Abstract
Xenotropic murine leukemia virus-related virus (XMRV) is a novel human gammaretrovirus discovered in association with human prostate tumors. XMRV was first identified in prostate stromal cells surrounding the tumors of patients carrying a mutation in the HPC1 gene locus. To determine the tropism of XMRV in cell culture, we tested the ability of XMRV to spread and replicate in various prostate and nonprostate cell lines. We found that although the expression of XMRV viral proteins and the spread of infectious virus were minimal in a variety of cell lines, XMRV displayed robust expression and infection in LNCaP prostate tumor cells. The transcriptional activity of the XMRV long terminal repeat (LTR) was found to be higher than the Moloney murine leukemia virus LTRs in both LNCaP and WPMY-1 (simian virus 40-transformed prostate stromal cells). The U3 promoter of XMRV and a glucocorticoid response element (GRE) within the U3 were required for the transcriptional activity in LNCaP cells. Coexpression of the androgen receptor and stimulation with dihydrotestosterone stimulated XMRV-LTR-dependent transcription in 293T cells, and the GRE was required for this activity. These data suggest that XMRV may replicate more efficiently in LNCaP cells in part due to the transcriptional environment in LNCaP cells.
Nearly 35% of familial prostate cancer patients carry a germ line mutation (R462Q) in the HPC1 gene locus (15). This locus encodes the protein RNase L, which is expressed and activated upon virus infection and degrades single-stranded viral and cellular RNA, thus blocking replication of the infecting virus and inducing apoptosis (1, 16). The association of prostate cancers with this variant of RNase L raised the possibility that mutant individuals were more susceptible to an unknown tumor virus (2, 15). Total polyadenylated RNA from prostate tumors that were either heterozygous or homozygous for the mutant RNase L allele was isolated and hybridized to a DNA microarray (Virochip) containing oligomers of ∼950 viral genomes (19). Seven of eleven tumors that carried at least one allele of the RNase L mutation were positive for the novel retrovirus. Isolation and sequencing of the virus from three different prostate cancer patients revealed nucleotide similarities to xenotropic murine leukemia viruses (MLVs), and the virus was named xenotropic MLV-related virus (XMRV) (19). The genome structure of XMRV is typical of gamma retroviruses. The env gene encodes a glycoprotein homologous to the MLV envelope protein that mediates virus binding to the xenotropic receptor, XPR1, on the surface of cells (4). In contrast to more complex retroviruses such as lentiviruses, XMRV does not encode any accessory genes, nor does it encode any host-derived oncogenes (3). Fluorescence in situ hybridization and immunohistochemistry revealed that a small number of stromal cells surrounding the tumor, but not tumor cells themselves, were positive for XMRV nucleotide sequences and viral proteins, suggesting that XMRV maintains a low level of infection in these tumors and that direct oncogenesis by XMRV might not play a role in prostate tumorigenesis (19).
Recent studies have demonstrated the affinity of XMRV for prostate cells. XMRV was produced at high titers from ∼10 integrated copies within the prostate carcinoma cell line 22Rv1 (11). Another study has confirmed the presence of XMRV-infected cells within the prostate but differs significantly from the original report describing XMRV. XMRV was found in 23% of all prostate cancers without correlation to the RNase L R462Q mutant allele. Significantly, malignant prostate epithelial cells were infected with XMRV at a higher rate compared to stromal cells, thus leaving open the possibility of direct oncogenesis by XMRV (14). Amyloidogenic fragments known as semen-derived enhancer of virus infection (SEVI) from prostatic acid phosphatase increased XMRV infectivity at the level of virus entry. XMRV nucleic acid was also found in prostatic secretions of prostate cancer patients, suggesting a possible mechanism of transmission (9).
XMRV has been shown to be sensitive to the antiviral actions of interferon (IFN) (4), a well-characterized antiviral mechanism against pathogenic infections (13). The DU145 prostate cell line treated with IFN-β prior to XMRV infection was more resistant to a spreading infection than cells without IFN (4). LNCaP prostate cells were permissive for XMRV infection in the presence or absence of IFN and were four times more supportive of virus infection than DU145 cells. The role that RNase L plays in regulating XMRV is still unclear: DU145 cells with a modest small interfering RNA knockdown of RNase L showed slower rather than enhanced replication of XMRV, and there was no change in replication with or without IFN treatment (4). Moreover, it is also unknown what effect the R429Q mutation in RNase L plays in the general response against viral infection. The threefold decrease in catalytic activity associated with this mutation may not profoundly change the susceptibility of the cells (2).
In the present study, we determined the ability of infectious XMRV to replicate in cell lines derived from various tissues. Of the cell lines tested, XMRV replicated most efficiently in the LNCaP cell line of prostate origin. To explore why these prostate cells are more permissive for XMRV replication, we analyzed the transcriptional activity of the XMRV long terminal repeat (LTR) in permissive and nonpermissive cell lines. Consistent with the tropism of XMRV replication, an increased transcriptional activity was seen in LNCaP and WPMY-1 prostate cell lines. The U3 region of XMRV and a glucocorticoid response element (GRE) was specifically required for this activity. The data presented here suggest that LNCaP prostate cells provide a transcriptional environment that supports efficient replication and spread of XMRV.
