Abstract
The role of human leukocyte antigen (HLA) class I supertypes in controlling human immunodeficiency virus type 1 (HIV-1) infection in African Americans has not been established. We examined the effects of the HLA-A and HLA-B alleles and supertypes on the outcomes of HIV-1 clade B infection among 338 African American women and adolescents. HLA-B58 and -B62 supertypes (B58s and B62s) were associated with favorable HIV-1 disease control (proportional odds ratio [POR] of 0.33 and 95% confidence interval [95% CI] of 0.21 to 0.52 for the former and POR of 0.26 and 95% CI of 0.09 to 0.73 for the latter); B7s and B44s were associated with unfavorable disease control (POR of 2.39 and 95% CI of 1.54 to 3.73 for the former and POR of 1.63 and 95% CI of 1.08 to 2.47 for the latter). In general, individual alleles within specific B supertypes exerted relatively homogeneous effects. A notable exception was B27s, whose protective influence (POR, 0.58; 95% CI, 0.35 to 0.94) was masked by the opposing effect of its member allele B*1510. The associations of most B supertypes (e.g., B58s and B7s) were largely explained either by well-known effects of constituent B alleles or by effects of previously unimplicated B alleles aggregated into a particular supertype (e.g., B44s and B62s). A higher frequency of HLA-B genotypic supertypes correlated with a higher mean viral load (VL) and lower mean CD4 count (Pearson's r = 0.63 and 0.62, respectively; P = 0.03). Among the genotypic supertypes, B58s and its member allele B*57 contributed disproportionately to the explainable VL variation. The study demonstrated the dominant role of HLA-B supertypes in HIV-1 clade B-infected African Americans and further dissected the contributions of individual class I alleles and their population frequencies to the supertype effects.
African Americans in the United States have been affected disproportionately by the human immunodeficiency virus type 1 (HIV-1) epidemic. They accounted for only 13% of the U.S. population but for 47% of AIDS cases diagnosed in the United States in 2006 (14, 17). An HIV-1 vaccine would be of enormous benefit to this subpopulation. One strategy for HIV-1 vaccine development seeks to capitalize on the recognition and destruction of HIV-1-infected cells by cytotoxic T lymphocytes (CTLs) (32, 33, 35).
CTL recognition is critically dependent on binding, presentation, and cell surface display of a variety of antigenic peptides (epitopes) by extremely polymorphic human leukocyte antigen (HLA) molecules. As the relative importance of increasing numbers of HIV-1 epitopes for individual HLA class I molecules has been recognized (9, 16), the feasibility of developing a vaccine tailored to every epitope and HLA specificity has come to seem more remote. Conceptualization of epitope specificity in terms of broad groupings (supertypes) of HLA molecules may provide a rational but simpler approach to this challenge.
Several efforts have succeeded at consolidating the huge spectrum of individual HLA class I alleles into four HLA-A and five HLA-B supertype categories (37-39), based on the ability of different HLA class I molecules to present similar epitopes. Unlike individual allele frequencies, which vary greatly across ethnic groups, all nine supertypes (comprising most, but not all, HLA-A and HLA-B alleles) are present in all human populations. This more uniform representation of allele groups may confer an advantage in the form of balancing selection (38).
HLA alleles within one supertype that share epitope binding specificities might be expected to demonstrate similar associations with HIV-1 outcomes or vaccine response; conversely, alleles within a supertype that differ substantially in function might be expected to show differential responses to natural infection or vaccines designed on the basis of supertype (2). Functional heterogeneity of alleles within the supertypes could be due to differences in the class I alleles themselves (e.g., variable epitope avidity or tolerance to viral mutations), in host background (genetic epistasis), or in the virus (e.g., clade-related epitope specificities or viral escape).
Previous work has detected associations between the HLA class I supertypes and HIV-1 outcomes for Caucasians with clade B infections (34, 44) and for native Africans with clade A or C infections (23, 28). We are unaware of studies among African Americans in the context of clade B HIV-1 infection or of any systematic attempt to tease apart the independent contributions of supertypes and their individual class I alleles to HIV-1 outcomes. Here we document the frequencies of the HLA class I alleles and supertypes in the Reaching for Excellence in Adolescent Care and Health (REACH) and HIV Epidemiologic Research Study (HERS) cohorts, and we report the relative effects of those alleles and supertypes on the degree of HIV-1 disease control.
MATERIALS AND METHODS
Subjects.
