Abstract
The detection of viroid-derived small RNAs (vd-sRNAs) similar to the small interfering RNAs (siRNAs, 21 to 24 nucleotides [nt]) in plants infected by nuclear-replicating members of the family Pospiviroidae (type species, Potato spindle tuber viroid [PSTVd]) indicates that they are inducers and targets of the RNA-silencing machinery of their hosts. RNA-dependent RNA polymerase 6 (RDR6) catalyzes an amplification circuit producing the double-stranded precursors of secondary siRNAs. Recently, the role of RDR6 in restricting systemic spread of certain RNA viruses and precluding their invasion of the apical growing tip has been documented using RDR6-silenced Nicotiana benthamiana (NbRDR6i) plants. Here we show that RDR6 is also engaged in regulating PSTVd levels: accumulation of PSTVd genomic RNA was increased in NbRDR6i plants with respect to the wild-type controls (Nbwt) early in infection, whereas this difference decreased or disappeared in later infection stages. Moreover, in situ hybridization revealed that RDR6 is involved in restricting PSTVd access in floral and vegetative meristems, thus providing firm genetic evidence for an antiviroid RNA silencing mechanism. RNA gel blot hybridization and deep sequencing showed in wt and RDR6i backgrounds that PSTVd sRNAs (i) accumulate to levels paralleling their genomic RNA, (ii) display similar patterns with prevailing 22- or 21-nt plus-strand species, and (iii) adopt strand-specific hot spot profiles along the genomic RNA. Therefore, the surveillance mechanism restraining entry of some RNA viruses into meristems likely also controls PSTVd access in N. benthamiana. Unexpectedly, deep sequencing also disclosed in NbRDR6i plants a profile of RDR6-derived siRNA dominated by 21-nt plus-strand species mapping within a narrow window of the hairpin RNA stem expressed transgenically for silencing RDR6, indicating that minus-strand siRNAs silencing the NbRDR6 mRNA represent a minor fraction of the total siRNA population.
Viroids are a unique class of replicons that in their minimal genome—composed exclusively of a circular, non-protein-coding single-stranded RNA (ssRNA) of only 246 to 401 nucleotides (nt), which folds into a compact secondary structure—store enough genetic information to initiate the infection of certain plants, to usurp their gene expression machinery for producing progenies that spread systemically, and to induce frequently specific diseases (14, 15, 16, 22, 23, 81). The available evidence indicates that viroids replicate in the nucleus (family Pospiviroidae) or in the chloroplast (family Avsunviroidae) through a rolling-circle mechanism involving RNA intermediates, some of which form double-stranded-RNA (dsRNA) complexes (3, 4, 9, 21, 53). In the type species of the family Pospiviroidae, Potato spindle tuber viroid (PSTVd) (55), the infecting monomeric circular RNA, the mc plus strand (Fig. 1), is reiteratively transcribed into oligomeric minus-strand RNA intermediates that are then transcribed into oligomeric plus-strand RNAs and finally cleaved and ligated into the mc plus-strand forms (3).
FIG. 1.
Primary and rod-like secondary structure of minimal free energy predicted for the plus strand of PSTVd variant NB (61, 64). The relative positions of the terminal left (TL), pathogenic (P), central (C), variable (V) and terminal right (TR) domains (39) are indicated. The reference PSTVd intermediate variant (NC_002030) shows the following nucleotidic changes with respect to the reference PSTVd NB variant: C46→G, U47→C, U201→G, U259→C, A313→U, and C317→U.
Most eukaryotes, and particularly plants, have evolved a gene-silencing defensive response against viruses based on the detection of viral dsRNAs or highly structured ssRNAs that are processed by one or more members (Dicer-like [DCL]) of the RNase III family into the so-called small interfering RNAs (siRNAs) of 21 to 24 nt (2, 30, 60). The siRNAs are subsequently incorporated into the RNA-induced silencing complex (RISC) and guide its Argonaute (AGO) core, an RNase H-like enzyme, for degradation or translation arrest of their cognate RNAs (31, 54, 58). In addition to the primary siRNAs directly derived from the trigger, RNA silencing is maintained by one or more cellular RNA-dependent RNA polymerases (RDRs) which, primed by these siRNAs or just sensing aberrant features in certain RNAs, use them as template for the synthesis of dsRNAs that are DCL-processed to secondary siRNAs.
Distinct plant RDRs have been implicated in virus defense (6, 13, 51, 83), prominent among which are RDR1 and, particularly, RDR6. Also related to antiviral defense in plants, and to systemic silencing in transgenic plants, is a yet-uncharacterized RNA-silencing signal that spreads via the plasmodesmata and the phloem (20, 38, 40, 57, 80, 84). RDR6 has been shown to be involved in long-distance silencing through the vasculature and in long-range cell-to-cell signaling but not in short-range cell-to-cell signaling (10 to 15 cells), and recent data indicate that this enzyme is required for maintenance but not for initiation of posttranscriptional gene silencing (83). To counteract antiviral RNA silencing, most plant viruses encode protein suppressors that may stimulate virus accumulation in the initially infected cells, promote intercellular movement within the inoculated leaves, assist virus spread through the phloem, and exacerbate symptoms in systemically infected leaves (12).
Downregulation of RDR6 in Nicotiana benthamiana through RNA interference (NbRDR6i plants) induces hypersusceptibility to certain viruses like Potato virus X (PVX), with hypersusceptibility being coupled with enhanced viral invasion of the growing tip (73). Furthermore, RDR6 is required not for production or movement of the silencing signal but rather for synthesis of dsRNA precursors of the secondary siRNAs, which mediate antiviral RNA silencing and preclude invasion of the shoot apical meristem (SAM) by PVX and perhaps other viruses (73). On the other hand, the efficiencies of virus-induced gene silencing and RNA-directed DNA methylation in the same NbRDR6i transgenic line are compromised for PVX and a potyvirus despite accumulating high levels of viral siRNAs; this reduced efficiency is unrelated to the siRNA size and (for PVX) independent of the virus-encoded silencing suppressor, suggesting that viral primary siRNAs produced in the absence of RDR6 may not be good silencing effectors and that RDR6 is required to generate secondary siRNAs that drive a more effective antiviral response (82). In an independent work with another NbRDR6i transgenic line (66), RDR6 was shown to be involved in protecting differentiated and apical plant tissues from invasion by distinct RNA plant viruses, including PVX. The effects of reducing the expression of RDR6 are stimulated by rising the temperature but lessened by virus-encoded silencing suppressors, with the final outcome depending on the relative intensity of these two factors (66).
Infection by viroids from both families occurs with the accumulation of 21- to 24-nt viroid-derived small RNAs (vd-sRNAs) with the characteristic properties of the siRNAs associated with RNA silencing, as revealed by RNA gel blot hybridization (5, 36, 43, 46, 47, 59, 74), low-scale sequencing (35, 41, 44, 77), and deep sequencing (17, 52). Moreover, mechanically codelivered or agroinoculated dsRNAs interfere with viroid infection in a sequence-specific manner (as also do, at least in one case, in vitro-generated homologous vd-sRNAs codelivered mechanically) (5). Altogether, these results strongly suggest that viroids also induce the RNA-silencing machinery of their hosts and, considering the sequence-specific nature of the observed effects, that these replicons are targeted by RISC, which provides sequence specificity to the RNA-silencing machinery. Regarding their origin, primary vd-sRNAs may result from DCL-mediated processing of the highly structured genomic ssRNAs and dsRNAs arising from replication by a host DNA-dependent RNA polymerase forced to transcribe RNA templates, with secondary RNAs arising from dsRNAs generated by an RDR(s) in concert with a DCL(s). Therefore, in contrast with virus dsRNAs, a fraction of which are produced by virus-encoded polymerases, viroid dsRNAs are exclusively produced by host-encoded enzymes.
In situ hybridization of sections of PSTVd-infected N. benthamiana and tomato has revealed the presence of the viroid in the vascular tissues and in other cells in the subapical region but not in the SAM (63, 90). These observations suggest that the surveillance mechanism that restricts the entry of certain RNA viruses into the SAM (24) also limits the PSTVd access to this plant compartment. Here we report evidence indicating that RDR6 is engaged in this mechanism. Specifically, accumulation of the PSTVd genomic RNA and vd-sRNAs was increased in leaves of NbRDR6i plants with respect to their wild-type counterparts (Nbwt) during the initial infection stages, whereas the difference decreased or disappeared in later infection stages. Moreover, in situ hybridization showed that PSTVd invades floral and vegetative meristems of NbRDR6i plants. These results have been complemented with deep sequencing examination of the vd-sRNAs from Nbwt and NbRDR6i plants in an attempt to define their role, as well as that of the RDR6-derived siRNAs in NbRDR6i plants.
MATERIALS AND METHODS
Plant material and viroid source.
Apical leaves and shoot and floral apices were from Nicotiana benthamiana plants grown from seeds of the wild type and of a transgenic line in which RDR6 was silenced by RNA interference (RNAi). Plants, maintained in a glasshouse (25 to 23°C day and 20 to 18°C night), were mock or viroid inoculated in the first true leaves with an RNA preparation from tissue infected by PSTVd (NB strain; GenBank accession number AJ634596.1).
