Abstract
The Epstein-Barr virus efficiently infects human B cells. The EBV genome is maintained extrachromosomally and replicates synchronously with the host's chromosomes. The latent origin of replication (oriP) guarantees plasmid stability by mediating two basic functions: replication and segregation of the viral genome. While the segregation process of EBV genomes is well understood, little is known about its chromatin association and nuclear distribution during interphase. Here, we analyzed the nuclear localization of EBV genomes and the role of functional oriP domains FR and DS for basic functions such as the transformation of primary cells, their role in targeting EBV genomes to distinct nuclear regions, and their association with epigenetic domains. Fluorescence in situ hybridization visualized the localization of extrachromosomal EBV genomes in the regions adjacent to chromatin-dense territories called the perichromatin. Further, immunofluorescence experiments demonstrated a preference of the viral genome for histone 3 lysine 4-trimethylated (H3K4me3) and histone 3 lysine 9-acetylated (H3K9ac) nuclear regions. To determine the role of FR and DS for establishment and subnuclear localization of EBV genomes, we transformed primary human B lymphocytes with recombinant mini-EBV genomes containing different oriP mutants. The loss of DS results in a slightly increased association in H3K27me3 domains. This study demonstrates that EBV genomes or oriP-based extrachromosomal vector systems are integrated into the higher order nuclear organization. We found that viral genomes are not randomly distributed in the nucleus. FR but not DS is crucial for the localization of EBV in perichromatic regions that are enriched for H3K4me3 and H3K9ac, which are hallmarks of transcriptionally active regions.
To maintain genetic stability, proliferating cells must pass on their genomes as exact copies to their daughter cells. Viruses that establish latent infections have developed various strategies to ensure genetic stability as they integrate their genomes into the host's chromosomes or infect nonproliferating cells. Only a limited number of viruses with extrachromosomally maintained genomes establish a latent infection in proliferating cells. The gammaherpesvirus Epstein-Barr virus (EBV) is a 170-kbp circular double-stranded DNA molecule that resides as a nucleosome-packaged episome in latently infected B cells. The cis-acting origin of plasmid replication (oriP) is essential for DNA replication, nuclear retention, and viral gene regulation (30, 56). These functions are dependent on the EBV-encoded nuclear antigen EBNA1 (33, 58). EBNA1 has a modular structure: its C-terminal DNA binding and dimerization domain recognizes consensus motifs located within oriP. A mutational analysis of the N terminus led to the identification of two regions (amino acids [aa] 40 to 89 and aa 329 to 379) capable of linking DNA elements, which have therefore been termed linking regions (37). Later studies demonstrated that these domains tether EBV genomes to metaphase chromosomes (23, 24, 36, 47, 54, 55). oriP-bearing plasmids are distributed by a piggyback mechanism and not by a centromeric system, but these plasmids are symmetrically partitioned to sister chromatids with a confidence of 88% (23, 38). Furthermore, the linking regions also support DNA replication by recruiting the origin recognition complex (ORC) in an RNA-dependent manner (5, 14, 35, 41, 44, 45).
oriP is 1.8 kbp in size and is a paradigm for a mammalian autonomously replicating system (52, 56). The dual function of oriP also reflects its bipartite structure. The dyad symmetry element (DS) is the viral replicator and mediates the replication functions discussed above. The family of repeats (FR) consists of an array of 20 imperfect 30-bp repeats, each containing one EBNA1 binding site. In conjunction with EBNA1, FR tethers the EBV genomes to the host's chromosomes to ensure the stable maintenance of oriP plasmids, which segregate with a plasmid loss rate of 3 to 5% per generation (27, 32). The precise architecture of DS is important for its replication function. However, the interplay between FR and DS of oriP has not been fully elucidated yet. The sequences between DS and FR can be either deleted or, to a certain degree, extended without affecting replication competence, although the copy number of the oriP plasmids is reduced (43). The spatial limits of DS and FR have not been addressed in the context of the virus, but plasmids bearing DS and lacking FR replicate in an EBNA1-dependent manner. They are not stably maintained regardless of their ability to replicate, indicating that the integrity of oriP is important for certain functions of EBV (20, 21, 48, 57).
Several studies have analyzed the symmetrical segregation mechanism of EBV genomes and oriP plasmids using various in vitro and in vivo techniques (12, 23, 38, 47). While the contribution of EBNA1 to the segregation process is reasonably well understood (24, 29, 34, 47), very little is known about the nuclear localization of EBV genomes and EBNA1 with respect to the higher nuclear structure. In the last years it became increasingly clear that the nucleus is a complex network of distinct domains (49), creating interacting functional territories (7, 8). A preferred nuclear localization environment has not been determined for extrachromosomal viruses like EBV, and it is not clear whether or not such localization correlates with an epigenetic pattern at or near oriP. Using a combination of immunofluorescence techniques, fluorescence in situ hybridization (FISH), and confocal microscopy, we demonstrate that EBV genomes localize in perichromatic regions of the host cell's nucleus. The interphase nucleus is not a uniform landscape of chromatin but a complex network of chromosome regions (8), protein clusters (49), and interchromatin compartments. The interchromatin domains serve as traveling channels, giving the nucleus structure and function (1). The border between the higher-order chromatin and interchromatin compartments is the structurally defined perichromatin (see Fig. S1 in the supplemental material). The perichromatin is characterized by its open chromatin structure and its functional importance, because it is highly accessible for the replication and transcription machineries as well as for chromatin-modifying proteins (9).
Our experiments indicate that EBV genomes reside preferentially in histone 3 lysine 4-trimethylated (H3K4me3) as well as H3K9-acetylated (H3K9ac) domains. These histone modifications are linked to activation of transcription. A partial overlap with H3 trimethylated at lysine 27 (H3K27me3)-enriched foci was detected, which is found in repressed euchromatic genes and pericentric heterochromatin. No association with the heterochromatic H3K9me3 modification was observed. This pattern was also detected at oriP using chromatin immunoprecipitation (ChIP) experiments. EBV genomes and EBNA1 colocalize, but EBV genomes do not overlap with transcriptional centers, replication foci, or any other functional compartments of the nucleus. Using the mini-EBV genomes containing 41% of the EBV genome, we questioned how translocation and deletion of FR and DS affect transformation of primary human B cells, copy number, nuclear localization, and the epigenetic environment of the mini-EBV genomes. The mini-EBV system encompasses 71 kbp of noncontiguous EBV DNA sequences cloned into a prokaryotic F-factor plasmid and allows the generation of mutants of essential components in Escherichia coli (25). In this system, FR and DS can be analyzed in the context of DNA sequences that naturally flank oriP or are at a different position of the EBV genome. These experiments indicate that FR is essential for the nuclear localization of EBV and that DS has only a minor impact on the epigenetic environment.
MATERIALS AND METHODS
Generation of mini-EBV mutants.
To generate the various oriP mini-EBV strains, the recombination plasmid p2891 was generated by conventional cloning technology. p2891 contains nucleotides 5,357 to 12,290 of the B95.8 genome, including oriP and the auxiliary element Rep* (nucleotides 7,315 to 9,673). oriP was flanked by loxP sites inserted at positions 7,309 and 9,674. A frt-flanked selectable marker encoding resistance against kanamycin was positioned next to the loxP at the coordinate 7,309. p2891 was used to generate the mini-EBV p2908 by insertional mutagenesis in E. coli DH10B as described in detail (13, 15, 39). The recombination plasmids for the ΔFR and ΔDS deletion mutants, p2892 and p2893, contain the same flanking regions as p2891. FR was deleted using EcoRI (position 7,316) and MluI (position 8,314), and DS was removed using HpaI (position 8,994) and EcoRV (position 9,314). The basic recombination plasmid p2088 used to integrate FR and DS at an ectopic site contains nucleotides 86,110 to 93,008 of the B95.8 strain. p2088 contains a frt-flanked kanamycin-encoding resistance cassette used for selection, which was cloned between NheI (90,101) and HindIII (90,792) sites. Both oriP elements were cloned into the BstEII site at nucleotide 89,146. The FR fragment encompasses the EcoRI/MluI sequences between 7,316 and 8,314, generating p2900. The 114-bp DS element was PCR amplified and cloned into the BstEII site (89,146), generating p2735.
For homologous recombination in E. coli, p2891, p2892, and p2893 were cleaved with BsrG1 and MunI (nucleotide coordinates 5,564 and 11,268 of the B95.8 genome, respectively) to provide the EBV flanking regions for targeted insertional mutagenesis in DH10B as described previously (19). DH10B, containing p2800 (wild-type oriP, lacking the neomycin gene of p1478.A), was transformed with p2650, which is encoded by the recA gene and carries a ts origin. This strain was grown at 30°C and prepared for the transformation with the linearized recombination plasmids carrying the different oriP mutations (loxP-flanked oriP, ΔFR, and ΔDS, respectively). After electroporation, the strains were grown on LB plates containing chloramphenicol and kanamycin for 4 to 6 h at 30°C and then overnight at 42°C. Individual colonies were purified at least twice on double-selection plates. DNAs from single colonies were analyzed with restriction enzymes. One clone of each mutant was selected, and large-scale plasmid DNA was prepared. To delete the kanamycin resistance gene, the mini-EBV mutants were transformed into DH5α containing a plasmid encoding the flp recombinase. Candidates were confirmed with restriction enzyme analysis and termed p2908 (wild-type oriP), p2909 (ΔFR), and p2910 (ΔDS). To generate the oriP deletion mutant p2906, p2908 was transformed into a DH5α strain containing the cre recombinase (6). Mini-EBV p2912eFR was generated by transfecting linearized p2900 (digested with XbaI [position 86,110] and SalI [position 93,008]) into DH10B carrying p2909ΔFR and p2650 as described above. Similarly, p2913eDS was generated using p2910ΔDS and the recombination plasmid p2735 also linearized with XbaI and SalI.