MATERIALS AND METHODS
Cell culture and virus.
All cell lines were maintained at 37°C and 5% CO2 and supplemented with 10% fetal bovine serum (Invitrogen). LNCaP cells (human prostate epithelial tumor cells, a gift of the Gelmann lab, Columbia University) were maintained in RPMI 1640 (Invitrogen). PC-3 cells (human prostate epithelial tumor cells; American Type Culture Collection [ATCC]) were maintained in Eagle modified essential medium (Invitrogen). WMPY-1 cells are prostate stromal cell immortalized with simian virus 40 (SV40) T antigen and were maintained in Dulbecco modified Eagle medium (DMEM). DU145 cells (human prostate carcinoma cells; ATCC) were maintained in FK12 media (Invitrogen). 2fTGH (human fibrosarcoma; gift of Horvath lab, Northwestern University), HeLa (cervical carcinoma; ATCC), TE671 (human rhabdomyosarcoma), Rat2 (rat fibroblast), and 293T (human embryonic kidney) cells were maintained in DMEM (Invitrogen). NIH 3T3 cells (ATCC) were maintained in DMEM supplemented with 10% bovine calf serum (Invitrogen). XMRV virus particles were generated by transfecting LNCaP cells with 5 μg of proviral DNAs. For XMRV spreading infection assays, LNCaP cell culture medium was harvested 8 days postinfection, passed through a 0.45-μm-pore-size filter, and stored at −80°C. Polybrene (8 μg/ml) was added during virus harvesting. The relative concentration of XMRV in supernatants was determined by measuring the reverse transcriptase (RT) activity in the cell culture media of harvested stocks (7). To generate pseudotyped reporter virions, the plasmid Moloney MLV (MoMLV)-XMRV Env, along with pFB-Luc (firefly luciferase with a MoMLV packaging signal), was cotransfected into 293T cells and supernatants were harvested after filtration through a 0.45-μm-pore-size filter.
Plasmids and reagents.
The full-length genome of XMRV (patient VP62) was obtained from D. Ganem (University of California, San Francisco). XMRV halves AM 2-9 and AO H4 (19) were joined by introducing a novel MluI site and performing overlapping PCR, which generated one amino acid change: glycine to alanine at position 385 of RT (gift of I. R. Singh, University of Utah). The genome was ligated into the pCR2-TOPO cloning vector. To generate the provirus, the U3 region was amplified and ligated to 5′ R region. This proviral construct was also ligated into the pCR2-TOPO cloning vector and utilized for all subsequent experiments. pcDNA3.1(+) was utilized as a control plasmid for generating mock virus stocks and as a control for mock provirus transfection. MoMLV and XMRV LTR (U3-R-U5-Gag start site) DNAs were amplified by PCR and cloned in between NdeI and HindIII sites by sequence ligation-independent cloning (SLIC) into the plasmid pRL-null (plasmid encoding a promoterless Renilla luciferase) and used as a reporter gene. The XM1 reporter plasmid in which the MoMLV U3 was swapped for the XMRV U3 was generated by SLIC cloning in the same manner as the full-length LTRs. The mutant reporter, mGRE, was generated in the same fashion with the nucleotides 192 and 193 (adenine) of the full-length LTRs both being changed to cytosine. The androgen receptor (AR) expression and dihydrotestosterone (DHT) was a gift from Liang-Nian Song of the Gelmann lab (Columbia University [17]). pQXCIP-dsRed (dsRed expression driven by the cytomegalovirus [CMV] promoter) and pCDNA-GFP-PT (green fluorescent protein [GFP] expression driven by the CMV promoter) were utilized as markers for transfection efficiency of 293T or LNCaP cells. pNCA-XMRV Env encodes a full-length MoMLV provirus with the XMRV Env in place of the MoMLV Env. The plasmid pFB-Luc was used as a MoMLV marker for single-round infection assays.
Preparation of cell lysates and immunoblotting.
Transfected cells were lysed with NP-40 lysis buffer (150 mM NaCl, 0.5% NP-40, 50 mM Tris [pH 8.0], 0.5 mM EDTA) supplemented with protease inhibitor cocktail (Roche) at 4°C for 30 min. Equal amounts of protein from clarified extracts were added to protein loading buffer, boiled for 5 min, and subjected to SDS-PAGE. Proteins were transferred to nitrocellulose and immunoblotted with antisera against MoMLV CA (goat polyclonal antibody that cross-reacts with XMRV Gag and CA), rabbit polyclonal GFP (ab290; Abcam), or mouse monoclonal β-Actin (A-1978; Sigma) antibody.
XMRV spreading assays.