RNA extraction, fractionation, and RNA gel blot hybridization.
Total nucleic acid preparations, extracted from apical leaves with phenol-chloroform and recovered by ethanol precipitation (7), were fractionated by denaturing PAGE on either 5% gels (for mc and ml PSTVd RNAs) or 17% gels (for plant and vd-sRNAs), stained with ethidium bromide, and transferred to Hybond-N+ (Roche Diagnostics) membranes that were hybridized with radioactively labeled full-length riboprobes for detecting PSTVd plus and minus strands, and with a 501-nt riboprobe (from positions 2279 to 2780; GenBank accession number AY722008) for detecting NBRDR6-sRNAs. Riboprobes were obtained by in vitro transcription under the control of the promoter of the T7 RNA polymerase. Hybridization was at 70°C in the presence of 50% formamide (for mc and ml PSTVd RNAs) or at 35°C with PerfectHyb Plus hybridization buffer (Sigma) for the sRNAs. Equal loading was confirmed by the fluorescence emitted by the 5S rRNA after ethidium bromide staining and UV irradiation.
In situ hybridization.
Longitudinal sections of shoot and floral apices collected 30 to 35 dpi were fixed with p-formaldehyde, dehydrated, embedded in paraffin, and sectioned to 7 mm, and in situ hybridization was performed according to reference 37 with some modifications (69). In brief, digoxigenin (DIG)-labeled riboprobes were added to the hybridization solution containing 50% formamide, and the hybridization and the three subsequent washes were performed at 70°C. After treatment for 1 h with blocking solution, the sections were incubated with alkaline phosphatase-conjugated anti-DIG antibody, and following three final washes, the alkaline phosphatase was detected by the nitroblue tetrazolium and 5-bromo-4-chloro-3-indolyl phosphate procedure according to the supplier (Roche Diagnostics). A bright-field microscope (E600; Nikon) was used for sample visualization and photography.
Deep sequencing of host sRNAs and vd-sRNAs.
sRNAs were eluted from the denaturing polyacrylamide gels using as size markers oligonucleotides of 20 to 30 nt, recovered by ethanol precipitation, and subjected to (i) single-strand ligation of the 3′ adapter, (ii) single-stranded ligation of the 5′ adapter, (iii) polyacrylamide gel electrophoresis (PAGE) purification to select the 65- to 80-nt fragments, (iv) reverse transcription and PCR amplification to generate the DNA colony template library, (v) PAGE purification, (vi) quality control of the DNA colony template library by cloning an aliquot into a TOPO plasmid and capillary sequencing of a few clones, (vii) library purification, estimation of the concentration by fluorescence, and dilution at 10 nM, (viii) flow cell preparation on the cluster station, and (ix) high-throughput DNA sequencing on an Illumina EAS269-GAII genome analyzer (Fasteris SA, Plan-les-Ouates, Switzerland). The four bar-coded samples were analyzed in a single read channel.
Sequence analysis of the sRNAs.
Data were analyzed on the GA Pipeline to convert images into sequences, without mapping of the reads, using ELAND. The resulting sequences were examined for the presence of the adapters, and after their trimming, they were sorted into separate files according to their length. For further analysis, the sequences between 20 and 24 nt were pooled, and each set of sequences was analyzed by BLAST (1) against the nucleotide sequence of the PSTVd NB strain and the RDR6 of N. benthamiana. No mismatch was allowed, and the circularity of the viroid genome was taken into consideration. A set of Perl scripts was developed to filter, analyze, and visualize the mapping data, searching for specific distribution patterns and phasing.
RESULTS
Accumulation of the PSTVd genomic RNA is increased in leaves of NbRDR6i plants.
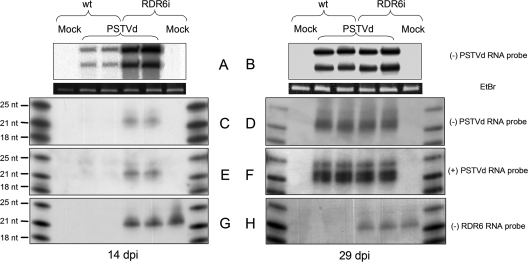
To investigate the potential role of RDR6 in defense against PSTVd, we first determined the relative accumulation of the monomeric circular (mc) and linear (ml) viroid plus-strand RNAs in wild-type N. benthamiana (Nbwt plants) and in a transgenic line in which RDR6 is silenced by RNA interference with a hairpin construct (NbRDR6i plants) (73). Quantitative reverse transcription PCR (RT-PCR) analyses have shown that the NbRDR6 transcript levels in this line are reduced to 4% of the level in a line carrying an unrelated RNAi construct, whereas expression of the NbRDR1 and NbRDR2 are essentially unaffected (73). Furthermore, by denaturing PAGE and RNA gel blot hybridization with an NbRDR6-specific riboprobe, we detected the presence of RDR6-siRNAs of 21 nt in the NbRDR6i plants but not in their Nbwt counterparts (Fig. 2G and H), thus showing that the hairpin transcript was targeted by the RNA-silencing machinery (see below for further evidence supporting this view). However, parallel attempts by agarose gel electrophoresis and RNA gel blot hybridization failed to reveal the NbRDR6 transcript in NbRDR6i and Nbwt plants (data not shown), indicating low steady-state levels of this transcript not only in the former but also in the latter, in contrast with a previous report in which a strong hybridization signal was detected in Nbwt plants (66). We do not know the reasons for this discrepancy, but, in support of our claim, there are indications that RDR6 is expressed poorly in plants (86) and, specifically, in Nbwt plants (J. Burgyan, personal communication).
FIG. 2.
Accumulation of PSTVd RNAs in Nbwt and NbRDR6i plants at early and late infection stages. (A to H) Total RNAs were separated by denaturing PAGE in 5% gels (A and B) and 17% gels (C to H) and analyzed by RNA gel blot hybridization with strand-specific PSTVd riboprobes (A to F) or with a riboprobe for detecting NbRDR6 sequences (G and H). Loading was equalized by the intensity of 5S RNA. The upper and lower bands in panels A and B correspond to the monomeric circular and linear viroid plus-strand RNAs. The sizes of the RNA markers (which were radioactively labeled at their 5′ termini) are on the left, and the specificity and polarity of the riboprobes are on the right. The 21-nt marker was composed of ribo- and deoxyribonucleotides and, therefore, migrated faster than expected for a marker of the same size composed of only ribonucleotides.
NbRDR6i and Nbwt plants were mechanically inoculated with a nucleic acid preparation from tissue infected by PSTVd (NB variant) (Fig. 1). Analysis by denaturing PAGE and RNA gel blot hybridization with a riboprobe for detecting PSTVd plus strands revealed that the PSTVd mc and ml plus-strand RNAs accumulated in apical noninoculated leaves, collected 14 days postinoculation (dpi), at a significantly higher level in NbRDR6i than in Nbwt plants (Fig. 2A), suggesting that RDR6 is involved in a defensive pathway restricting early systemic invasion of the viroid. However, the difference disappeared when the apical leaves were collected 29 dpi, indicating that the viroid overcomes the defensive pathway in later infection stages (Fig. 2B). The experiment was repeated three times, using five Nbwt and five NbRDR6i plants per experiment, with similar results.
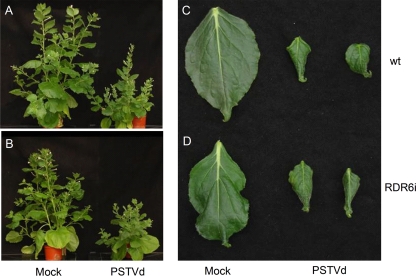
NbRDR6i and Nbwt plants infected with the PSTVd-NB variant showed stunting and curled leaves of reduced size with respect to their mock-inoculated controls, resembling the symptoms observed previously in Nbwt plants infected by the severe PSTVd-A3 strain (47). On the other hand, phenotypic differences between mock-inoculated NbRDR6i and Nbwt plants were undetectable or limited to a mild stunting of the former (Fig. 3). These results differ from those of a recent report in which Nbwt plants infected with Hop stunt viroid (HSVd) showed moderate stunting compared with their noninoculated controls, whereas HSVd-infected NbRDR6i plants did not (28); therefore, in contrast with the HSVd situation, we have not observed that PSTVd-induced symptoms are dependent on RDR6 activity.
FIG. 3.
PSTVd-induced phenotypes in Nbwt and NbRDR6i plants. (A to D) Stunting (A and B) and reduced leaf size (C and D) displayed by Nbwt (A and C) and NbRDR6i (B and D) plants inoculated with an in vitro transcript of the PSTVd-NB strain with respect to their mock-inoculated controls. Pictures were taken 48 dpi.
PSTVd sRNAs levels parallel those of their genomic RNAs in Nbwt and NbRDR6i plants.