Cell culture.
A39 is a lymphoblastic B-cell line generated from human primary B lymphocytes with EBV virions containing the mini-EBV 1478.A (25). All lymphoblastic cell lines (LCLs) generated in this study were maintained in RPMI 1640 medium supplemented with 10% fetal calf serum (Biochrom). Raji cells were maintained in RPMI 1640 medium supplemented with 10% fetal calf serum (Biochrom). HEK293-EBV+ cells were established by transfecting the human embryonic kidney HEK293 cell line with a recombinant full-length circular EBV genome. HEK293-EBV+ cells were selected and maintained in Dulbecco modified Eagle medium (DMEM) supplemented with 10% fetal calf serum (Biochrom) and 80 μg/ml hygromycin (PAA Laboratories).
Mounting cells on coverslips prior to immunofluorescence staining.
Coverslips (diameter, 18 mm) were covered with poly-l-lysine solution (0,1%, Sigma-Aldrich), rinsed with water, and air dried under the lamina flow. Soluble cells (Raji; LCLs) were transferred to a 15-ml tube and centrifuged for 10 min at 1,250 rpm. Cells were resuspended in the appropriate medium to a concentration of 1 × 106 cells/ml. Coverslips were rinsed with phosphate-buffered saline (PBS) (Biochrom), and 300 μl of cell suspension was spread on a coverslip and incubated at 37°C for 15 min. Adherent HEK293 cells and its derivates were directly seeded onto glass coverslips and cultivated overnight.
Hypertonic treatment.
For the analysis of nuclei with condensed chromatin, cells were incubated for 2 min at 37°C in the appropriate medium supplemented with 10% 20× PBS (final osmolarity: 570 mOsm) (1). These cells were fixed directly after the hypertonic treatment.
Preparation of fixed cells for FISH.
For FISH analysis, cells were fixed with 2% paraformaldehyde (Merck) for 10 min and permeabilized with 0.05% Triton X-100 (Sigma-Aldrich) in PBS three times for 5 min, followed by one PBS-0.5% Triton X-100 wash and three 5-min incubations in 0.05% Triton X-100-PBS at room temperature. DNA was nicked by incubating the cells with 0.1 M HCl for 6 min at room temperature. Cells were subsequently treated with 200 μg/ml RNase A, equilibrated with 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate), and stored in 50% formamide-2× SSC for 24 h.
Immunofluorescence staining prior to iFISH.
Cells for immuno-FISH (iFISH) were fixed with 2% paraformaldehyde (Merck) for 10 min and permeabilized three times for 5 min in 0.15% Triton X-100 (Sigma-Aldrich). Cells were blocked three times for 10 min with blocking solution (1% bovine serum albumin [BSA], 0.15% lysine) and subsequently incubated with the primary antibody in fetal calf serum overnight at 4°C. Cells were then washed twice with 0.15% Triton X-100-PBS, once for 5 min and once for 10 min, followed by an additional blocking step for 7 min. The secondary antibody was diluted 1:200 in blocking solution and applied for 45 min at room temperature. The cells were washed with 0.15% Triton X-100-PBS twice for 5 min at room temperature. To prepare the hybridization step, the coverslips were postfixed with 1% paraformaldehyde and washed twice with PBS supplemented with 20 mM glycine. After equilibration in 2× SSC, cells were stored in 50% formamide-2× SSC for 24 h.
Generation of EBV DNA probes.
DNA probes for in situ hybridizations were obtained by labeling of cMABA-SalA-cosmids with the hapten digoxigenin dUTPs using nick translation (42). The probes were precipitated with ethanol and resuspended in deionized formamide.
FISH/iFISH.
Coverslips were hybridized using 200 ng of labeled EBV probes for 2 min at 75°C. After hybridization, the probes were transferred to 37°C for 3 days. Unbound probes were removed by incubating the coverslips three times for 5 min in 2× SSC at 37°C followed by three washing steps for 5 min with 0.1× SSC at 60°C. The coverslips were blocked for 45 min with blocking solution (4% BSA in 4× SSC-0.2% Tween 20) at room temperature. The probes were detected with goat anti-digoxigenin-Cy3 antibody (dilution, 1:200 in blocking solution). After incubation for 1 h at room temperature, cells were washed with 4× SSC-0.2% Tween 20 and counterstained with DAPI (4′,6-diamidino-2-phenylindole). The completely stained coverslips were mounted on glass slides with Vectashield (Vector Laboratories) and sealed with nail varnish.
Microscopy/image analysis.
Images were acquired with a Leica TCS SP2 confocal laser scanning microscope fitted with a 63× 1.4 HCX Plan Apo λblue objective. Image-series for three-dimensional (3D) reconstruction were taken at 0.25-μm intervals. The acquired digital images were deconvoluted, rendered, and evaluated with Huygens Essential Suite 3.2 (Scientific Volume Imaging). Colocalization events were analyzed with signal intensity scans (line scans) and Li′s approach calculation (2).
Antibodies.
Primary antibodies used were as follows: EBNA1, rat monoclonal antibody (MAb) clone 1H4 (18); BrdU, mouse MAb (catalog no. 11170376001, Roche); SC35, mouse MAb (catalog no. S4045, Sigma); LaminB1, rabbit polyclonal antibody (pAb) (catalog no. ab16048, Abcam); H3K4me3, rabbit pAb (catalog no. ab8580, Abcam); H3K9me3, rabbit pAb (catalog no. ab8898, Abcam); H3K27me3, rabbit pAb (catalog no. 17-622, Upstate); H3K9ac, rabbit pAb (catalog no. ab10812, Abcam); Snf2h, rabbit pAb (catalog no. ab3749, Abcam); and HP1α, rabbit pAb (catalog no. ab9057, Abcam). Secondary antibodies were as follows: digoxigenin Cy3, mouse MAb (catalog no. 200-162-156, Dianova); anti-mouse Alexa488 (catalog no. A11029, Invitrogen); and anti-rabbit Alexa488 (catalog no. A1103, Invitrogen).
RESULTS
EBV genomes are located in perichromatic regions.
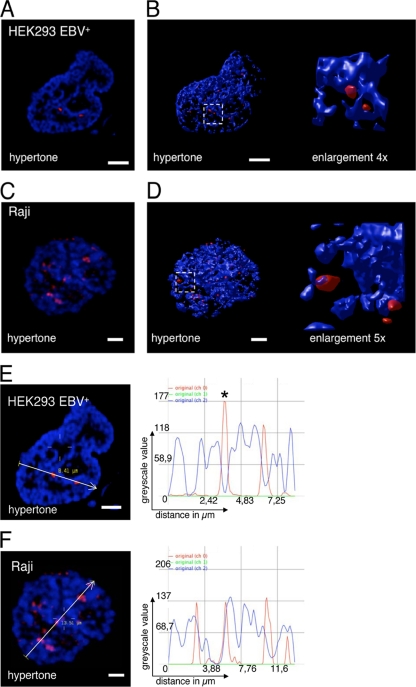
EBV is maintained as a nucleosome-coated extrachromosomal genome in the host cell nucleus (16, 53). During mitosis it is tethered to the host cell chromatin by its viral transactivator EBNA1 (47). The preferred interphase localization of EBV genomes with respect to nuclear subdomains is unknown. The interphase nucleus is not a uniform landscape of chromatin but a complex network of chromosome-dense regions (8), protein clusters (49), and interchromatin compartments. By hypertonic treatment of cells, the chromatin rapidly condenses, revealing the interchromatin compartment in a reversible manner (1). Cells fixed during hypertonic treatment can be subsequently analyzed regarding the chromatic, perichromatic, or interchromatic localization of the studied protein (immunofluorescence [IF]) or DNA (fluorescence in situ hybridization [FISH]). To determine where EBV genomes are located, we analyzed EBV-positive Raji cells and HEK293 cells stably transfected with recombinant EBV genomes (HEK293-EBV+) in FISH experiments of isotonically and hypertonically treated cells. Signals that are associated with but are not part of the condensed chromatin regions qualify for perichromatic localization. Our experiments clearly demonstrate that the EBV genomes of HEK293-EBV+ and Raji cells localize in perichromatic regions of the host cell nucleus (Fig. 1). Immuno-FISH images taken from the studied cells displayed EBV signals that were attached to but not part of the condensed chromatin, protruding into the interchromatic domain (Fig. 1A and C). The captured images were deconvoluted using Huygens software and were 3D reconstructed to enable a comprehensive localization of the signals (Fig. 1B and D). 3D reconstructions of 23 Raji and 42 HEK293 EBV+ cells were used to quantify the localization of EBV genomes (Table 1).
