Abstract
Mouse bioassay remains the gold standard for determining proof of infectivity, strain type, and infectious titer estimation in prion disease research. The development of an approach using ex vivo cell-based assays remains an attractive alternative, both in order to reduce the use of mice and to hasten results. The main limitation of a cell-based approach is the scarcity of cell lines permissive to infection with natural transmissible spongiform encephalopathy strains. This study combines two advances in this area, namely, the standard scrapie cell assay (SSCA) and the Rov9 and MovS6 cell lines, which both express the ovine PrP VRQ allele, to assess to what extent natural and experimental ovine scrapie can be detected ex vivo. Despite the Rov9 and MovS6 cell lines being of different biological origin, they were both permissive and resistant to infection with the same isolates of natural sheep scrapie as detected by SSCA. Rov9 subclones that are 20 times more sensitive than Rov9 to SSBP/1-like scrapie infection were isolated, but all the subclones maintained their resistance to isolates that failed to transmit to the parental line. The most sensitive subclone of the Rov9 cell line was used to estimate the infectious titer of a scrapie brain pool (RBP1) and proved to be more sensitive than the mouse bioassay using wild-type mice. Increasing the sensitivity of the Rov9 cell line to SSBP/1 infection did not correlate with broadening susceptibility, as the specificity of permissiveness and resistance to other scrapie isolates was maintained.
Prion diseases are a group of neurodegenerative diseases affecting humans and animals, including scrapie in sheep and goats and bovine spongiform encephalopathy (BSE) in cattle. A feature of prion diseases and, in particular, of scrapie, is the existence of different strains (6) which influence pathology and is most probably related to the conformation of the pathogenic form of the prion protein (PrPSc). The susceptibility of sheep to scrapie is determined by the PrP genotype; codons 136, 154, and 171 determine relative resistance and susceptibility, with amino acids valine (V), arginine (R), and glutamine (Q) at these positions (known as VRQ) being considered the sheep PrP allele most susceptible to classical scrapie (3).
An array of diagnostic tests exist for prion diseases, aimed at the detection of the disease-associated protease-resistant form of the naturally occurring PrPC protein, termed PrPSc or PrPres after partial protease digestion. However, the level of detectable PrPSc does not quantitatively correlate with prion infectivity (2) and the current biochemical analysis of PrPSc cannot always determine the strain (6, 7).
Mouse bioassay remains the gold standard for determining proof of infectivity, strain type, and infectious titer estimate in ruminant transmissible spongiform encephalopathy (TSE) research. Conventional mouse bioassays using wild-type mice are generally slow (>150 days, and considerably longer, >600, days for obtaining infectious titer information) and require multiple mice to be dosed (typically 6 or more) at each dilution of infectious material. Therefore, the development of an approach using ex vivo cell-based assays remains an ethically and economically desirable alternative. Using cell lines permissive to mouse-passaged scrapie strains, Klöhn et al. have developed a cell-based assay for measuring de novo infection and the titer of mouse-passaged scrapie (18).
The main limitation of adopting a cell-based approach is the scarcity of cell lines permissive to infection with natural TSE strains (for a review, see references 31 and 34), as the majority of permissive cell lines can only be infected with rodent-adapted strains of scrapie and BSE (4, 9, 16, 20, 23, 24, 29, 33, 36). While there are currently no cell lines reported to be permissive to bovine BSE or human TSE diseases, there are cell lines which express ovine PrP that have been shown to be permissive to natural scrapie infection (1, 35). There is also one fibroblast-like deer cell line that is able to propagate chronic wasting disease (27).
Two of the sheep scrapie-susceptible cell lines are the MovS6 cell line (1), a Schwann cell line derived from the tg301 transgenic mouse, and the Rov9 cell line (35), based on a stably transfected rabbit kidney epithelial cell line (RK13) that does not express endogenous PrP. Both express the VRQ allele of ovine PrP, the latter upon induction with doxycycline (35). These cell lines were found to be permissive to infection with a PrP genotype-matched VRQ homozygous scrapie field case, and de novo PrPSc maintained its phenotype when used as an inoculum in mouse bioassays (1, 35). Using fluorescence-activated cell sorting, Falanga et al. isolated Rov9 subclones that produce higher levels of PrPC and PrPSc than the parental cell line when infected (11).
The primary objective of this study was to assess the permissiveness of the Rov9 and MovS6 cell lines to a panel of scrapie isolates from a range of sheep breeds with a range of PrP genotypes. Second, subcloning of the Rov9 cell line was undertaken in an attempt to identify subclones with greater sensitivity and more diverse permissibility to ovine scrapie isolates.
MATERIALS AND METHODS
Cell lines.
The MovS6 and Rov9 cell lines have been described previously (1, 35). The MovS6 cell line was cultured in 75% Dulbecco's modified Eagle's medium (DMEM; Sigma) and 25% F-12 Ham medium (Sigma) supplemented with 10% fetal calf serum and antibiotics. Infections in MovS6 cells were performed with Optimem (Gibco) as a replacement for DMEM. Rov9 cells were routinely cultured in Eagle's minimal essential medium (EMEM; Gibco) supplemented with 10% fetal calf serum, 2% HEPES, and antibiotics (penicillin, streptomycin, and mycostatin). All Rov9 infections were performed using Optimem (Gibco) as a replacement for EMEM. Cells were cultured at 37°C in a humidified 5% CO2 atmosphere in a cell culture incubator.
SSCA.
For the standard scrapie cell assay (SSCA), cells were infected following the published protocol from Klöhn et al. (18), briefly described here. Approximately 20,000 cells per well were plated out in 96-well plates, Rov9 cells were cultured in the presence of 1 μg/ml doxycycline (Sigma) to induce PrPC expression for 48 h prior to the addition of infectious brain homogenate, and doxycycline was added to the medium for subsequent culturing of the Rov9 cells in order to maintain PrPC expression. Cells were cultured for a further 3 days before being passaged every 3 days at a 1:6 dilution into fresh 96-well plates and then on the fourth day were transferred to 96-well enzyme-linked immunospot assay (ELISPOT) plates (Multiscreen-IP filter plates; Millipore). The plates were dried, and a modified SSCA was performed. Alterations to the published protocol included increased proteinase K (PK) concentrations (4 and 10 μg/ml for MovS6 and Rov9 cells, respectively) and the use of 6H4 (Prionics) as a primary antibody for detection of PrPres in the plates. Following immunodetection, infected cells were visualized with an alkaline phosphate conjugate substrate kit (Bio-Rad). Positive cells (spots) were counted using a Zeiss KS-ELISPOT imaging system running WellScan software (Imaging Associates).
