Abstract
The current vaccine against smallpox is an infectious form of vaccinia virus that has significant side effects. Alternative vaccine approaches using recombinant viral proteins are being developed. A target of subunit vaccine strategies is the poxvirus protein A33, a conserved protein in the Chordopoxvirinae subfamily of Poxviridae that is expressed on the outer viral envelope. Here we have determined the structure of the A33 ectodomain of vaccinia virus. The structure revealed C-type lectin-like domains (CTLDs) that occur as dimers in A33 crystals with five different crystal lattices. Comparison of the A33 dimer models shows that the A33 monomers have a degree of flexibility in position within the dimer. Structural comparisons show that the A33 monomer is a close match to the Link module class of CTLDs but that the A33 dimer is most similar to the natural killer (NK)-cell receptor class of CTLDs. Structural data on Link modules and NK-cell receptor-ligand complexes suggest a surface of A33 that could interact with viral or host ligands. The dimer interface is well conserved in all known A33 sequences, indicating an important role for the A33 dimer. The structure indicates how previously described A33 mutations disrupt protein folding and locates the positions of N-linked glycosylations and the epitope of a protective antibody.
Poxviruses are large DNA viruses that infect a wide range of hosts. The smallpox virus devastated human populations until its eradication 3 decades ago. Other poxviruses are emerging, such as monkeypox virus, which also infects humans and causes disease (61). The smallpox vaccine is a model of vaccine efficacy, but how the vaccine induces protection is not well understood. Knowledge of how the vaccine produces protection will also likely be important for efforts to produce vaccines that are effective against other pathogens. Though highly successful in the population overall, the smallpox vaccine uses an infectious strain of vaccinia virus and has a significant level of serious side effects (6). One focus of current research is to develop vaccines using recombinant poxvirus proteins that are as protective as the live virus vaccine but produce fewer complications. A more detailed structural characterization of the protein antigens that are important for conferring protection will improve our knowledge of how the smallpox vaccine works and lead to a better understanding of poxvirus biology.
There are two morphologically distinct forms of poxviruses: the mature virion (MV) and the enveloped virion (EV) (53). The MV, also known as the intracellular mature virion (IMV), is found inside the infected cell (62, 65). Enveloped virions are formed from MVs that have been wrapped by modified Golgi or endosomal membranes (53). MVs are thought to be responsible for host-to-host proliferation of the virus, while the EVs mediate virus spread within a host (40, 49). EVs that are attached to the cell surface, also termed cell-associated enveloped virions (CEV), are thought to propagate viral infection to neighboring cells (65). EVs that are released from the cell surface, also termed extracellular enveloped virus (EEV), mediate longer-range dissemination in the host (65). The outer membranes of the MV and EV forms each have a distinct assemblage of proteins. Candidate subunit vaccines have been shown to require proteins from both virus forms to be most effective (17, 27, 28).
A33 is a type II integral membrane glycoprotein found on the surface of the EV form of the virus and is also expressed on the host cell membrane (62). A33 is a disulfide-bonded homodimer with both N- and O-linked glycosylation (57, 62). Deletion of the A33R gene in vaccinia virus results in a small-plaque phenotype, defects in actin tail formation, and inefficient cell-to-cell spread in cell culture (63). Evidence implicates A33 in the spreading of virus from cell to cell by a mechanism that is antibody resistant (40). Antibodies against A33 in cell culture prevent the formation of comet-shaped viral plaques, which are assayed in an overlay of plaques with liquid and are indicative of cell-to-cell spreading of the EV (2, 17). A33 has been shown to interact through its cytoplasmic and transmembrane regions with the EV proteins A36 (18, 64, 72, 76) and B5 (58, 64). Vaccination with A33 is protective in a number of animal models of poxvirus infection as a component of protein subunit vaccination (16, 17, 19, 77), DNA vaccination (19, 27-29), or a combination of the two methods (24). Despite the inclusion of A33 in vaccination studies, the functions of A33 in the virus are unclear.
Members of the C-type lectin-like domain (CTLD) superfamily of proteins are found in organisms ranging from bacteria to humans (13, 78). The first crystal structures of carbohydrate-binding domains with this fold gave rise to the name “lectin” for the family, but many family members do not bind carbohydrates. The classification of a domain as being C type lectin-like is currently based on similarities in protein sequence and fold (74). CTLDs have been shown to bind noncarbohydrate small molecules, lipids, proteins, and other structures, such as ice (13, 78). One group of type II transmembrane proteins that contain CTLDs is comprised of the natural killer (NK)-cell receptors of the innate immune system (78). The NK-cell receptors are composed of dimers of CTLDs, which are the only dimers out of the several hundred C-type lectin-like structures that are known. Protein binding by NK-cell receptors occurs on a surface formed by the “long loop” and nearby residues that are on the opposite side of the dimer from the N and C termini (60). A second group of proteins contains a monomeric CTLD, termed a “Link module” CTLD, that binds glycosaminoglycans but lacks the “long loop” region that is present in almost all other CTLDs (78).
To gain a better understanding of the structure and possible functions of A33 and to further its development in vaccines, we determined the X-ray crystal structure of the A33 ectodomain from vaccinia virus. Based on the structure and sequence of A33, the carbohydrate-binding site of the canonical CTLD is not present. The structure revealed A33 to have dimers of CTLDs. Comparison of A33 with other CTLDs, including dimers from NK-cell receptors and monomers from Link modules, indicates that A33 contains an unusual CTLD that likely binds ligands of host or virus origins.
MATERIALS AND METHODS
A33 expression and purification.
We expressed several A33 proteins that differed in their N termini but extended to the full-length C terminus at residue 185. The first A33 length variant encoded residues 82 to 185 and was generated by PCR using the 5′ primer GGGAATTCTAAGGAGGATATTCATATGTCTACTCATAGAAAGGTTGCGTCTAG, the 3′ primer GGGGATCCTTAGTTCATTGTTTTAACACAAAAATACTTTC, and viral DNA from the vaccinia virus Western Reserve (WR) strain. A33 is the A33R gene product. In the WR strain of vaccinia virus, A33R is given the locus tag WR156. PCR products were cloned into the vector pNAN using the NdeI and NotI restriction sites (67). The A33 protein (residues 82 to 185) formed crystals, but electrospray ionization mass spectrometry of A33(82-185) in stock solution showed that the mass of A33 had become smaller, presumably by proteolytic cleavage by Escherichia coli proteases remaining in the preparation. A shorter A33 (residues 90 to 185) molecule was generated using the 5′ primer GGGAATTCTAAGGAGGATATTCATATGAGCACTACACAATATGATCACAAAGAAAG and the same 3′ primer as the 82-185 construct. The L118M mutation was made with the 5′ primer CAGACTACCAGATGTTCTCGGATGC and 3′ primer GCATCCGAGAACATCTGGTAGTCTG. The L140M mutation was introduced by the 5′ primer CCGATGTCATGATTACCTGGCTCATTG and 3′ primer CAATGAGCCAGGTAATCATGACATCGG. The K123A mutation was generated with the 5′ primer CAGATGTTCTCGGATGCTGCGGCAAATTGCACTGCGG and 3′ primer CCGCAGTGCAATTTGCCGCAGCATCCGAGAACATCTG. The K123C mutation was generated with the 5′ primer GGATGCTTGCGCAAATTGCACTGCGG and 3′ primer GCAGTGCAATTTGCGCAAGCATCCGAG. The E149C mutation was generated with the 5′ primer CTCATTGATTATGTTTGCGATACATGGGG and 3′ primer CCCCATGTATCGCAAACATAATCAATGAG. All plasmids and mutations were verified by DNA sequencing.
Expression and refolding of A33 were carried out using methods similar to those used for producing the L1 (66) and G4 (67) proteins. A33 was expressed as inclusion bodies in E. coli strain BL21(DE3)-RIL-X (Stratagene, La Jolla, CA) at 37°C in 6 liters of selenomethionine (SeMet) medium (Molecular Dimensions, Apopka, FL) containing 100 μg/ml ampicillin and supplemented with 50 mg per liter of l-SeMet (Sigma, St. Louis, MO). When the optical density of the culture at 600 nm reached 0.6, A33 expression was induced with 0.5 mM isopropyl-β-d-thiogalactopyranoside for 4 to 6 h. Cells were harvested by centrifugation at 4,000 × g and resuspended in 150 ml of lysis buffer (100 mM Tris-HCl [pH 8.0], 150 mM NaCl, 1% NP-40, 0.1% [wt/vol] sodium deoxycholate) with one protease inhibitor tablet per 50 ml (Complete protease inhibitor cocktail; Roche Diagnostics, Indianapolis, IN). Cells were lysed by freezing and thawing in the presence of lysozyme (1 mg/ml).