We analyzed the population frequencies of HLA-A and HLA-B supertypes and their effects on HIV-1 disease control among the combined subsets of 338 African American adolescents and women from the REACH and HERS cohorts. The details of the study design and data structure of each entire cohort have been described elsewhere (31, 41, 45). African American race/ethnicity was self-reported by all study participants in a questionnaire with four race/ethnic categories (i.e., African American, Caucasian, Hispanic, and Native American). In brief, the REACH cohort is a prospective cohort of HIV-1-infected and high-risk uninfected adolescents from 13 U.S. cities. Of the 355 seropositive participants (71% African American, 75% female, and 91% with CD4+ T-cell count of ≥200 cells/ml) accrued by March 2001, 227 participants, selected for their outcomes during the treatment-free follow-up intervals, were included in the genetic analysis (66% African American, 75% female, median age of 18 years, and 93% with CD4 count of ≥200 cells/ml) (45). Hence, our subset of 161 African Americans well represented the entire REACH cohort, according to both demographic (71% female, with a median age of 18 years) and clinical (93% with CD4 count of ≥200 cells/ml) characteristics.
The HERS cohort is a larger multicenter prospective cohort including 871 HIV-infected women (60.3% African American, median age of 35 years, and 83% with CD4 count of ≥200 cells/ml) (41). Genetic analysis was performed on 286 eligible women (62% African American, median age of 34 years, and 86% with CD4 count of ≥200 cells/ml), selected for their HIV-1 outcomes during the treatment-free follow-up intervals. From these 286 women, 177 with complete HLA data and complete viral load (VL) and CD4 count data during the treatment-free interval were included in this study. These 177 individuals were similar to the global HERS cohort by age (median age, 35 years) and immunologic profile (86% with CD4 count of ≥200 cells/ml).
HLA class I typing.
HLA class I alleles were typed by PCR with sequence-specific primers, using a commercial kit (Pel-Freez/Invitrogen, Brown Deer, WI). A higher resolution of the HLA class I alleles was achieved by reference-strand conformation analysis, followed by sequence-based typing for ultimate resolution of remaining ambiguities in allele definitions (43).
Supertype assignment.
HLA class I alleles were grouped into four HLA-A and five HLA-B supertypes according to the framework of Sette and Sidney (38, 39), which served as a benchmark for most previous research on the role of HLA class I supertypes in HIV-1 infection.
HIV-1 outcomes.
Assessments of HIV-1 RNA VL in REACH and HERS participants were performed by nucleic acid sequence-based amplification (NASBA) (NucliSens; Organon Teknika, Durham, NC) and Quantiplex branched-chain DNA (b-DNA) assay (Chiron Corp., Emeryville, CA), respectively. Values determined by NASBA for the REACH cohort were predictably higher than those determined by b-DNA assay for the HERS cohort (29). CD4 counts were quantified by an AIDS Clinical Trials Group standard flow cytometry protocol. At least three consecutive VL and CD4 count measurements were collected for all study participants over 3- to 6-month intervals of antiretroviral treatment-free follow-up. Based on combinations of average values of VLs and CD4 counts over those intervals, the subjects were further categorized into 76 HIV-1 disease “controllers” (VL of <1,000 copies/ml and CD4 count of >450 cells/ml), 93 “noncontrollers” (VL of >16,000 copies/ml and CD4 count of <450 cells/ml), and 169 “intermediates” (any other combination of VL and CD4 count). Threshold viral loads used to categorize subjects were based on their division into quartiles: 1,000 copies/ml was the threshold for the lowest quartile and 16,000 copies/ml was the threshold for the highest quartile (rounded to the nearest 1,000 integer).
Statistical methods.
Ordinal categories of HIV-1 disease control were analyzed by Cochran-Mantel-Haenszel χ2 test and proportional-odds logistic regression. Although the latter approach was based on the assumption of constant change in the effect of a genetic marker(s) from one disease control category to another, it successfully captured the differences in both VL and CD4 count, as reflected by disease control categories. The associations between the genetic markers and continuous repeated CD4 counts were tested by linear growth models (also known as random-coefficient models) (40). The effects of the HLA class I supertypes and individual alleles on CD4 count were tested for associations with significant changes throughout consecutive patient visits. In the absence of interaction between HLA class I supertype and time, the estimates of supertype effect corresponded to average differences in VL or CD4 count within the follow-up, for as long as 12 months. These estimates were further adjusted for participation in one or the other cohort, patient age at baseline, and other HLA and non-HLA covariates, as applicable. The best model fit was achieved with an unstructured correlation matrix based on the lowest Akaike information criterion (5). Fixed effects from mixed models were used to estimate the associations between the HLA markers and continuous HIV-1 outcomes. By extending the supertype analysis to the genotype level, we introduced a hierarchical data structure with up to 15 hypothetical supertype combinations for HLA-B and up to 10 for HLA-A, according to the following formula:
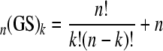 |
where n reflects the spectrum of HLA-A (n = 4) or HLA-B (n = 5) supertypes and k = 2, corresponding to paternal and maternal HLA-A or -B supertypes present in each individual, hereafter referred as genotypic supertypes (GS).
Pearson's correlation coefficient (r) was used to assess the linear relationship between the GS frequencies and HIV-1 outcomes. We used multilevel analysis with random-coefficient linear regression (40) to investigate the difference in VLs between the GS. In these single multilevel models, both the higher-level (i.e., supertype) and lower-level (i.e., individual class I alleles and their population frequencies) predictors were analyzed together. These models were used to quantify percent changes in explainable viral load variation, as predicted by each individual HLA class I marker.