FIG. 1.
EBV episomes are localized in perichromatic regions of the nucleus. Immuno-FISH image of HEK293-EBV+ (A) and Raji (C) cells displaying a DAPI DNA counterstain (blue; channel 2) and a fluorescence in situ hybridization of EBV genomic DNA (red). 3D reconstruction of HEK293-EBV+ (B) and Raji (D) treated with hypertonic buffer with an enlarged section. EBV DNA is detected near the condensed chromatin, which defines the perichromatic localization. EBV DNA neither colocalizes with the chromatic domain nor exists unassociated with the chromatin. Enlargements are indicated by white-lined squares. 3D reconstructions were used to quantify the perichromatic localization of EBV genomes. Scale bar = 2 μm. Signal intensity scans for EBV (red; channel 0) and DNA counterstain (blue; channel 1) along the indicated line and direction in HEK293-EBV+ (E) and Raji (F) confirm the perichromatic localization. Localization of EBV is not observed in the peaks of the DNA counterstain but is observed in the shoulders of DAPI-stained regions. Line scans represent one selected confocal plane, which might result in unambiguous results. The asterisk indicates a signal that seems unassociated in this particular confocal plane but is clearly associated in an adjacent plane. See also Fig. S2 in the supplemental material for details. Scale bar = 2 μm.
TABLE 1.
Localization analysis of the different cell linesa
| Type of localization | % with result for cell line |
|||||
|---|---|---|---|---|---|---|
| Raji (n = 23) | HEK293 EBV+ (n = 42) | 2908 wt-oriP (n = 26) | 2910ΔDS (n = 28) | 2912eFR (n = 29) | 2913eDS (n = 30) | |
| Perichromatic localization | 95.7 | 85 | 84.6 | 96.4 | 69.6 | 93.3 |
| Chromatic localization | 0 | 0 | 0 | 0 | 0 | 0 |
| Interchromatic localization | 4.3 | 0 | 0 | 0 | 0 | 0 |
| Cytosolic localization | 0 | 0 | 0 | 0 | 15.2 | 0 |
| No signal | 0 | 15.0 | 15.4 | 3.6 | 15.2 | 6.7 |
EBV genomes localize predominantly in the perichromatin. Cells showing no signal for EBV-DNA were included in the statistics to take the number of cells into account that have lost the EBV genome. Signals qualify for chromatic localization if signals for EBV-FISH and DAPI signals completely overlap, which was not observed (see also Fig. S2 in the supplemental material). EBV signals score for perichromatin if the FISH signal localizes at the border of the chromatin-dense region and a partial overlap of both signals is observed. There was no association of both signals in the interchromatin.
Because EBV genomes are three-dimensional specimens, only 3D reconstructions allow the unbiased analysis of acquired images. However, measuring the signal intensity distribution in line scans facilitates the visualization in two dimensions. One representative confocal plane was selected as an example (Fig. 1E and F; see Fig. S2 in the supplemental material). Signal intensity scans of the DAPI stain and EBV signals confirm the perichromatic localization, which is characterized by a partial overlap of the DAPI and FISH signals for EBV; the red EBV signals are located at the shoulders of the blue DAPI line (Fig. 1; see Fig. S2 in the supplemental material). We did not observe a colocalization of chromatin-dense regions and EBV-FISH signals and only very rarely an interchromatic localization typified by nonassociation of both signals (see Fig. S2D in the supplemental material). These findings indicate that the EBV genomes are associated to perichromatic regions of interphase chromosomes.
EBV genomes colocalize with EBNA1 but not with other nuclear functions.
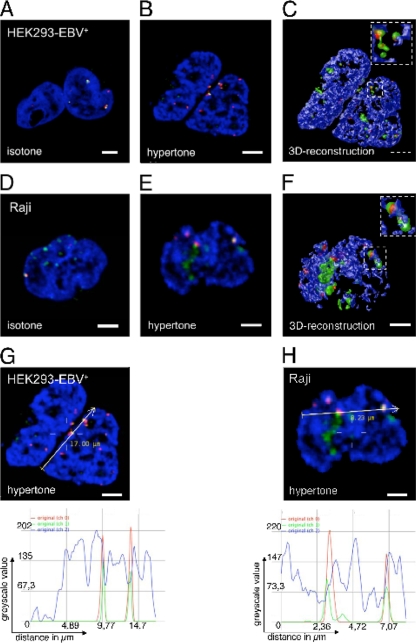
It is known that EBV genomes colocalize with the viral transactivator EBNA1 in interphase nuclei (23), but the distinct localization within a nucleus was unknown. To study the colocalization of EBNA1 and EBV genomes in more detail, we used a combination of immunofluorescence and FISH experiments. Isotonically treated HEK293-EBV+ genomes (Fig. 2A) and EBV genomes of Raji cells (Fig. 2D) displayed colocalizing signals for EBV DNA and EBNA1. The number of EBNA1 clusters (green signals) was higher than that of EBV hybridization signals (red spots). This reflected the higher copy number of EBNA1 proteins in relation to EBNA1 binding sites within oriP (48 sites per oriP) (51). The abundant EBNA1 protein was bound to the host cell chromatin. The majority of EBV and EBNA1 signals colocalized, and no EBV signal was detected without EBNA1, confirming that the chromatin association mediated by EBNA1 is essential for viral genome stability. Both analyzed cell lines, HEK293-EBV+ and Raji, displayed a perichromatic localization of EBV/EBNA1 signals, which became obvious in hypertonically treated cells (Fig. 2B and E) and was visualized after 3D reconstruction analysis (Fig. 2C and F). The colocalization became even more evident in the signal intensity scans of HEK293-EBV+ cells (Fig. 2G) and Raji cells (Fig. 2H). The ratio of the EBV/EBNA1 correlation is summarized in Table 2.
FIG. 2.
EBV and EBNA1 colocalize in perichromatic regions. (A to C) HEK293-EBV+ cells stained for EBV genomic DNA (red), EBNA1 (green), and DNA (blue) by combinational immunofluorescence and fluorescence in situ hybridization. EBV and EBNA1 colocalize in regions adjacent to condensed chromatin domains of the nucleus. This perichromatic localization is revealed by the hypertonic treatment (B) and 3D reconstruction (C). Scale bar = 2 μm. (D to F) Raji cells after combined immunofluorescence and fluorescence in situ hybridization. EBV genomic DNA (red), EBNA1, (green) and DNA (blue); EBV and EBNA1 colocalize in perichromatic regions of the nucleus. The perichromatic localization is obvious after hypertonic treatment (E) and 3D reconstruction (F). Enlargements are indicated by white-lined squares. Scale bar = 2 μm. (G) Signal intensity scan of EBNA1 (green; channel 1 of the microscope), EBV (red; channel 0), and DNA counterstain (blue; channel 2) along the indicated line in HEK293-EBV+ cells. EBV peaks colocalize with peaks for EBNA1. Colocalization does not occur in the peaks of the DNA counterstain but next to it, indicating perichromatic localization. (H) Signal intensity scans for EBNA1 (green), EBV (red), and DNA counterstain (blue) along the indicated line in Raji cells. EBV peaks colocalize with peaks for EBNA1. Scale bar = 2 μm.
TABLE 2.
Colocalization analysis of the different cell linesa
| Genome | % with result for cell line |
|||||
|---|---|---|---|---|---|---|
| Raji (n = 43) | HEK293-EBV+ (n = 20) | 2908 wt-oriP (n = 30) | 2910ΔDS (n = 46) | 2912eFR (n = 24) | 2913eDS (n = 26) | |
| EBNA1 only | 19.7 | 17.7 | 32.8 | 29.5 | 67.9 | 26.1 |
| EBV and EBNA1 colocalized | 80.3 | 82.3 | 67.2 | 70.5 | 32.1 | 73.9 |
EBV genomes colocalize with the viral transactivator EBNA1. The relative percentages of EBNA1 foci and FISH signals colocalizing with EBNA1 are shown. Due to the high expression levels of EBNA1, the number of EBNA1 foci exceeds the number of EBV foci thus generating EBNA1 foci that show no signal for EBV.