The A33 inclusion bodies were purified by multiple washes using centrifugation and resuspension. After the final wash, the pellet was dissolved in 6 M guanidine-HCl with 2 mM dithiothreitol, and insoluble material was removed by centrifugation at 14,000 × g for 15 min. A33 was refolded by rapid dilution in buffer containing 100 mM Tris-HCl (pH 8.0), 800 mM arginine-HCl, 0.5 mM oxidized glutathione, 5 mM reduced glutathione, and one Complete protease inhibitor cocktail tablet. After 24 h, the refolded protein was dialyzed at least three times against 10 volumes of 10 mM Tris-HCl (pH 8.0). The protein was filtered through a 0.22-μm membrane, concentrated in centrifugal devices (Centricon-70; Millipore, Billerica, MA), and purified by size exclusion chromatography (Superdex 75; GE Healthcare, Fairfield, CT). The Superdex 75 column was calibrated with standard proteins (catalog no. 151-1901; Bio-Rad, Hercules, CA). Finally, A33 was dialyzed into 10 mM Tris-HCl (pH 8.0) and concentrated. The extent of SeMet incorporation was determined by mass spectrometry.
Crystallization and structure determination.
Crystals of A33 were obtained by the hanging drop vapor diffusion method at room temperature by mixing 1 μl of protein solution (10 mg/ml) and 1 μl of reservoir solution (0.5 ml) containing 100 mM sodium acetate (pH 4.3), 20% polyethylene glycol 8000, and 10% isopropanol. The crystals were cryo-preserved for X-ray diffraction in a solution of 100 mM sodium acetate (pH 4.3), 30% polyethylene glycol 8000, and 15% glycerol. Similar crystallization conditions with modified A33 proteins gave rise to four other crystal forms (see Table 2).
TABLE 2.
Structural parameters and crystal cell constants of the five A33 crystals
| Space group | Resolution (Å) | Solvent content (%) | Matthews coeff. | Content asym. unit | Angle(s) between monomers | a (Å) | b (Å) | c (Å) | α (°) | β (°) | γ (°) |
|---|---|---|---|---|---|---|---|---|---|---|---|
| C2 old | 2.6 | 30.7 | 1.77 | Dimer | 179.5° | 125.9 | 32.4 | 38.5 | 90 | 92.3 | 90 |
| P21212 | 3.0 | 32.1 | 1.81 | Dimer | 177.9° | 130.8 | 32.7 | 37.4 | 90 | 90 | 90 |
| P212121 | 2.8 | 22.0 | 1.58 | Dimer | 179.3° | 29.2 | 44.2 | 107.7 | 90 | 90 | 90 |
| P21 | 2.3 | 34.3 | 1.87 | Two dimers | 179.6°, 180° | 41.8 | 57.6 | 68.9 | 90 | 95.6 | 90 |
| C2 new | 2.1 | 38.6 | 2.00 | Dimer | 179.8° | 80.8 | 56.2 | 41.6 | 90 | 110.7 | 90 |
The structure of A33 was determined using diffraction data obtained from the “C2 new” crystal. The A33 protein that crystallized in the “C2 old” space group has the wild-type sequence. The “P21212” sequence contains the mutations K123C, L118M, and L140M. The “P212121” sequence contains the mutations E149C, L118M, and L140M. The “P21” and “C2 new” sequences contain the mutations K123A, L118M, and L140M. coeff., coefficient; asym., asymmetric.
X-ray data were collected at the peak, edge, and high remote Se energies with the intention of obtaining phases by a multiple-wavelength anomalous diffraction experiment. X-ray data were integrated and scaled using the HKL2000 (55) and XDS (32) software programs. When the SOLVE software program (70) was used with various combinations of the three SeMet data sets, the best electron density maps were obtained by using the high-energy remote data set alone in a single-wavelength anomalous diffraction experiment. Density modification and automated model building were performed using the RESOLVE software program (68, 69) (see Table 1). The asymmetric unit of the crystal contained two A33 monomers, but noncrystallographic symmetry was not used during structure solution, model building, or refinement. Cycles of manual model building using the O graphics software program (31) and refinement using the CNS software program (7) improved the electron density map and allowed the tracing of residues 96 to 184 of one monomer and 98 to 185 of the other monomer. Three residues (166 to 168) were removed from both monomers in the final model because of poor electron density. Setting of the occupancies of the Se and Cɛ atoms of SeMet side chains to less than 1.0 was guided by Fo-Fc difference electron density maps in order to model the loss of those atoms due to radiation damage.
TABLE 1.
X-ray data and refinement statistics
| Parametera | Value or identifierb |
|---|---|
| Space group | C2 |
| Resolution (Å) | 50-2.10 (2.18-2.10) |
| X-ray source | 22-ID (SER-CAT) |
| Wavelength (Å) | 0.97625 |
| Completeness (%) | 98.2 (96.2) |
| Avg redundancy | 3.9 (3.6) |
| Rsym (%) | 4.8 (30.4) |
| <I>/<σI> | 26.9 (4.4) |
| Phasing (figure of merit): | |
| SOLVE | 0.44 |
| RESOLVE | 0.79 |
| Ramachandran (%): | |
| Favored | 85.4 |
| Allowed | 14.6 |
| Mean B value (Å2) for protein | 40.9 |
| Main chain | 39.4 |
| Side chain | 42.4 |
| Mean B value of all atoms | 40.8 |
| No. of protein atoms | 1,343 |
| No. of waters | 42 |
| No. of reflections refined/no. free | 9573/500 |
| Rwork/Rfree (%) | 21.8/25.9 (25.7/26.4) |
Rsym = ΣhΣi|Ih,i − 〈Ih〉|/ΣhΣi|Ih,i Where Ih,i is the ith measurement of intensity for reflection h and 〈Ih〉 is the average intensity of that reflection; Rwork/Rfree = Σh|Fp − Fc|/Σh|Fp, where Fp is the observed structure factor and Fc is the calculated structure factor amplitude for reflection h for the working set and free set, respectively.
Values in parentheses are for the highest-resolution bin.
The placement of the A33 model in the other crystal forms was accomplished by molecular replacement. A monomer from the refined structure of A33 was used as a search model after the removal of residues 161 to 174 from the loop and residues 183 to 185 from the C terminus. The molecular replacement and rigid body refinement of the best solutions were carried out using the PHASER software program (50). The rigid-body refined models were examined without any further refinement.
Protein model geometry and side-chain rotamer conformations for Asn, Gln, and His were examined using the Procheck software program (39) and corrected using the Molprobity program (47). PISA was used to calculate buried surface areas (37). Shape complementarity statistics were calculated using the SC software program (41). The DALI (26), SSM (36), and FFAS (30) servers were used to search the structural and sequence databases. LSQMAN (35) and PISA were used to structurally superimpose A33 on other structures. Depictions of molecules were generated with PyMOL (11). Sequence alignments and figures were made with the Multalin (10), ClustalW (38), and Jalview (9) software programs. Selection and comparison of poxvirus sequences were performed using software and databases at the Poxvirus Bioinformatics Resource Center (www.poxvirus.org) (42).
Protein structure accession number.
Coordinates for the refined A33 structure and structure factors for the five data sets have been deposited in the Research Collaboratory for Structural Bioinformatics Protein Data Bank (accession code 3k7b).
RESULTS
Based on its sequence, A33 is predicted to be a type II integral membrane protein with a short N-terminal cytoplasmic region, an uncleaved signal sequence-transmembrane region spanning the outer membrane of the EV, and an ectodomain on the surface of the virus or infected cell (Fig. 1) (62). The ectodomain is predicted to be comprised of residues 58 to 185, which includes five cysteines (62). Secondary structure prediction algorithms predicted that the first regular secondary structure in the A33 sequence begins around residue 82. An expressed A33 protein consisting of residues 82 to 185 formed crystals after 3 weeks of incubation. Mass spectrometry analysis of the 82-185 protein indicated that it had been shortened by proteolysis during storage to residues 90 to 185. When the shorter A33 protein (residues 90 to 185) was expressed and purified, it formed crystals in 1 day.