The multiple imputation procedure (1) with the Markov chain Monte Carlo technique (36) was used to account for a few missing repeated outcome measures (24 VL and 38 CD4 count values). The normality assumptions for all continuous variables were validated via the Kolmogorov-Smirnov test (SAS 9.1.3; SAS, Cary, NC), and log10 VL transformation was performed to achieve normality in the VL distribution.
The thresholds for designating alleles at HLA-A and HLA-B as “infrequent” were based on the median allele frequency (5.3% and 4.4%, respectively) in each population (2N). SAS 9.1.3, SAS/Genetics (Statistical Analysis Software Institute, Cary, NC), GraphPad Prism 4.0 (La Jolla, CA), and PopGene 1.32 (http://www.ualberta.ca/∼fyeh/index.htm) were used to analyze the data. Allele and supertype carriage frequencies were calculated by dividing the number of subjects carrying a particular allele by the total number of subjects (1N), whereas population genetic marker (i.e., allele or supertype) frequencies were calculated by dividing the total number of individual marker copies by the number of chromosomes (2N).
RESULTS
Descriptive statistics.
The average age of the population was 27 years (median, 25 years; range, 13 to 54 years) (Table 1). Females (n = 307) accounted for 91% of the total study population (n = 338). The average population mean and median CD4 counts for all study subjects at all visits were 515 cells/ml and 476 cells/ml (range, 0 to 1,461 cells/ml), respectively. Both the mean and median log10 VL values were 3.6 copies/ml (range, 1.0 to 6.7 log10 copies/ml). Overall and cohort-specific distributions of log10 VL and CD4 count measurements conformed to normality (Kolmogorov-Smirnov P > 0.1).
TABLE 1.
Demographic, immunologic, virologic, and genetic features of African Americans in the individual and combined REACH (1996-2001) and HERS (1993-2000) cohorts
| Variable | Value |
||
|---|---|---|---|
| REACH cohort | HERS cohort | Combined cohorts | |
| Age (yr) (median [range])a | 18 (13-21) | 35 (21-54) | 25 (13-54) |
| Gender (% females)a | 81 | 100 | 91 |
| No. of CD4+ T cells/ml (median [range]) | 477 (0-1,461) | 475 (11-1,416) | 476 (0-1,461) |
| Viral load (log10 copies/ml) (median [range])a | 3.8 (1.0c-6.7) | 3.5 (1.4-5.6) | 3.6 (1-6.7) |
| HLA class I supertypes (n [% of patients with supertype])b | |||
| A1s | 29 (9) | 37 (11) | 66 (10) |
| A2s | 78 (24) | 83 (24) | 161 (24) |
| A24s | 89 (28) | 103 (29) | 192 (28) |
| A3s | 126 (39) | 131 (37) | 257 (38) |
| B7s | 146 (45) | 152 (43) | 298 (44) |
| B27s | 56 (17) | 56 (16) | 112 (16) |
| B44s | 70 (22) | 69 (20) | 139 (21) |
| B58s | 45 (14) | 68 (19) | 113 (17) |
| B62s | 5 (2) | 9 (2) | 14 (2) |
P < 0.05 for differences in between the individual cohorts.
HLA-A and -B supertype distributions are based on their population (2N) frequencies.
Corresponding to 1/2 log10VL for plasma HIV-1 RNA level of <80 copies/ml.
A somewhat higher median log10 VL was observed for REACH (3.8 log10 copies/ml) than for HERS (3.5 log10 copies/ml) participants, whereas median CD4 counts for these two subcohorts were nearly identical (477 versus 475 cells/ml). There was no significant difference among the subcohorts in the prevalence (1N) of either HLA-A or -B supertypes (all P values were >0.2).
The HLA-A3 supertype (A3s) had the highest population (2N) frequency (38%) and the highest carriage (1N) frequency (63.3%). On the other hand, A1s was the least frequent of all HLA-A supertypes, with population and carriage frequencies of 9.8% and 18.3%, respectively. Among the HLA-B supertypes, B7s had a population frequency of 44.1% and a carrier frequency of 68%, in contrast to B62s, which had the lowest population and carriage frequencies (2.1% and 4.1%, respectively).
Supertypes and HIV-1 control.
For none of the four HLA-A supertypes did the population frequencies or proportions of carriers differ significantly across the HIV-1 disease progression categories (all P values were >0.2) (Table 2). Inconsistencies in favorable and unfavorable associations with disease progression were detected for most of the HLA-A supertype alleles. The patterns remained unchanged after excluding two alleles that appeared to be associated with protection in our analysis (P < 0.01): A*32, an allele that has been associated with protection elsewhere (13, 21, 44), was excluded from A1s; and A*74, associated with favorable HIV-1 outcomes here only, was excluded from A3s.