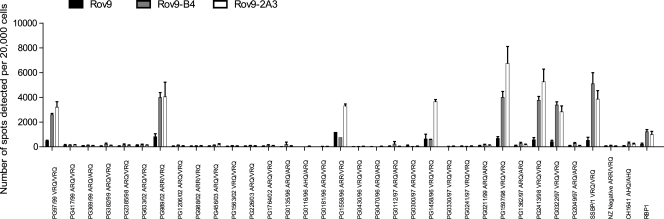
To accurately assess the total cell number per well, duplicate ELISPOT plates were prepared with a 1:10 dilution of cells and dried in the same way as described above. Cells were stained with Trypan blue solution and counted using the Zeiss KS-ELISPOT imaging system. All results are presented as the number of infected cells per 20,000 cells for ease of comparison; however, in most cases the number of cells assayed per well was ∼5,000 cells. In some cases, the number of infected cells detected by the SSCA with the Rov9-2A3 subclone is an underestimate: due to the large number of infected cells present, the individual spots merge and the ELISPOT plate reader is unable to count the individual infected cells, resulting in undercounting of infected cells (see Fig. 3). However, at lower concentrations of inocula, the number of infected cells is decreased and differences between the sensitivities of the subclones become apparent (see Fig. 5a). In all cases, the wells have been inspected under the microscope to confirm infected cells; this is especially important for samples that are borderline positive with regard to the negative controls. In some cases, wells appear positive by spot numbers but, upon microscopic inspection, the spots detected are drying defects within the well or the effects of the edge of the well.
FIG. 3.
Permissibility of Rov9, Rov9-B4, and Rov9-2A3 cell lines to a range of ovine scrapie isolates (a 10−3 dilution). The number of infected cells (spots) was determined using the SSCA. The cutoff values for determining samples to be positive or negative for infection, calculated from the average number of spots detected in the New Zealand-derived negative controls plus 3 times the standard deviation, are 107, 223, and 105 spots per 20,000 cells plated in the Rov9, Rov9-B4, and Rov9-2A3 cells, respectively. In some cases, the number of infected cells detected for the most sensitive subclones is an underestimate that is due to the ELISPOT plate reader being unable to separate the large numbers of individual spots. Error bars show standard errors of the means.
FIG. 5.
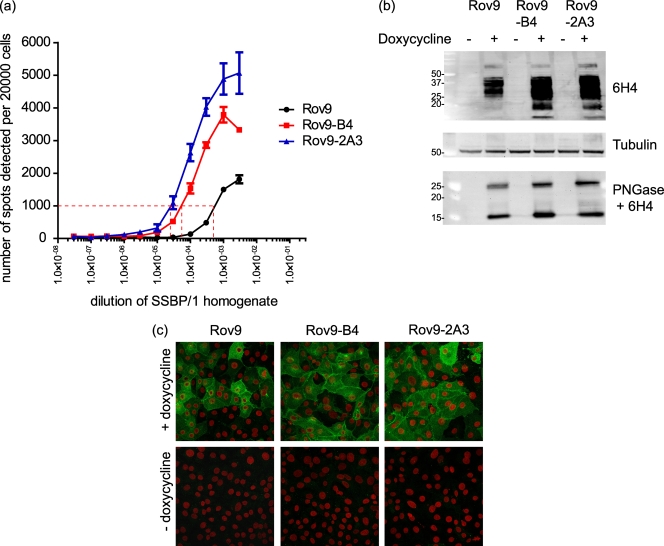
Comparison of parental Rov9 cells and the most SSBP/1-permissive first- and second-generation subclones. (a) Dose-response graph showing the average number of infected cells (spots) detected by the SSCA when cells were challenged with increasingly dilute SSBP/1. x axis is a logarithmic scale. The dashed red line represents 1,000 infected cells. Error bars show standard errors of the means. (b) Western blot of PrPC expression levels in Rov9 cells and the Rov9-B4 and -2A3 subclones. Amounts of 10 μg of total protein from induced (+ doxycycline) and uninduced (− doxycycline) cells were used. Lysates were PNGase treated to deglycosylate the PrPC so that the total amount of PrPC could be compared without the interference of glycosylated PrP. Molecular masses of size markers are shown on the left in kDa. (c) Immunofluorescent staining of PrPC in Rov9 and the two subclones Rov9-B4 and Rov9-2A3. Cells were either induced or uninduced for 48 h prior to staining for PrPC, which is colored green. Nuclei are falsely colored red.
To aid comparison of the subclones, the concentration of the inoculum required to infect 1,000 cells out of 20,000 cells plated has been calculated. This is an arbitrary value similar to that used by Mahal et al. (21), but whereas Mahal et al., used 300 infected cells per 20,000, we have chosen 1,000 cells.
Scrapie inocula.
Two experimental scrapie isolates, SSBP/1 from a VRQ/VRQ sheep and CH1641 from an AHQ/AHQ sheep (14); a brain pool (RBP1) (30); New Zealand-derived uninfected controls; and field case samples of scrapie were obtained from the Veterinary Laboratories Agency (Table 1). All material was brain frontal cortex tissue except RBP1, which is a pool of whole brains from 17 sheep of different PrP genotypes. All brain samples were homogenized in sterile phosphate-buffered saline (PBS) (10% wt/vol) and stored at −80°C; they were diluted in cell culture medium to appropriate dilutions prior to being added to cells.
TABLE 1.