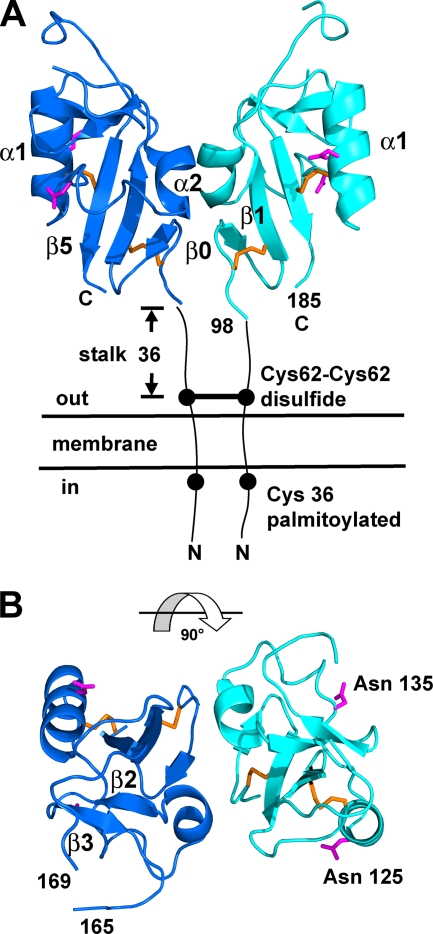
FIG. 1.
A33 contains a dimer of C-type lectin-like domains (CTLDs). (A) A ribbon representation depicts the two monomers of A33, residues 98 to 185, on the membrane surface (“out”). The C100-C109 and C126-C180 disulfide bonds (orange) and the Asn side chains of the two N-linked glycosylation sites (magenta) are shown on each A33 CTLD monomer. The intermolecular disulfide bond between the C62 residues of each monomer and the palmitoylated C36 residues on the virion or cytosol side (“in”) of the membrane are shown. The β2 and β3 β-strands are labeled in panel B for clarity. The 36 residues from Cys 62 to the first ordered residue in the crystal, Glu 98, comprise the stalk between the membrane and A33. (B) View of A33 after a 90° rotation from the view in panel A. The predicted N-linked glycosylation sites (Asn 125 and Asn 135) are labeled. The three residues (166, 167, and 168) between residues 165 and 169 in both monomers were not modeled (see Methods).
Mutations and manipulations used to determine the A33 structure.
X-ray data collected from several heavy atom-soaked crystals led to estimated phases that did not yield interpretable electron density. This was due to the moderate resolution of the X-ray data, coupled with inadequate heavy atom binding in the acidic pH of crystallization and the low solvent content of the crystals. Our second approach for obtaining a bound heavy atom was to mutate K123 and E149 one at a time to cysteine in order to bind a mercury atom to the sulfur atom of the new cysteine at neutral pH prior to crystallization. Since only one methionine (M184) is present in residues 90 to 185 of the wild-type sequence, we also mutated L118 and L140 to methionines so as to use the anomalous scattering of the Se atoms of SeMet. Mass spectrometry analysis of the K123C mutant A33 protein indicated that C123 was covalently bonded to a glutathione molecule from the refolding buffer. Under crystallization conditions similar to those for the wild-type protein, crystals appeared that exhibited a new morphology. Though larger, these crystals did not diffract as well as the crystals with the wild-type A33 sequence.
Suspecting that the glutathione adduct on the new cysteine at residue 123 had altered the packing within the crystal, we mutated K123 to alanine with the intention of changing the molecular packing in a way that would result in a better diffracting crystal. The K123A protein crystallized in two new space groups (C2 and P21) that exhibited markedly improved diffraction. Phases and clear electron density were successfully obtained in the new C2 space group using the Se atoms of the three SeMet residues with the single-wavelength anomalous diffraction method (Table 1). In the new C2 space group, molecules from neighboring asymmetric units contact each other at the molecular surface containing the alanine at position 123. This well-diffracting molecular packing of the protein in the crystal is not possible with a Lys or Cys at position 123.
Structure of A33.
The 2.1-Å crystal structure of vaccinia virus A33 reveals a domain that is structurally similar to CTLDs based on a search of the structural database (Fig. 1). The A33 CTLD forms dimers with an overall “butterfly” shape (Fig. 1A). Each A33 monomer has five β-strands in two β-sheets and two α-helices numbered from the N to C termini as β0, β1, α1, α2, β2, β3, and β5 (Fig. 1), in the numbering convention used for mannose binding protein (73), the first structurally determined CTLD. In A33, one β-sheet is composed of the β0, β1, and β5 strands, and the second β-sheet is composed of only two β-strands, β2 and β3. A33 is unique in not having a β4 strand that pairs with the β3 strand of the second β-sheet, as in all other CTLDs with known structures. Residues 1 to 97 of A33 make up the intracellular, transmembrane, and stalk portions of A33 (Fig. 1).
There are six cysteines in the full-length A33 sequence. Four of these were observed in electron density forming the two disulfide bonds C126-C180 and C100-C109 (Fig. 1). CTLDs have been found to contain one (48), two (73), three (46), five (54), or no disulfide bonds (52). In all CTLDs with disulfide bonds, the bond that links a cysteine in the α1 helix to a cysteine in the β5 strand is conserved. In A33, this is the C126-C180 disulfide bond. Another conserved disulfide bond, lacking in A33 but found in most CTLDs, ties the β3 strand to the loop that follows the β4 strand. The second A33 disulfide bond, C100-C109, is largely conserved in CTLDs that have three disulfide bonds and have been termed “long-form” CTLDs (13). As in the “long-form” CTLDs, the A33 C100-C109 bond connects the loop before the β0 strand to the β1 strand. Of the two other cysteines in A33, C62 is predicted to be located outside the membrane and involved in the interdomain disulfide bond between the two monomers (Fig. 1). The other cysteine, C36, is predicted to be inside the membrane and to be palmitoylated (21, 57) (Fig. 1).
There are two predicted N-linked glycosylation sites in vaccinia virus A33, at residues N125 and N135 (Fig. 1B). Both sites are located away from the dimer interface. The N125 site is on the α1 helix adjacent to the conserved C109. The N135 site is in the connecting region between the α1 and α2 helices. Gel electrophoresis mobility differences after enzymatic deglycosylation of A33 and after virus growth in the presence of glycosylation inhibitors indicate that the N-linked sites are used in vaccinia virus (57). Variola virus and monkeypox virus A33 orthologs lack the N125 site, despite their 95% sequence identity with vaccinia virus A33.
The A33 ectodomain is a unique C-type lectin-like domain.
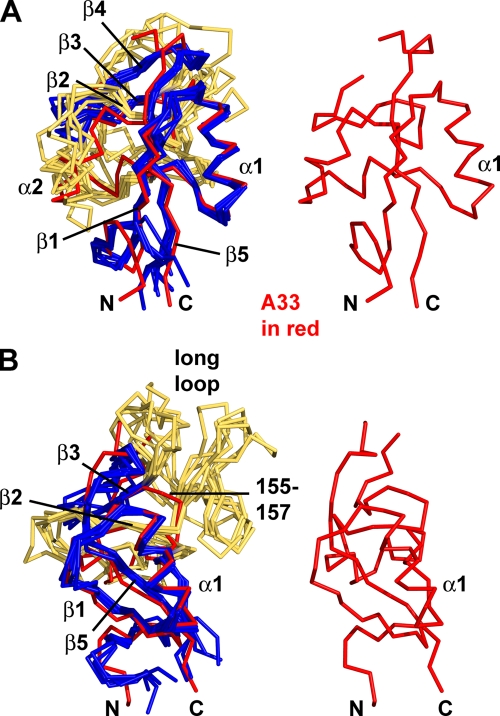
One α-helix and two β-sheets are conserved in almost all C-type lectin-like proteins (Fig. 2). Upon superimposition of several CTLD structures, close structural similarity is observed in the α1 helix and in the β-strands. Overlaying an A33 monomer and a selected CTLD results in root mean squared deviations (RMSD) of 2.0 to 2.9 Å over the positions of 65 to 75 Cα pairs (36). A hallmark of CTLDs is that the C-terminal β-strand (β5 in A33) pairs with an N-terminal β-strand (β1 in A33). This results in the N and C termini of CTLDs being close to each other. The α2 helix is almost always present but takes a distinct path in each CTLD as it connects α1 and β2 (Fig. 2A). For the comparison shown in Fig. 2, any of the several hundred CTLDs that have been structurally determined would yield a similar superimposition.