TABLE 2.
HLA class I supertypes and HIV-1 control in African Americans from the REACH (1996-2001) and HERS (1993-2000) cohorts
| HLA class I supertype | No. (%) of patientsa |
P valueb | POR (95% CI)c | ||
|---|---|---|---|---|---|
| Controllers (n = 76) | Intermediates (n = 169) | Noncontrollers (n = 93) | |||
| A1s | 16 (21.1) | 30 (17.8) | 16 (17.2) | 0.5 | 0.9 (0.5-1.4) |
| No A*32 | 10 (14.3) | 25 (15.2) | 16 (17.2) | 0.6 | 1.1 (0.7-2.1) |
| A2s | 31 (40.8) | 78 (46.2) | 36 (38.7) | 0.7 | 0.9 (0.6-1.4) |
| A24s | 36 (47.4) | 78 (46.2) | 48 (51.6) | 0.6 | 1.1 (0.8-1.7) |
| A3s | 52 (68.4) | 106 (62.7) | 56 (60.2) | 0.3 | 0.8 (0.5-1.2) |
| No A*74 | 36 (60.0) | 81 (56.3) | 51 (58.0) | 0.9 | 0.9 (0.6-1.5) |
| B7s | 40 (52.6) | 115 (68.1) | 75 (80.7) | <0.001 | 2.4 (1.5-3.7) |
| No B*35 | 31 (46.3) | 91 (62.8) | 56 (75.7) | <0.001 | 2.3 (1.5-3.7) |
| No B*35 or B*53 | 25 (41.0) | 48 (47.1) | 32 (64.0) | 0.02 | 1.8 (1.1-3.1) |
| No B*35, B*53, or B22d | 25 (41.0) | 46 (46.0) | 31 (63.3) | 0.02 | 1.8 (1.1-3.0) |
| No B*35, B*53, B22,d or B*81 | 20 (35.7) | 42 (43.8) | 31 (63.3) | <0.01 | 2.1 (1.3-3.6) |
| B27s | 25 (32.9) | 53 (31.4) | 23 (24.7) | 0.2 | 0.8 (0.5-1.2) |
| No B*1510 | 24 (32.0) | 40 (25.6) | 14 (16.7) | 0.03 | 0.6 (0.4-0.9) |
| No B*1510 or B*14 | 13 (20.3) | 28 (19.4) | 12 (14.6) | 0.36 | 0.8 (0.4-1.4) |
| B44s | 17 (22.4) | 70 (41.4) | 38 (40.9) | 0.02 | 1.6 (1.1-2.5) |
| No B*44 | 10 (14.5) | 44 (30.8) | 24 (30.4) | 0.04 | 1.7 (1.0-2.7) |
| No B*44 or B*45 | 4 (6.4) | 30 (23.3) | 14 (20.3) | 0.05 | 1.8 (1.0-3.2) |
| B58s | 42 (55.3) | 42 (24.9) | 19 (20.4) | <0.0001 | 0.3 (0.2-0.5) |
| No B*57 | 17 (33.3) | 31 (19.6) | 17 (18.7) | 0.07 | 0.6 (0.4-1.1) |
| No B*57 or B*5801 | 9 (20.9) | 20 (13.6) | 8 (9.7) | 0.09 | 0.6 (0.3-1.1) |
| B62s | 7 (9.2) | 6 (3.6) | 1 (1.1) | 0.01 | 0.3 (0.1-0.7) |
Disease control categories were based on the average favorable (controllers; VL of <1,000 copies/ml and CD4 count of >450 cells/ml), unfavorable (noncontrollers; VL of >16,000 copies/ml and CD4 count of <450 cells/ml), and remaining (intermediates) VL and CD4 count combinations.
Assessed by rank score Cochran-Mantel-Haenszel test for trend.
POR, proportional odds ratio.
B22 serogroup includes B*54 to B*56.
In contrast to HLA-A supertypes, most of the HLA-B supertypes demonstrated significant associations with either control (B58s and B62s) or lack of control (B7s and B44s) in the univariate analyses. Stepwise restriction of B7s (proportional odds ratio [POR], 2.39; 95% confidence interval [95% CI], 1.54 to 3.73) carriers to subjects without alleles reported to accelerate disease progression (B*35, B*53, and B22 [B*54 to B*56]) reduced the strength of association, but the higher frequency of B7s among noncontrollers remained significant (P = 0.02).
As in previous studies of HIV-1-infected native Africans (23) and North Americans from the SCOPE cohort (8), here too the small number of B*81 carriers (n = 14) within the B7s supertype tended to differ from carriers of other B7s alleles in demonstrating a trend toward disease control (6.6% B*81 carriage in controllers versus 2.2% in noncontrollers; P = 0.15). In the present study, after exclusion of B*81 carriers, the remainder of those with B7s showed an even stronger association with poor disease control (P = 0.005). Similar to B7s, B44s was associated with noncontrol (POR, 1.63; 95% CI, 1.1 to 2.5), even after excluding B*44 and B*45 alleles (P < 0.05). B58s (POR, 0.33; 95% CI, 0.21 to 0.52) was associated with control that was heavily determined by B*57 alleles (POR, 0.11; 95% CI, 0.05 to 0.22). However, neither the strong protective effect observed elsewhere for B*5801 (22, 27) nor the strong deleterious effect observed elsewhere for B*5802 (22, 23, 27) was observed in our study.