Characteristics of sheep from which scrapie field samples, experimental scrapie, and negative-control samples were obtained for use as inoculaa
| Inoculum | Sex | Age | Breed or source | PrP genotype(s) |
|---|---|---|---|---|
| PG0097/99 | F | 6 y | British Friesland | VRQ/VRQ |
| PG0417/99 | F | 5 y | Swaledale | ARQ/VRQ |
| PG0369/99 | F | 4 y | Mule | ARQ/ARQ |
| PG0382/99 | F | 3 y 1 mo | Suffolk cross | ARQ/VRQ |
| PG2385/98 | F | 3 y 11 mo | Charollais | ARQ/VRQ |
| PG0633/02 | F | 4 y | Charollais × Suffolk | ARQ/VRQ |
| PG0989/02 | F | 8 y | Easicare | ARR/VRQ |
| PG1206/02 | F | 3 y | Brecknock Hill Cheviot | ARQ/VRQ |
| PG0988/02 | F | 7 y | Easicare | ARR/VRQ |
| PG0456/03 | F | 3 y | Swaledale | ARQ/VRQ |
| PG1563/02 | M | 2 y 8 mo | Bleu de Maine | VRQ/VRQ |
| PG0226/03 | F | 5 y | Swaledale | ARQ/VRQ |
| PG1764/02 | F | 3 y 8 mo | British Friesland | ARQ/VRQ |
| PG0135/96 | F | 5 y | Finn Dorset cross | ARQ/ARQ |
| PG0116/96 | F | 3 y 1 mo | Finn Dorset | AHQ/ARQ |
| PG0181/96 | F | 2 y 9 mo | Swaledale | ARQ/VRQ |
| PG1558/96 | F | 4 y 6 mo | Welsh Mountain | ARR/VRQ |
| PG0430/96 | F | 2 y 2 mo | Swaledale | VRQ/VRQ |
| PG0470/96 | F | 3 y | Suffolk | ARQ/ARQ |
| PG0112/97 | F | 6 y | Mule | ARQ/ARQ |
| PG0300/97 | F | 3 y 10 mo | Mule | ARQ/VRQ |
| PG1458/96 | F | 2 y 5 mo | Welsh Mountain × Wiltshire | VRQ/VRQ |
| PG0330/97 | F | 3 y | Bleu de Maine | VRQ/VRQ |
| PG0241/97 | F | 3 y | Swaledale | VRQ/VRQ |
| PG0211/99 | F | 5 y | Mule | ARQ/ARQ |
| PG1597/98 | F | 2 y 7 mo | Crossbreed | VRQ/VRQ |
| PG1362/97 | M | 3 y | Suffolk | ARQ/ARQ |
| PG2413/98 | F | 2 y 2 mo | Polled Dorset | VRQ/VRQ |
| PG0322/97 | F | 2 y | Welsh Mountain cross | VRQ/VRQ |
| PG2049/97 | F | 8 y | Suffolk cross | ARQ/ARQ |
| PG1475/04 (SSBP/1) | M | 1 y 4 mo | Cheviot | VRQ/VRQ |
| PG1271/05 (CH1641) | M | 2 y 3 mo | Cheviot | AHQ/AHQ |
| PG1531/01 (NZ negative) | M | 4 y | Bleu de Maine | ARR/VRQ |
| RBP1 | Pool from of 17 scrapie-infected sheep | 5 × ARQ/VRQ, 1 × ARR/VRQ, 4 × ARQ/ARQ, 1 × ARQ/AHQ, 6 × VRQ/VRQ |
Scrapie samples from those animals whose data are highlighted in bold font were able to infect all the cell lines and subclones thereof that were tested.
Subcloning by limiting dilution.
Rov9 cells were counted and plated out in 96-well plates with an average of one cell per 3 wells. After clonal cells were established, they were transferred to new plates and tested for permissiveness to infection using the SSCA.
Western blotting.
For PrPC analysis, Rov9 cells and subclones were grown in 25-cm2 flasks in the presence of 1 μg/ml doxycycline for 48 h. For infected cells, cells were grown in 24-well plates and inoculated with scrapie-infected brain; after 3 days the cells were passaged 1:6 into a fresh 24-well plate. At 3-day intervals, cells were passaged 1:1 into 6-well plates, transferred to 25-cm2 flasks, and finally transferred to 75-cm2 flasks before collection once confluent. Prior to collection, 20,000 cells from each infection were plated out for SSCA analysis to confirm infection. Cells were lysed in 1 ml radioimmunoprecipitation buffer (50 mM Tris, pH 8.0, 150 mM NaCl, 0.5% sodium deoxycholate, 0.1% SDS, 1% NP-40, 1 mM EDTA). Lysates were treated with benzonase (25 U/ml of lysates; Sigma) to digest DNA. Cell lysates were clarified by centrifugation, and protein concentration determined by bicinchoninic acid (BCA) protein assay (Pierce). For PrPC analysis, 100 μg of protein was precipitated in 1 ml cold methanol at −20°C for 1 h before being washed and resuspended in 50 μl loading buffer and boiled for 5 min. Five microliters (equivalent to 10 μg of total protein) was loaded onto each lane of a gel. For detecting PrPres in infected cell lysates, 500 μg of total protein was digested with PK (20 μg/ml for 60 min at 37°C, Roche) before centrifugation at 47,000 × g for 30 min, and the pellet was resuspended in 25 μl loading dye (Prionics) and loaded onto a polyacrylamide gel (Novex 12% Bis-Tris gels). After separation by electrophoresis, proteins were transferred to polyvinylidene difluoride (PVDF) membrane (Millipore) and detected by antibodies against PrP (6H4; Prionics) or tubulin (Santa Cruz Biotechnology, Inc.). Proteins were visualized by using CDP-Star.
For Western blotting of scrapie inoculum samples, 20 μl of 10% brain homogenate was mixed with an equal volume of homogenization buffer (Prionics). The homogenate was digested with 37.5 μg/ml PK for 1 h at 37°C before the addition of 2 mM phenylmethylsulfonyl fluoride (PMSF; Sigma), to stop digestion, and 40 μl loading buffer. Samples were boiled, 25-μl amounts run on 12% Bis-Tris gels (Novex), and Western blots performed as described above for cell lysates.
Deglycosylation of PrPC.
For deglycosylating PrPC, lysates were digested with peptide-N-glycosidase F (PNGase). Proteins in 10 μl of cell lysate were precipitated with 7 volumes of ice-cold methanol for 2 h at −20°C and collected by centrifugation at 13,000 rpm for 20 min. The protein pellet was resuspended in 2 μl of denaturing buffer (New England Biolabs) and 18 μl water and heated to 100°C for 10 min. Two microliters Nonidet P40, 2 μl G7 buffer, and 0.8 μl PNGase (all reagents from New England Biolabs) were added to the resuspended pellet. Samples were briefly vortexed for mixing and incubated at 37°C overnight. Digested proteins were reprecipitated with 7 volumes of ice-cold methanol for 2 h at −20°C and collected by centrifugation at 13,000 rpm for 20 min. Precipitates were resuspended in loading dye and separated by electrophoresis as described above. Densitometry measurements were performed using Labworks software (UVP Bioimaging Systems).
Immunofluorescence confocal microscopy.
Uninfected Rov9 cells were grown on coverslips in 24-well plates in the presence or absence of doxycycline for 48 h and then fixed in 4% formaldehyde solution in PBS. Cells were permeabilized with 0.1% Triton X-100 in PBS for 2 min before immunofluorescent staining with an antibody targeted against PrP (6H4; Prionics) and an IgG secondary anti-mouse antibody to which Alexa 488 was conjugated (Molecular Probes). Nuclei were counterstained with Hoechst 33342 (Molecular Probes). Coverslips were mounted in ProLong Gold (Molecular Probes), and images were captured using a Leica TCS SP2 confocal microscope.