FIG. 2.
Overlays of A33 and five other CTLD structures in stick representations. (A) A33 (red) is overlaid with five other CTLDs (blue and yellow, at left) and is shown alone (at right and in panel B) to orient the reader. The other CTLDs are colored blue where they exhibit the more conserved portions of a C-type lectin-like fold and are colored yellow where they are not as conserved. Conserved secondary structure elements are labeled from N to C termini: β1, α1, α2, β2, β3, β4, and β5. Note that A33 does not contain a β4 strand. The β2 strand of A33 (red) and the β2 strands of the other CTLDs (“β2,” colored yellow for clarity) also superimpose closely. The orientation here is similar to the orientation of the A33 monomer on the right in Fig. 1A. CTLDs that have been found to be homodimers were chosen for comparison. (B) The view is rotated toward the viewer and to the left about 45° in order to see the “long loop.” Each of the five CTLDs has the “long loop” connecting the β2 and β3 strands, but A33 does not. Instead, A33 has a short connecting loop (residues 155 to 157). The legend to Fig. 3 identifies the five structures from which monomer CTLDs were extracted and superimposed on the A33 monomer.
CTLDs that bind carbohydrates usually have conserved motifs, EPN or EPD, of residues that interact with the carbohydrate ligand. The residues EPN are involved in mannose binding and the EPD residues are involved in galactose binding with the central proline in a cis conformation that allows side chain carbonyl groups of the adjacent residues to coordinate a Ca2+ atom (13, 78). Neither sequence motif is found in A33, and there is no evidence that A33 binds Ca2+. The two prolines at positions 134 and 158 in the A33 CTLD are both trans isomers, and each is adjacent to a nonpolar residue, LPN and NPI, which does not have side chain carbonyl groups to coordinate Ca2+.
Almost all known CTLDs have a β-sheet composed of the β2, β3, and β4 strands located immediately N-terminal to the β5 strand at the C terminus. But A33 is different and to our knowledge unique. Starting at the β5 strand and examining the A33 structure in the N-terminal direction, there is a loop extending 15 residues from T160 to E174 that connects the β5 strand to the β3 strand (Fig. 2). Three of these residues (166, 167, and 168) were not modeled in the A33 structure due to insufficient electron density supporting their positions, which is likely due to the mobility of these residues in the crystal. Without a β4 strand in A33, the 15-residue loop forms the connection between the β3 and β5 strands.
Connecting the β2 and β3 strands, A33 has an unusually short three-residue loop (residues 155 to 157) (Fig. 2B). In most other CTLDs, this same β2-β3 connecting loop is 30 to 40 residues in length and is termed the “long loop” (78). The “long loop” is involved in the many binding activities of CTLDs (Fig. 2B). Short β2-β3 connecting loops that are similar to those in A33 are found in the Link module (3, 25), invasin (23), and intimin (48) CTLDs. All three types of CTLDs are monomers and, like the NK-cell receptors, are close structural matches to the A33 monomer but with low sequence identities to A33. When A33 is superimposed on the Link module of CD44 (PDB code 2jcp [3]), the RMSD is 2.7 Å over 72 Cα pairs, and when superimposed on the Link module from tumor necrosis factor-stimulated gene 6 (TSG-6; pdb code 2pf5 [25]), the RMSD is 2.5 Å over 67 Cα pairs. Structure-based sequence identities of A33 and the Link modules are 13% and 15%, respectively. Superimposing the A33 monomer and invasin (PDB code 1cwv [23]) yields an RMSD of 2.1 Å over 71 Cα pairs but a sequence identity of 10%. The comparison between A33 and intimin (PDB code 1f02 [48]) yields an RMSD of 2.6 Å over 70 Cα pairs with a sequence identity of 17%. Link modules, invasins, and intimins have not been observed as dimers (78).
A33 dimer of CTLDs resembles dimeric CTLDs of natural killer (NK)-cell receptors.
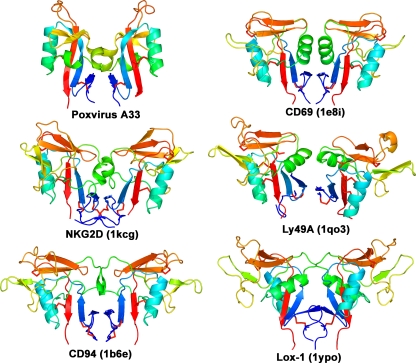
When the structural database is queried using the A33 dimer as the search molecule, the molecules that are identified as similar are the dimeric NK-cell receptors of the immune system that contain CTLDs (Fig. 3). These NK-cell receptors are CD69, NKG2D, Ly49A, CD94, Lox-1, and killer cell lectin-like receptor G1 (KLRG1), though not all are found exclusively on natural killer cells (78). Structure-based sequence identities between A33 and these NK-cell receptors are 19, 13, 21, 13, 12, and 13%, respectively. When the A33 dimer is structurally aligned to the NK-cell receptors, the RMSD range from 3.0 to 4.0 Å over 131 to 150 Cα pairs.
FIG. 3.
The A33 homodimer of CTLDs is structurally similar to CTLD dimers found in only a few other proteins. A ribbon depiction of the A33 dimer (Poxvirus A33) is shown at the upper left. Other known homodimers of CTLDs are from the immune system and are colored similarly for comparison: CD69 (PDB code 1e8i [46]), NKG2D (1kcg [59]), Ly49A (1qo3 [71]), CD94 (1b6e [5]), and Lox-1 (1ypo [56]). The KLRG1 dimer (3ff8 [45]) is also similar (not shown). The molecules are rainbow colored in a gradient from the N terminus (dark blue) to the C terminus (red). Disulfide bonds are shown in red. All proteins in this figure are type II transmembrane proteins.
The NK-cell receptors are classified as group V in the classification scheme for CTLDs (13, 14, 78). Members of group V are dimeric type II integral membrane proteins and have a stalk between the transmembrane segment and the CTLD. The A33 stalk encompasses the 36 residues from the interdomain C62-C62 disulfide bond to the beginning of the CTLD (Fig. 1A). The A33 stalk is comparable in length to the stalks seen in members of the NK-cell receptors in group V CTLDs and is likely to be similarly nonglobular in structure (78).
The dimer of A33.
When recombinant A33 was subjected to size exclusion chromatography during protein production for crystallization, it migrated as a single population of molecules at a molecular mass consistent with a dimer (data not shown). We did not observe an A33 monomer. Since recombinant A33 (residues 90 to 185) did not include C62, recombinant A33 forms stable dimers without the intersubunit disulfide bond. Reported light scattering results using orthologous recombinant A33 from ectromelia virus, which causes mousepox, and A33 from vaccinia virus also indicate that A33 is a dimer in solution (16).
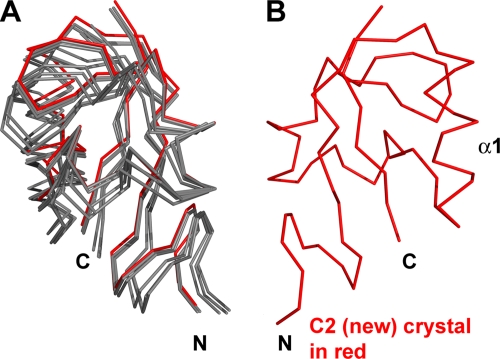
We examined five crystals containing six independent ways that A33 molecules packed to form crystals. In each packing, A33 was exclusively dimeric (Fig. 4). In four of the five crystals, noncrystallographic symmetry related the two monomers that composed one dimer in each asymmetric unit. The fifth crystal (P21) (Table 2) contained four monomers per asymmetric unit. Two monomers formed a dimer and were related by noncrystallographic symmetry. The two other monomers formed dimers with two other A33 monomers in neighboring asymmetric units and thus were related by crystallographic symmetry (Table 2). The angle of rotation that would be required to overlay one A33 monomer on the other monomer in the dimer changes up to 2° between space groups (Table 2). The A33 dimer, though stable without a disulfide bond, has a degree of flexibility in that the monomers can move relative to each other (Fig. 4).