Although relatively few subjects carried B62s (n = 14), it was also associated with disease control (POR, 0.26; 95% CI, 0.1 to 0.7). B27s (POR, 0.77; 95% CI, 0.5 to 1.2) showed no overall association with disease status and only a minimal protective effect after exclusion of two individual alleles with opposing associations: favorable for B*14 (POR, 0.3; 95% CI, 0.1 to 0.7) and unfavorable for B*1510 (POR, 2.2; 95% CI, 1.0 to 5.0).
Supertypes and continuous HIV-1 outcomes.
We detected no significant differences in the VL or CD4 count slopes across sequential patient visit intervals based on the possession of any HLA-A or HLA-B supertype. Hence, fixed supertype effects on these two outcomes were estimated from the multivariable models accounting for patient age, cohort membership, and the HLA class I alleles with widely recognized effects on disease control (Table 3). In general, our findings from the VL analysis and, to a lesser extent, the CD4 count analysis, paralleled those in the categorical assessment of disease control. Although differences in CD4 count by HLA-B supertype were not statistically significant, the trends for the associations with CD4 count paralleled the effects of the supertypes on VL. No such parallel effects on VL and CD4 count were detected for HLA-A supertypes.
TABLE 3.
Multivariable analysis of HLA class I supertypes in relation to HIV-1 outcomes among African Americans from the REACH (1996-2001) and HERS (1993-2000) cohortsa
| HLA class I supertype | Log10 VL |
CD4 count |
||||||
|---|---|---|---|---|---|---|---|---|
| Adjustedb |
Adjustedc |
Adjustedd |
Adjustede |
|||||
| ΔVL | P value | ΔVL | P value | ΔCD4 count | P value | ΔCD4 count | P value | |
| A1s | +0.06 | 0.73 | +0.001 | 0.99 | +47 | 0.33 | +67 | 0.17 |
| A2s | −0.1 | 0.34 | −0.14 | 0.18 | −16 | 0.78 | −9 | 0.87 |
| A24s | +0.05 | 0.6 | −0.01 | 0.86 | +3 | 0.93 | +28 | 0.4 |
| A3s | −0.01 | 0.91 | −0.006 | 0.96 | −14 | 0.7 | −27 | 0.45 |
| B7s | +0.28 | 0.02 | +0.22 | 0.05 | −81 | 0.02 | −52 | 0.13 |
| B27s | −0.2 | 0.15 | −0.25 | 0.05 | +4 | 0.92 | +24 | 0.49 |
| B44s | +0.33 | <0.01 | +0.39 | <0.001 | −54 | 0.11 | −50 | 0.13 |
| B58s | −0.26 | 0.04 | −0.17 | 0.17 | +32 | 0.39 | +39 | 0.29 |
| B62s | −0.65 | 0.01 | −0.49 | 0.03 | +82 | 0.27 | +53 | 0.44 |
Estimates correspond to the fixed effects of average log10 VL and CD4 count changes within all patient visits. Both the estimates and P values were derived from mixed linear models, with imputed missing values for log10 VL (24 observations) and CD4 count (38 observations). ΔVL and ΔCD4 count estimates correspond to the β coefficients from mixed linear models.
Adjusted for cohort, age, patient visit, and alleles associated with log10 VL from uni- and bivariate analyses (A*32 and A*36 for A1s, A*74 for A3s, B*3501, B*53, and B*81 for B7s, B*14 and B*1510 for B27s, B*45 for B44s, and B*57 for B58s).
Adjusted for cohort, age, patient visit, and significant alleles within and outside the particular supertype, as applicable (A*32, A*36, A*74, B*3501, B*53, B*14, B*1510, B*45, B*57, and B*81).
Adjusted for patient visit, cohort, age, and alleles associated with CD4 from uni- and bivariate analyses (A*32 and A*36 for A1s, A*02 for A2s, A*74 and A*03 for A3s, B*3503, B*53, and B*81 for B7s, B*14 and B*27 for B27s, B*45 for B44s, and B*57 for B*58s).
Adjusted for patient visit, cohort, age, and significant alleles from within and outside the supertype, as applicable (A*02, A*03, A*32, A*36, A*74, B*14, B*27, B*3503, B*45, B*53, and B*57).