Flow cytometry.
Rov9 cells and subclones were grown in 25-cm2 flasks in the presence or absence of 1 μg/ml doxycycline for 48 h. Cells were detached by incubation with 3 ml Versene (Gibco) for 30 min at 37°C. Detached cells were washed in PBS and labeled with SAF32 anti-PrP antibody (Spi-Bio) (1 μg/106 cells for 1 h). Cells were washed twice in wash buffer (PBS, 0.5% bovine serum albumin, 0.1% sodium azide, 0.002% EDTA) and incubated for an hour at room temperature with a phycoerythrin-conjugated secondary antibody (BD Pharminogen). Cells were washed twice and resuspended in 0.5 ml PBS. Cells were analyzed using a Beckman Coulter Epics XL Flow Cytometer. Ten thousand events were recorded for each sample and analyzed using Expo32 ADC software.
Titer estimation with SCEPA.
Thirty-two wells for each of 7 dilutions of homogenate and 32 wells of New Zealand-derived negative control were plated, and the scrapie cell endpoint assay (SCEPA) performed (22). This differs from the standard scrapie cell assay in that after the initial 3-day incubation period with the inoculum, 50% of the medium is changed and the cells are cultured for a further 3 days. The cells are then passaged three times at a 1:3 dilution every 2 days, followed by three passages at a 1:6 dilution every 3 days before the ELISPOT assay is performed as described for the SSCA. Titers were estimated using the Reed and Muench 50% endpoint calculation method (28). A well is deemed to be positively infected if the number of spots detected by SCEPA is greater than the average of the results for the 32 wells of New Zealand-derived scrapie-negative isolate plus 3 times the standard deviation.
RESULTS
Transmission of sheep scrapie to Rov9 and MovS6 cells.
To compare the susceptibilities of the Rov9 and MovS6 cell lines to natural cases of scrapie, both cell lines were challenged with a panel of 30 field isolates of scrapie and de novo infection assessed with the standard scrapie cell assay (SSCA) (18). As the Rov9 and MovS6 cell lines express high levels of PrPC, the proteinase K (PK) concentration was optimized to ensure complete digestion of PrPC, ensuring that only PK-resistant PrP (PrPSc)-expressing cells were detected in the SSCA and preventing false-positive results (data not shown).
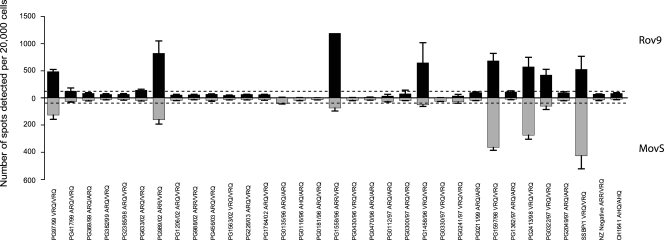
Rov9 and MovS6 cells were challenged with brain homogenates from 30 field cases of scrapie, 2 experimental scrapie isolates (SSBP/1 and CH1641) (10, 12), and a New Zealand-derived scrapie-negative case, described in Table 1.
The average numbers of infected cells per 20,000 cells determined by SSCA are plotted in Fig. 1. The Rov9 and MovS6 cells are permissive to the same 7 natural scrapie samples (PG97/99, PG989/02, PG1558/96, PG1458/96, PG1597/98, PG2413/98, and PG0322/97) and the SSBP/1 experimental isolate (Fig. 1). A greater number of infected cells was seen in the Rov9 cell line than in the MovS6 cell line.
FIG. 1.
Permissibility of Rov9 and MovS6 cell lines to infection with natural and experimental sheep scrapie isolates. The average number of de novo infected cells (spots) detected by SSCA in 4 duplicate wells of Rov9 (upper panel, black) and MovS6 (lower panel, gray) cell lines after exposure to a 10−3 dilution of each scrapie inoculum is shown, and the PrP genotype is given for each isolate. The New Zealand-derived negative control represents a scrapie-negative isolate used to calculate the background level of nonspecific spots. The dashed lines represent the average number of spots detected in the negative control plus 3 times the standard deviation; below this line, the samples are considered uninfected. Error bars show standard errors of the means. Note the difference in y axis scale for the two cell lines.
Five of the 7 field case samples and the SSBP/1 isolate that produced de novo infection are from VRQ homozygous sheep, and the other two samples are from ARR/VRQ heterozygous animals. Among the PrP genotypes of the 23 natural scrapie isolates that failed to infect the cell lines as determined by SSCA, 7 were ARQ/ARQ, 10 ARQ/VRQ, 1 AHQ/ARQ, 1 ARR/VRQ, and 4 VRQ/VRQ (Fig. 1). The CH1641 experimental isolate, from an AHQ/AHQ sheep, also failed to transmit infectivity to the cell lines.
Subcloning of Rov9 cells and screening for increased permissiveness to infection.
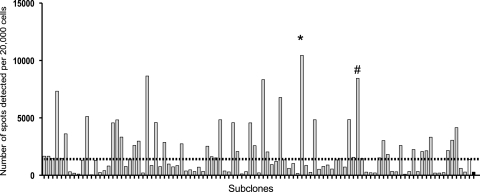
Using cell lines permissive to murine prion strains, it has been shown that sensitivity to scrapie infection can be altered through subcloning (18, 21). To further explore this finding, the Rov9 cell line was subcloned by limiting dilution and 50 first-generation subclones were obtained. These 50 subclones were screened for permissiveness to infection using SSBP/1 and CH1641 scrapie isolates as inocula. Using the SSCA, 11 subclones were found to be more permissive to infection with SSBP/1 than the parental Rov9 cells. No subclone was found to be permissive to infection with CH1641. The Rov9-B4 subclone was the most sensitive to SSBP/1 infection, and therefore, Rov9-B4 was subjected to a second round of subcloning by limiting dilution. Ninety-nine Rov9 second-generation subclones were obtained from the Rov9-B4 subclone and challenged with the SSBP/1 isolate. Thirty-five of the 99 Rov9 second-generation subclones were more permissive to SSBP/1 than the Rov9-B4 subclone (Fig. 2). The Rov9-1G10 second-generation subclone was found to be the most sensitive (highest number of infected cells per 20,000 cells) by SSCA; however, this clone did not grow consistently in culture and therefore the next most SSBP/1-sensitive subclone, Rov9-2A3, was chosen for further investigation in this study.