A total of 105 Cells were seeded onto six-well dishes and infected the day after with 100 μl of LNCaP culture supernatants containing XMRV viral particles. The cells were allowed to recover after 8 h of adsorption at 37°C with appropriate cell media. Samples were taken each day and subjected to RT assays as described previously (7) to monitor the release of viral particles into the culture supernatants. XMRV was isolated by ultracentrifugation of filter-sterilized (0.45-μm pore size) supernatants at 75,830 × g and 4°C for 2 h. Virus pellets were lysed in NP-40 lysis buffer supplemented with protease inhibitors and protein loading buffer. Samples were boiled, subjected to SDS-PAGE, and immunoblotted with antisera to the MoMLV CA protein.
Luciferase reporter gene assays.
Luciferase reporter gene assays were performed according to the manufacturer's protocol (Promega). 293T, LNCaP, or WPMY-1 cells were transfected with different reporter genes and lysed with passive lysis buffer 24 h posttransfection. For treatment with DHT, cells were exposed to 10 μM DHT at the time of transfection, and all experiments were performed using charcoal-stripped serum. Luciferase activity was measured from triplicate samples using a POLARstar Omega plate reader (BMG Labtech). All conditions represent the average values from triplicate samples, normalized to cotransfected firefly luciferase (pcDNA4.0-Fluc).
RESULTS
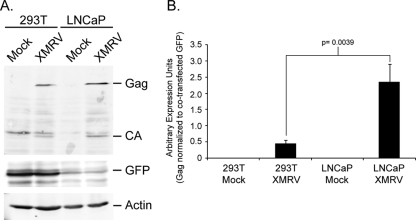
To generate infectious virus particles, we obtained the XMRV full-length genome isolated from patient designated VP62 (19). The provirus was transfected into both 293T or LNCaP prostate cells and lysates of transfected cells were tested for XMRV Gag and CA accumulation 2 days posttransfection (Fig. 1). Both 293T and LNCaP lysates contained nearly the same amount of steady-state Gag and CA proteins. The transfection efficiency of LNCaP cells is extremely poor, as indicated by the levels of exogenously expressed GFP. To rule out the possibility that GFP was expressed poorly in these cells, expression vectors encoding either dsRed or GFP were transfected, and fluorescent cells were quantitated by fluorescence-activated cell sorting (FACS) analysis. Although the number of transfected LNCaP cells was less, the fluorescence intensity of dsRed- or GFP-positive cells in both 293T and LNCaP cells were the same, indicating the marker was expressed equally well in transfected cells (data not shown). Normalization of Gag proteins to cotransfected GFP levels suggests that LNCaP cells express a higher amount of Gag compared to 293T cells (Fig. 1B).
FIG. 1.
LNCaP cells express a higher level of Gag and CA compared to 293T cells. (A) Cells were transfected with XMRV DNAs and a plasmid encoding a CMV promoter directing the expression of exogenous GFP. Cell lysates were prepared 2 days posttransfection and characterized for expression of Gag, CA, and GFP levels by SDS-PAGE. (B) Normalization of Gag protein levels to coexpressed exogenous GFP. This experiment was performed in triplicate with the panel A being one representative of the three experiments and panel B showing the quantification of Gag and GFP from all three experiments. P values were obtained by using a two-tailed Student t test of the data presented.
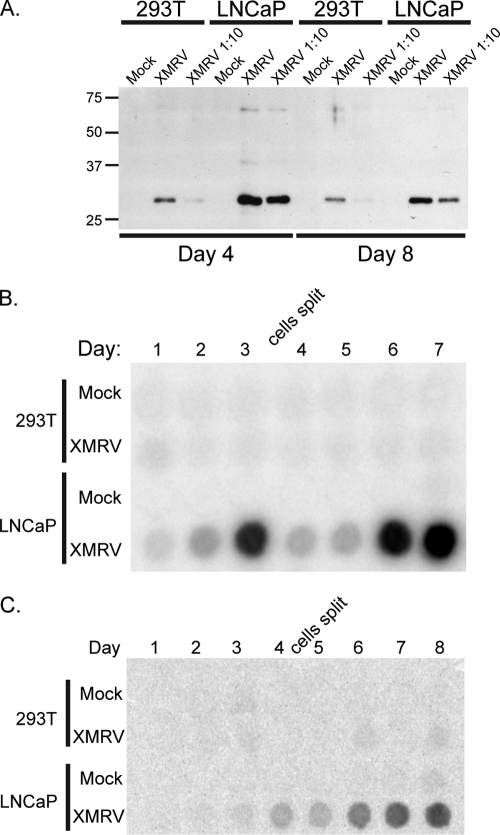
To determine whether the XMRV particles are infectious, LNCaP culture media containing XMRV virus particles were applied to 293T or LNCaP cells (Fig. 2A), and these infected cultures were monitored for virus release into the media by analyzing CA accumulation in the supernatants. At 4 and 8 days postinfection, distinctly higher levels of CA at steady state were found in the culture media from infected LNCaP cells compared to 293T cells, suggesting that XMRV spreads more efficiently in LNCaP cells. Virion spread and release into the media were also examined by measuring the activity of RT in the cell culture supernatants (Fig. 2B). LNCaP cells were supportive of an XMRV spreading infection, with peak RT activity detected at day 3, and then continued at day 7, after the cells were reseeded. RT activity in 293T cell culture supernatants, however, was not detected, indicating a lack of replication. Although 293T cells were poor producers of XMRV virus particles, we tested whether the virions from 293T culture media were infectious by applying supernatants to naive LNCaP or 293T cells and monitoring RT activity. Although 293T cells did not support efficient XMRV replication and spread, LNCaP cells rescued the few virus particles produced from 293T cells and could then initiate a spreading XMRV infection (Fig. 2C).