After all adjustments for the effects of individual alleles highlighted above, B7s (+0.22 Δlog10 VL; P = 0.05) and B44s (+0.39 Δlog10 VL; P < 0.001) remained associated with higher VLs. B62s (−0.49 Δlog10 VL; P = 0.03) and B27s (−0.25 Δlog10 VL; P = 0.05) remained associated with lower VLs. Again in contrast to the case for HLA-B, with analysis adjusted for the effects of individual contributing alleles, there were no significant associations between HLA-A supertypes and continuous HIV-1 outcomes.
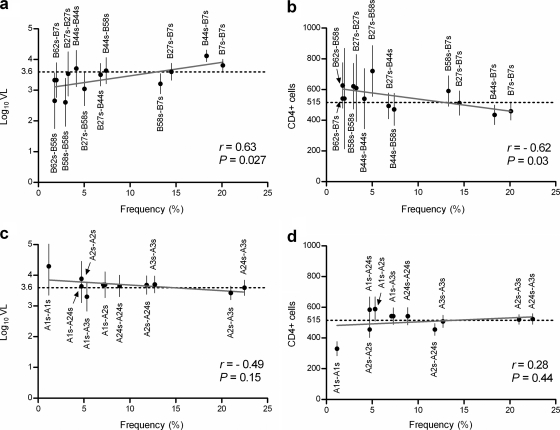
Genotypic supertypes (GS) at the HLA-B, but not HLA-A, loci were differentially associated with HIV-1 outcomes (Fig. 1). Specifically, GS comprising favorable combinations of maternal and paternal B58s, B62s, and B27s showed effects contrasting with those of unfavorable combinations of B7s and B44s. Those GS at the B but not the A locus that were associated with lower VLs and higher CD4 counts also tended to occur at lower population frequencies (r > 0.6; P < 0.05) (Fig. 1). The correlation between the supertype frequencies and viral load was comparable between the two subcohorts (r = 0.7 and P = 0.01 for REACH and r = 0.62 and P = 0.025 for HERS).
FIG. 1.
Frequencies of HLA class I genotypic supertypes and their correlations with HIV-1 outcomes among African Americans from the REACH (1996-2001) and HERS (1993-2000) cohorts. In contrast to the apparent correlations (Pearson's r < 0.05) between frequencies of HLA-B genotypic supertypes and virological (a) and immunological (b) outcomes, frequencies of HLA-A supertypes (c and d) have no clear correlation with HIV-1 outcomes. Predicted values constitute regression lines superimposed on the graphs. Horizontal dotted lines correspond to the average population VL or CD4 count, and vertical bars correspond to 95% confidence intervals for the mean values.
Having demonstrated independent associations of infrequent HLA-B alleles with VL (unpublished data), we investigated specific contributions of these B alleles and their population frequencies to the heterogeneity in VLs among B locus GS (Table 4). We found that the percent reductions in explainable VL variation attributed to B58s (53%) overall and to B*57 (45%) in particular were substantially higher than the contributions of other individual (5 to 10%) or infrequent (9%) B alleles in aggregate. To evaluate our inferences against the background of previous work with other populations, we further partitioned all B locus GS into two major clusters, as proposed by Trachtenberg et al. for HIV-1-infected Caucasian males (44). We found that 57% of the VL variation was explained by the cluster of GS including B58s-B7s, B58s-B44s, B27s-B44s, B58s-B27s, B58s-B58s, and B58s-B62s. Our analysis thus supported a dominant model for B58s and further highlighted the major contribution of B*57 alleles to the explainable variation in VLs across the GS.
TABLE 4.
Factors associated with HIV-1 load heterogeneity across genotypic supertypes among African Americans from the REACH (1996-2001) and HERS (1993-2000) cohortsa
| HLA class I marker | Intercept | VL variationc (%) |
|---|---|---|
| Baseline modelb | 0.126 | |
| Level 2 markers | ||
| B7s | 0.107 | 15 |
| B27s | 0.139 | −10 |
| B44s | 0.094 | 25 |
| B58s | 0.059 | 53 |
| B62s | 0.114 | 10 |
| Level 1 markers | ||
| B*14 | 0.117 | 7 |
| B*1510 | 0.139 | −10 |
| B*35 | 0.12 | 5 |
| B*45 | 0.116 | 8 |
| B*53 | 0.117 | 7 |
| B*57 | 0.069 | 45 |
| Infrequent HLA-B allelesd | 0.115 | 9 |
All estimates were derived from random-effect multilevel models.
Adjusted for individual cohort.
Corresponding to percentage of explainable log10 VL variation and quantified as the percent difference in variation of VL attributed to each HLA marker included in the model [(baseline intercept − intercept) × 100/baseline intercept], with positive values indicating a decrease and negative values indicating an increase in VL variation (e.g., B27s or B*1510).
Based on HLA-B population frequency distribution below the median (4.4%).