FIG. 4.
The A33 dimer interface exhibits limited flexibility. The “B” monomers from each of the six A33 models were superimposed (red monomer at right). The relative positions of the “A” monomers at left (gray) are seen to vary among the six kinds of packing (see Table 2). The P21 crystal had two A33 dimers in the asymmetric unit. The A33 model from the “C2 (new)” crystal in which the A33 structure was determined is colored red.
Formation of the A33 dimer buries a total solvent-exposed surface area of 970 Å2 or approximately 9% of the total surface area of 10,820 Å2 for the two monomers (37). As a measure of the quality of the fit of the interface between the two A33 monomers, a shape complementarity statistic was calculated, where a perfect fit between monomers would yield a score of 1.0. The shape complementarity of the A33 dimer interface is 0.80, which is higher than typical values for protein oligomers, which range from 0.70 to 0.76 (41).
Poxvirus A33 dimer interface is conserved.
A33 orthologs have been identified in each of the genera of the Chordopoxvirinae subfamily (Fig. 5A), except in the Avipoxvirus genus. A33 orthologs also appear to be absent in the Entomopoxvirinae subfamily and in the unassigned crocodilepox virus (1). An A33 ortholog is present in the genome of squirrelpox virus, which is also unassigned to a genus (51) (see the legend to Fig. 5). A33 orthologs from viruses within the same genus have relatively conserved sequences. For example, the orthologous A33 genes of members of the Orthopoxvirus genus are nearly identical in sequence (not shown). In contrast, orthologous A33 sequences from different genera in the Chordopoxvirinae subfamily have a lower sequence similarity (Fig. 5A).
FIG. 5.
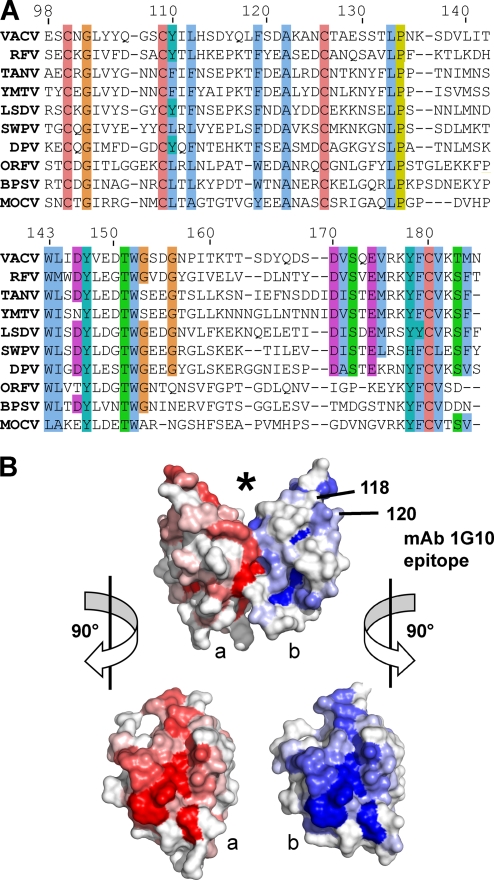
The A33 dimer interface is conserved in the Chordopoxvirinae subfamily. (A) The top line is the sequence of A33 from vaccinia virus (VACV) in the structure described herein. The A33 sequence is aligned with the sequences of nine A33 orthologs: RFV (rabbit fibroma virus), TANV (tanapox virus), YMTV (yaba monkey tumor virus), LSDV (lumpy skin disease virus), SWPV (swinepox virus), DPV (deerpox virus), ORFV (orf virus), BPSV (bovine papular stomatitis virus), and MOCV (molluscum contagiosum virus). Sequence similarities have been shaded with the ClustalX color scheme. Cysteines for the C100-C109 and C126-C180 disulfides are completely conserved. (B) Surface representation of A33 with the molecule in the same view as in Fig. 1. The location of the putative ligand binding site is marked (*). The location of the surface implicated in the MAb 1G10 epitope and contributed by residues 118 and 120 is indicated on the “b” monomer (blue). Monomers “a” and “b” have been shaded red and blue, respectively, accordingly to the sequence conservation at that position in the alignment in panel A. A bright red or blue patch means that a high percentage of the sequences in panel A have the same residue as does vaccinia virus A33 at that position. The alignment in panel A was used as input to the ProtSkin server (12). Residues 118 and 120 of the “a” monomer (red) are not visible in this view. GenBank geninfo identifiers (gi) are as follows: VACV, 66275953; RFV, 9633931; TANV, 157939746; YMTV, 38229285; LSDV, 15150561; SWPV, 18640205; DPV, 62637509; ORFV, 32167503; BPSV, 41018861; MOCV, 9629074. The sequence of the squirrelpox virus A33 ortholog (SQV) (88769957) is not shown for clarity, due to an eight-residue insertion between 141 and 142 and a six-residue insertion between residues 160 and 161.
Aligning the sequences of the predicted CTLDs of A33 orthologs from different genera reveals conserved residues (Fig. 5A). In addition to the cysteines of the CTLD, several residues are completely conserved: G102, A122, L133, P134, W143, Y147, T151, and W152. At other positions, residues of the vaccinia virus A33 sequence are replaced with similar residues. When the percent identity at each aligned residue is mapped onto a surface representation of vaccinia virus A33, it is clear that those residues that interact across the dimer interface are the most conserved (Fig. 5B).
Locating antibody epitopes on A33.
Several monoclonal antibodies (MAb) that bind to the ectodomain of A33 have been described (8, 27, 57). The MAbs 4, 66, and 105 precipitate A33 molecules from vaccinia virus EV in the range of 23 to 28 kDa, which was shown to result from the several glycosylation states of A33 (57). Although MAb 4 recognizes similarly sized proteins (A33 orthologs) in several other orthopoxviruses, MAb 4 does not bind to ectromelia virus proteins (62). By transfection of vaccinia virus DNA fragments into cells infected with ectromelia virus and screening for restored MAb 4 binding, the vaccinia virus A33R gene and protein were identified as the target for MAb 4 (62).
Anti-A33 chimpanzee/human MAbs 6C, 12C, and 12F have been produced from a phage library that displays chimpanzee Fab antibody fragments (8). The MAbs competed with each other for binding to A33, and their epitopes were mapped to A33 residues 99 to 185, which comprise the CTLD of the current work (8). A33 molecules (residues 99 to 185) that were further truncated at the N or C terminus were not expressed in bacteria (8). The prediction from the current work is that deletions at the N or C terminus of the A33 CTLD would disrupt the β-sheet formed between the N- and C-terminal β-strands and would thus prevent protein folding.
The anti-A33 MAb 1G10 was shown to be protective in mice challenged with vaccinia virus (27, 49). MAb 10F10 and MAb 1G10 compete for binding to vaccinia virus A33, implying that their epitopes likely overlap. MAb 10F10 also binds to monkeypox virus A33, but MAb 1G10 does not (20). The monkeypox virus A33 ortholog has only a few sequence differences from vaccinia virus A33. As reported, mutations that replaced monkeypox virus amino acid residues with the analogous vaccinia virus residues yielded a mutant monkeypox virus A33 protein to which 1G10 bound, thereby mapping the location of the 1G10 epitope to residues 118 and 120 (20) (Fig. 5B). enzyme-linked immunosorbent assay (ELISA) data using overlapping A33 peptides further supported the location of the 1G10 epitope. Sera from immunized, protected rhesus macaque monkeys recognized peptides that identified a 31-residue region that included residues 118 and 120 (20, 24). Mapping of residues 118 (Leu) and 120 (Ser) on the vaccinia A33 crystal structure reveals that the 1G10 epitope lies on the edge of the A33 monomer and above the dimer interface (Fig. 5B). Antibody binding to the epitope would occlude portions of the top of the CTLD and the new surface formed by dimerization. Analogously to both the Link modules and the NK-cell receptors, which use the top of the CTLD to bind ligands, A33 ligand binding would be predicted to be disrupted by antibody binding. It is possible that the bivalent interaction of an antibody molecule could bind the same A33 dimer, since the distance between the two 1G10 epitopes on the monomers is about 40 Å.