FIG. 2.
Permissibility of second-generation Rov9 subclones to a 10−4 dilution of SSBP/1 detected by SSCA. Each gray bar represents the average number of de novo infected cells (spots) detected by SSCA for an individual subclone; the white bar represents the Rov9-B4 first-generation subclone; and the black bar represents the parental Rov9 cells. The dashed black line represents the number of spots detected with the first-generation subclone Rov9-B4. *, Rov9-1G10 subclone; #, Rov9-2A3 subclone.
Comparison of Rov9 subclones with the parental cell line.
The most SSBP/1-sensitive first- and second-generation subclones, Rov9-B4 and Rov9-2A3, were challenged with the 30 natural and 2 experimental (SSBP/1 and CH1641) scrapie isolates. In addition, the cells were challenged with the scrapie brain pool, RBP1 (30). The data presented in Fig. 3 show that although the number of infected cells detected by the SSCA is increased, in some cases by more than 10-fold, in the second-generation subclone compared to the number in the parental Rov9 cells, the range of samples that resulted in de novo infection remains unchanged.
To confirm the presence of de novo PrPres in the infected cell lines, Western blotting was performed on PK-digested lysates of Rov9-2A3 cells challenged with 19 of the field case isolates that were assessed by SSCA (Fig. 4a). The presence of PrPres in the cell lysate correlates with the de novo infection results obtained for the same scrapie inocula (PG97/99, PG1558/96, PG989/02, PG1458/86, PG1597/98, PG2413/98, PG0322/97, SSBP/1, and RBP1) in the SSCA (Fig. 3).
FIG. 4.
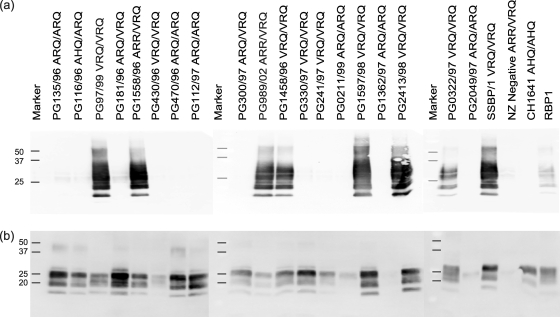
Western blot analysis of PrPres in cell lysates of scrapie-challenged Rov9-2A3 cells (a) and ovine inocula (b). Molecular masses of size markers are shown in kDa.
The amount of detectable PrPSc present in the inocula does not seem to be a determining factor in successful de novo infection, as there is no correlation between PrPres levels and success of infection (compare Fig. 4a and b). For example, field cases PG330/97 and PG2413/98 are both from VRQ/VRQ sheep and have similar levels of PrPres on the Western blot (Fig. 4b); however, only PG2413/98 is able to produce de novo infection in the cell lines.
The estimated infectious titers of SSBP/1 in the Rov9 parental cells and two of the subclones, Rov9-B4 and Rov9-2A3, were calculated using the scrapie cell endpoint assay (SCEPA). The SCEPA is a modification of the original SSCA that allows for more sensitive detection than the standard assay (22). Cells were challenged with increasingly dilute SSBP/1 inocula (10−2 to 10−8 dilution), and the SCEPA performed. From the data shown in Fig. 5a, the dilutions of SSBP/1 required to produce 1,000 (1 in 20) infected cells were calculated as 5.5 × 10−4, 5 × 10−5, and 2.5 × 10−5 for Rov9, Rov9-B4, and Rov9-2A3 cells, respectively. Therefore, more than 20 times the amount of SSBP/1 was required to infect the same number of cells in the Rov9 cell line as in the Rov9-2A3 cells. The infectious titers (50% infectious dose [ID50]) of SSBP/1 calculated using the method of Reed and Muench (28) ranged from 105.18 in the Rov9 parental cell line to 106.49 ID50/ml in the most sensitive subclone, Rov9-2A3. The difference in sensitivity of these cells is reflected in the infectious titer estimates, where a higher infectious titer is given for the subclones.
To elucidate potential reasons why the Rov9 subclones were more sensitive to some scrapie isolates than the parental Rov9 cells, various aspects of their PrPC expression were studied.
First, PrPC expression levels were assessed by Western blot, where the more sensitive subclones were seen to have increased PrPC levels (Fig. 5b). Densitometry measurements of the PNGase-digested full-length PrPC (27-kDa band) show 37 and 54% increases in expression in the Rov9-B4 and Rov9-2A3 subclones, respectively, compared to the expression level in the Rov9 cells. The 17-kDa band, which is possibly the C1 degradation product of the full-length PrPC (8), shows 62 and 16% increases in expression in the Rov9-B4 and Rov9-2A3 subclones, respectively, compared to its expression level in the Rov9 cells.
Second, the subcellular location of PrPC was assessed by immunofluorescent staining and confocal microscopy. In the parental Rov9 cells and the Rov9-B4 and Rov9-2A3 subclones, the PrPC staining is predominantly localized on the plasma membrane, as shown by the strong staining at the boundaries between cells (Fig. 5c). The only difference observed appears to be the number of cells staining positively for PrPC, with a greater number of PrPC-expressing cells found in the subclones than in the Rov9 cells. This increased number of cells with positive expression may account for the increased total PrPC levels seen in the Western blots (Fig. 5b).
To quantify whether there are indeed more PrPC-expressing cells in the subclones than in the parental cells, the cell surface PrPC was fluorescently labeled and cells subjected to flow cytometry. Approximately 76% of the Rov9 cells stained positively for cell surface PrPC, slightly less than for Rov9-B4, at 81%, and Rov9-2A3 cells, at 88% (data not shown). These results confirm the findings seen with the immunofluorescent staining and confocal microscopy.
SCEPA titer estimation of a well-characterized scrapie sample and comparison to mouse bioassay.
The Ripley brain pool 1 (RBP1) is a scrapie pool produced from 17 scrapie-infected brains from sheep with a range of PrP genotypes. By mouse bioassay in RIII mice, the infectious titer of the RBP1 was calculated to be 103.96 ID50/gram of tissue (30). The infectious titer of RBP1 in Rov9-2A3 cells, calculated using the 50% endpoint estimation of Reed and Muench (28), was 104.99 ID50/gram of tissue, more than 1 log higher than the wild-type mouse bioassay titer estimate.