DISCUSSION
In our study of clade B HIV-1 infections among African Americans, HLA-B but not HLA-A supertypes were associated with disease control. Our analysis further demonstrated the importance of individual alleles at the B locus compared with those at the A locus. Associations of B7s and B44s were consistently disadvantageous for disease control overall and separately for VL and CD4 count. The latter are the most prevalent HLA-B supertypes in both African Americans and Caucasians (34, 44). Our findings corroborated the few previous studies on the effects of HLA-B supertypes broadly in Caucasians (34, 44) and of B44s specifically in native Africans (28). The consistently favorable overall associations of B58s with disease control and with VL and the favorable association of B62s also agreed with published findings.
Associations between supertypes and HIV-1 outcomes have been demonstrated in previous studies of Caucasians with clade B infection (34, 44) and of native Africans with clade A or C infection (23, 28). Caucasians in one study showed an unfavorable effect of B7s and a favorable effect of B58s on HIV-1 control (44); in another study, Caucasians with the relatively infrequent B58s, B27s, and B62s supertypes showed enhanced CTL responses, while the more frequent B7s and B44s supertypes were associated with diminished CTL responses (34). Among native Africans, carriers of A2s, A24s, B27s, and B58s, but not B44s, had lower VLs and more vigorous CTL responses to total Gag, Gag p24, and Nef (28).
Whether the effect of a particular supertype is driven by one, several, or all of the individual class I alleles currently assigned to that supertype has received little attention. In one population of European ancestry (44), the advantage of B27s alleles persisted after excluding carriers with favorable B*27 alleles and the disadvantage of B7s alleles persisted after removing unfavorable B*54-56 (6) and B*35 (12) alleles. These findings suggested analogous outcomes associated with the seemingly cross-reacting alleles within each particular supertype. A large proportion of our African Americans with B7s remained “noncontrollers” of disease, even after exclusion of participants with B*35-Px and B*35-Py, both of which have appeared disadvantageous in at least one other HIV-1 clade B-infected North American population besides ours (3).
The pattern of association with B7s described above has an exception: B*81 appeared to predispose individuals to more favorable HIV-1 outcomes here and elsewhere (8, 23).
As a largely African allele, B*81 was only recently assigned to B7s (39), based on predictions from its three-dimensional protein structure of a preference for proline at position 2 of the binding epitope (7). Important functional differences between various B7s alleles that present cross-reacting epitopes were previously detected among clade C-infected Africans (24). Any of several mechanisms may explain this within-supertype allelic heterogeneity, including frequency, magnitude, immunodominance, or avidity of allele-specific CTL responses, as well as distinct T-cell receptor (TCR) usage by the individual alleles. In this study, B*81 accounted for only 6% of B7s alleles, and overall the remainder were disadvantageous.
As a whole, B44s (largely represented by B*44, B*45, and B*49) also appeared to be disadvantageous for disease control. All individual B44s alleles tended toward an association with poorer control (data not shown). These findings have extended the recognition of B44s as disadvantageous for multiple human ethnic groups and viral clades.
In contrast, the strongly protective B62s supertype was the least prevalent in both cohorts. The latter supertype includes B*13, an allele implicated repeatedly in protection against HIV-1 (4, 18, 42). Interestingly, only 2 of 14 African Americans with B62s carried B*13 in our study; 9 carried B*52, an allele not previously associated with prognosis of HIV-1 infection. Since both B*52 and B*13 appear to share an affinity for glutamine at position 2 of binding epitopes, B*52 may successfully target an immunodominant CTL epitope(s) that cross-reacts with B*13 (e.g., epitope R19 in Nef) (18). The low B62s population frequency, perhaps along with protective Bw4-80Ile specificity (10, 30) of its two common members, may have contributed to its favorable effect.
The observed relationships between supertype population frequencies and HIV-1 outcomes in the REACH and HERS cohorts separately and in aggregate are consistent with earlier findings (34, 44). On the other hand, infrequent individual HLA-B alleles accounted in aggregate for only a modest portion of explainable VL variation across supertypes at the genotypic level; the magnitude of this effect was similar to that seen with more common individual HLA-B alleles. Because any advantage conferred by infrequent individual HLA-B alleles likely reflects past negative selective pressure by more than just a single pathogen, infrequent HLA-B alleles might not be expected to exert great control of HIV-1 disease. Despite this lower magnitude of effect on HIV-1 outcomes, infrequent class I alleles remained consistent in their association with lower VLs (−0.26 Δlog10 VL; P < 0.005) in the multivariable analysis restricted to the carriers of the two most frequent HLA-B supertypes (i.e., B7s and B44s).
The disproportionate contribution of a single B58s allele (B*57) to the VL heterogeneity among genotypic supertypes highlights the uniqueness of B*57 alleles in conferring highly effective natural immunity to HIV-1. In our study population, the number of carriers of the African B*5703 allele (n = 28) outnumbered those with the Caucasian B*5701 allele (n = 8), but the protective associations were quite similar. In additional exploratory analysis, the trend toward an association between B*1516 and -17 and lower VLs was in the direction predicted by other work (11). Our earlier investigation of B58s alleles among clade A- and C-infected native Africans demonstrated contrasting effects of B*57 and B*58 alleles on HIV-1 control (23). Here we found no appreciable effect of either B*5802, established as unfavorable, or B*5801, established as favorable.
