruglyak. 1995. Genetic dissection of complex traits: guidelines for interpreting and reporting linkage results. Nat. Genet. 11:241-247. [DOI] [PubMed] [Google Scholar]
- 36.Lofgren, J., M. Ramet, M. Renko, R. Marttila, and M. Hallman. 2002. Association between surfactant protein A gene locus and severe respiratory syncytial virus infection in infants. J. Infect. Dis. 185:283-289. [DOI] [PubMed] [Google Scholar]
- 37.Lukacs, N. W., M. L. Moore, B. D. Rudd, A. A. Berlin, R. D. Collins, S. J. Olson, S. B. Ho, and R. S. Peebles, Jr. 2006. Differential immune responses and pulmonary pathophysiology are induced by two different strains of respiratory syncytial virus. Am. J. Pathol. 169:977-986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Manly, K. F., R. H. Cudmore, Jr., and J. M. Meer. 2001. Map Manager QTX, cross-platform software for genetic mapping. Mamm. Genome 12:930-932. [DOI] [PubMed] [Google Scholar]
- 39.McCurdy, L. H., and B. S. Graham. 2003. Role of plasma membrane lipid microdomains in respiratory syncytial virus filament formation. J. Virol. 77:1747-1756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.McDowell, S. A., K. Gammon, C. J. Bachurski, J. S. Wiest, J. E. Leikauf, D. R. Prows, and G. D. Leikauf. 2000. Differential gene expression in the initiation and progression of nickel-induced acute lung injury. Am. J. Respir. Cell Mol. Biol. 23:466-474. [DOI] [PubMed] [Google Scholar]
- 41.Mejias, A., S. Chavez-Bueno, A. M. Rios, J. Saavedra-Lozano, M. Fonseca Aten, J. Hatfield, P. Kapur, A. M. Gomez, H. S. Jafri, and O. Ramilo. 2004. Anti-respiratory syncytial virus (RSV) neutralizing antibody decreases lung inflammation, airway obstruction, and airway hyperresponsiveness in a murine RSV model. Antimicrob. Agents Chemother. 48:1811-1822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mohapatra, S. S., and S. Boyapalle. 2008. Epidemiologic, experimental, and clinical links between respiratory syncytial virus infection and asthma. Clin. Microbiol. Rev. 21:495-504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Parnes, C., J. Guillermin, R. Habersang, P. Nicholes, V. Chawla, T. Kelly, J. Fishbein, P. McRae, M. Goessler, A. Gatti, J. A. Calcagno, C. Eki, K. A. Harris, J. Joyave, K. McFarland, P. Protter, M. Sullivan, A. Stanford, N. Lovett, M. Ortiz, S. Rojas, S. Cyrus, J. Cyrus, S. Cohen, D. Buchin, L. Riordan, M. Zuniga, R. Shah, C. Minard, A. Quintin, G. Douglas, J. van Houten, S. Freutner, S. Chartrand, P. Nowatzke, J. Romero, T. Rhodes, M. Benoit, E. Walter, L. Walker, L. DeBonnett, M. Cross, T. Free, S. Martin, K. Shank, B. Guedes, L. A. Atkinson, G. J. Halpin, K. Rouse, I. Hand, D. Geiss, J. R. Marshall, L. Burleson, J. Boland, K. Seybold, V. Hunter, S. Unfer, J. Schmucker, M. Gley, M. Marcus, P. Thompson, P. Milla, C. Young, R. Zanni, V. Zinno, A. Fetter-Zarzeka, A. Busey, M. A. Sokunbi, S. Airington, N. Richard, V. Muraligopal, S. Lewis, F. T. Weber, B. P. Giordano, D. Linehan, J. Roach, R. Davis, A. A. Rzepka, T. Booth, D. Smeltzer, J. Walsh, E. Arispe, R. Rowley, C. Bolling, T. Botts, K. Haskett, D. Raby, E. Batiz, A. Gelfand, L. Farrell, S. Butler, L. Colby, P. Schochet, J. Bentler, D. Hirsch, L. Wilkinson, A. Aaronson, E. Bennett, J. Wingate, D. Quinn, et al. 2003. Palivizumab prophylaxis of respiratory syncytial virus disease in 2000-2001: results from The Palivizumab Outcomes Registry. Pediatr. Pulmonol. 35:484-489. [DOI] [PubMed] [Google Scholar]
- 44.Pastey, M. K., T. L. Gower, P. W. Spearman, J. E. Crowe, Jr., and B. S. Graham. 2000. A RhoA-derived peptide inhibits syncytium formation induced by respiratory syncytial virus and parainfluenza virus type 3. Nat. Med. 6:35-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Perez-Yarza, E. G., A. Moreno, P. Lazaro, A. Mejias, and O. Ramilo. 2007. The association between respiratory syncytial virus infection and the development of childhood asthma: a systematic review of the literature. Pediatr. Infect. Dis. J. 26:733-739. [DOI] [PubMed] [Google Scholar]
- 46.Petersen, N. T., N. Hoiby, C. H. Mordhorst, K. Lind, E. W. Flensborg, and B. Bruun. 1981. Respiratory infections in cystic fibrosis patients caused by virus, chlamydia and mycoplasma—possible synergism with Pseudomonas aeruginosa. Acta Paediatr. Scand. 70:623-628. [DOI] [PubMed] [Google Scholar]