DISCUSSION
In this study, we have adapted the SSCA protocol for use with Rov9 and MovS6 cell lines and subclones thereof, and the assay has been successfully used for the detection of de novo infectivity and infectious titer estimation of ovine scrapie.
Using this method, the range of sheep scrapie samples able to infect the cell lines examined in this study was found to be limited. Out of the 30 natural scrapie and 2 experimental scrapie isolates used to challenge the MovS6 and Rov9 cells, the same 7 natural scrapie isolates and SSBP/1 resulted in de novo PrPres in all the cell lines, while the remaining 23 natural isolates and CH1641 did not transmit to any of the cell lines.
There is evidence that different cell lines are able to propagate different strains of mouse-adapted scrapie (21). Furthermore, Mahal et al., have shown that for mouse-passaged TSE strains, cell lines of different origin have different levels of sensitivity (21). For example, all four of the cell lines they used (2 subclones of neuroblastoma [N2a-PK1 and R33], a fibroblast cell line [L929], and a central nervous system-derived cell line [CAD]) were permissive to infection with 22L, but only three cell lines were permissive to infection with RML, 2 were permissive to infection with ME7, and one cell line was permissive to infection with the 301C strain. In our study, in spite of using cell lines from different origins, namely, Rov9 cells, derived from a rabbit kidney epithelial cell line, and the MovS6 cell line, which is a murine Schwann cell line, no differences in permissibility to the different isolates were seen. However, the Rov9 cell line was consistently more sensitive to all the transmissible ovine scrapie isolates than the MovS6 cell line.
The amount of PrPres present in the scrapie inocula did not correlate with de novo infection results. Western blotting of the original inocula used to challenge the cell lines shows a range of PrPres levels; in fact, the isolate with the highest levels of PrPres is an ARQ/VRQ heterozygous sheep (PG181/96) (Fig. 4b) which is unable to infect the cell lines. Of the VRQ/VRQ isolates tested, those that are able to infect the cell lines do not have higher levels of PrPres than those isolates that are unable to infect the cells, and the glycoform profiles are the same (Fig. 4a and b).
All of the 7 natural scrapie isolates that were able to infect the cells (5 VRQ/VRQ and 2 VRQ/ARR) plus the SSBP/1 isolate (VRQ/VRQ) were from sheep that carry at least one copy of the VRQ PrP allele, the same allele expressed by the cell lines. However, not all scrapie isolates from VRQ heterozygous or homozygous sheep were able to infect the cells. Therefore, although some PrP homology between inoculum and cell line may be required, it is not sufficient to ensure de novo infection.
There was no discernible difference in the ages of the sheep that had the transmissible and nontransmissible VRQ/VRQ isolates (Table 1); however, the four nontransmissible VRQ/VRQ isolates are, notably, from the Swaledale and Bleu de Maine breeds. The significance of this observation cannot be determined due to the low numbers of samples analyzed. It is possible that the nontransmitting isolates, particularly the VRQ/VRQ isolates, are a different scrapie strain than the transmitting isolates of the same PrP genotype. It is well known that different strains of natural sheep scrapie exist, as determined by lesion profiles (13) and incubation periods in mice. There are very limited amounts of ovine scrapie material of known strains available for testing; however, 17 of the scrapie isolates assayed here are currently being strain typed by mouse bioassay. Once known, the strain information will be used to elucidate whether the permissiveness to infection of the cells tested here is determined by strain.
Studies of mice have shown that scrapie strain can affect whether an isolate is transmissible to a cell line. In a study by Mahal et al. (22), the 4 cell lines used in a cell panel assay and the 4 mouse-adapted scrapie strains used were all from Prnpa mice, and therefore, permissiveness or resistance to infection was thought to be due to strain. The authors considered that strains of prions with the same PrP protein could be differentiated by glycosylation, the requirement for different cell-specific factors (i.e., small RNAs), structure-specific chaperones, or a cell-specific fibril cleavage activity.
Subcloning of the Rov9 cell line was undertaken to try to improve the sensitivity of the parental line to infection and in an attempt to increase the range of scrapie isolates that resulted in de novo PrPres detection. While the first undertaking was successful, the second was not. All 50 first-generation and 99 second-generation sibling clones were screened with 2 experimental scrapie isolates (CH1641 and SSBP/1) which differ in terms of PrP genotypes of inocula, biochemical characteristics of PrPSc, and known transmissibility to sheep (12, 15, 17, 32).
As previously described, SSBP/1 infected the Rov9 and MovS6 cell lines and the Rov9 subclones to various degrees. The CH1641 scrapie isolate did not infect any of the cell lines examined in this study. The CH1641 isolate has a lower apparent PrPres molecular mass than other sheep scrapie isolates and, in this respect, is more similar to ovine BSE. Like ovine BSE, CH1641 is not easily transmitted to VRQ/VRQ sheep (12); therefore, it is perhaps not surprising that the VRQ-expressing cell lines used in this study were not permissive to infection with this isolate. However, although rare, natural cases of CH1641-like scrapie detected in VRQ/VRQ sheep have been reported (19).
The process of selecting the Rov9 subclones evaluated in this study may have biased the results such that only subclones which are highly permissive to SSBP/1 and SSBP/1-like scrapie samples were selected. Screening of all the Rov9 subclones with other natural scrapie cases, in particular the VRQ/VRQ nontransmitting isolates, may result in identifying additional permissive subclones.
The finding in our study that sibling Rov9 clones have different sensitivities to SSBP/1-like scrapie isolates is not unexpected, as a similar effect has been noted in other cell lines (4, 18, 22). The fraction of cells expressing PrPC may be a contributory factor in the increased sensitivity in the subclones, and the PrPC levels in the most sensitive subclones are elevated compared to the levels in the parental Rov9 cell line. However, previous studies have shown that cells expressing increased levels of PrPC are not necessarily the most sensitive to infection (23), so this may not completely account for the increased sensitivity seen in the subclones.
The distribution of PrPC throughout the cell does not appear to vary greatly between the subclones and the parental cells, with the gross subcellular location revealed by confocal microscopy being indistinguishable between the parental cells and the subclones. The majority of PrPC staining is located on the plasma membrane, and a small fraction in intracellular organelles, most likely the Golgi body. Using flow cytometry, we have shown that the number of cells expressing cell surface PrPC increased from 76% in the Rov9 cells to 88% in the Rov9-2A3 clone, and this may have an influence on infection, as more cells would be available for infection. A study by Paquet et al. (25) has shown that Rov9 cells need to be expressing PrPC at the time of exposure to prions for infection to be successful.