To get a deeper insight of the underlying mechanism, we also examined the accumulation pattern of the vd-sRNAs by denaturing PAGE and RNA gel blot hybridization with radioactive riboprobes for detecting plus and minus PSTVd strands. No qualitative (size) differences could be discerned between those from leaves of NbRDR6i and Nbwt plants: in line with previous results (5, 36, 41, 59, 74), at least two populations of 21- or 22-nt and 24-nt vd-sRNAs were detected with both riboprobes, with smaller ones being predominant (Fig. 2C to F). From a quantitative standpoint, accumulation of the vd-sRNAs always paralleled that of the genomic PSTVd mc and ml plus-strand RNAs: they were less abundant in Nbwt than in NbRDR6i leaves collected 14 dpi (Fig. 2, compare panels C and E with A), as well as in leaves collected 14 dpi with respect to those collected 29 dpi (Fig. 2, compare panel C with panel D and panel E with panel F).
This apparent correlation between the levels of the PSTVd genomic and vd-sRNAs in both NbRDR6i and Nbwt plants reinforces the idea that most of the latter are vd-RNAs from Dicer-mediated processing of the viroid genomic RNAs or of some dsRNAs resulting from replication or from the activity of a RDR distinct from RDR6. However, the enhanced accumulation of the genomic PSTVd mc and ml plus-strand RNAs in early infection stages of NbRDR6i plants supports the view that a quantitatively minor population of RDR6-derived secondary vd-sRNAs loads RISC and temporarily attenuates buildup of the viroid progeny in Nbwt plants. Eventually, active viroid replication and accumulation overcome this barrier, and most of the vd-sRNAs result from DCL-mediated processing of the genomic RNAs or of some dsRNAs.
PSTVd invades meristems of NbRDR6i plants.
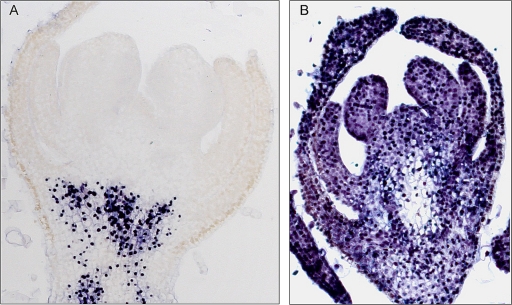
To explore the possibility that RDR6 might have some role in restricting PSTVd invasion of floral and vegetative meristems, longitudinal sections of flower and shoot apices from PSTVd-infected NbRDR6i and Nbwt plants, as well as from mock-inoculated controls, were analyzed by in situ hybridization with strand-specific digoxigenin-labeled PSTVd riboprobes. To prevent cross-hybridization between the strand-specific riboprobes and viroid RNAs of the same polarity—a side effect of the high self-complementarity of viroid RNAs—hybridizations were performed at 70°C in the presence of 50% formamide, conditions that have been previously shown to minimize the problem (69). PSTVd plus strands were detected in developing flower organs of NbRDR6i plants (Fig. 4B) but not in the corresponding organs of their Nbwt counterparts (Fig. 4A). The intense hybridization signals are the result of the nuclear (nucleolar) localization of the most abundant PSTVd mc plus-strand RNA (32, 62). Results were less clear with the probe for detecting PSTVd minus strands (data not shown), most likely because of their lower accumulation in vivo with respect to the PSTVd plus strands (3, 21).
FIG. 4.
RDR6 Prevents PSTVd invasion of developing flowers in N. benthamiana. In situ hybridizations with a digoxigenin-labeled riboprobe specific for plus PSTVd strands show that the viroid does not invade the floral meristem of an Nbwt plant (A) in contrast with the complete invasion of the floral meristem of an NbRDR6i plant (B). The concentrated hybridization signals are the result of the nuclear localization of PSTVd.
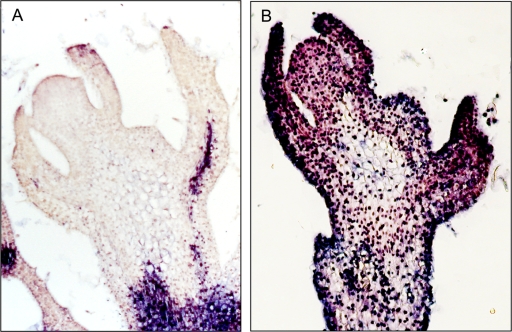
In line with these observations, PSTVd plus strands were also detected in the SAM of NbRDR6i plants (Fig. 5B) but not in Nbwt controls (Fig. 5A). Judging from the intensity of the hybridization signals in NbRDR6i plants, PSTVd penetrated partially into some vegetative meristems (see Fig. S1 in the supplemental material), while in others the viroid reached the upper layers of the SAM (Fig. 5B). In some experiments, vegetative meristems of NbRDR6i plants failed to show detectable hybridization signals, suggesting the involvement of additional factors presumably related to development; this assumption is supported by previous results showing that another type of PSTVd trafficking, specifically, from bundle sheath to mesophyll, is under developmental control (61). No hybridization signals were observed in vegetative and floral meristems from mock-inoculated controls.
FIG. 5.
RDR6 prevents SAM invasion of PSTVd in N. benthamiana. In situ hybridizations with a digoxigenin-labeled riboprobe specific for plus PSTVd strands show that the viroid does not invade the SAM of an Nbwt plant (A), in contrast with the complete SAM invasion in an NbRDR6i plant (B). The concentrated hybridization signals are the result of the nuclear localization of PSTVd.
These findings (i) confirm and extend previous in situ hybridization studies in which PSTVd was not detected either in developing or mature flower organs (except in sepals) or in the SAM of Nbwt and tomato plants (63, 89, 90), although viroid levels below the detection limit of the technique cannot be excluded, (ii) suggest that RNA trafficking into sink organs is differentially regulated, in line with previous results (89, 90), and that PSTVd access into floral and vegetative meristems is restricted by a surveillance system in which RDR6 is involved, and (iii) show that PSTVd invasion of the SAM (in NbRDR6i plants) is not associated with symptom exacerbation. The possibility that meristem invasion could be a transient phenomenon, directly or indirectly linked with the early increased PSTVd accumulation in vegetative tissues of NbRDR6i plants, seems unlikely because samples for in situ hybridization were taken 30 to 35 dpi, when the viroid had reached similar accumulation in Nbwt and NbRDR6i plants (Fig. 2A and B).
Deep sequencing reveals multiple PSTVd sRNAs in Nbwt and NbRDR6i plants.
To get a better understanding of the role of RDR6, we performed a deep sequencing study of four preparations of gel-purified sRNAs from young expanding leaves from mock-inoculated or PSTVd-infected Nbwt and NbRDR6i plants; leaves were collected 14 days postinoculation (dpi), when RNA gel blot hybridization revealed conspicuous differences in the genomic and small viroid RNAs between Nbwt and NbRDR6i plants (Fig. 2A to F). Each sRNA preparation was linked to a bar-coded adapter in order to sequence the four libraries in the same channel, thus avoiding any undesired bias and obtaining four independent datasets that could be further compared. Adapters were synthesized assuming that the vd-sRNAs have the characteristic 5′ phosphomonoester and 3′ hydroxyl termini generated by DCLs (42, 60). Although this is the prevailing view for plant siRNAs (82), with some data from vd-sRNAs supporting it (44), the possibility that a minor fraction of the vd-sRNAs may have other 5′ termini—as recently reported for secondary siRNAs in Caenorhabditis elegans (56, 75)—cannot be dismissed.
Of approximately 7,710,000 reads obtained by the high-throughput sequencing, 97.6% were clearly attributable to the four bar-coded samples (the fraction of each sample in the pooled data set varied from 23 to 26%, with an average of 24.4% and a standard deviation of 1.2%) and adopted a profile with two prominent 21- and 24-nt peaks (excluding the adapters and the file with empty inserts) (see Table S1 in the supplemental material). Intriguingly, while the 21-nt RNAs dominate in the two NbRDR6i samples, the 24-nt RNAs are the most prevalent in the two Nbwt samples. In contrast, viroid infection has minor effects in the sRNA populations (see Fig. S2 in the supplemental material). A comprehensive account of the characteristics of the sRNAs from Nbwt and NbRDR6i plants will be reported elsewhere.
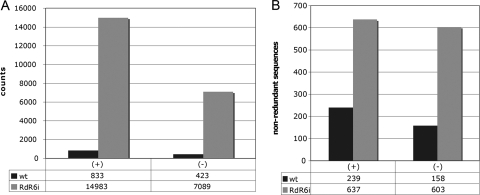
Searching the reads of the infected Nbwt and NbRDR6i samples (1,866,590 and 2,047,373, respectively) for PSTVd sRNAs of 20 to 24 nt matching perfectly the parental sequence resulted in 1,256 and 22,072 counts, respectively (Fig. 6). This difference most likely reflected the initial PSTVd sRNA concentration, which was lower in the Nbwt preparation than in the NbRDR6i one, as revealed by RNA gel blot hybridization with PSTVd-specific riboprobes (Fig. 2C to F).
FIG. 6.
Multiple PSTVd sRNAs in Nbwt and NbRDR6i plants. The histograms compare the total counts (A) and number (B) of nonredundant (unique) plus- and minus-strand PSTVd sRNAs (20 to 24 nt) obtained by deep sequencing from PSTVd-infected Nbwt and NbRDR6i samples.
Similar patterns with prevailing 22- and 21-nt plus-strand PSTVd sRNA in Nbwt and NbRDR6i plants.