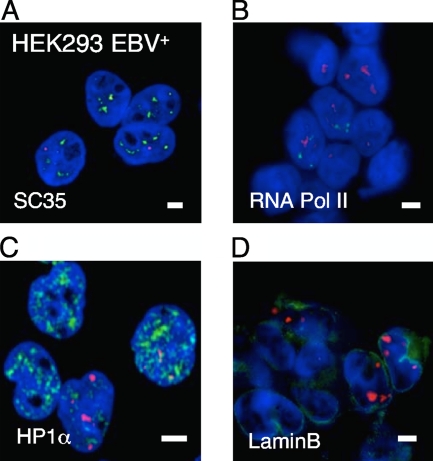
The perichromatin is considered to be the region where DNA synthesis, transcription, and DNA modification take place (9). We next analyzed whether EBV genomes were predominantly located in specific regions of nuclear activity. Toward this end, we again combined FISH and immunofluorescence. The colocalization experiments were carried out in HEK293-EBV+ cells due to their lower background in immunofluorescence experiments. SC35 is a splicing factor and localizes in characteristic speckles in the interchromatin compartment. An anti-SC35 antibody was used to visualize these domains (Fig. 3A), and an RNA-PolII-specific antibody was used to stain transcriptionally active perichromatic regions (Fig. 3B). Heterochromatic regions were stained with an HP1α antibody (Fig. 3C). A laminB1 antibody was employed to visualize the nuclear meshwork, which is important for the organization of nuclear microdomains (Fig. 3D). An Snf2h-specific antibody was utilized to detect chromatin domains undergoing chromatin remodeling and BrdU incorporation for sites of DNA synthesis (data not shown). Only a small percentage of the different immunofluorescence signals colocalized with EBV DNA (Fig. 3 and data not shown), which might be explained by the dynamic nature of the different processes. These results suggested that the stable maintenance of EBV genomes is dependent only on the EBNA1-mediated tethering to perichromatic regions of the host.
FIG. 3.
Localization of EBV genomes in subnuclear compartments of interphase nuclei. HEK293-EBV+ cells were fixed and FISH was used to visualize the viral genome with an EBV-specific probe. The DNA was counterstained with DAPI. A potential colocalization with different subnuclear markers was analyzed with IF techniques. (A) Immunofluorescence image for the splicing protein SC35 (red), EBV genomes (green), and DNA counterstain (blue) in HEK293-EBV+ cells. EBV and SC35 signals show no significant colocalization. (B) Immunofluorescence image for RNA-Pol II (red), EBV genomes (green), and DNA counterstain (blue) in HEK293-EBV+ cells. EBV and RNA-Pol II signals show no significant colocalization. (C) No specific colocalization of EBV DNA with the heterochromatin protein HP1α (red) was observed. (D) A potential colocalization of EBV with the nuclear meshwork was determined with a LaminB1-specific antibody (red). Scale bar = 2 μm.
EBV genomes colocalize with specific epigenetic markers.
Previous studies analyzed the histone modifications at oriP and parts of different EBV genomes (10, 59). These results suggested that the latent origin is specifically enriched with histone modifications that correlate with active transcription such as histone H3 and H4 acetylation as well as H3K4 trimethylation (H3K4me3). In a combination of FISH and immunofluorescence (IF) with antibodies specific for different methylation marks (Fig. 4; see Fig. S3 in the supplemental material), we found that in Raji cells EBV genomes always localized in perichromatic regions that were enriched with H3K4me3 and lysine 9-acetylated H3 (P < 10−6; Fig. 4A and Table 2). We used the Wilcoxon rank sum test, which assesses two independent samples. The EBV DNA showed no specific preference for a colocalization with H3K27me3 (P < 10−5). Furthermore, no association with H3K9me3 was observed (P to avoid a colocalization, <10−6). These results were in agreement with results of ChIP experiments, which indicated an enrichment of H3K4me3 and H3 acetylation at and near oriP in Raji cells (10). A comparable nuclear localization was observed for an LCL transformed with the recombinant full-size EBV genome 2089 (LCL-EBV+; Fig. 4B, Table 3). These findings clearly demonstrated that the higher order nuclear architecture influences the local epigenetic pattern. The subnuclear localization of the viral genome correlated with the local epigenetic pattern at oriP, indicating that the association of EBV DNA in H3K4me3- and H3K9ac-enriched nuclear regions might cause the epigenetic pattern at oriP. ChIP experiments confirmed that oriP is enriched with histone H3 trimethylated at lysines 4 and 27 (Fig. 4C). In line with a recent report, H3K9me3 was not associated with oriP (10). As a reference site, we used the Q promoter, which shows no specific enrichment of the analyzed patterns, which is consistent with a report from Day et al. (10).
FIG. 4.
EBV genomes associate with specific histone modifications. The localization of different EBV genomes in epigenetic regions was determined with a combination of immunofluorescence techniques. The EBV genome of Raji cells (A) and an LCL transformed with a full-length EBV genome (B) were visualized by FISH using an EBV-specific probe (red). Colocalization with histone 3 trimethylated at lysine 4 (H3K4me3; first panel), histone 3 trimethylated at lysine 9 (H3K9me3; second panel), histone 3 trimethylated at lysine 27 (H3K27me3; third panel), and histone 3 acetylated at lysine 9 (H3K9ac; fourth panel) is shown in green. 3D reconstructions of the Raji cells are shown in Fig. S3 in the supplemental material. Scale bar = 2 μm. (For signal intensity scans of panel A, see Fig. S3 in the supplemental material). (C) ChIP experiments of HEK293 cells transfected with wild-type oriP. Cells were cross-linked for 8 min at room temperature with 1% formaldehyde. Sonicated chromatin (200 μg) was immunoprecipitated with 2.5 μg of the indicated antibody. Coprecipitated DNA was analyzed with oriP-, oriLyt-, and Q-promoter specific primer pairs as described previously (45) and quantified in relation to the amount of the input chromatin (y axis).
TABLE 3.
Quantification of the colocalization analysis of EBV genomes and epigenetic markersa
| Cell line and modification | No. of cells evaluated | % (± SD) of EBV clusters: |
||
|---|---|---|---|---|
| Partially colocalizing with epigenetic mark | Attached to epigenetic mark | Not associated with epigenetic mark | ||
| Raji cells | ||||
| H3K4me3 | 47 | 57.5 (± 26.9) | 42.5 (± 26.9) | 0 |
| H3K9me3 | 50 | 8.9 (± 17.5) | 29.6 (± 24.1) | 62.1 (± 24.4) |
| H3K27me3 | 49 | 32.3 (± 25.1) | 32.6 (± 20.9) | 35 (± 27.3) |
| H3K9ac | 25 | 78.5 (± 14.9) | 21.5 (± 14.1) | 0 |
| LCL-EBV+ | ||||
| H3K4me3 | 20 | 63.2 (± 13.5) | 36.8 (± 13.5) | 0 |
| H3K9me3 | 24 | 14.6 (± 14.8) | 37.9 (± 14.6) | 47.6 (± 17.8) |
| H3K27me3 | 21 | 20.9 (± 17.5) | 54.5 (± 14.0) | 25.5 (± 14.2) |
| H3K9ac | 20 | 85.5 (± 22.5) | 14.5 (± 22.5) | 0 |
| LCL2908wt-oriP | ||||
| H3K4me3 | 24 | 72.8 (± 20.6) | 27.2 (± 20.6) | 0 |
| H3K9me3 | 23 | 28.7 (± 11.5) | 31.2 (± 9.7) | 40.9 (± 7.6) |
| H3K27me3 | 39 | 49.4 (± 18.1) | 33.3 (± 16.3) | 17.3 (± 14.9) |
| LCL2910ΔDS | ||||
| H3K4me3 | 27 | 67.9 (± 16.0) | 32.1 (± 16.0) | 0 |
| H3K9me3 | 22 | 26.8 (± 16.4) | 36.4 (± 9.7) | 36.8 (± 18.2) |
| H3K27me3 | 27 | 56.6 (± 15.9) | 41.25 (± 15.6) | 3.1 (± 8.7) |
The analyzed cell line and respective histone modifications are indicated in the stub. For abbreviations, see the legends to Fig. 4 and 7. The percentages of overlapping and directly adjacent voxels are indicated, as well as the percentage of nonassociated signals for EBV and the modification. The statistical relevance of the association was calculated using the Wilcoxon rank sum test. For details, see text.
Only FR is essential for episomal maintenance and B-cell transformation.
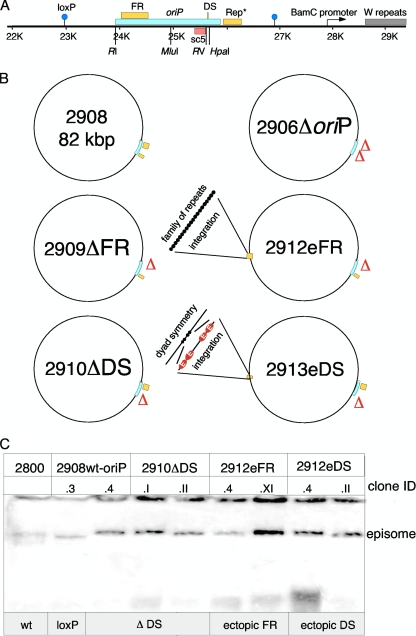
It is generally accepted that the extrachromosomal maintenance of plasmids and EBV genomes is dependent on oriP. The family of repeats (FR) functions as a segregation element of oriP, whereas the dyad symmetry element (DS) acts as a replicator that can be deleted in the context of larger constructs (52, 56). However, it is currently unclear whether both FR and DS have an impact on the nuclear localization of EBV genomes and if they are required for the transformation process of human primary B lymphocytes. To address these questions, we generated a series of oriP mutants in E. coli in the context of mini-EBV genomes that were subsequently used to generate viral particles in a helper cell line. Mini-EBV plasmids with wild-type oriP contain all information required for the proliferation of in vitro transformed human B cells (25), while virus genes that mediate the productive phase of EBV are missing. The mini-EBV system was chosen instead of full-size EBV genomes to study the specific contributions of oriP only. Full-size EBV genomes contain multiple replication initiation sites that have been mapped outside oriP (32, 40). These sites are missing in the mini-EBV genome. The mini-EBV mutant p2908 contains a loxP-flanked wild-type oriP as in the original mini-EBV p1478.A (Fig. 5A and B) (45). In p2910ΔDS the viral replicator was deleted. We altered the spatial integrity by transferring DS to a BstEII site located downstream of the EBNA3 gene (position 89,146 of B95.8) to generate p2913eDS (p2913 ectopic DS). In parallel, we deleted the FR in p2909ΔFR at the oriP locus and placed FR also into the BstEII site to generate p2912eFR (p2912 ectopic FR). The oriP deletion mutant p2906ΔoriP was used as a negative control.