Furthermore, PrPC expression is required for efficient cell-to-cell transmission of prions (26). The subclone cell populations in our study are clearly not homogenous in this respect, as not all cells were expressing PrPC upon induction. The homogeneity may have been lost during the subsequent culturing through some degree of genetic drift. Alternatively, the phase of the cell cycle may affect inducible expression, resulting in not all cells expressing detectable PrPC at the same time, as has been shown for the cytomegalovirus promoter (5).
Improving the sensitivity of cell lines to scrapie infection is an essential requirement since, if they are to be used to estimate infectious titer, the cell-based bioassay needs to be at least as sensitive as the mouse bioassay. The titer estimation of the RBP1 isolate using Rov9-2A3 cells and the SCEPA approach determined in this study demonstrated that the Rov9-2A3 bioassay was 1 log more sensitive than the mouse bioassay using a wild-type mouse line.
In conclusion, this study has shown that the SSCA sensitively detects de novo infection in a subset of natural scrapie cases, including a 10−6 dilution of high-titer brain tissue (estimated to contain 106.49 ID50/gram of tissue). The SSCA efficiently detected de novo infection in two cell lines and proved to be more sensitive in its titer estimation than the mouse bioassay. Subcloning of cell lines was successfully applied to the Rov9 cells, and subclones more sensitive to natural scrapie infection were isolated. However, increasing the sensitivity of the Rov9 cell line to SSBP/1 infection did not correlate with broadening susceptibility, as the specificity of permissiveness and resistance to other scrapie isolates was maintained. Furthermore, neither high levels of detectable PrPSc nor PrP protein homology between inoculum and cell line were sufficient to ensure successful transmission of infectivity, suggesting that other factors contributing to the diversity of isolates, such as the scrapie strain, influence transmission ex vivo. With further development of existing cell lines and identification of new cell lines permissive to infection with scrapie, a cell-based approach could allow for a cheaper, faster, and more ethical way to assess infectivity, estimate titer, and determine the strain of ovine scrapie isolates.
Acknowledgments
This work was supported by Defra, UK, project no. SE2003.
We are grateful to R. Lockey, S. Bellworthy, S. Ryder, and H. Simmons for the RBP1 tissue and titer estimation by mouse bioassay and to Peter Klöhn for advice on the SSCA and SCEPA.
Footnotes
Published ahead of print on 23 December 2009.
REFERENCES
- 1.Archer, F., C. Bachelin, O. Andreoletti, N. Besnard, G. Perrot, C. Langevin, A. Le Dur, D. Vilette, A. Baron-Van Evercooren, J. L. Vilotte, and H. Laude. 2004. Cultured peripheral neuroglial cells are highly permissive to sheep prion infection. J. Virol. 78:482-490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barron, R. M., S. L. Campbell, D. King, A. Bellon, K. E. Chapman, R. A. Williamson, and J. C. Manson. 2007. High titers of transmissible spongiform encephalopathy infectivity associated with extremely low levels of PrPSc in vivo. J. Biol. Chem. 282:35878-35886. [DOI] [PubMed] [Google Scholar]
- 3.Baylis, M., C. Chihota, E. Stevenson, W. Goldmann, A. Smith, K. Sivam, S. Tongue, and M. B. Gravenor. 2004. Risk of scrapie in British sheep of different prion protein genotype. J. Gen. Virol. 85:2735-2740. [DOI] [PubMed] [Google Scholar]
- 4.Bosque, P. J., and S. B. Prusiner. 2000. Cultured cell sublines highly susceptible to prion infection. J. Virol. 74:4377-4386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brightwell, G., V. Poirier, E. Cole, S. Ivins, and K. W. Brown. 1997. Serum-dependent and cell cycle-dependent expression from a cytomegalovirus-based mammalian expression vector. Gene 194:115-123. [DOI] [PubMed] [Google Scholar]
- 6.Bruce, M. E. 1993. Scrapie strain variation and mutation. Br. Med. Bull. 49:822-838. [DOI] [PubMed] [Google Scholar]
- 7.Bruce, M. E., A. Boyle, S. Cousens, I. McConnell, J. Foster, W. Goldmann, and H. Fraser. 2002. Strain characterization of natural sheep scrapie and comparison with BSE. J. Gen. Virol. 83:695-704. [DOI] [PubMed] [Google Scholar]
- 8.Chen, S. G., D. B. Teplow, P. Parchi, J. K. Teller, P. Gambetti, and L. Autilio-Gambetti. 1995. Truncated forms of the human prion protein in normal brain and in prion diseases. J. Biol. Chem. 270:19173-19180. [DOI] [PubMed] [Google Scholar]
- 9.Courageot, M. P., N. Daude, R. Nonno, S. Paquet, M. A. Di Bari, A. Le Dur, J. Chapuis, A. F. Hill, U. Agrimi, H. Laude, and D. Vilette. 2008. A cell line infectible by prion strains from different species. J. Gen. Virol. 89:341-347. [DOI] [PubMed] [Google Scholar]
- 10.Dickinson, A. G. 1976. Scrapie in sheep and goats. Front. Biol. 44:209-241. [PubMed] [Google Scholar]
- 11.Falanga, P. B., M. C. Blom-Potar, P. Bittoun, M. E. Goldberg, and M. Hontebeyrie. 2006. Selection of ovine PrP high-producer subclones from a transfected epithelial cell line. Biochem. Biophys. Res. Commun. 340:309-317. [DOI] [PubMed] [Google Scholar]
- 12.Foster, J. D., and A. G. Dickinson. 1988. The unusual properties of CH1641, a sheep-passaged isolate of scrapie. Vet. Rec. 123:5-8. [DOI] [PubMed] [Google Scholar]
- 13.Fraser, H., and A. G. Dickinson. 1968. The sequential development of the brain lesion of scrapie in three strains of mice. J. Comp. Pathol. 78:301-311. [DOI] [PubMed] [Google Scholar]
- 14.Gielbert, A., L. A. Davis, A. R. Sayers, J. Hope, A. C. Gill, and M. J. Sauer. 2009. High-resolution differentiation of transmissible spongiform encephalopathy strains by quantitative N-terminal amino acid profiling (N-TAAP) of PK-digested abnormal prion protein. J. Mass Spectrom. 44:384-396. [DOI] [PubMed] [Google Scholar]