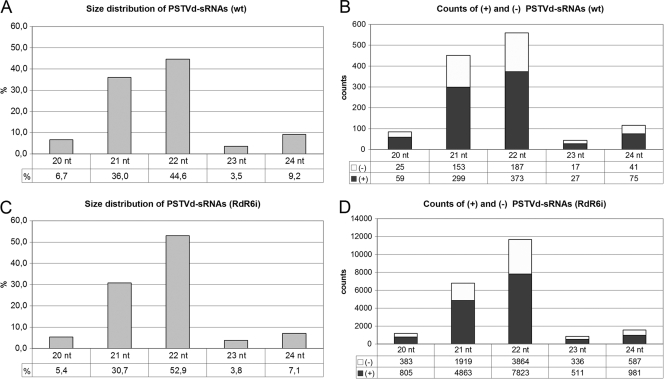
Examination of the counts corresponding to PSTVd sRNAs revealed in the Nbwt sample a distribution in which approximately 45% were of 22 nt and 36% of 21 nt, with significantly lower proportions of RNAs of 24 nt (9%), 20 nt (7%), and 23 nt (3.5%) (Fig. 7). A parallel analysis of the NbRDR6i sample showed an essentially similar size distribution (Fig. 7), suggesting that the PSTVd sRNAs from both samples are generated by the same DCLs.
FIG. 7.
Similar size distribution of PSTVd sRNAs in Nbwt and NbRDR6i plants. The histograms compare in Nbwt (A and B) and NbRDR6i (C and D) the size distribution of 20- to 24-nt PSTVd sRNA counts (A and C) and the total counts of plus- and minus-strand PSTVd sRNAs (B and D).
With regard to polarity, about 65 and 35% of the total PSTVd sRNA counts in the Nbwt sample were of plus and minus polarity, respectively, with a similar ratio being observed for the counts corresponding to the predominant 22- and 21-nt components (Fig. 7). Data derived from the NbRDR6i sample were comparable (Fig. 7), indicating that the absence of RDR6 does not affect the balance between the accumulating plus- and minus-strand PSTVd sRNAs. This moderate excess of the plus-strand PSTVd sRNAs is far below that of the sum of the unit-length and longer-than-unit plus PSTVd strands with respect to the sum of their minus-strand counterparts (3, 4), arguing against a direct precursor-product relationship between the PSTVd sRNAs and the genomic (and multimeric) PSTVd RNAs.
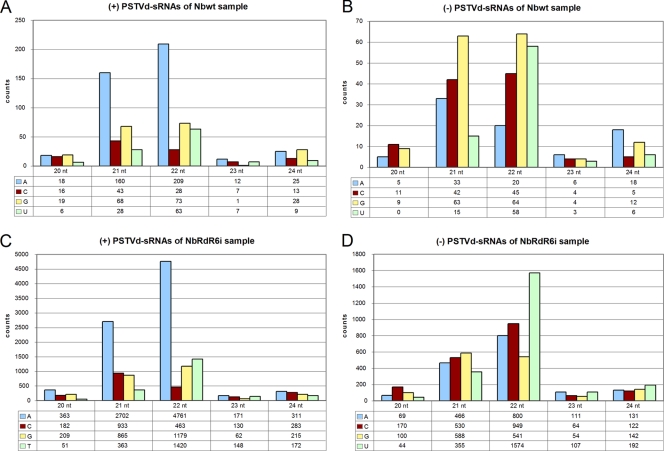
PSTVd sRNAs of 21 and 22 nt differ in their 5′ nucleotides.
Analysis of the 5′-terminal position of the most abundant PSTVd sRNAs revealed an intriguing bias: in both Nbwt and NbRDR6i samples, the 21- and 22-nt species of plus polarity present a dominant A, while in the minus polarity the 21-nt species has a dominant G and the 22-nt species a dominant or codominant U (Fig. 8). The 5′-terminal position of PSTVd sRNAs thus depends on the polarity rather than on the size; RDR6 silencing affected particularly the profile of the minus-strand 22-nt PSTVd sRNAs, G being the most (34%) and least (14%) prevalent in Nbwt and NbRDR6i plants, respectively.
FIG. 8.
Biased distribution of the 5′ nucleotide in plus- and minus-strand PSTVd sRNAs. The histograms compare in Nbwt (A and B) and NbRDR6i (C and D) the total counts corresponding to PSTVd sRNAs (20 to 24 nt) of plus polarity (A and C) and minus polarity (B and D) with different 5′ termini.
Irrespective of the redundancy of the specific 5′ termini of the PSTVd sRNA and their allocation along the genomic plus- and minus-strand RNAs (see below), these data are indicative of a disproportionate distribution, because PSTVd is rich in G+C (29). Potential factors shaping this distribution include the concurrent action of distinct DCLs and auxiliary proteins, the preference of certain AGOs for the 5′ termini of their guide sRNAs (see below), and their differential targeting by one or more exoribonucleases.
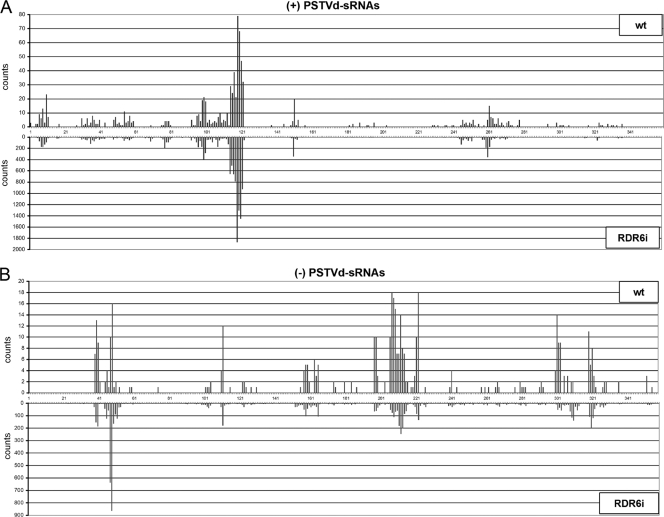
Mapping of the PSTVd sRNAs along the genomic plus- and minus-strand RNAs reveals specific hot spot profiles.
Examination of the PSTVd sRNA frequency showed that their 5′ termini mapped at many positions of the genomic plus- and minus-strand RNAs, with a large fraction of the counts merging in specific regions (hot spots). Some features are worth mentioning. First, the hot spot patterns formed by the plus-strand PSTVd sRNAs are to a large extent similar in the Nbwt and NbRDR6i samples (although some sRNAs, at positions 180 to 200 and 300 to 340, are present only in the less abundant Nbwt population), as are those generated by their minus-strand counterparts (although the PSTVd sRNAs mapping at positions 47 and 48 are overrepresented in the NbRDR6i population) (Fig. 9); these observations thus attest to the small contribution of the RDR6-derived PSTVd sRNAs and, indirectly, to the reproducibility of the sequencing approach. However, the hot spot patterns of plus- and minus-strand PSTVd sRNAs differ, indicating that the potential factors determining these profiles (see above) operate differentially on both viroid strands. Second, the most prominent hot spot of plus-strand PSTVd sRNAs (Fig. 9, positions 110 to 120), does not correspond to a region particularly structured in the rod-like conformation (Fig. 1) or in an alternative folding adopted during thermal denaturation (68); this alternative folding contains the so-called hairpin I (HPI), which facilitates adoption of a conserved double-stranded structure presumably directing the in vivo cleavage of oligomeric plus strands, but HPI covers positions 80 to 110 (26). Therefore, the PSTVd genomic plus-strand RNA, by itself, does not fully explain the observed vd-sRNA profile; the PSTVd genomic minus-strand RNA, in the form of multimeric strands paired to plus strands (3), also does not account for the vd-sRNA profile. Third, the hot spot profiles of the 20- to 24-nt plus-strand PSTVd sRNAs are very similar, as are those of the 20- to 24-nt minus-strand PSTVd sRNAs (see Fig. S3 in the supplemental material), indicating that these profiles are determined by the RNA substrates rather than by the specific DCLs acting upon them; a similar situation was recently reported for some plant viruses (18). Fourth, although a search identified some perfectly complementary vd-sRNAs with two 3′-protruding nucleotides in each strand, no clear phasing became evident. And fifth, the plus-strand vd-sRNAs mapping at the pathogenic (P) domain and at certain positions of the central (C) domain associated with pathogenesis (63, 72) accumulated with low frequency (Fig. 1), a result with implications for the mechanism of symptom induction (see below).
FIG. 9.
Similar hot spot profiles of plus- and minus-strand PSTVd sRNAs in Nbwt and NbRDR6i plants. Location and frequency in the genomic PSTVd RNA of the NB variant of the 5′ termini of the plus-strand (A) and minus-strand (B) PSTVd sRNA counts from Nbwt and NbRDR6i samples. Note that the scale of counts is different in the two polarities and that the same numbers are used in the plus polarity (5′→3′ orientation is from left to right) and in the minus polarity (5′→3′ orientation is from right to left).