FIG. 2.
XMRV spreads efficiently in LNCaP cells but not 293T cells. (A) Eight-day culture media from LNCaP cells that were transfected with the XMRV provirus were adsorbed onto either naive 293T or LNCaP cells. Virus spreading was monitored over 8 days by immunoblotting supernatants against XMRV CA. XMRV 1:10 culture medium was diluted 10-fold at the time of infection. (B) Same as in panel A, but with monitoring of RT activity over 7 days. On the third day, the cells were diluted and reseeded to allow accumulation of XMRV virus particles. (C) Same as in panel A, but culture media from 293T cells that were transfected with the XMRV provirus were adsorbed onto either naive 293T or LNCaP cells. The results shown are representative of five different experiments.
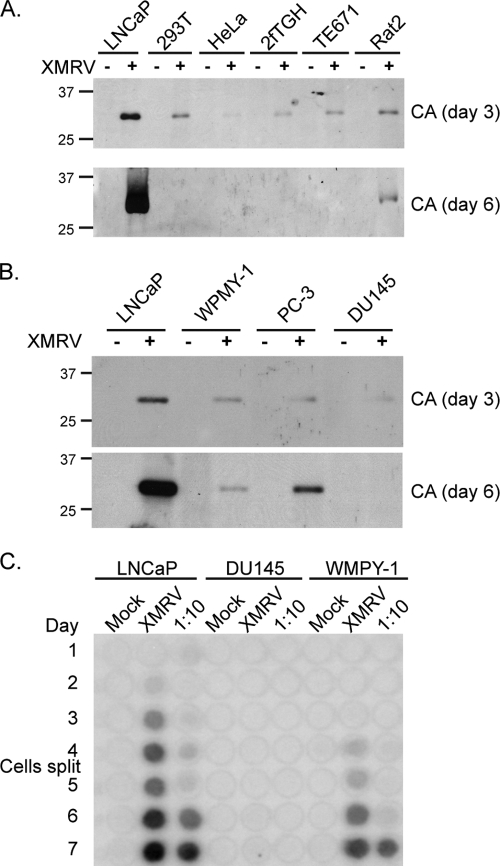
To further assess the cell line tropism of XMRV, we infected seven different cell lines with XMRV harvested from LNCaP cell culture supernatants. XMRV infection was conducted on LNCaP, 293T, HeLa (human cervical carcinoma), 2fTGH (human fibrosarcoma), TE671 (human rhabdomyosarcoma), and Rat2 (rat fibroblast) cells, and subsequent CA release into the culture media was examined (Fig. 3A). Three days postinfection, CA could be detected in the media supernatants from all cell lines, but LNCaP supernatants contained the highest levels. After 6 days, high levels of CA were observed in culture supernatants from LNCaP cells but not from the other cell lines, indicating that XMRV infection and spread was most efficient in LNCaP prostate cells. Next, we tested XMRV replication in three other prostate cell lines: DU145 prostate carcinoma cells, WPMY-1 prostate stromal cells immortalized with T-antigen, and PC-3 prostate carcinoma cells. As before, XMRV supernatants were applied to naive cells and release of CA in the culture media was measured (Fig. 3B). Compared to other prostate cell lines, LNCaP cells again were the most permissive for XMRV replication and spread, as monitored by CA production. RT release into the media was detected in LNCaP, DU145, and WPMY-1 cells (Fig. 3C), with LNCaP cells supporting the most robust spreading infection.
FIG. 3.
XMRV spreading in LNCaP cells. (A) Same as in Fig. 2, but different cell lines were infected with XMRV and levels of CA were measured by immunoblotting at 3 and 6 days postinfection. 293T, human embryonic kidney cells; HeLa, human cervical carcinoma; 2fTGH, human fibrosarcoma; TE671, human rhabdomyosarcoma; Rat2, rat fibroblast. (B) Same as in panel A, but three prostate cell lines were tested: WPMY-1 (prostate stromal immortalized with SV40 T-antigen), DU145 (epithelial prostate carcinoma), and PC-3 (prostate epithelial). (C) RT assay measuring XRMV spread in three different prostate cell lines: LNCaP, DU145, and WPMY-1 cells. The results shown are representative of three different experiments.