- 47.Prince, G. A., R. L. Horswood, J. Berndt, S. C. Suffin, and R. M. Chanock. 1979. Respiratory syncytial virus infection in inbred mice. Infect. Immun. 26:764-766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Prows, D. R., H. G. Shertzer, M. J. Daly, C. L. Sidman, and G. D. Leikauf. 1997. Genetic analysis of ozone-induced acute lung injury in sensitive and resistant strains of mice. Nat. Genet. 17:471-474. [DOI] [PubMed] [Google Scholar]
- 49.Puthothu, B., J. Forster, J. Heinze, A. Heinzmann, and M. Krueger. 2007. Surfactant protein B polymorphisms are associated with severe respiratory syncytial virus infection, but not with asthma. BMC Pulm. Med. 7:6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Puthothu, B., M. Krueger, J. Forster, J. Heinze, M. Weckmann, and A. Heinzmann. 2007. Interleukin (IL)-18 polymorphism 133C/G is associated with severe respiratory syncytial virus infection. Pediatr. Infect. Dis. J. 26:1094-1098. [DOI] [PubMed] [Google Scholar]
- 51.Sartor, M., J. Schwanekamp, D. Halbleib, I. Mohamed, S. Karyala, M. Medvedovic, and C. R. Tomlinson. 2004. Microarray results improve significantly as hybridization approaches equilibrium. Biotechniques 36:790-796. [DOI] [PubMed] [Google Scholar]
- 52.Sartor, M. A., C. R. Tomlinson, S. C. Wesselkamper, S. Sivaganesan, G. D. Leikauf, and M. Medvedovic. 2006. Intensity-based hierarchical Bayes method improves testing for differentially expressed genes in microarray experiments. BMC Bioinform. 7:538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Seaton, G., C. S. Haley, S. A. Knott, M. Kearsey, and P. M. Visscher. 2002. QTL Express: mapping quantitative trait loci in simple and complex pedigrees. Bioinformatics 18:339-340. [DOI] [PubMed] [Google Scholar]
- 54.Shay, D. K., R. C. Holman, R. D. Newman, L. L. Liu, J. W. Stout, and L. J. Anderson. 1999. Bronchiolitis-associated hospitalizations among US children, 1980-1996. JAMA 282:1440-1446. [DOI] [PubMed] [Google Scholar]
- 55.Smyth, G. K. 2004. Linear models and empirical Bayes methods for assessing differential expression in microarray experiments. Stat. Appl. Genet. Mol. Biol. 3:Article3. [DOI] [PubMed] [Google Scholar]
- 56.Stark, J. M., S. A. McDowell, V. Koenigsknecht, D. R. Prows, J. E. Leikauf, A. M. Le Vine, and G. D. Leikauf. 2002. Genetic susceptibility to respiratory syncytial virus infection in inbred mice. J. Med. Virol. 67:92-100. [DOI] [PubMed] [Google Scholar]
- 57.Tang, Y. W., and B. S. Graham. 1997. T cell source of type 1 cytokines determines illness patterns in respiratory syncytial virus-infected mice. J. Clin. Invest. 99:2183-2191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Taylor, G., E. J. Stott, M. Hughes, and A. P. Collins. 1984. Respiratory syncytial virus infection in mice. Infect. Immun. 43:649-655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Thompson, W. W., D. K. Shay, E. Weintraub, L. Brammer, N. Cox, L. J. Anderson, and K. Fukuda. 2003. Mortality associated with influenza and respiratory syncytial virus in the United States. JAMA 289:179-186. [DOI] [PubMed] [Google Scholar]
- 60.Venkatakrishnan, A., A. A. Stecenko, G. King, T. R. Blackwell, K. L. Brigham, J. W. Christman, and T. S. Blackwell. 2000. Exaggerated activation of nuclear factor-kappaB and altered IkappaB-beta processing in cystic fibrosis bronchial epithelial cells. Am. J. Respir. Cell Mol. Biol. 23:396-403. [DOI] [PubMed] [Google Scholar]
- 61.Weber, A. J., G. Soong, R. Bryan, S. Saba, and A. Prince. 2001. Activation of NF-kappaB in airway epithelial cells is dependent on CFTR trafficking and Cl− channel function. Am. J. Physiol. Lung Cell. Mol. Physiol. 281:L71-L78. [DOI] [PubMed] [Google Scholar]
- 62.Welliver, R. C. 2003. Review of epidemiology and clinical risk factors for severe respiratory syncytial virus (RSV) infection. J. Pediatr. 143:S112-S117. [DOI] [PubMed] [Google Scholar]
- 63.Wesselkamper, S. C., L. M. Case, L. N. Henning, M. T. Borchers, J. W. Tichelaar, J. M. Mason, N. Dragin, M. Medvedovic, M. A. Sartor, C. R. Tomlinson, and G. D. Leikauf. 2005. Gene expression changes during the development of acute lung injury: role of transforming growth factor beta. Am. J. Respir. Crit. Care Med. 172:1399-1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhang, B., D. Schmoyer, S. Kirov, and J. Snoddy. 2004. GOTree Machine (GOTM): a web-based platform for interpreting sets of interesting genes using Gene Ontology hierarchies. BMC Bioinform. 5:16. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.