DISCUSSION
Here we describe the crystal structure of the CTLD domain from the vaccinia virus A33 protein, which is currently a focus of protein and DNA vaccine development. The CTLD domain of A33 shows little sequence similarity with other proteins and has not, to our knowledge, been identified as a CTLD in previously published reports. The unusually small CTLD of 95 residues, lacking the β4 strand, a disulfide bond, and the “long loop,” has contributed to the difficulty of making a positive identification of the A33 CTLD from its sequence alone.
Two other vaccinia virus proteins, A34 and A40, have been predicted to contain CTLDs and to be type II membrane proteins. A34 is located on the EV membrane, as is A33, but A34 is not known to be a dimer and lacks a cysteine residue that could make a predicted disulfide bond between two CTLD-containing monomers (4, 15). A40 is present on the surface of the infected cell but not on virions. Like A33, A40 has a cysteine in the predicted stalk region and migrates as a dimer in nonreduced SDS-PAGE (75).
In CTLDs, the first and last β-strands are adjacent to each other in a β-sheet. This pairing brings the N and C termini of the domain close together. Modifications of the N and C termini, such as truncations or mutations, could disrupt the pairing of the β1 and β5 strands. The A33 structure predicts that interfering with β-strand pairing would prevent folding of the CTLD and would likely yield a phenotype similar to that for deletion of the entire extracellular CTLD. Katz et al. reported three independent vaccinia virus A33 mutants with C-terminal truncations of at least 35 residues (34). The A33 mutants were obtained by screening for mutant vaccinia viruses that would release more virus than the parental virus (34). When the A33 mutants were transferred to a wild-type virus, the phenotype of increased release of extracellular soluble virus by the A33 mutant viruses was retained (33). When the A33 gene itself is experimentally deleted, the phenotype is also an increased release of virus in vitro (63). Viruses bearing truncated A33 genes also had a markedly diminished virulence in mice (22, 33).
The identification of a CTLD in A33 implicates a role in binding a ligand, since CTLD-containing proteins use the CTLD to bind ligands (78). A33 is structurally similar to the group of NK receptors that bind proteins and to the group of Link module-containing proteins that bind glycosaminoglycans. For NK receptors, the two monomers of the homodimer act together to bind proteins. Structures of such complexes of dimeric receptors with their protein ligands have been determined (43-45, 59, 71). In these structures, the surface that mediates ligand binding is on the side of the dimer that is opposite to the N and C termini of the CTLD. The binding surface includes significant contributions from the “long loop” of the canonical CTLD and the β-sheet that consists of the β2, β3, and β4 strands (74).
One well-characterized Link module is that of CD44, which binds hyaluronan, a glycosaminoglycan (3). In the CD44 crystal structure, the bound hyaluronan extends across the top of the CTLD, making contacts with the β-sheet that consists of the β2, β3, and β4 strands. Without a “long loop,” the way that the Link module of CD44 binds hyaluronan is different from how the NK-cell receptors bind proteins. At a location similar to that observed in the crystal structures of NK receptors and Link module domains, A33 dimerization forms a V-shaped surface that may bind to ligands (Fig. 1A and 5B). Contributing to such a putative ligand interaction surface at the top of A33 is the small β-sheet composed of the β2 and β3 strands and the loop between the β3 and β5 strands (Fig. 1A). The mobility of this unliganded loop as observed in the electron density would be consistent with a binding partner or ligand for A33 that would stabilize the loop. The binding of protective antibodies near this region at the top of the CTLD supports the possibility of an A33 ligand bound by its CTLD.
Acknowledgments
We thank Joseph Golden, Jay Hooper, Greg Snyder, Michael March, and the members of the Structural Biology Section for helpful discussions. We thank Bernard Moss for discussions and vaccinia virus WR DNA, Mark Garfield for mass spectrometry, and the staff of the SER-CAT and SBC-CAT beam lines for assistance with X-ray data collection. X-ray data were collected at the Southeast Regional (SER-CAT) and Structural Biology Center Collaborative Access Team (SBC-CAT) beam lines at the Advanced Photon Source, Argonne National Laboratory.
Use of the Advanced Photon Source beam lines was supported by the U.S. Department of Energy, Office of Science, Office of Basic Energy Sciences, under contract no. W-31-109-Eng-38. This research was supported by the Intramural Research Program of the NIH, NIAID.
Footnotes
Published ahead of print on 23 December 2009.
REFERENCES
- 1.Afonso, C. L., E. R. Tulman, G. Delhon, Z. Lu, G. J. Viljoen, D. B. Wallace, G. F. Kutish, and D. L. Rock. 2006. Genome of crocodilepox virus. J. Virol. 80:4978-4991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Appleyard, G., A. J. Hapel, and E. A. Boulter. 1971. An antigenic difference between intracellular and extracellular rabbitpox virus. J. Gen. Virol. 13:9-17. [DOI] [PubMed] [Google Scholar]
- 3.Banerji, S., A. J. Wright, M. Noble, D. J. Mahoney, I. D. Campbell, A. J. Day, and D. G. Jackson. 2007. Structures of the CD44-hyaluronan complex provide insight into a fundamental carbohydrate-protein interaction. Nat. Struct. Mol. Biol. 14:234-239. [DOI] [PubMed] [Google Scholar]
- 4.Blasco, R., J. R. Sisler, and B. Moss. 1993. Dissociation of progeny vaccinia virus from the cell membrane is regulated by a viral envelope glycoprotein: effect of a point mutation in the lectin homology domain of the A34R gene. J. Virol. 67:3319-3325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boyington, J. C., A. N. Riaz, A. Patamawenu, J. E. Coligan, A. G. Brooks, and P. D. Sun. 1999. Structure of CD94 reveals a novel C-type lectin fold: implications for the NK cell-associated CD94/NKG2 receptors. Immunity 10:75-82. [DOI] [PubMed] [Google Scholar]
- 6.Bray, M. 2003. Pathogenesis and potential antiviral therapy of complications of smallpox vaccination. Antiviral Res. 58:101-114. [DOI] [PubMed] [Google Scholar]
- 7.Brunger, A. T. 2007. Version 1.2 of the Crystallography and NMR system. Nat. Protoc. 2:2728-2733. [DOI] [PubMed] [Google Scholar]
- 8.Chen, Z., P. Earl, J. Americo, I. Damon, S. K. Smith, F. Yu, A. Sebrell, S. Emerson, G. Cohen, R. J. Eisenberg, I. Gorshkova, P. Schuck, W. Satterfield, B. Moss, and R. Purcell. 2007. Characterization of chimpanzee/human monoclonal antibodies to vaccinia virus A33 glycoprotein and its variola virus homolog in vitro and in a vaccinia virus mouse protection model. J. Virol. 81:8989-8995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Clamp, M., J. Cuff, S. M. Searle, and G. J. Barton. 2004. The Jalview Java alignment editor. Bioinformatics 20:426-427. [DOI] [PubMed] [Google Scholar]
- 10.Corpet, F. 1988. Multiple sequence alignment with hierarchical clustering. Nucleic Acids Res. 16:10881-10890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.DeLano, W. L. 2002. The PyMOL molecular graphics system. http://www.pymol.org.