- 15.Goldmann, W., N. Hunter, G. Smith, J. Foster, and J. Hope. 1994. PrP genotype and agent effects in scrapie: change in allelic interaction with different isolates of agent in sheep, a natural host of scrapie. J. Gen. Virol. 75(Pt. 5):989-995. [DOI] [PubMed] [Google Scholar]
- 16.Iwamaru, Y., T. Takenouchi, K. Ogihara, M. Hoshino, M. Takata, M. Imamura, Y. Tagawa, H. Hayashi-Kato, Y. Ushiki-Kaku, Y. Shimizu, H. Okada, M. Shinagawa, H. Kitani, and T. Yokoyama. 2007. Microglial cell line established from prion protein-overexpressing mice is susceptible to various murine prion strains. J. Virol. 81:1524-1527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jeffrey, M., L. Gonzalez, A. Chong, J. Foster, W. Goldmann, N. Hunter, and S. Martin. 2006. Ovine infection with the agents of scrapie (CH1641 isolate) and bovine spongiform encephalopathy: immunochemical similarities can be resolved by immunohistochemistry. J. Comp. Pathol. 134:17-29. [DOI] [PubMed] [Google Scholar]
- 18.Klöhn, P. C., L. Stoltze, E. Flechsig, M. Enari, and C. Weissmann. 2003. A quantitative, highly sensitive cell-based infectivity assay for mouse scrapie prions. Proc. Natl. Acad. Sci. U. S. A. 100:11666-11671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lezmi, S., S. Martin, S. Simon, E. Comoy, A. Bencsik, J. P. Deslys, J. Grassi, M. Jeffrey, and T. Baron. 2004. Comparative molecular analysis of the abnormal prion protein in field scrapie cases and experimental bovine spongiform encephalopathy in sheep by use of Western blotting and immunohistochemical methods. J. Virol. 78:3654-3662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Maas, E., M. Geissen, M. H. Groschup, R. Rost, T. Onodera, H. Schatzl, and I. M. Vorberg. 2007. Scrapie infection of prion protein-deficient cell line upon ectopic expression of mutant prion proteins. J. Biol. Chem. 282:18702-18710. [DOI] [PubMed] [Google Scholar]
- 21.Mahal, S. P., C. A. Baker, C. A. Demczyk, E. W. Smith, C. Julius, and C. Weissmann. 2007. Prion strain discrimination in cell culture: the cell panel assay. Proc. Natl. Acad. Sci. U. S. A. 104:20908-20913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mahal, S. P., C. A. Demczyk, E. W. Smith, Jr., P. C. Klohn, and C. Weissmann. 2008. Assaying prions in cell culture: the standard scrapie cell assay (SSCA) and the scrapie cell assay in end point format (SCEPA). Methods Mol. Biol. 459:49-68. [DOI] [PubMed] [Google Scholar]
- 23.Nishida, N., D. A. Harris, D. Vilette, H. Laude, Y. Frobert, J. Grassi, D. Casanova, O. Milhavet, and S. Lehmann. 2000. Successful transmission of three mouse-adapted scrapie strains to murine neuroblastoma cell lines overexpressing wild-type mouse prion protein. J. Virol. 74:320-325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nishida, N., S. Katamine, and L. Manuelidis. 2005. Reciprocal interference between specific CJD and scrapie agents in neural cell cultures. Science 310:493-496. [DOI] [PubMed] [Google Scholar]
- 25.Paquet, S., N. Daude, M. P. Courageot, J. Chapuis, H. Laude, and D. Vilette. 2007. PrPc does not mediate internalization of PrPSc but is required at an early stage for de novo prion infection of Rov cells. J. Virol. 81:10786-10791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Paquet, S., C. Langevin, J. Chapuis, G. S. Jackson, H. Laude, and D. Vilette. 2007. Efficient dissemination of prions through preferential transmission to nearby cells. J. Gen. Virol. 88:706-713. [DOI] [PubMed] [Google Scholar]
- 27.Raymond, G. J., E. A. Olsen, K. S. Lee, L. D. Raymond, P. K. Bryant III, G. S. Baron, W. S. Caughey, D. A. Kocisko, L. E. McHolland, C. Favara, J. P. Langeveld, F. G. van Zijderveld, R. T. Mayer, M. W. Miller, E. S. Williams, and B. Caughey. 2006. Inhibition of protease-resistant prion protein formation in a transformed deer cell line infected with chronic wasting disease. J. Virol. 80:596-604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Reed, L. J., and H. Muench. 1938. A simple method of estimating fifty percent endpoints. Am. J. Hygiene 27:493-497. [Google Scholar]
- 29.Rubenstein, R., R. I. Carp, and S. M. Callahan. 1984. In vitro replication of scrapie agent in a neuronal model: infection of PC12 cells. J. Gen. Virol. 65(Pt. 12):2191-2198. [DOI] [PubMed] [Google Scholar]
- 30.Ryder, S. J., G. E. Dexter, L. Heasman, R. Warner, and S. J. Moore. 2009. Accumulation and dissemination of prion protein in experimental sheep scrapie in the natural host. BMC Vet. Res. 5:9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Solassol, J., C. Crozet, and S. Lehmann. 2003. Prion propagation in cultured cells. Br. Med. Bull. 66:87-97. [DOI] [PubMed] [Google Scholar]
- 32.Stack, M. J., M. J. Chaplin, and J. Clark. 2002. Differentiation of prion protein glycoforms from naturally occurring sheep scrapie, sheep-passaged scrapie strains (CH1641 and SSBP1), bovine spongiform encephalopathy (BSE) cases and Romney and Cheviot breed sheep experimentally inoculated with BSE using two monoclonal antibodies. Acta Neuropathol. 104:279-286. [DOI] [PubMed] [Google Scholar]
- 33.Vella, L. J., R. A. Sharples, V. A. Lawson, C. L. Masters, R. Cappai, and A. F. Hill. 2007. Packaging of prions into exosomes is associated with a novel pathway of PrP processing. J. Pathol. 211:582-590. [DOI] [PubMed] [Google Scholar]
- 34.Vilette, D. 2008. Cell models of prion infection. Vet. Res. 39:10. [DOI] [PubMed] [Google Scholar]
- 35.Vilette, D., O. Andreoletti, F. Archer, M. F. Madelaine, J. L. Vilotte, S. Lehmann, and H. Laude. 2001. Ex vivo propagation of infectious sheep scrapie agent in heterologous epithelial cells expressing ovine prion protein. Proc. Natl. Acad. Sci. U. S. A. 98:4055-4059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vorberg, I., A. Raines, B. Story, and S. A. Priola. 2004. Susceptibility of common fibroblast cell lines to transmissible spongiform encephalopathy agents. J. Infect. Dis. 189:431-439. [DOI] [PubMed] [Google Scholar]