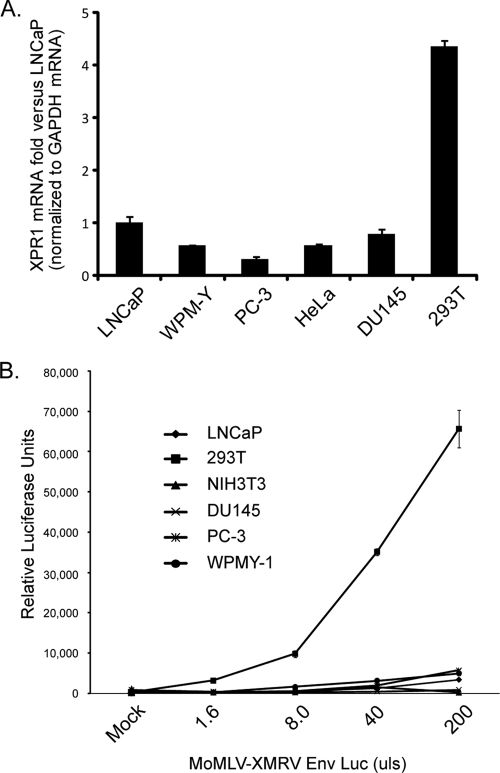
Next, we determined whether XMRV replicates efficiently in LNCaP cells due to increased viral entry at the level of the cellular receptor, XPR1. Analysis of XPR1 endogenous mRNA levels in different cell lines (Fig. 4A) demonstrated that whereas all of the cell lines tested expressed relatively small amount of XPR1 transcripts (data not shown), 293T cells had the highest XPR1 transcript levels. MoMLV particles that were psuedotyped with XMRV Env and contained a packaged luciferase reporter gene was used to infect cells and analyze virus entry in a single round of infection (Fig. 4B). Interestingly, although most cell lines, including LNCaP cells, exhibited low luciferase activities, 293T cells, which are largely refractory to XMRV spread, supported a robust degree of virus entry. These data suggests the amount of expressed receptor and viral entry is not a determining factor in the spread of XMRV in LNCaP cells.
FIG. 4.
293T cells mediate a higher amount of viral entry at the level of receptor. (A) Total mRNA isolated from LNCaP, WPMY-1, PC-3, HeLa, DU145, 293T, and 2fTGH cells were reverse transcribed to obtain cDNA. Triplicate RT products were amplified by using primers for XPR1 or GAPDH (glyceraldehyde-3-phosphate dehydrogenase) by quantitative PCR and SYBR green reaction mix. Standard curves for both XPR1 and GAPDH were created and XPR1 values were normalized to GAPDH transcript levels. The data are represented as the fold difference against LNCaP XPR1 mRNA levels where LNCaP XPR1 = 1. (B) A total of 2.5 × 105 cells of each cell type were infected with MoMLV virions that contain the XMRV Env and a packaged firefly luciferase reporter (MoMLV-XMRV Env Luc). To obtain data from a single round of infection, lysates were prepared 24 h postinfection and analyzed for luciferase activity. Each sample was normalized to total protein levels, and all analyses were performed in triplicate.
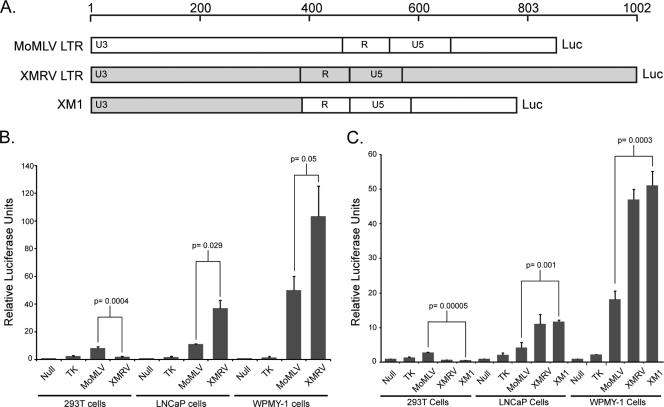
The intracellular accumulation of XMRV Gag and CA to higher levels in LNCaP cells suggested the possibility that the promoter of XMRV displayed an enhanced level of transcription in these cells. We generated luciferase reporters that were fused to the LTRs of both MoMLV and XMRV, in which the fusion point was the translation initiation site of Gag (Fig. 4A). We then tested the transcriptional output of these reporter genes by transfecting LNCaP, WPMY-1, or 293T cells with these DNAs and measuring luciferase activity (Fig. 4B). The transcriptional activity of the MoMLV LTR was higher than the herpes simplex virus type 1 (HSV-1) TK promoter in all cell lines tested, but the pattern of XMRV LTR activity in different cells was distinct. The XMRV LTR transcriptional activity was lower than that of MoMLV in 293T cells but higher in both LNCaP and WPMY-1 prostate cells.