- 12.Deprez, C., R. Lloubes, M. Gavioli, D. Marion, F. Guerlesquin, and L. Blanchard. 2005. Solution structure of the E.coli TolA C-terminal domain reveals conformational changes upon binding to the phage g3p N-terminal domain. J. Mol. Biol. 346:1047-1057. [DOI] [PubMed] [Google Scholar]
- 13.Drickamer, K. 1999. C-type lectin-like domains. Curr. Opin. Struct. Biol. 9:585-590. [DOI] [PubMed] [Google Scholar]
- 14.Drickamer, K., and A. J. Fadden. 2002. Genomic analysis of C-type lectins. Biochem. Soc. Symp. 2002:59-72. [DOI] [PubMed] [Google Scholar]
- 15.Duncan, S. A., and G. L. Smith. 1992. Identification and characterization of an extracellular envelope glycoprotein affecting vaccinia virus egress. J. Virol. 66:1610-1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fang, M., H. Cheng, Z. Dai, Z. Bu, and L. J. Sigal. 2006. Immunization with a single extracellular enveloped virus protein produced in bacteria provides partial protection from a lethal orthopoxvirus infection in a natural host. Virology 345:231-243. [DOI] [PubMed] [Google Scholar]
- 17.Fogg, C., S. Lustig, J. C. Whitbeck, R. J. Eisenberg, G. H. Cohen, and B. Moss. 2004. Protective immunity to vaccinia virus induced by vaccination with multiple recombinant outer membrane proteins of intracellular and extracellular virions. J. Virol. 78:10230-10237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Frischknecht, F., V. Moreau, S. Rottger, S. Gonfloni, I. Reckmann, G. Superti-Furga, and M. Way. 1999. Actin-based motility of vaccinia virus mimics receptor tyrosine kinase signalling. Nature 401:926-929. [DOI] [PubMed] [Google Scholar]
- 19.Galmiche, M. C., J. Goenaga, R. Wittek, and L. Rindisbacher. 1999. Neutralizing and protective antibodies directed against vaccinia virus envelope antigens. Virology 254:71-80. [DOI] [PubMed] [Google Scholar]
- 20.Golden, J. W., and J. W. Hooper. 2008. Heterogeneity in the A33 protein impacts the cross-protective efficacy of a candidate smallpox DNA vaccine. Virology 377:19-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Grosenbach, D. W., S. G. Hansen, and D. E. Hruby. 2000. Identification and analysis of vaccinia virus palmitylproteins. Virology 275:193-206. [DOI] [PubMed] [Google Scholar]
- 22.Gurt, I., I. Abdalrhman, and E. Katz. 2006. Pathogenicity and immunogenicity in mice of vaccinia viruses mutated in the viral envelope proteins A33R and B5R. Antiviral Res. 69:158-164. [DOI] [PubMed] [Google Scholar]
- 23.Hamburger, Z. A., M. S. Brown, R. R. Isberg, and P. J. Bjorkman. 1999. Crystal structure of invasin: a bacterial integrin-binding protein. Science 286:291-295. [DOI] [PubMed] [Google Scholar]
- 24.Heraud, J. M., Y. Edghill-Smith, V. Ayala, I. Kalisz, J. Parrino, V. S. Kalyanaraman, J. Manischewitz, L. R. King, A. Hryniewicz, C. J. Trindade, M. Hassett, W. P. Tsai, D. Venzon, A. Nalca, M. Vaccari, P. Silvera, M. Bray, B. S. Graham, H. Golding, J. W. Hooper, and G. Franchini. 2006. Subunit recombinant vaccine protects against monkeypox. J. Immunol. 177:2552-2564. [DOI] [PubMed] [Google Scholar]
- 25.Higman, V. A., C. D. Blundell, D. J. Mahoney, C. Redfield, M. E. Noble, and A. J. Day. 2007. Plasticity of the TSG-6 HA-binding loop and mobility in the TSG-6-HA complex revealed by NMR and X-ray crystallography. J. Mol. Biol. 371:669-684. [DOI] [PubMed] [Google Scholar]
- 26.Holm, L., and C. Sander. 1996. Mapping the protein universe. Science 273:595-603. [DOI] [PubMed] [Google Scholar]
- 27.Hooper, J. W., D. M. Custer, C. S. Schmaljohn, and A. L. Schmaljohn. 2000. DNA vaccination with vaccinia virus L1R and A33R genes protects mice against a lethal poxvirus challenge. Virology 266:329-339. [DOI] [PubMed] [Google Scholar]
- 28.Hooper, J. W., D. M. Custer, and E. Thompson. 2003. Four-gene-combination DNA vaccine protects mice against a lethal vaccinia virus challenge and elicits appropriate antibody responses in nonhuman primates. Virology 306:181-195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hooper, J. W., E. Thompson, C. Wilhelmsen, M. Zimmerman, M. A. Ichou, S. E. Steffen, C. S. Schmaljohn, A. L. Schmaljohn, and P. B. Jahrling. 2004. Smallpox DNA vaccine protects nonhuman primates against lethal monkeypox. J. Virol. 78:4433-4443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jaroszewski, L., L. Rychlewski, Z. Li, W. Li, and A. Godzik. 2005. FFAS03: a server for profile-profile sequence alignments. Nucleic Acids Res. 33:W284-W288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jones, T. A., J. Y. Zou, S. W. Cowan, and M. Kjeldgaard. 1991. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A 47:110-119. [DOI] [PubMed] [Google Scholar]
- 32.Kabsch, W. 1993. Automatic processing of rotation diffraction data from crystals of initially unknown symmetry and cell constants. J. Appl. Crystallogr. 26:795-800. http://www.mpimf-heidelberg.mpg.de/∼kabsch/xds/. [Google Scholar]
- 33.Katz, E., B. M. Ward, A. S. Weisberg, and B. Moss. 2003. Mutations in the vaccinia virus A33R and B5R envelope proteins that enhance release of extracellular virions and eliminate formation of actin-containing microvilli without preventing tyrosine phosphorylation of the A36R protein. J. Virol. 77:12266-12275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Katz, E., E. Wolffe, and B. Moss. 2002. Identification of second-site mutations that enhance release and spread of vaccinia virus. J. Virol. 76:11637-11644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kleywegt, G. J., J. Y. Zou, M. Kjeldgaard, and T. A. Jones. 2001. Around O, p. 353-356, 366-367. In M. G. Rossmann and E. Arnold (ed.), International tables for crystallography, vol F: crystallography of biological macromolecules. Kluwer Academic Publishers, Dordrecht, The Netherlands.
- 36.Krissinel, E., and K. Henrick. 2004. Secondary-structure matching (SSM), a new tool for fast protein structure alignment in three dimensions. Acta. Crystallogr. D. Biol. Crystallogr. 60:2256-2268. http://www.ebi.ac.uk/msd-srv/ssm/. [DOI] [PubMed] [Google Scholar]
- 37.Krissinel, E., and K. Henrick. 2007. Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 372:774-797. [DOI] [PubMed] [Google Scholar]
- 38.Larkin, M. A., G. Blackshields, N. P. Brown, R. Chenna, P. A. McGettigan, H. McWilliam, F. Valentin, I. M. Wallace, A. Wilm, R. Lopez, J. D. Thompson, T. J. Gibson, and D. G. Higgins. 2007. Clustal W and Clustal X version 2.0. Bioinformatics 23:2947-2948. [DOI] [PubMed] [Google Scholar]
- 39.Laskowski, R. A., M. W. MacArthur, D. S. Moss, and J. M. Thornton. 1993. PROCHECK: a program to check the stereochemical quality of protein structures. J. Appl. Crystallogr. 26:283-291. [Google Scholar]
- 40.Law, M., R. Hollinshead, and G. L. Smith. 2002. Antibody-sensitive and antibody-resistant cell-to-cell spread by vaccinia virus: role of the A33R protein in antibody-resistant spread. J. Gen. Virol. 83:209-222. [DOI] [PubMed] [Google Scholar]
- 41.Lawrence, M. C., and P. M. Colman. 1993. Shape complementarity at protein/protein interfaces. J. Mol. Biol. 234:946-950. [DOI] [PubMed] [Google Scholar]
- 42.Lefkowitz, E. J., C. Upton, S. S. Changayil, C. Buck, P. Traktman, and R. M. Buller. 2005. Poxvirus Bioinformatics Resource Center: a comprehensive Poxviridae informational and analytical resource. Nucleic Acids Res. 33:D311-D316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li, P., G. McDermott, and R. K. Strong. 2002. Crystal structures of RAE-1beta and its complex with the activating immunoreceptor NKG2D. Immunity 16:77-86. [DOI] [PubMed] [Google Scholar]
- 44.Li, P., D. L. Morris, B. E. Willcox, A. Steinle, T. Spies, and R. K. Strong. 2001. Complex structure of the activating immunoreceptor NKG2D and its MHC class I-like ligand MICA. Nat. Immunol. 2:443-451. [DOI] [PubMed] [Google Scholar]