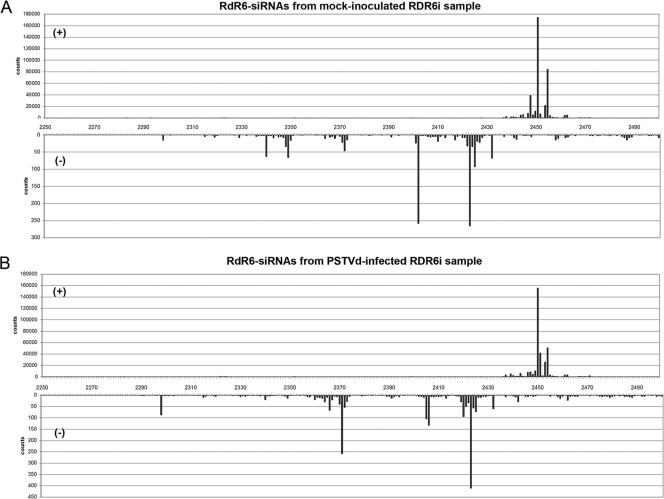
Deep sequencing discloses an uneven profile of RDR6 hairpin-derived siRNAs in NbRDR6i plants.
To complement the RNA gel blot hybridization results showing the accumulation of NbRDR6 siRNAs of 21 nt in NbRDR6i plants (Fig. 2), we searched the deep sequencing datasets from mock-inoculated and PSTVd-infected NbRDR6i samples for the presence, size, and distribution of hairpin-derived NbRDR6 siRNAs. Our results (Fig. 10; also, see Fig. S4 in the supplemental material) show that (i) the plus-strand siRNAs are considerably more abundant that their minus-strand counterparts, (ii) the 21-nt and, to a lesser extent, the 22-nt species are the most prevalent in the siRNA populations of both polarities, (iii) the siRNAs adopt a hot spot distribution, but while the 5′ termini of plus-strand siRNAs map within a very narrow window (of approximately 25 nt) of the hairpin stem, the minus-strand siRNAs cover a broader region of this stem, and thus, siRNAs arising from the intron or from regions of the NbRDR6 transcript not targeted by the hairpin construct were not retrieved, and (iv) the siRNA profile is not appreciably affected by PSTVd infection. Collectively, these data indicate that all NbRDR6 siRNAs derive from the stem of the hairpin construct and, more specifically, that only restricted portions of this stem are effective for siRNA accumulation. Additionally, given that the hot spot profiles of the 21- and 22-nt plus-strand siRNAs are very similar, as are those of the 21- and 22-nt minus-strand siRNAs (see Fig. S4 in the supplemental material), these profiles do not appear to be dictated by the DCL4 and DCL2 that are involved in their respective genesis (which appear to act hierarchically) (25), but rather, as with the vd-sRNAs (see Fig. S3 in the supplemental material), they are intrinsic attributes of the RNA substrate.
FIG. 10.
PSTVd infection does not alter the hot spot profiles of RDR6-siRNAs in NbRDR6i plants. Location and frequency in the RDR6 sequence of the 5′ termini of the hairpin-derived siRNAs of plus and minus polarity (upper and lower panels, respectively) in mock-inoculated (A) and PSTVd-infected (B) NbRDR6i plants are shown. Note that the scale of counts is different in both polarities and that the 5′ termini of the plus-strand siRNAs map at a very narrow window of the hairpin stem, while those of the minus-strand siRNAs extend over a much broader region. The hairpin stem covers a 243-bp portion of the NbRDR6 sequence (approximately between positions 2250 and 2500) (73).
DISCUSSION
Although a key role in viroid pathogenesis has been proposed for vd-sRNAs, which, acting like microRNAs or trans-acting RNAs, would target endogenous mRNAs for inactivation (27, 59, 85), no direct evidence for this intriguing hypothesis is yet available. Even if it is still an open possibility, some data argue against it. First, the postulated endogenous mRNAs have not been identified so far. Second, low-scale sequencing has revealed that vd-sRNAs arising from the P and C domains associated with pathogenesis in PSTVd (Fig. 1) and Citrus exocortis viroid (CEVd)—another member of the genus Pospiviroid—are relatively scarce in infected tissues (35, 44), and deep sequencing has produced similar results for PSTVd (at least regarding the most abundant plus-strand vd-sRNAs [this work]) and for the vd-sRNAs arising from the pathogenic determinant of one member of the family Avsunviroidae (17); low-abundance vd-sRNAs, however, might be functionally relevant. And third and most important, in contrast with previous results reporting that tomato plants expressing a noninfectious PSTVd hairpin RNA developed symptoms similar to those of PSTVd infection (85), a reexamination of some of these plants has failed to confirm the symptoms despite the accumulation of abundant hairpin-derived siRNAs (74). Therefore, vd-sRNAs might play other alternative functions and (i) regulate viroid titer (by loading RISC and targeting the genomic viroid RNA for inactivation), as inferred from in planta experiments (5), although in transfected protoplasts this RNA appears RISC-resistant (35), and (ii) provide specificity to viroid cross-protection (with the siRNAs from the first inoculated strain targeting the RNA of the challenge strain for inactivation) (5, 22).
In line with previous results for some RNA viruses (66, 73), our present data provide firm genetic evidence that NbRDR6, which mediates an amplification circuit leading to the generation of dsRNA precursors of secondary siRNAs, is also involved in an RNA silencing-based plant defense mechanism against PSTVd, a viroid that replicates in the nucleus. RDR6 negatively modulates PSTVd accumulation in differentiated leaf tissues at early infection stages—when the viroid load is low—and also prevents the incoming PSTVd from invading floral and vegetative meristems, possibly because they concurrently receive a viroid-specific systemic silencing signal that triggers a defensive response. In this context, the lack in viroid RNAs of 5′ cap and 3′ poly(A) tail, which are characteristic features of aberrant RNAs (83), may promote binding and activation of RDR6 to produce additional viroid-specific dsRNAs. However, despite our present results and others along the same line reported previously (89, 90), we presume that minor amounts of PSTVd should be able to invade floral organs in late developmental stages because this RNA is transmitted through both botanical seed and pollen in potato and tomato (55), which belong to the same family. Furthermore, RT-PCR has revealed the presence of PSTVd in all floral parts of tomato (76).
On the other hand, the entry of some viruses into the SAM of plants ectopically expressing virus-encoded RNA silencing suppressors suggests that meristem exclusion may be governed by an RNA silencing-based surveillance system (24), and that viral invasion may depend on the balance between the relative strengths of the host RNA silencing defense and the virus suppressor (66). Yet recent results indicate that PVX-induced gene silencing is independent of the presence of the virus suppressor (82). This must also be the case for viroid-induced gene silencing, because protein suppressors are not encoded by viroids, which alternatively might suppress RNA silencing by sequestering for their replication enzymes involved in the biogenesis of the host siRNAs and microRNAs, as has been reported for an RNA virus (79). Furthermore, in contrast to PSTVd, Peach latent mosaic viroid (PLMVd) (33), a member of the family Avsunviroidae, has recently been detected in the SAM (69), showing that it can bypass the RNA surveillance system and raising the question of whether this behavior is related to its replication and accumulation in plastids, where no silencing has been reported. Hence, like viruses, viroids may have evolved strategies to overcome this system in order to be seed or pollen transmitted.
To get a deeper insight of the function of RDR6 in the genesis of vd-sRNAs, we analyzed the PSTVd sRNAs retrieved by deep sequencing from the infected Nbwt and NbRDR6i samples 14 dpi. Consistent with the intensity of RNA gel blot hybridization signals (Fig. 2), the PSTVd sRNAs were significantly more abundant in the NbRDR6i preparation, but their size and polarity were similar in both samples, with the 22-nt plus-strand and to a lesser extent 21-nt plus-strand RNAs being prevalent in the population (Fig. 7). This profile is in good agreement with low-scale sequencing data for the vd-sRNAs recovered from tomato infected by PSTVd (35) and CEVd (44) but differs somewhat from the vd-sRNA profile from another study with PSTVd-infected tomato in which the predominant species were 21- and 22-nt minus-strand RNAs (41) and from the vd-sRNA profile from PLMVd-infected peach with prevalent 21-nt plus- and minus-strand RNAs (17).
Considering that PSTVd replicates in the nucleus (32, 62) and that plus strands accumulate to much higher levels than minus strands (3), and assuming for the N. benthamiana DCLs the same cellular and molecular properties as their Arabidopsis homologues (42, 60), DCL1 appears to be a candidate for producing the 21-nt plus-strand PSTVd sRNAs because it mediates nuclear processing of miRNA precursors with a secondary structure resembling that of the genomic PSTVd plus-strand RNA (29). Additionally, DCL2 and DCL4—which act hierarchically in antiviral defense (10, 25, 65)—most likely mediate the genesis of the 22-nt PSTVd sRNA and part of the 21-nt PSTVd sRNA, respectively, dicing (i) the nuclear genomic PSTVd plus-strand RNA or the replicative dsRNA intermediates or (ii) the genomic plus-strand RNA during its cytoplasmic translocation to infect adjacent cells or the PSTVd-dsRNAs produced by an RDR, like RDR6, which operates concerted with DCL4 (34). However, because of the small quantitative contribution of RDR6 to vd-RNA accumulation in later infection stages (Fig. 2D and F) and because the nuclear RDR2—which acts in concert with DCL3—also seems to play a minor role in view of the weak hybridization signals and low frequency of the 24-nt PSTVd sRNAs (Fig. 2 and 7), the involvement of other RDRs appears to be a feasible possibility. RDR1, which was the first RDR characterized biochemically and genetically (70, 71), is stimulated by viroid infection in tomato and has been implicated in the production of viral siRNAs from Tobacco rattle virus (19) and a mutant of Cucumber mosaic virus (13) in Arabidopsis. Yet N. benthamiana is likely to be RDR1 deficient (87).