Most, if not all, of the transcriptional activity of the retroviral LTR typically originates from cis-acting DNA elements within the U3 region that recruit various cellular transcription factors. Whether the XMRV U3 was responsible for the enhancement of XMRV transcription in LNCaP and WPMY-1 cells was tested by generating a chimeric luciferase reporter gene in which the MoMLV U3 was replaced with the U3 of XMRV (Fig. 5A) and tested for its transcriptional activity (Fig. 5C). The chimeric reporter gene (XM1) behaved exactly like the full-length XMRV LTR: transcriptional activity was lower than MoMLV LTR in 293T cells but higher in LNCaP and WMPY-1 prostate cells. Together, these data indicate that the U3 region of the XMRV LTR promotes transcription more efficiently in LNCaP and WMPY-1 prostate cell lines.
FIG. 5.
XMRV LTR exhibits a higher transcriptional activity in LNCaP and WPMY-1 prostate cells. (A) Schematic diagram of MoMLV and XMRV LTR luciferase reporter gene constructs. The 5′ LTR of both MoMLV and XMRV were fused to Renilla luciferase at the translational start site for Gag. XM1, chimeric reporter gene where the U3 region of MoMLV was swapped for the U3 of XMRV. (B) Reporter genes in panel A were transfected into 293T, LNCaP, or WPMY-1 cells, and the luciferase activity was measured 24 h later. Null, luciferase with no promoter; TK, HSV-1 thymidine kinase promoter upstream of Renilla luciferase. All samples were assayed in triplicate and normalized to cotransfected firefly luciferase. The data are represented as the fold difference compared to the null control (null = 1). (C) Same as in panel B, but the XM1 reporter gene was included, demonstrating that the XMRV U3 is required for the observed transcriptional specificity in LNCaP and WMPY-1 cells. P values were obtained by using a two-tailed Student t test of the data presented. This experiment was repeated three times.
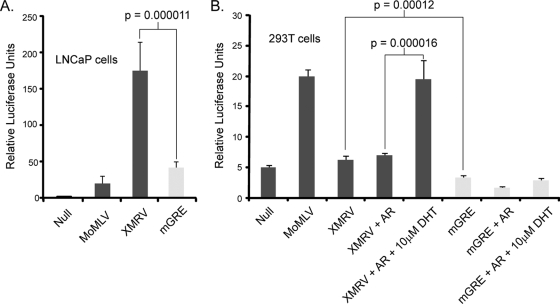
Xenotropic MLVs contain a GRE within their U3 region which is conserved within the XMRV LTR. To determine whether the XMRV GRE binding site plays a role in transcription, the GRE was mutated (GAACAGATGG to GCCCAGATGG; mGRE), and the promoter activity of the mutant LTR (mGRE) was determined by luciferase assays (Fig. 6). Significantly, the mutant mGRE reporter exhibited a fourfold reduction in LNCaP cells, suggesting that this DNA element may play a role in transcription from the XMRV LTR (Fig. 6A). We next tested whether the AR can activate the XMRV-LTR reporter gene in 293T cells (Fig. 6B). Coexpression of AR per se had no effect on the transcriptional activity of the XMRV LTR; however, after coexpression of AR and stimulation with DHT, an AR agonist, the activity increased to that of the MoMLV LTR. DHT and AR coexpression had no effect on the transcriptional activity of the MoMLV reporter gene (data not shown). Importantly, mutation of the GRE site abolished activity with or without DHT stimulation and AR coexpression. Coexpression with glucocorticoid receptor (GR) and treatment with dexamethasone, a GR agonist, had no effect on the XMRV reporter gene (data not shown).
FIG. 6.
The GRE site within the XMRV LTR is required for transcriptional activity. (A) 293T cells were transfected with the LTR reporter genes depicted in Fig. 4A and with a XMRV LTR containing a mutant within the GRE site (mGRE; GAACAGATGG to GCCCAGATGG). Luciferase activity was measured 24 h later and normalized to cotransfected firefly luciferase. All samples were assayed in triplicate and were normalized to cotransfected firefly luciferase. (B) Same as in panel A, but LNCaP cells were transfected. (C) Same as in panel A, but the XMRV LTR was cotransfected with the AR and treated with DHT for 24 h after lysis. mGRE was included to demonstrate that the mutation does not respond to AR expression and DHT stimulation. P values were obtained through a two-tailed Student t test of the data presented. This experiment was repeated twice.
DISCUSSION
This study examines the tropism of XMRV in different culture cell lines. We found that expression of Gag and CA from the provirus of XMRV is more efficient in LNCaP cells than in 293T cells. Similarly, LNCaP cells supported XMRV viral spread, whereas 293T cells did not. The few virus particles that did arise from 293T cells could be rescued by transfer to LNCaP cells; thus, virions from 293T cells were not grossly defective. Of the prostate and non-prostate cell lines tested, LNCaP cells are the most efficient host cell line for spread of infectious XMRV. We analyzed the transcriptional output from the XMRV LTR and found that the U3 region provides a higher transcriptional activity than the constitutively highly active MoMLV LTR in LNCaP and WPMY-1 cells but lower activity than the MoMLV LTR in 293T cells. These data suggest that the U3 promoter region of the XMRV LTR plays a significant role in transcriptional activity in LNCaP cells.