FIG. 5.
Maps of mini-EBV plasmids. (A) The latent origin, schematically shown at the top, is flanked by loxP sites (blue circles) for cloning purposes. The minimal oriP encompasses the family of repeats (FR) and the dyad symmetry element (DS). Rep* is a 298-bp fragment that can partially replace the DS element if multimerized on a plasmid (28). The oriP-specific PCR fragment sc5 (red box) was utilized for quantification experiments. The restriction enzymes used for the deletion of FR (EcoRI [RI] and MluI) and DS (EcoRV [RV] and HpaI) are indicated at their respective positions. (B) Different oriP mutants were generated in the context of the mini-EBV genome and used for immortalization experiments. Depicted are oriP (blue box) and its two functional elements DS and FR (yellow boxes). Starting from p2908, three deletion mutants were generated: p2910ΔDS is lacking DS, p2909ΔFR is lacking the family of repeats, and p2906ΔoriP is lacking the entire oriP, including the auxiliary element Rep*. p2913eDS and p2912eFR carrying the respective oriP element integrated at an ectopic site, positioned 35 kbp apart from the oriP locus. (C) The Gardella gel technique was used to determine the episomal status of LCL subclones containing the different mini-EBV mutants (17). 2800 is identical to the basic mini-EBV p1478. A but lacking the neomycin resistance marker (25, 26). 2908 carries a wild-type oriP that is flanked by loxP sites used for the generation of 2912eFR and 2913eDS. 2 × 106 to 5 × 106 cells of each individual cell clone were lysed in the wells of a Gardella gel and probed with a radiolabeled plasmid recognizing the prokaryotic backbone of the mini-EBV genomes and oriP (17). Two clones of each established LCL were analyzed (clone ID).
At first we tested whether the different oriP mutants could transform human primary B lymphocytes. The generation of mini-EBV virus progeny is dependent on the lytic origin, oriLyt, and the packaging signals (terminal repeats) in cis. Both elements are part of the mini-EBV genomes (11, 61). The helper cell line HEK293-TR− provides all viral factors required in trans, thereby supporting the lytic amplification and packaging of mini-EBV mutants (13). To induce the lytic phase, an expression plasmid for the lytic inducer BZLF1 was cotransfected with the different mini-EBV mutants. Subsequently, the supernatants were used for the infection of human primary B lymphocytes isolated from the adenoids of 3- to 4-year-old children. Infected cells were cultivated without fibroblast feeder cells. No transformed cell lines were obtained for the p2906ΔoriP and p2909ΔFR mutants lacking oriP and FR, respectively. Transformed cell lines grew out from infections with p2908wt-oriP, p2910ΔDS, p2912eFR, and p2913eDS virions. For these mutants the transformation efficiency was similar, although the ΔDS-lymphoblastoid cell line (LCL 2910ΔDS) grew slightly slower than the other LCLs. Southern blot hybridization and PCR analysis confirmed the correctness of the different oriP variants of the transformed cell lines (see Fig. S4 in the supplemental material). A Gardella gel analysis verified the extrachromosomal status of the mini-EBV genomes (Fig. 5C). All further experiments were performed with the cell lines LCL2908.3, LCL2910.II, LCL2912.XI, and LCL2913.4.
The integrity of oriP is important for achieving higher copy numbers.
The infection experiments with virus particles generated with different oriP mutants indicated that the integrity of oriP in the context of the mini-EBV is not essential for the episomal stability of the viral genome and the transformation process. FR-dependent stable episomal maintenance correlated with the transformation of B cells, whereas DS was not essential for this process. The integrity of oriP, however, might be important for the establishment of higher copy numbers of the EBV genome. To determine the distribution of the different cell lines, we used FISH, quantitative PCR, and Southern blotting. The episomal stability of the different mini-EBV genomes was confirmed and quantified after the cell lines were cultivated for 3 months (data not shown) and 6 months (Fig. 6A). The wild-type LCL2908wt-oriP contains approximately 5 to 10 copies, which is comparable to the result for LCL A39 (45). On average, LCLs with oriP mutants have two episomes per cell as estimated by quantitative PCR and Southern blot hybridization (Fig. 6A). The separation of FR and DS as well as the deletion of DS resulted in a reduced number of extrachromosomal plasmids. The copy number of the LCL2910 was slightly lower than the copy number in LCLp2912eFR and LCL2913eDS. Once established, the copy number was stable over months.
FIG. 6.
Integrity of oriP affects copy number. (A) The different mini-EBV mutants were analyzed by the Gardella gel technique in order to confirm the episomal status of the viral genomes 6 months posttransfection. Quantitative PCR and Southern blotting determined the copy numbers of the different mini-EBV variants. For Gardella gel analysis, the indicated amount of cells was lysed for the individual lanes (×105 cells). Two different amounts of Raji cells were included for standardization, resulting in 10 × 106 and 30 × 106 EBV genomes, respectively (51). We used an oriP-bearing plasmid as a hybridization probe that recognizes the prokaryotic backbone of the mini-EBV genomes as well as oriP fragments. The copy numbers of the different mutants were determined using the AIDA Image Analyser software (Raytest) (third row). The resulting absolute copy number is given in row two. In parallel, the copy number was revealed by quantitative PCR generating a standard curve from a series of 10-fold dilutions from purified mini-EBV genomes (row four). The mean values and standard deviations of results of three independent experiments are shown in row five (copies/cell; Q-PCR). (B) Examples of FISH experiments that indicate different populations in the 2908wt-oriP and 2910ΔDS cell lines. 3D stacks of fluorescence images were projected according to the maximum intensity of the acquired volume pixels along the z axis. 2908wt-oriP cells showed an average of 5.5 and 12.4 signals per cell. 2910ΔDS cells illustrated an average of 1.8 and 5.5 signals per cell. Scale bar = 2 μm. The data are summarized in Table 3.
Nanbo et al. have recently reported that in EBV-positive LCL721 cells heterogeneous populations of copy numbers exist within one culture (38). To determine whether this is a general phenomenon and if this is dependent on DS, we used FISH to analyze the establishment of different populations in LCL2908wt-oriP and LCL2910ΔDS. In our FISH experiments we also observed 5.5 and 12.4 signals per cell for LCL2908wt-oriP and 1.8 and 5.5 signals per cell for LCL2910ΔDS (Fig. 6B and Table 3). These findings confirmed with an independent approach that the presence of DS is important for the establishment of higher copy numbers. In line with results reported by Nanbo et al., we also observed two heterogeneous populations, one having approximately twice as many mini-EBV genomes as the other (38). In conclusion, separating the core elements of oriP by moving either FR or DS to a different location did not affect their functionality with respect to genome stability and transformation of primary B cells, but it does reduce the copy number of the mini-EBV genomes.
The integrity of oriP does not alter the nuclear localization of mini-EBV genomes.
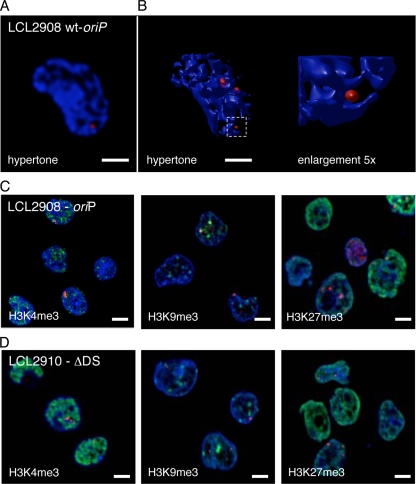
To determine the impact of oriP integrity on the nuclear localization of mini-EBV genomes, immuno-FISH experiments were performed with 2908wt-oriP- (Fig. 7A; B), 2910ΔDS-, and 2913eDS-transformed B cells as well as HEK293 cells transfected with 2912eFR (see Fig. S5 in the supplemental material). A quantitative evaluation of the acquired images classified the signals according to their chromatic or perichromatic localizations (Table 1). The majority of cells infected with different EBV genomes showed hybridization signals characteristic for perichromatic localizations. Unfortunately, the fluorescent background of the LCL2912eFR was on average too high to allow the quantification of sufficient cells. Therefore, we analyzed HEK293 cells transfected with 2912eFR. For this reason, some cells displayed a cytosolic signal of the EBV genome (Table 1). However, all nuclear 2912eFR episomes were located in the perichromatin. Subsequent analyses of the mini-EBV cell lines carrying the different oriP mutants also revealed a perichromatic localization of colocalizing EBV and EBNA1 (see Fig. S5 in the supplemental material). Again, not all EBNA1 proteins colocalized with EBV genomes in perichromatic regions, which was due to the molar excess of EBNA1 protein. The data summarized in Table 2 indicated that approximately 80% of the detected EBV and EBNA1 colocalized with full-size EBV genomes and approximately 70% colocalized in the different mini-EBV-infected LCLs. The transiently transfected HEK293-p2912eFR showed a lower number of colocalizing spots.