The absence of a clear relationship between the hot spot profile of PSTVd sRNAs and structural features of the genomic plus- and minus-strand RNAs, also observed in previous low-scale sequencing studies (35, 41), also does not favor the idea that these RNAs are the predominant DCL substrates but rather invokes the participation of one or more RDRs. The hot spot profile of vd-sRNAs may also be conditioned by host proteins protecting specific domains of their precursors from DCL attack (8, 45), as well as by the differential stability of the vd-sRNA strands because of their HEN1-mediated 3′ methylation (67, 88) and specific binding through their 5′-terminal nucleotide to distinct AGO members forming the RISC core (48, 49). PSTVd sRNAs may thus resemble virus-derived small RNAs, which recent evidence indicates act as genuine siRNAs and program RISC for degradation of their cognate viral RNAs (54, 58). In this context, AGO1 and AGO7 target viral RNAs in tissue infected by Turnip crinkle virus (65), resistance to Cucumber mosaic virus (CMV) is impaired in AGO1 mutants (50), and AGO2 and AGO5 bind CMV-derived sRNAs (78).
Supporting this lack of correlation between the secondary structure of the RNA substrate upon which DCLs act and the resulting sRNAs, we recovered from NbRDR6i plants an uneven profile of NbRDR6 siRNAs mostly of 21- and 22-nt and exclusively derived from very specific regions of the hairpin stem, which is presumed to fold into a perfect double-stranded structure. However, they neither present a clear phasing nor correspond to duplex siRNAs directly generated by DCLs—most likely DCL4 and DCL2—thus indicating that other factors greatly influence the final siRNA profile. In the only (to our knowledge) previous work reporting the low-scale sequencing of the siRNAs derived from a hairpin construct, they also displayed an unexpected profile (74). Moreover, recent deep sequencing of the sRNAs from the chalcone synthase A (CHS-A) gene in cosuppressed transgenic petunia has also revealed an unanticipated pattern with two extremely abundant siRNAs that guide prominent cleavage events in CHS-A mRNA (11). Therefore, dissecting the underlying factors and understanding how they operate may help to improve the efficiency of RNA interference mediated by hairpin constructs, because our results indicate that the siRNAs active in silencing the NbRDR6 mRNA, the minus-strand siRNAs, represent a minor fraction of the total population. Furthermore, the absence of NbRDR6 siRNAs derived from other regions of the NbRDR6 mRNA (apart from that forming the hairpin stem) is consistent with the role of RDR6 in generating the dsRNA templates for secondary siRNAs, a role in which it cannot be apparently supplanted by other RDRs.
In summary, our results showing that RDR6 delays accumulation of PSTVd in differentiated tissues and precludes invasion of floral and vegetative meristems of N. benthamiana provide additional support for the notion that viroids, like viruses, are inducers and targets (and perhaps even suppressors) of the RNA-silencing machinery of their hosts.
Supplementary Material
Acknowledgments
This research was supported by grants to R.F. (Ministerio de Ciencia e Innovación of Spain, BFU2008-03154/BMC, and Generalidad Valenciana, ACOMP/2009/151), to F.D.S. (Dipartimento Agroalimentare of the CNR of Italy, A. Leone and D. Mariotti 2008 award for advanced research in agriculture), and to A.-E.M.A. (Generalidad Valenciana, GVPRE/2008/219).
We are grateful to D. C. Baulcombe for providing NdRDR6i and Nbwt seeds, to M. Wassenegger for the NB variant of PSTVd, and to A. Ahuir for excellent technical assistance.
Footnotes
Published ahead of print on 16 December 2009.
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 2.Bernstein, E., A. A. Caudy, S. M. Hammond, and G. J. Hannon. 2001. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature 409:363-366. [DOI] [PubMed] [Google Scholar]
- 3.Branch, A. D., B. J. Benenfeld, and H. D. Robertson. 1988. Evidence for a single rolling circle in the replication of potato spindle tuber viroid. Proc. Natl. Acad. Sci. U. S. A. 85:9128-9132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Branch, A. D., and H. D. Robertson. 1984. A replication cycle for viroids and other small infectious RNAs. Science 223:450-455. [DOI] [PubMed] [Google Scholar]
- 5.Carbonell, A., A. E. Martínez de Alba, R. Flores, and S. Gago. 2008. Double-stranded RNA interferes in a sequence-specific manner with infection of representative members of the two viroid families. Virology 371:44-53. [DOI] [PubMed] [Google Scholar]
- 6.Dalmay, T., A. Hamilton, S. Rudd, S. Angell, and D. C. Baulcombe. 2000. An RNA-dependent RNA polymerase gene in Arabidopsis is required for posttranscriptional gene silencing mediated by a transgene but not by a virus. Cell 101:543-553. [DOI] [PubMed] [Google Scholar]
- 7.Dalmay, T., L. Rubino, J. Burgyan, A. Kollár, and M. Russo. 1993. Functional analysis of cymbidium ringspot virus genome. Virology 194:697-704. [DOI] [PubMed] [Google Scholar]
- 8.Daròs, J. A., and R. Flores. 2002. A chloroplast protein binds a viroid RNA in vivo and facilitates its hammerhead-mediated self-cleavage. EMBO J. 21:749-759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Daròs, J. A., J. F. Marcos, C. Hernández, and R. Flores. 1994. Replication of avocado sunblotch viroid: Evidence for a symmetric pathway with two rolling circles and hammerhead ribozyme processing. Proc. Natl. Acad. Sci. U. S. A. 91:12813-12817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Deleris, A., J. Gallego-Bartolomé, J. Bao, K. D. Kasschau, J. C. Carrington, and O. Voinnet. 2006. Hierarchical action and inhibition of plant Dicer-like proteins in antiviral defense. Science 313:68-71. [DOI] [PubMed] [Google Scholar]
- 11.De Paoli, E., A. Dorantes-Acosta, J. Zhai, M. Accerbi, D.-H. Jeong, S. Park, B. C. Meyers, R. A. Jorgensen, and P. J. Green. 2009. Distinct extremely abundant siRNAs associated with cosuppression in petunia. RNA. 15:1965-1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Díaz-Pendón, J. A., and S. W. Ding. 2008. Direct and indirect roles of viral suppressors of RNA silencing in pathogenesis. Annu. Rev. Phytopathol. 46:303-326. [DOI] [PubMed] [Google Scholar]
- 13.Díaz-Pendón, J. A., F. Li, W. X. Li, and S. W. Ding. 2007. Suppression of antiviral silencing by cucumber mosaic virus 2b protein in Arabidopsis is associated with drastically reduced accumulation of three classes of viral small interfering RNAs. Plant Cell 19:2053-2063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Diener, T. O. 2003. Discovering viroids—a personal perspective. Nature Rev. Microbiol. 1:75-80. [DOI] [PubMed] [Google Scholar]
- 15.Ding, B. 2009. The biology of viroid-host interactions. Annu. Rev. Phytopathol. 47:105-131. [DOI] [PubMed] [Google Scholar]
- 16.Ding, B., and A. Itaya. 2007. Viroid: a useful model for studying the basic principles of infection and RNA biology. Mol. Plant Microbe Interact. 20:7-20. [DOI] [PubMed] [Google Scholar]
- 17.Di Serio, F., A. Gisel, B. Navarro, S. Delgado, A. E. Martínez de Alba, G. Donvito, and R. Flores. 2009. Deep sequencing of the small RNAs derived from two symptomatic variants of a chloroplastic viroid: implications for their genesis and for pathogenesis. PLoS One 4:e7539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Donaire, L., Y. Wang, D. Gonzalez-Ibeas, K. F. Mayer, M. A. Aranda, and C. Llave. 2009. Deep-sequencing of plant viral small RNAs reveals effective and widespread targeting of viral genomes. Virology 392:203-214. [DOI] [PubMed] [Google Scholar]
- 19.Donaire, L., D. Barajas, B. Martínez-García, L. Martínez-Priego, I. Pagan, and C. Llave. 2008. Structural and genetic requirements for the biogenesis of tobacco rattle virus-derived small interfering RNAs. J. Virol. 82:5167-5177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fagard, M., and H. Vaucheret. 2000. Systemic silencing signal(s). Plant Mol. Biol. 43:285-293. [DOI] [PubMed] [Google Scholar]
- 21.Feldstein, P. A., Y. Hu, and R. A. Owens. 1998. Precisely full length, circularizable, complementary RNA: an infectious form of potato spindle tuber viroid. Proc. Natl. Acad. Sci. U. S. A. 95:6560-6565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Flores, R., C. Hernández, A. E. Martínez de Alba, J. A. Daròs, and F. Di Serio. 2005. Viroids and viroid-host interactions. Annu. Rev. Phytopathol. 43:117-139. [DOI] [PubMed] [Google Scholar]