- 45.Li, Y., M. Hofmann, Q. Wang, L. Teng, L. K. Chlewicki, H. Pircher, and R. A. Mariuzza. 2009. Structure of natural killer cell receptor KLRG1 bound to E-cadherin reveals basis for MHC-independent missing self recognition. Immunity 31:35-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Llera, A. S., F. Viedma, F. Sanchez-Madrid, and J. Tormo. 2001. Crystal structure of the C-type lectin-like domain from the human hematopoietic cell receptor CD69. J. Biol. Chem. 276:7312-7319. [DOI] [PubMed] [Google Scholar]
- 47.Lovell, S. C., I. W. Davis, W. B. Arendall III, P. I. de Bakker, J. M. Word, M. G. Prisant, J. S. Richardson, and D. C. Richardson. 2003. Structure validation by Calpha geometry: phi, psi and Cbeta deviation. Proteins 50:437-450. [DOI] [PubMed] [Google Scholar]
- 48.Luo, Y., E. A. Frey, R. A. Pfuetzner, A. L. Creagh, D. G. Knoechel, C. A. Haynes, B. B. Finlay, and N. C. Strynadka. 2000. Crystal structure of enteropathogenic Escherichia coli intimin-receptor complex. Nature 405:1073-1077. [DOI] [PubMed] [Google Scholar]
- 49.Lustig, S., C. Fogg, J. C. Whitbeck, R. J. Eisenberg, G. H. Cohen, and B. Moss. 2005. Combinations of polyclonal or monoclonal antibodies to proteins of the outer membranes of the two infectious forms of vaccinia virus protect mice against a lethal respiratory challenge. J. Virol. 79:13454-13462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.McCoy, A. J., R. W. Grosse-Kunstleve, P. D. Adams, M. D. Winn, L. C. Storoni, and R. J. Read. 2007. Phaser crystallographic software. J. Appl. Crystallogr. 40:658-674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.McInnes, C. J., A. R. Wood, K. Thomas, A. W. Sainsbury, J. Gurnell, F. J. Dein, and P. F. Nettleton. 2006. Genomic characterization of a novel poxvirus contributing to the decline of the red squirrel (Sciurus vulgaris) in the UK. J. Gen. Virol. 87:2115-2125. [DOI] [PubMed] [Google Scholar]
- 52.McMahon, S. A., J. L. Miller, J. A. Lawton, D. E. Kerkow, A. Hodes, M. A. Marti-Renom, S. Doulatov, E. Narayanan, A. Sali, J. F. Miller, and P. Ghosh. 2005. The C-type lectin fold as an evolutionary solution for massive sequence variation. Nat. Struct. Mol. Biol. 12:886-892. [DOI] [PubMed] [Google Scholar]
- 53.Moss, B. 2007. Poxviridae: the viruses and their replication, p. 2905-2946. In D. M. Knipe and P. M. Howley (ed.), Fields virology, vol. 2. Lippincott Williams & Wilkins, Philadelphia, PA. [Google Scholar]
- 54.Mullen, M. M., K. M. Haan, R. Longnecker, and T. S. Jardetzky. 2002. Structure of the Epstein-Barr virus gp42 protein bound to the MHC class II receptor HLA-DR1. Mol. Cell 9:375-385. [DOI] [PubMed] [Google Scholar]
- 55.Otwinowski, Z., and W. Minor. 1997. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276:307-326. [DOI] [PubMed] [Google Scholar]
- 56.Park, H., F. G. Adsit, and J. C. Boyington. 2005. The 1.4 angstrom crystal structure of the human oxidized low density lipoprotein receptor lox-1. J. Biol. Chem. 280:13593-13599. [DOI] [PubMed] [Google Scholar]
- 57.Payne, L. G. 1992. Characterization of vaccinia virus glycoproteins by monoclonal antibody precipitation. Virology 187:251-260. [DOI] [PubMed] [Google Scholar]
- 58.Perdiguero, B., and R. Blasco. 2006. Interaction between vaccinia virus extracellular virus envelope A33 and B5 glycoproteins. J. Virol. 80:8763-8777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Radaev, S., B. Rostro, A. G. Brooks, M. Colonna, and P. D. Sun. 2001. Conformational plasticity revealed by the cocrystal structure of NKG2D and its class I MHC-like ligand ULBP3. Immunity 15:1039-1049. [DOI] [PubMed] [Google Scholar]
- 60.Radaev, S., and P. D. Sun. 2003. Structure and function of natural killer cell surface receptors. Annu. Rev. Biophys. Biomol. Struct. 32:93-114. [DOI] [PubMed] [Google Scholar]
- 61.Rimoin, A. W., N. Kisalu, B. Kebela-Ilunga, T. Mukaba, L. L. Wright, P. Formenty, N. D. Wolfe, R. L. Shongo, F. Tshioko, E. Okitolonda, J. J. Muyembe, R. W. Ryder, and H. Meyer. 2007. Endemic human monkeypox, Democratic Republic of Congo, 2001-2004. Emerg. Infect. Dis. 13:934-937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Roper, R. L., L. G. Payne, and B. Moss. 1996. Extracellular vaccinia virus envelope glycoprotein encoded by the A33R gene. J. Virol. 70:3753-3762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Roper, R. L., E. J. Wolffe, A. Weisberg, and B. Moss. 1998. The envelope protein encoded by the A33R gene is required for formation of actin-containing microvilli and efficient cell-to-cell spread of vaccinia virus. J. Virol. 72:4192-4204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Rottger, S., F. Frischknecht, I. Reckmann, G. L. Smith, and M. Way. 1999. Interactions between vaccinia virus IEV membrane proteins and their roles in IEV assembly and actin tail formation. J. Virol. 73:2863-2875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Smith, G. L., A. Vanderplasschen, and M. Law. 2002. The formation and function of extracellular enveloped vaccinia virus. J. Gen. Virol. 83:2915-2931. [DOI] [PubMed] [Google Scholar]
- 66.Su, H. P., S. C. Garman, T. J. Allison, C. Fogg, B. Moss, and D. N. Garboczi. 2005. The 1.51-angstrom structure of the poxvirus L1 protein, a target of potent neutralizing antibodies. Proc. Natl. Acad. Sci. U. S. A. 102:4240-4245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Su, H. P., D. Y. Lin, and D. N. Garboczi. 2006. The structure of G4, the poxvirus disulfide oxidoreductase essential for virus maturation and infectivity. J. Virol. 80:7706-7713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Terwilliger, T. C. 2000. Maximum-likelihood density modification. Acta Crystallogr. D Biol. Crystallogr. 56:965-972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Terwilliger, T. C. 2003. Automated main-chain model building by template matching and iterative fragment extension. Acta. Crystallogr. D Biol. Crystallogr. 59:38-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Terwilliger, T. C., and J. Berendzen. 1999. Automated MAD and MIR structure solution. Acta. Crystallogr. D Biol. Crystallogr. 55:849-861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Tormo, J., K. Natarajan, D. H. Margulies, and R. A. Mariuzza. 1999. Crystal structure of a lectin-like natural killer cell receptor bound to its MHC class I ligand. Nature 402:623-631. [DOI] [PubMed] [Google Scholar]
- 72.Ward, B. M., A. S. Weisberg, and B. Moss. 2003. Mapping and functional analysis of interaction sites within the cytoplasmic domains of the vaccinia virus A33R and A36R envelope proteins. J. Virol. 77:4113-4126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Weis, W. I., R. Kahn, R. Fourme, K. Drickamer, and W. A. Hendrickson. 1991. Structure of the calcium-dependent lectin domain from a rat mannose-binding protein determined by MAD phasing. Science 254:1608-1615. [DOI] [PubMed] [Google Scholar]
- 74.Weis, W. I., M. E. Taylor, and K. Drickamer. 1998. The C-type lectin superfamily in the immune system. Immunol. Rev. 163:19-34. [DOI] [PubMed] [Google Scholar]
- 75.Wilcock, D., S. A. Duncan, P. Traktman, W. H. Zhang, and G. L. Smith. 1999. The vaccinia virus A40R gene product is a nonstructural, type II membrane glycoprotein that is expressed at the cell surface. J. Gen. Virol. 80:2137-2148. [DOI] [PubMed] [Google Scholar]
- 76.Wolffe, E. J., A. S. Weisberg, and B. Moss. 2001. The vaccinia virus A33R protein provides a chaperone function for viral membrane localization and tyrosine phosphorylation of the A36R protein. J. Virol. 75:303-310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Xiao, Y., L. Aldaz-Carroll, A. M. Ortiz, J. C. Whitbeck, E. Alexander, H. Lou, H. L. Davis, T. J. Braciale, R. J. Eisenberg, G. H. Cohen, and S. N. Isaacs. 2007. A protein-based smallpox vaccine protects mice from vaccinia and ectromelia virus challenges when given as a prime and single boost. Vaccine 25:1214-1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Zelensky, A. N., and J. E. Gready. 2005. The C-type lectin-like domain superfamily. FEBS J. 272:6179-6217. [DOI] [PubMed] [Google Scholar]