FIG. 7.
The integrity of oriP does not alter the nuclear localization of mini-EBV genomes. (A) Immuno-FISH image of LCL2908wt-oriP cells displaying a DAPI DNA counterstain (blue) and a fluorescence in situ hybridization of EBV genomic DNA (red). (B) 3D-reconstruction LCL2908wt-oriP cells treated with hypertonic buffer. The enlargement depicts the area outlined by the white square. Mini-EBV DNA is detected near the condensed chromatin, which defines the perichromatic localization. Mini-EBV DNA neither colocalizes with the chromatic domain nor exists unassociated with the chromatin. Enlargements are indicated by the white-lined square. (For signal intensity scans of panel A, see Fig. S2 in the supplemental material). The mini-EBV genome p2908 (C) or the mini-EBV genome lacking DS (D) were visualized by FISH using an EBV-specific probe (red). Colocalization with histone 3 trimethylated at lysine 4 (H3K4me3; first panel), histone 3 trimethylated at lysine 9 (H2H9me3; second panel), and histone 3 trimethylated at lysine 27 (H3K27me3; third panel) are shown in green. Scale bar = 2 μm. (For signal intensity scans of panels C and D) see Fig. S6 in the supplemental material).
Neither the altered spatial integrity of oriP in p2912eFR and 2913eDS (translocated oriP elements) nor the deletion of DS in p2910ΔDS had an impact on the nuclear distribution of EBV genomes to perichromatic regions. This suggests that EBV genomes localize in this highly accessible domain, guaranteeing access to the transcription and replication proteins of the host's nucleus. The mini-EBV genomes are not dependent on a special cis-functional element other than the provided EBNA1-binding ability of the FR region. As such, the perichromatic localization is a functional interplay of FR and EBNA1.
Next, we compared the nuclear localization of LCL2908wt-oriP and LCL2910ΔDS (Fig. 7C and D and Table 3) epigenetic markers representative for transcriptionally active chromatin (H3K4me3) and heterochromatin (H3K9me3 and H3K27me3). Because both H3K9 acetylation and H3K4me3 mark transcriptionally active chromatin and the H3K4me3 antibody was more specific and reliable in immunofluorescence experiments, we concentrated on H3K4me3. Immuno-FISH experiments confirmed that the mini-EBV genome of the cell line LCL2908wt-oriP localized preferentially to the same epigenetic regions as cell lines carrying full-size EBV genomes with a slight shift toward H3K27me3 sites. Signal intensity scans are shown in Fig. S6 in the supplemental material. Interestingly, the mini-EBV genomes lacking DS colocalized in nearly all cases at H3K27me3 and H3K4me3 foci. Again, no specific association with H3K9me3 foci was observed. Taken together, the combination of immunofluorescence and FISH experiments clearly demonstrated that FR but not DS is crucial for the localization of EBV in perichromatic regions. All EBV genomes preferentially localized in transcriptionally active H3K4me3 regions but not in heterochromatic H3K9me3 regions. Furthermore, our experiments indicate that DS-dependent replication has an impact on the localization of the viral genome in H3K27me3 foci.
DISCUSSION
Efficient once-per-cell-cycle replication and faithful segregation of genomes to daughter cells are the major hallmarks of the EBV system (30) and are required for the stable episomal maintenance of EBV genomes. This is achieved by oriP's bipartite structure and function. The family of repeats (FR) consists of 20 imperfect 30-bp repeats of the binding motif for the viral transactivator EBNA1. Separated from FR by 1 kb lies the dyad symmetry element (DS) that consists of 2 × 2 binding sites for EBNA1. Binding of EBNA1 to FR is needed for transcriptional activation, chromatin tethering, and faithful segregation of the genomes to daughter cells. Binding to DS induces a once-per-cell-cycle replication starting at the DS element. The mechanisms behind each of these processes have been studied separately in the past (29, 34), but the interdependency of oriP's FR region and DS element has not been analyzed in detail.
The role of FR and DS in establishment and maintenance of episomal EBV genomes.
In the study presented here we show that the integrity, the wild-type configuration, and the spacing of the two functional elements of oriP are not essential for its function. As expected, deletion of FR leads to the loss of the growth-transforming capacity of primary B cells, highlighting its importance in oriP function. In contrast, deletion of DS does not significantly interfere with the growth transformation of human primary B cells but results in a decreased copy number of viral genomes in the cells. In the context of mini-EBV genomes, FR and DS can be separated by 40 kbp and still maintain their individual functions with respect to tethering, segregation, and replication. We suggest that the spatial integrity of oriP and the replicator DS are required to establish higher copy numbers, although the spatial integrity of oriP is not essentially needed for oriP function per se. These findings extend the model that the efficiency of plasmid establishment in part reflects the efficiency of the initiation of DNA synthesis (31).
The FISH measurements showed a broad distribution of copy numbers, with two main values for each cell line, one being approximately twice as high as the other (Fig. 6B; Table 4). This phenomenon, which is consistent with results reported by Nanbo et al. (38), was also observed for the oriP mutants 2912eFR and 2913eDS (data not shown), indicating that the maintenance of copy numbers is independent from spatial configuration of the viral origin or the presence of DS, although an unaltered oriP establishes higher copy numbers. The spatial separation of DS and FR in 2912eFR and 2913eDS does not significantly alter the replication and tethering functions of both oriP elements. Although we did not study the enhancer function of FR, the effect of FR as a transcriptional enhancer for the BamC promoter seems to be independent from its distance from DS, confirming FR as a bona fide enhancer. Both mutants also suggest that the oriP plasmids analyzed by Reisman et al., containing large additional sequences between FR and DS, were probably lost because of plasmid instability rather than a lack of functionality of FR or DS (43).
TABLE 4.
Distribution of p2908 and p2919ΔDS signalsa
| LCL | Total cell count | Avg no. of signals/cell (no. of cells counted) |
|
|---|---|---|---|
| Lower copy no. | Higher copy no. | ||
| p2908wt-oriP | 34 | 5.5 ± 1.3 (22) | 12.4 ± 1.1 (12) |
| p2910ΔDS | 58 | 1.8 ± 0.8 (33) | 5.5 ± 1.3 (25) |
EBV genomes of asynchronously growing LCLs 2908wt-oriP and 2910ΔDS were visualized by FISH and counted. The average number of cells displaying the lower copy number is shown in the middle panel, the one with the higher copy number on the right.
The combination of our infection experiments and Gardella gel analyses indicates that not DS but FR is crucial for the efficient establishment of growth-transformed LCLs that maintain the EBV genomes as extrachromosomal genomes. Three lines of evidence support this thesis. (i) Only extrachromosomal genomes can be isolated from cells and rescued into E. coli, as has been shown for mini-EBVs (26). (ii) The integration of viral genomes is a rare and very inefficient process. EBNA1-deficient EBV genomes growth transform human primary B cells an at least 10,000 times lower rate than EBNA1+-EBV genomes (22). (iii) We did not obtain any growth-transformed LCLs of FR-deficient and oriP-deficient mini-EBV mutants in infection experiments. These findings confirm that the EBNA1-FR interaction is essential for efficient transformation and extrachromosomal maintenance of EBV-based genomes. Although we cannot rule out a very infrequent integration of EBV genomes, the frequency would be too low to be detected by the utilized imaging analyses.
EBV genomes colocalize with EBNA1 in perichromatic regions.
We aimed to understand how EBV genomes are extrachromosomally maintained. Toward this end, we needed to define in detail how EBV genomes interact with the different nuclear compartments. In the last few years, highly detailed and divergent models of nuclear domains, chromosome territories, and functional subcompartments have emerged (1, 3, 4, 8, 9). We hypothesized that EBV genomes prefer specific nuclear domains or compartments. To verify this model, we analyzed the localization of the EBV genomes in interphase nuclei and their colocalization with specific landmarks of the nuclear landscape. Furthermore, it was of interest whether or not a preferred localization might be dependent on EBNA1 interacting with FR and/or DS.