- 23.Flores, R., J. A. Daròs, and C. Hernández. 2000. The Avsunviroidae family: viroids with hammerhead ribozymes. Adv. Virus Res. 55:271-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Foster, T. M., T. J. Lough, S. J. Emerson, R. H. Lee, J. L. Bowman, R. L. Forster, and W. J. Lucas. 2002. A surveillance system regulates selective entry of RNA into the shoot apex. Plant Cell 14:1497-1508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fusaro, A. F., L. Matthew, N. A. Smith, S. J. Curtin, J. Dedic-Hagan, G. A. Ellacott, J. M. Watson, M. B. Wang, C. Brosnan, B. J. Carroll, and P. M. Waterhouse. 2006. RNA interference-inducing hairpin RNAs in plants act through the viral defence pathway. EMBO Rep. 7:1168-1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gas, M. E., C. Hernández, R. Flores, and J. A. Daròs. 2007. Processing of nuclear viroids in vivo: an interplay between RNA conformations. PLoS Pathog. 3:e182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gómez, G., G. Martínez, and V. Pallás. 2009. Interplay between viroid-induced pathogenesis and RNA silencing pathways. Trends Plant Sci. 14:264-269. [DOI] [PubMed] [Google Scholar]
- 28.Gómez, G., G. Martínez, and V. Pallás. 2008. Viroid-induced symptoms in Nicotiana benthamiana plants are dependent on RDR6 activity. Plant Physiol. 148:414-423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gross, H. J., H. Domdey, C. Lossow, P. Jank, M. Raba, H. Alberty, and H. L. Sänger. 1978. Nucleotide sequence and secondary structure of potato spindle tuber viroid. Nature 273:203-208. [DOI] [PubMed] [Google Scholar]
- 30.Hamilton, A. J., and D. C. Baulcombe. 1999. A species of small antisense RNA in posttranscriptional gene silencing in plants. Science 286:950-952. [DOI] [PubMed] [Google Scholar]
- 31.Hammond, S. M., E. Bernstein, D. Beach, and G. J. Hannon. 2000. An RNA-directed nuclease mediates post-transcriptional gene silencing in Drosophila cells. Nature 404:293-296. [DOI] [PubMed] [Google Scholar]
- 32.Harders, J., N. Lukacs, M. Robert-Nicoud, J. M. Jovin, and D. Riesner. 1989. Imaging of viroids in nuclei from tomato leaf tissue by in situ hybridization and confocal laser scanning microscopy. EMBO J. 8:3941-3949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hernández, C., and R. Flores. 1992. Plus and minus RNAs of peach latent mosaic viroid self-cleave in vitro through hammerhead structures. Proc. Natl. Acad. Sci. U. S. A. 89:3711-3715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Howell, M. D., N. Fahlgren, E. J. Chapman, J. S. Cumbie, C. M. Sullivan, S. A. Givan, K. D. Kasschau, and J. C. Carrington. 2007. Genome-wide analysis of the RNA-DEPENDENT RNA POLYMERASE6/DICER-LIKE4 pathway in Arabidopsis reveals dependency on miRNA- and tasiRNA-directed targeting. Plant Cell 19:926-942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Itaya, A., X. Zhong, R. Bundschuh, Y. Qi, Y. Wang, R. Takeda, A. R. Harris, C. Molina, R. S. Nelson, and B. Ding. 2007. A structured viroid RNA is substrate for Dicer-like cleavage to produce biologically active small RNAs but is resistant to RISC-mediated degradation. J. Virol. 81:2980-2994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Itaya, A., A. Folimonov, Y. Matsuda, R. S. Nelson, and B. Ding. 2001. Potato spindle tuber viroid as inducer of RNA silencing in infected tomato. Mol. Plant Microbe. Interact. 14:1332-1334. [DOI] [PubMed] [Google Scholar]
- 37.Jackson, D. P. 1992. In situ hybridization in plants, p. 163-174. In D. J. Bowles, S. J. Gurr, and M. J. McPherson (ed.), Molecular plant pathology: a practical approach. Oxford University Press, Oxford.
- 38.Kalantidis, K., H. T. Schumacher, T. Alexiadis, and J. M. Helm. 2008. RNA silencing movement in plants. Biol. Cell 100:13-26. [DOI] [PubMed] [Google Scholar]
- 39.Keese, P., and R. H. Symons. 1985. Domains in viroids: evidence of intermolecular RNA rearrangements and their contribution to viroid evolution. Proc. Natl. Acad. Sci. U. S. A. 82:4582-4586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lindbo, J. A., L. Silva-Rosales, W. M. Proebsting, and W. G. Dougherty. 1993. Induction of a highly specific antiviral state in transgenic plants: implications for regulation of gene expression and virus resistance. Plant Cell 5:1749-1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Machida, S., N. Yamahata, H. Watanuki, R. A. Owens, and T. Sano. 2007. Successive accumulation of two size classes of viroid-specific small RNA in potato spindle tuber viroid-infected tomato plants. J. Gen. Virol. 88:3452-3457. [DOI] [PubMed] [Google Scholar]
- 42.Margis, R., A. F. Fusaro, N. A. Smith, S. J. Curtin, J. M. Watson, E. J. Finnegan, and P. M. Waterhouse. 2006. The evolution and diversification of Dicers in plants. FEBS Lett. 580:2442-2450. [DOI] [PubMed] [Google Scholar]
- 43.Markarian, N., H. W. Li, S. W. Ding, and J. S. Semancik. 2004. RNA silencing as related to viroid induced symptom expression. Arch. Virol. 149:397-406. [DOI] [PubMed] [Google Scholar]
- 44.Martín, R., C. Arenas, J. A. Daròs, A. Covarrubias, J. L. Reyes, and N. H. Chua. 2007. Characterization of small RNAs derived from citrus exocortis viroid (CEVd) in infected tomato plants. Virology 367:135-146. [DOI] [PubMed] [Google Scholar]
- 45.Martínez de Alba, A. E., R. Sagesser, M. Tabler, and M. Tsagris. 2003. A bromodomain-containing protein from tomato specifically binds potato spindle tuber viroid RNA in vitro and in vivo. J. Virol. 77:9685-9694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Martínez de Alba, A. E., R. Flores, and C. Hernández. 2002. Two chloroplastic viroids induce the accumulation of the small RNAs associated with post-transcriptional gene silencing. J. Virol. 76:13094-13096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Matousek, J., P. Kozlová, L. Orctová, A. Schmitz, K. Pesina, O. Bannach, N. Diermann, G. Steger, and D. Riesner. 2007. Accumulation of viroid-specific small RNAs and increase of nucleolytic activities linked to viroid-caused pathogenesis. Biol. Chem. 388:1-13. [DOI] [PubMed] [Google Scholar]
- 48.Mi, S., T. Cai, Y. Hu, Y. Chen, E. Hodges, F. Ni, L. Wu, S. Li, H. Zhou, C. Long, S. Chen, G. J. Hannon, and Y. Qi. 2008. Sorting of small RNAs into Arabidopsis argonaute complexes is directed by the 5′ terminal nucleotide. Cell 133:116-127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Montgomery, T. A., M. D. Howell, J. T. Cuperus, D. Li, J. E. Hansen, A. L. Alexander, E. J. Chapman, N. Fahlgren, E. Allen, and J. C. Carrington. 2008. Specificity of ARGONAUTE7-miR390 interaction and dual functionality in TAS3 trans-acting siRNA formation. Cell 133:128-141. [DOI] [PubMed] [Google Scholar]
- 50.Morel, J. B., C. Godon, P. Mourrain, C. Beclin, S. Boutet, F. Feuerbach, F. Proux, and H. Vaucheret. 2002. Fertile hypomorphic ARGONAUTE (ago1) mutants impaired in posttranscriptional gene silencing and virus resistance. Plant Cell 14:629-639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mourrain, P., C. Beclin, T. Elmayan, F. Feuerbach, C. Godon, J. B. Morel, D. Jouette, A. M. Lacombe, S. Nikic, N. Picault, K. Remoue, M. Sanial, T. A. Vo, and H. Vaucheret. 2000. Arabidopsis SGS2 and SGS3 genes are required for post-transcriptional gene silencing and natural virus resistance. Cell 101:533-542. [DOI] [PubMed] [Google Scholar]
- 52.Navarro, B., V. Pantaleo, A. Gisel, S. Moxon, T. Dalmay, G. Bisztray, F. Di Serio, and J. Burgyán. 2009. Deep sequencing of viroid-derived small RNAs from grapevine provides new insights on the role of RNA silencing in plant-viroid interaction. PLoS One 4:e7686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Navarro, J. A., J. A. Daròs, and R. Flores. 1999. Complexes containing both polarity strands of avocado sunblotch viroid: Identification in chloroplasts and characterization. Virology 253:77-85. [DOI] [PubMed] [Google Scholar]
- 54.Omarov, R. T., J. J. Cioperlik, and H. B. Sholthof. 2007. RNAi-associated ssRNA-specific ribonucleases in tombusvirus P19 mutant-infected plants and evidence for a discrete siRNA-containing effector complex. Proc. Natl. Acad. Sci. U. S. A. 104:1714-1719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Owens, R. A. 2007. Potato spindle tuber viroid: The simplicity paradox resolved? Mol. Plant Pathol. 8:549-560. [DOI] [PubMed] [Google Scholar]