The U3 regions of xenotropic MLVs are conserved and contained transcriptional elements that regulate transcription of the integrated provirus (18, 20). Transcription factors such as NF-1, E-box proteins, and C/EBP coordinate with other factors to activate transcription signals to RNA Polymerase II. Interestingly, the mouse xenotropic MLVs contain two GREs that are conserved in XMRV. Cells that constitutively express the AR, such as hormone-responsive prostate cells and LNCaP cells, are thus predicted to be susceptible to AR-dependent agonists and could stimulate transcription of XMRV. In support of this notion, coexpression of the AR along with the XMRV LTR reporter in 293T cells, and stimulation with DHT, increased transcription. Mutation of one of the conserved GRE sequences abolished the transcriptional activity observed with DHT stimulation, suggesting that AR and steroid responses do indeed play a role in XMRV replication. Characterization of endogenous AR expression reveals that LNCaP cells express the AR, whereas DU145 and PC-3 cells do not (data not shown), suggesting that the reduced replication in these cell lines may be due to the absence of AR. Indeed, a study by Dong and Silverman (5) demonstrated that stimulating LNCaP cells with DHT increased the transcriptional activity of an XMRV reporter gene, whereas the AR antagonists Casodex and Flutinamide blocked this activity.
Alternative explanations for the enhanced replication of XMRV in LNCaP cells also exist. It is possible, for example, that the cellular receptor for XMRV, XPR1, may be expressed at a higher level in LNCaP cells than other cell types tested in the present study. However, quantitative PCR analysis revealed that 293T cells express fourfold-higher levels of XPR1 mRNA transcripts than LNCaP cells, suggesting that XPR1 does not account for the weak spreading infection of XMRV in 293T cells despite an increase in viral entry. We also considered the possibility that the lack of RNase L expression in LNCaP cells may be responsible for the permissiveness. To address this possibility, we depleted RNase L by using RNA interference in 293T cells. Despite >95% reduction in RNase L protein levels, XMRV replication was not enhanced (data not shown). However, it remains possible that production of IFN may limit the spread of XMRV. Increasing amounts of IFN exposure in an IFN signaling-competent cell line reduced XMRV replication in a dose-dependent manner (4). LNCaP cells are known to be deficient in JAK1 and have impaired IFN signaling, which may account for the robust spreading in this cell line (6). Experiments in which IFN antiviral signaling is restored in LNCaP cells will ultimately resolve this possibility. Another possibility might be that the lack of expression of an XMRV-restrictive factor in LNCaP cells provides a boost to viral replication. However, we consider this unlikely since the expression of APOBEC3G and Tetherin, both potent inhibitors of retrovirus replication (8, 10), are poorly expressed in 293T cells, and this suggests some other factor may play a role in XMRV spread. Importantly, XMRV was also found in the plasma (peripheral blood mononuclear cells, B cells, and T cells) in a high percentage (67%) of patients with chronic fatigue syndrome, suggesting that XMRV has a broader host range than previously appreciated (12).
We show here that XMRV can replicate efficiently in LNCaP prostate cells and induce the release of high levels of virus. This may in part be due to an enhanced transcriptional environment within LNCaP cells that allows for production of more viral proteins and subsequent budding of viral particles. The increase in transcriptional activity of the XMRV LTR is totally attributable to the U3 region and requires a GRE sequence element within the U3. The data presented further indicate that XMRV transcription can be enhanced by steroids, suggesting that XMRV may show selectivity for hormone responsive cell types, including the prostate.
Acknowledgments
This study was supported by PHS grant R37 CA 30488 from the National Cancer Institute. J.J.R. was supported by an institutional training grant T32 CA 009503-21A1 from the NCI. J.J.R. is an Associate and S.P.G. is an Investigator of the Howard Hughes Medical Institute.
We thank the Horvath laboratory for their generosity with cells and reagents. We also thank J. R. Hogg and D. Wolf for editorial advice.
The authors have paid a fee to allow immediate free access to this article.
Footnotes
Published ahead of print on 16 December 2009.