A combination of FISH and immunofluorescence demonstrated that EBV genomes and EBNA1 colocalize in all cells in the perichromatic region of the host cell nucleus (Fig. 2; see Fig. S5 in the supplemental material). Again, this distribution was not dependent on the spatial organization of oriP and the presence of DS. The perichromatic region is the major site for the different nuclear machineries and highly active regarding transcription, replication, and chromatin remodeling activity (8, 9). The localization of EBV in these highly active regions guarantees easy access to the host cells machineries, although we did not observe a significant colocalization with these active centers or processes (Fig. 3). The binding of EBV to mitotic chromosomes is conferred by EBNA1 (23, 24, 46, 47). We now provide detailed data on the EBNA1-dependent tethering of EBV to interphase host cell chromosomes in perichromatic regions. Furthermore, we observed that more EBNA1 foci than EBV clusters exist in each cell, generating binding platforms that might be used by newly synthesized EBV plasmids during cell cycle progression (Table 2). Interestingly, EBNA1 does not form distinct foci when expressed in the absence of EBV (data not shown), indicating that EBV DNA might induce a clustering of EBNA1. How this is accomplished is yet unknown. The analysis of the different oriP mutants in the context of the mini-EBV genome confirms that the perichromatic localization is entirely dependent on FR, the oriP element that mediates chromatin association.
The epigenetic profiling of oriP in different EBV genomes indicated elevated histone H3K4me3 and H3K9ac levels at the region surrounding oriP in ChIP experiments (10, 59). These marks increase the plasticity of chromatin and correlate with a permissive transcriptional status. All wild-type oriP-containing cell lines, Raji, mini-EBV genome LCL2908, and HEK203 EBV+, localize in nuclear regions characterized by these epigenetic markers. The LCL2910ΔDS also prefers these epigenetic regions, indicating that FR-dependent tethering and not DS-mediated replication directs EBV genomes to these domains. The observation that the epigenetic pattern of histones present at oriP corresponds with its preferred nuclear localization argues for an active interplay between oriP and the associated chromatin domain (Fig. 4). We detected no preference for heterochromatic H3K9me3 regions, an epigenetic mark also absent at histones associated with oriP. The iFISH analysis indicated a weak preference for H3K27me3 regions, an epigenetic mark that is also slightly enriched at oriP (Fig. 4 and 7). This association seems to be linked to DS, because the deletion of DS resulted in a slightly increased H3K27me3 association (Fig. 7). The molecular mechanism of the implementation of this histone modification via DS and its functional relevance remains open. However, it is conceivable that protein-protein interactions implemented via DS might exclude H3K27 foci. For example, Zhou et al. demonstrated that the loss of the telomere repeat factor (TRF2) binding to DS results in earlier replication timing of EBV genomes (60). This suggests that TRF2 delays replication timing at DS, which has an EBNA1-induced permissive chromatin configuration that normally replicates in early S phase. Alternatively, the preference of heterochromatic H3K27me3 localization might be caused by an altered, DS-independent origin usage and replication timing pattern.
To summarize, we observe a synergistic effect generated by the correct spatial conformation of oriP's subcomponents that exceeds the sum of the individual functions. The findings that EBV genomes localize in perichromatic regions and in open epigenetic domains are similar to a different extrachromosomal replicon model, pEPI (50). As such, oriP-based replicons might function as a model system regulated by higher order nuclear structures. The modular design of oriP allows the design of artificial but directed extrachromosomal systems to study the influence of nuclear localization on replication timing, plasmid segregation, and the expression of genes encoded by the vector.
Supplementary Material
Acknowledgments
This work was supported by institutional grants and grants from the Deutsche Forschungsgemeinschaft (grants SPP1230, SFB646, and SFB/TR05).
We thank Stefanie Fülöp, Christoph-Erik Mayer, and Máté Ravasz for critical reading, Wolfgang Hammerschmidt for discussion, Thomas Cremer and Steffen Dietzel for antibodies and help with confocal microscopy, and Achim Tresch for the statistical analyses.
Footnotes
Published ahead of print on 23 December 2009.
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1.Albiez, H., M. Cremer, C. Tiberi, L. Vecchio, L. Schermelleh, et al. 2006. Chromatin domains and the interchromatin compartment form structurally defined and functionally interacting nuclear networks. Chromosome Res. 14:707-733. [DOI] [PubMed] [Google Scholar]
- 2.Bolte, S., and F. P. Cordelieres. 2006. A guided tour into subcellular colocalization analysis in light microscopy. J. Microsc. 224:213-232. [DOI] [PubMed] [Google Scholar]
- 3.Branco, M. R., and A. Pombo. 2006. Intermingling of chromosome territories in interphase suggests role in translocations and transcription-dependent associations. PLoS Biol. 4:e138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chambeyron, S., and W. A. Bickmore. 2004. Chromatin decondensation and nuclear reorganization of the HoxB locus upon induction of transcription. Genes Dev. 18:1119-1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chaudhuri, B., H. Xu, I. Todorov, A. Dutta, and J. L. Yates. 2001. Human DNA replication initiation factors, ORC and MCM, associate with oriP of Epstein-Barr virus. Proc. Natl. Acad. Sci. U. S. A. 98:10085-10089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cherepanov, P. P., and W. Wackernagel. 1995. Gene disruption in Escherichia coli: TcR and KmR cassettes with the option of Flp-catalyzed excision of the antibiotic-resistance determinant. Gene 158:9-14. [DOI] [PubMed] [Google Scholar]
- 7.Cremer, T., and C. Cremer. 2001. Chromosome territories, nuclear architecture and gene regulation in mammalian cells. Nat. Rev. Genet. 2:292-301. [DOI] [PubMed] [Google Scholar]
- 8.Cremer, T., M. Cremer, S. Dietzel, S. Muller, I. Solovei, et al. 2006. Chromosome territories—a functional nuclear landscape. Curr. Opin. Cell Biol. 18:307-316. [DOI] [PubMed] [Google Scholar]
- 9.Cremer, T., K. Kupper, S. Dietzel, and S. Fakan. 2004. Higher order chromatin architecture in the cell nucleus: on the way from structure to function. Biol. Cell 96:555-567. [DOI] [PubMed] [Google Scholar]
- 10.Day, L., C. M. Chau, M. Nebozhyn, A. J. Rennekamp, M. Showe, et al. 2007. Chromatin profiling of Epstein-Barr virus latency control region. J. Virol. 81:6389-6401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Deiss, L. P., and N. Frenkel. 1986. Herpes simplex virus amplicon: cleavage of concatemeric DNA is linked to packaging and involves amplification of the terminally reiterated a sequence. J. Virol. 57:933-941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Delecluse, H. J., S. Bartnizke, W. Hammerschmidt, J. Bullerdiek, and G. W. Bornkamm. 1993. Episomal and integrated copies of Epstein-Barr virus coexist in Burkitt lymphoma cell lines. J. Virol. 67:1292-1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Delecluse, H. J., D. Pich, T. Hilsendegen, C. Baum, and W. Hammerschmidt. 1999. A first-generation packaging cell line for Epstein-Barr virus-derived vectors. Proc. Natl. Acad. Sci. U. S. A. 96:5188-5193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dhar, S. K., K. Yoshida, Y. Machida, P. Khaira, B. Chaudhuri, et al. 2001. Replication from oriP of Epstein-Barr virus requires human ORC and is inhibited by geminin. Cell 106:287-296. [DOI] [PubMed] [Google Scholar]
- 15.Dirmeier, U., B. Neuhierl, E. Kilger, G. Reisbach, M. L. Sandberg, et al. 2003. Latent membrane protein 1 is critical for efficient growth transformation of human B cells by epstein-barr virus. Cancer Res. 63:2982-2989. [PubMed] [Google Scholar]
- 16.Dyson, P. J., and P. J. Farrell. 1985. Chromatin structure of Epstein-Barr virus. J. Gen. Virol. 66(part 9):1931-1940. [DOI] [PubMed] [Google Scholar]
- 17.Gardella, T., P. Medveczky, T. Sairenji, and C. Mulder. 1984. Detection of circular and linear herpesvirus DNA molecules in mammalian cells by gel electrophoresis. J. Virol. 50:248-254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Grasser, F. A., P. G. Murray, E. Kremmer, K. Klein, K. Remberger, et al. 1994. Monoclonal antibodies directed against the Epstein-Barr virus-encoded nuclear antigen 1 (EBNA1): immunohistologic detection of EBNA1 in the malignant cells of Hodgkin's disease. Blood 84:3792-3798. [PubMed] [Google Scholar]