- 56.Pak, J., and A. Fire. 2007. Distinct populations of primary and secondary effectors during RNAi in C. elegans. Science 315:241-244. [DOI] [PubMed] [Google Scholar]
- 57.Palauqui, J. C., T. Elmayan, J. M. Pollien, and H. Vaucheret. 1997. Systemic acquired silencing: transgene-specific post-transcriptional silencing is transmitted by grafting from silenced stocks to non-silenced scions. EMBO J. 16:4738-4745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pantaleo, V., G. Szittya, and J. Burgyán. 2007. Molecular bases of viral RNA targeting by viral small interfering RNA-programmed RISC. J. Virol. 81:3797-3806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Papaefthimiou, I., A. J. Hamilton, M. A. Denti, D. C. Baulcombe, M. Tsagris, and M. Tabler. 2001. Replicating potato spindle tuber viroid RNA is accompanied by short RNA fragments that are characteristic of post-transcriptional gene silencing. Nucleic Acids Res. 29:2395-2400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Qi, Y., A. M. Denli, and G. J. Hannon. 2005. Biochemical specialization within Arabidopsis RNA silencing pathways. Mol. Cell 19:421-428. [DOI] [PubMed] [Google Scholar]
- 61.Qi, Y., T. Pelissier, A. Itaya, E. Hunt, M. Wassenegger, and B. Ding. 2004. Direct role of a viroid RNA motif in mediating directional RNA trafficking across a specific cellular boundary. Plant Cell 16:1741-1752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Qi, Y., and B. Ding. 2003. Differential subnuclear localization of RNA strands of opposite polarity derived from an autonomously replicating viroid. Plant Cell 15:2566-2577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Qi, Y., and B. Ding. 2003. Inhibition of cell growth and shoot development by a specific nucleotide sequence in a noncoding viroid RNA. Plant Cell 15:1360-1374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Qi, Y., and B. Ding. 2002. Replication of potato spindle tuber viroid in cultured cells of tobacco and Nicotiana benthamiana: the role of specific nucleotides in determining replication levels for host adaptation. Virology 302:445-456. [DOI] [PubMed] [Google Scholar]
- 65.Qu, F., X. Ye, and T. J. Morris. 2008. Arabidopsis DRB4, AGO1, AGO7, and RDR6 participate in a DCL4-initiated antiviral RNA silencing pathway negatively regulated by DCL1. Proc. Natl. Acad. Sci. U. S. A. 105:14732-14737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Qu, F., X. Ye, G. Hou, S. Sato, T. E. Clemente, and T. J. Morris. 2005. RDR6 has a broad-spectrum but temperature-dependent antiviral defense role in Nicotiana benthamiana. J. Virol. 79:15209-15217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ramachandran, V., and X. M. Chen. 2008. Small RNA metabolism in Arabidopsis. Trends Plant Sci. 13:368-374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Riesner, D., K. Henco, U. Rokohl, G. Klotz, A. K. Kleinschmidt, H. Domdey, P. Jank, H. J. Gross, and H. Sänger. 1979. Structure and structure formation of viroids. J. Mol. Biol. 133:85-115. [DOI] [PubMed] [Google Scholar]
- 69.Rodio, M. E., S. Delgado, A. De Stradis, M. D. Gómez, R. Flores, and F. Di Serio. 2007. A viroid RNA with a specific structural motif inhibits chloroplast development. Plant Cell 19:3610-3616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Schiebel, W., T. Pelissier, L. Riedel, S. Thalmeir, R. Schiebel, D. Kempe, F. Lottspeich, H. L. Sänger, and M. Wassenegger. 1998. Isolation of an RNA-Directed RNA polymerase-specific cDNA clone from tomato. Plant Cell 10:2087-2101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Schiebel, W., B. Haas, S. Marinkovic, A. Klanner, and H. L. Sänger. 1993. RNA-directed RNA polymerase from tomato leaves. 1. Purification and physical-properties. J. Biol. Chem. 268:11851-11857. [PubMed] [Google Scholar]
- 72.Schnolzer, M., B. Haas, K. Ramm, H. Hofmann, and H. L. Sänger. 1985. Correlation between structure and pathogenicity of potato spindle tuber viroid (PSTV). EMBO J. 4:2181-2190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Schwach, F., F. E. Vaistij, L. Jones, and D. C. Baulcombe. 2005. An RNA-dependent RNA polymerase prevents meristem invasion by potato virus X and is required for the activity but not the production of a systemic silencing signal. Plant Physiol. 138:1842-1852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Schwind, N., M. Zwiebel, A. Itaya, B. Ding, M. B. Wang, G. Krczal, and M. Wassenegger. 2009. RNAi-mediated resistance to potato spindle tuber viroid in transgenic tomato expressing a viroid hairpin RNA construct. Mol. Plant Pathol. 10:459-469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Sijen, T., F. A. Steiner, K. L. Thijssen, and R. H. Plasterk. 2007. Secondary siRNAs result from unprimed RNA synthesis and form a distinct class. Science 315:244-247. [DOI] [PubMed] [Google Scholar]
- 76.Singh, R. P. 2006. Reassessment of the presence of viroid species of the genus Pospiviroid in infected floral parts, using reverse transcription-polymerase chain reaction and infectivity assays. Can. J. Plant Pathol. 28:242-249. [Google Scholar]
- 77.St-Pierre, P., F. Hassen, D. Thompson, and J. P. Perreault. 2009. Characterization of the siRNAs associated with peach latent mosaic viroid infection. Virology 383:178-182. [DOI] [PubMed] [Google Scholar]
- 78.Takeda, A., S. Iwasaki, T. Watanabe, M. Utsumi, and Watanabe, Y. 2008. The mechanism selecting the guide strand from small RNA duplexes is different among Argonaute proteins. Plant Cell Physiol. 49:493-500. [DOI] [PubMed] [Google Scholar]
- 79.Takeda, A., M. Tsukuda, H. Mizumoto, K. Okamoto, M. Kaido, K. Mise, and T. Okuno. 2005. A plant RNA virus suppresses RNA silencing through viral RNA replication. EMBO J. 24:3147-3157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Tournier, B., M. Tabler, and K. Kalantidis. 2006. Phloem flow strongly influences the systemic spread of silencing in GFP Nicotiana benthamiana plants. Plant J. 47:383-394. [DOI] [PubMed] [Google Scholar]
- 81.Tsagris, E. M., A. E. Martínez de Alba, M. Gozmanova, and K. Kalantidis. 2008. Viroids. Cell Microbiol. 10:2168-2179. [DOI] [PubMed] [Google Scholar]
- 82.Vaistij, F. E., and L. Jones. 2009. Compromised virus-induced gene silencing in RDR6-deficient plants. Plant Physiol. 149:1399-1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Voinnet, O. 2008. Use, tolerance and avoidance of amplified RNA silencing by plants. Trends Plant Sci. 13:317-328. [DOI] [PubMed] [Google Scholar]
- 84.Voinnet, O., P. Vain, S. Angell, and D. C. Baulcombe. 1998. Systemic spread of sequence-specific transgene RNA degradation is initiated by localised introduction of ectopic promoterless DNA. Cell 95:177-187. [DOI] [PubMed] [Google Scholar]
- 85.Wang, M. B., X. Y. Bian, L. M. Wu, L. X. Liu, N. A. Smith, D. Isenegger, R. M. Wu, C. Masuta, V. B. Vance, J. M. Watson, A. Rezaian, E. S. Dennis, and P. M. Waterhouse. 2004. On the role of RNA silencing in the pathogenicity and evolution of viroids and viral satellites. Proc. Natl. Acad. Sci. U. S. A. 101:3275-3280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Wassenegger, M., and G. Krczal. 2006. Nomenclature and functions of RNA-directed RNA polymerases. Trends Plant Sci. 11:142-151. [DOI] [PubMed] [Google Scholar]
- 87.Yang, S. J., S. A. Carter, A. B. Cole, N. H. Cheng, and R. S. Nelson. 2004. A natural variant of a host RNA-dependent RNA polymerase is associated with increased susceptibility to viruses by Nicotiana benthamiana. Proc. Natl. Acad. Sci. U. S. A. 101:6297-6302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Yang, Z., Y. W. Ebright, B. Yu, and X. Chen. 2006. HEN1 recognizes 21-24 nt small RNA duplexes and deposits a methyl group onto the 2′ OH of the 3′ terminal nucleotide. Nucleic Acids Res. 34:667-675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Zhu, Y., Y. Qi, Y. Xun, R. Owens, and B. Ding. 2002. Movement of potato spindle tuber viroid reveals regulatory points of phloem-mediated RNA traffic. Plant Physiol. 130:138-146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Zhu, Y., L. Green, Y. M. Woo, R. Owens, and B. Ding. 2001. Cellular basis of potato spindle tuber viroid systemic movement. Virology 279:69-77. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.