REFERENCES
- 1.Bisbal, C., and T. Salehzada. 2008. RNase L, a crucial mediator of innate immunity and other cell functions. Med. Sci. 24:859-864. (In French.) [DOI] [PubMed] [Google Scholar]
- 2.Casey, G., P. J. Neville, S. J. Plummer, Y. Xiang, L. M. Krumroy, E. A. Klein, W. J. Catalona, N. Nupponen, J. D. Carpten, J. M. Trent, R. H. Silverman, and J. S. Witte. 2002. RNASEL Arg462Gln variant is implicated in up to 13% of prostate cancer cases. Nat. Genet. 32:581-583. [DOI] [PubMed] [Google Scholar]
- 3.Cullen, B. R. 1992. Mechanism of action of regulatory proteins encoded by complex retroviruses. Microbiol. Rev. 56:375-394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dong, B., S. Kim, S. Hong, J. Das Gupta, K. Malathi, E. A. Klein, D. Ganem, J. L. Derisi, S. A. Chow, and R. H. Silverman. 2007. An infectious retrovirus susceptible to an IFN antiviral pathway from human prostate tumors. Proc. Natl. Acad. Sci. U. S. A. 104:1655-1660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dong, B., and R. H. Silverman. 2010. Androgen stimulates transcription and replication of xenotropic murine leukemia virus-related virus. J. Virol. 84:1648-1651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dunn, G. P., K. C. Sheehan, L. J. Old, and R. D. Schreiber. 2005. IFN unresponsiveness in LNCaP cells due to the lack of JAK1 gene expression. Cancer Res. 65:3447-3453. [DOI] [PubMed] [Google Scholar]
- 7.Goff, S., P. Traktman, and D. Baltimore. 1981. Isolation and properties of Moloney murine leukemia virus mutants: use of a rapid assay for release of virion reverse transcriptase. J. Virol. 38:239-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Harris, R. S., and M. T. Liddament. 2004. Retroviral restriction by APOBEC proteins. Nat. Rev. Immunol. 4:868-877. [DOI] [PubMed] [Google Scholar]
- 9.Hong, S., E. A. Klein, J. Das Gupta, K. Hanke, C. J. Weight, C. Nguyen, C. Gaughan, K. A. Kim, N. Bannert, F. Kirchhoff, J. Munch, and R. H. Silverman. 2009. Fibrils of prostatic acid phosphatase fragments boost infections with XMRV (xenotropic murine leukemia virus-related virus), a human retrovirus associated with prostate cancer. J. Virol. 83:6995-7003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jouvenet, N., S. J. Neil, M. Zhadina, T. Zang, Z. Kratovac, Y. Lee, M. McNatt, T. Hatziioannou, and P. D. Bieniasz. 2009. Broad-spectrum inhibition of retroviral and filoviral particle release by tetherin. J. Virol. 83:1837-1844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Knouf, E. C., M. J. Metzger, P. S. Mitchell, J. D. Arroyo, J. R. Chevillet, M. Tewari, and A. D. Miller. 2009. Multiple integrated copies and high-level production of the human retrovirus XMRV (xenotropic murine leukemia virus-related virus) from 22Rv1 prostate carcinoma cells. J. Virol. 83:7353-7356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lombardi, V. C., F. W. Ruscetti, J. Das Gupta, M. A. Pfost, K. S. Hagen, D. L. Peterson, S. K. Ruscetti, R. K. Bagni, C. Petrow-Sadowski, B. Gold, M. Dean, R. H. Silverman, and J. A. Mikovits. 2009. Detection of an infectious retrovirus, XMRV, in blood cells of patients with chronic fatigue syndrome. Science 326:585-589. [DOI] [PubMed] [Google Scholar]
- 13.Samuel, C. E. 1994. Interferon-induced proteins and their mechanisms of action. Hokkaido Igaku Zasshi 69:1339-1347. [PubMed] [Google Scholar]
- 14.Schlaberg, R., D. J. Choe, K. R. Brown, H. B. Thaker, and I. R. Singh. 2009. XMRV is present in malignant prostatic epithelium and is associated with prostate cancer, especially high-grade tumors. Proc. Natl. Acad. Sci. U. S. A. 106:16351-16356. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 15.Silverman, R. H. 2003. Implications for RNase L in prostate cancer biology. Biochemistry 42:1805-1812. [DOI] [PubMed] [Google Scholar]
- 16.Silverman, R. H. 2007. Viral encounters with 2′,5′-oligoadenylate synthetase and RNase L during the interferon antiviral response. J. Virol. 81:12720-12729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Song, L. N., M. Coghlan, and E. P. Gelmann. 2004. Antiandrogen effects of mifepristone on coactivator and corepressor interactions with the androgen receptor. Mol. Endocrinol. 18:70-85. [DOI] [PubMed] [Google Scholar]
- 18.Tomonaga, K., and J. M. Coffin. 1999. Structures of endogenous nonecotropic murine leukemia virus (MLV) long terminal repeats in wild mice: implication for evolution of MLVs. J. Virol. 73:4327-4340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Urisman, A., R. J. Molinaro, N. Fischer, S. J. Plummer, G. Casey, E. A. Klein, K. Malathi, C. Magi-Galluzzi, R. R. Tubbs, D. Ganem, R. H. Silverman, and J. L. DeRisi. 2006. Identification of a novel gammaretrovirus in prostate tumors of patients homozygous for R462Q RNASEL variant. PLoS Pathog. 2:e25. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 20.Verdin, E., and C. Van Lint. 1995. Internal transcriptional regulatory elements in HIV-1 and other retroviruses. Cell Mol. Biol. 41:365-369. [PubMed] [Google Scholar]