- 19.Gruffat, H., J. Batisse, D. Pich, B. Neuhierl, E. Manet, et al. 2002. Epstein-Barr virus mRNA export factor EB2 is essential for production of infectious virus. J. Virol. 76:9635-9644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Haase, S. B., and M. P. Calos. 1991. Replication control of autonomously replicating human sequences. Nucleic Acids Res. 19:5053-5058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Harrison, S., K. Fisenne, and J. Hearing. 1994. Sequence requirements of the Epstein-Barr virus latent origin of DNA replication. J. Virol. 68:1913-1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Humme, S., G. Reisbach, R. Feederle, H. J. Delecluse, K. Bousset, et al. 2003. The EBV nuclear antigen 1 (EBNA1) enhances B-cell immortalization several thousandfold. Proc. Natl. Acad. Sci. U. S. A. 100:10989-10994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kanda, T., M. Kamiya, S. Maruo, D. Iwakiri, and K. Takada. 2007. Symmetrical localization of extrachromosomally replicating viral genomes on sister chromatids. J. Cell Sci. 120:1529-1539. [DOI] [PubMed] [Google Scholar]
- 24.Kanda, T., M. Otter, and G. M. Wahl. 2001. Coupling of mitotic chromosome tethering and replication competence in Epstein-Barr virus-based plasmids. Mol. Cell. Biol. 21:3576-3588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kempkes, B., D. Pich, R. Zeidler, and W. Hammerschmidt. 1995. Immortalization of human primary B lymphocytes in vitro with DNA. Proc. Natl. Acad. Sci. U. S. A. 92:5875-5879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kempkes, B., D. Pich, R. Zeidler, B. Sugden, and W. Hammerschmidt. 1995. Immortalization of human B lymphocytes by a plasmid containing 71 kilobase pairs of Epstein-Barr virus DNA. J. Virol. 69:231-238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kirchmaier, A. L., and B. Sugden. 1995. Plasmid maintenance of derivatives of oriP of Epstein-Barr virus. J. Virol. 69:1280-1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kirchmaier, A. L., and B. Sugden. 1998. Rep*: a viral element that can partially replace the origin of plasmid DNA synthesis of Epstein-Barr virus. J. Virol. 72:4657-4666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Leight, E. R., and B. Sugden. 2000. EBNA-1: a protein pivotal to latent infection by Epstein-Barr virus. Rev. Med. Virol. 10:83-100. [DOI] [PubMed] [Google Scholar]
- 30.Lindner, S. E., and B. Sugden. 2007. The plasmid replicon of Epstein-Barr virus: mechanistic insights into efficient, licensed, extrachromosomal replication in human cells. Plasmid 58:1-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lindner, S. E., K. Zeller, A. Schepers, and B. Sugden. 2008. EBNA1's affinity for its origin of DNA synthesis is a determinant of the origins's replicative efficiency. J. Virol. 82:5693-5702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Little, R. D., and C. L. Schildkraut. 1995. Initiation of latent DNA replication in the Epstein-Barr virus genome can occur at sites other than the genetically defined origin. Mol. Cell. Biol. 15:2893-2903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lupton, S., and A. J. Levine. 1985. Mapping genetic elements of Epstein-Barr virus that facilitate extrachromosomal persistence of Epstein-Barr virus-derived plasmids in human cells. Mol. Cell. Biol. 5:2533-2542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mackey, D., and B. Sugden. 1999. Applications of oriP plasmids and their mode of replication. Methods Enzymol. 306:308-328. [DOI] [PubMed] [Google Scholar]
- 35.Mackey, D., and B. Sugden. 1999. The linking regions of EBNA1 are essential for its support of replication and transcription. Mol. Cell. Biol. 19:3349-3359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Marechal, V., A. Dehee, R. Chikhi-Brachet, T. Piolot, M. Coppey-Moisan, et al. 1999. Mapping EBNA-1 domains involved in binding to metaphase chromosomes. J. Virol. 73:4385-4392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Middleton, T., and B. Sugden. 1992. EBNA1 can link the enhancer element to the initiator element of the Epstein-Barr virus plasmid origin of DNA replication. J. Virol. 66:489-495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nanbo, A., A. Sugden, and B. Sugden. 2007. The coupling of synthesis and partitioning of EBV's plasmid replicon is revealed in live cells. EMBO J. 26:4252-4262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Neuhierl, B., R. Feederle, W. Hammerschmidt, and H. J. Delecluse. 2002. Glycoprotein gp110 of Epstein-Barr virus determines viral tropism and efficiency of infection. Proc. Natl. Acad. Sci. U. S. A. 99:15036-15041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Norio, P., C. L. Schildkraut, and J. L. Yates. 2000. Initiation of DNA replication within oriP is dispensable for stable replication of the latent Epstein-Barr virus chromosome after infection of established cell lines. J. Virol. 74:8563-8574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Norseen, J., A. Thomae, V. Sridharan, A. Aiyar, A. Schepers, et al. 2008. RNA-dependent recruitment of the origin recognition complex. EMBO J. 27:3024-3035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Polack, A., G. Hartl, U. Zimber, U. K. Freese, G. Laux, et al. 1984. A complete set of overlapping cosmid clones of M-ABA virus derived from nasopharyngeal carcinoma and its similarity to other Epstein-Barr virus isolates. Gene 27:279-288. [DOI] [PubMed] [Google Scholar]
- 43.Reisman, D., J. Yates, and B. Sugden. 1985. A putative origin of replication of plasmids derived from Epstein-Barr virus is composed of two cis-acting components. Mol. Cell. Biol. 5:1822-1832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ritzi, M., K. Tillack, J. Gerhardt, E. Ott, S. Humme, et al. 2003. Complex protein-DNA dynamics at the latent origin of DNA replication of Epstein-Barr virus. J. Cell Sci. 116:3971-3984. [DOI] [PubMed] [Google Scholar]
- 45.Schepers, A., M. Ritzi, K. Bousset, E. Kremmer, J. L. Yates, et al. 2001. Human origin recognition complex binds to the region of the latent origin of DNA replication of Epstein-Barr virus. EMBO J. 20:4588-4602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sears, J., J. Kolman, G. M. Wahl, and A. Aiyar. 2003. Metaphase chromosome tethering is necessary for the DNA synthesis and maintenance of oriP plasmids but is insufficient for transcription activation by Epstein-Barr nuclear antigen 1. J. Virol. 77:11767-11780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sears, J., M. Ujihara, S. Wong, C. Ott, J. Middeldorp, et al. 2004. The amino terminus of Epstein-Barr Virus (EBV) nuclear antigen 1 contains AT hooks that facilitate the replication and partitioning of latent EBV genomes by tethering them to cellular chromosomes. J. Virol. 78:11487-11505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shirakata, M., and K. Hirai. 1998. Identification of minimal oriP of Epstein-Barr virus required for DNA replication. J. Biochem. 123:175-181. [DOI] [PubMed] [Google Scholar]
- 49.Spector, D. L. 2001. Nuclear domains. J. Cell Sci. 114:2891-2893. [DOI] [PubMed] [Google Scholar]
- 50.Stehle, I. M., J. Postberg, S. Rupprecht, T. Cremer, D. A. Jackson, et al. 2007. Establishment and mitotic stability of an extra-chromosomal mammalian replicon. BMC Cell Biology 8:33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sternas, L., T. Middleton, and B. Sugden. 1990. The average number of molecules of Epstein-Barr nuclear antigen 1 per cell does not correlate with the average number of Epstein-Barr virus (EBV) DNA molecules per cell among different clones of EBV-immortalized cells. J. Virol. 64:2407-2410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wang, J., and B. Sugden. 2005. Origins of bidirectional replication of Epstein-Barr virus: models for understanding mammalian origins of DNA synthesis. J. Cell Biochem. 94:247-256. [DOI] [PubMed] [Google Scholar]
- 53.Wensing, B., and P. J. Farrell. 2000. Regulation of cell growth and death by Epstein-Barr virus. Microbes Infect. 2:77-84. [DOI] [PubMed] [Google Scholar]
- 54.Wu, H., D. F. Ceccarelli, and L. Frappier. 2000. The DNA segregation mechanism of Epstein-Barr virus nuclear antigen 1. EMBO Rep. 1:140-144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wu, H., P. Kapoor, and L. Frappier. 2002. Separation of the DNA replication, segregation, and transcriptional activation functions of Epstein-Barr nuclear antigen 1. J. Virol. 76:2480-2490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yates, J., N. Warren, D. Reisman, and B. Sugden. 1984. A cis-acting element from the Epstein-Barr viral genome that permits stable replication of recombinant plasmids in latently infected cells. Proc. Natl. Acad. Sci. U. S. A. 81:3806-3810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yates, J. L., S. M. Camiolo, and J. M. Bashaw. 2000. The minimal replicator of Epstein-Barr virus oriP. J. Virol. 74:4512-4522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yates, J. L., N. Warren, and B. Sugden. 1985. Stable replication of plasmids derived from Epstein-Barr virus in various mammalian cells. Nature 313:812-815. [DOI] [PubMed] [Google Scholar]
- 59.Zhou, J., C. M. Chau, Z. Deng, R. Shiekhattar, M. P. Spindler, et al. 2005. Cell cycle regulation of chromatin at an origin of DNA replication. EMBO J. 24:1406-1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhou, J., A. R. Snyder, and P. M. Lieberman. 2009. Epstein-Barr virus episome stability is coupled to a delay in replication timing. J. Virol. 83:2154-2162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zimmermann, J., and W. Hammerschmidt. 1995. Structure and role of the terminal repeats of Epstein-Barr virus in processing and packaging of virion DNA. J. Virol. 69:3147-3155. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.