Although B*27 alleles have repeatedly been associated with favorable outcomes (12, 19, 20), B27s did not show a clear trend toward an association in either direction, for two reasons: B*27 alleles are relatively rare in individuals of African ancestry, and the effects of two other major B27s alleles (B*14 and B*1510) offset each other. Alleles of B*14, with or without its closely linked Cw*08 allele, have been associated with protection against HIV-1 (19, 26). In addition, B*14 appears to be more frequent among subjects with immunodominant CTL responses toward Gag p24 epitope SG20 (15). B*1510 occurs more frequently among Africans than among Caucasians, but no prior epidemiologic association has been reported for this allele in the context of HIV-1. Our multilevel model (Table 4) also reflected a mixed picture for B27s; genotypic supertypes containing it accounted for a modest 10% increase in viral load heterogeneity. Our results support the suggested reallocation of B*1510 from B27s into the novel B39 supertype (25), which would leave the remaining B27s alleles showing uniformly favorable effects.
Our study had limitations. With 338 subjects, it was underpowered to study alleles at frequencies of <2%. Despite the sample size, it was the more frequent HLA-A supertypes that failed to show any evidence of association with disease control. Although the apparent heterogeneity of our two subcohorts (e.g., in median VL) might have precluded combining them, their close resemblance in the distribution of HLA markers (all with P values of >0.1) and in their median values for CD4 cell counts justified doing so. Moreover, the observed population differences in median VL were most likely due to the distinct methods for quantifying HIV-1 plasma RNA levels. Nevertheless, we further adjusted our analysis for individual cohort membership.
Our study was the largest to date to examine the relationship between HLA class I supertypes and HIV-1 outcomes in African Americans, the first to include a systematic assessment of the effects of individual class I alleles within each of the supertypes, and the first to analyze the contributions of individual class I alleles and their population frequencies to the differences in VLs among genotypic supertypes. Multilevel random-coefficient regression enabled us to address the hierarchical structure of the data in a flexible framework for analyzing the heterogeneity of VL, as predicted by both the HLA class I alleles (i.e., individual level) and their supertypes (i.e., group level).
The consistent demonstration of complementary influences of HLA-B supertypes and individual HLA-B alleles may focus attention on functional convergence in HLA polymorphism. Strong, consistent evidence for supertype effects could guide the development of multiepitope-based HIV-1 vaccines. Finally, the techniques introduced here to analyze the hierarchical relationships of HLA supertypes and their component alleles should be applicable to future studies of HLA in other infectious diseases.
Acknowledgments
The authors thank the investigators and staff (listed in J. Adolesc. Health 29(Suppl.):5-6, 2001) of the Adolescent Medicine HIV/AIDS Research Network (1994-2001) and the youth who participated in the REACH project for their valuable contributions. We are deeply grateful to all women participants of the HERS cohort.
REACH was supported by grant U01 HD32830 from the National Institute of Child Health and Human Development, with additional funding from the National Institute on Drug Abuse, National Institute of Allergy and Infectious Diseases, and National Institute of Mental Health. HERS was supported by the Centers for Disease Control and Prevention, the National Institute of Allergy and Infectious Diseases (NIAID), the National Institute on Drug Abuse, the Agency for Health Care Policy and Research, and the National Institutes of Health Office for Women's Research. Genetic analysis was supported by grant R01 AI071906 from the National Institute of Allergy and Infectious Diseases.
The HERS group consists of Robert S. Klein, from the Mount Sinai School of Medicine; Ellie Schoenbaum, Julia Arnsten, Robert D. Burk, Penelope Demas, and Andrea Howard, from Montefiore Medical Center and the Albert Einstein College of Medicine; Paula Schuman, Jack Sobel, and Wayne Lancaster, from the Wayne State University School of Medicine; Anne Rompalo, David Vlahov, and David Celentano, from the Johns Hopkins University School of Medicine; Charles Carpenter, Kenneth Mayer, Susan Cu-Uvin, Timothy Flanigan, Joseph Hogan, and Josiah Rich, from the Brown University School of Medicine; Lytt I. Gardner, Chad Heilig, Scott D. Holmberg, Denise J. Jamieson, Janet S. Moore, and Dawn K. Smith, from the Centers for Disease Control and Prevention; and Katherine Davenny, from the National Institute of Drug Abuse.
The findings and conclusions in this report are those of the authors and do not necessarily represent the views of the Centers for Disease Control and Prevention.
Footnotes
Published ahead of print on 23 December 2009.
REFERENCES
- 1.Allison, P. D. 2000. Multiple imputation for missing data: a cautionary tale. Sociol. Methods Res. 28:301-309. [Google Scholar]
- 2.Brander, C. 2004. HLA and HIV: implications for HIV vaccine design. ASHI Q. 2004:58-59. [Google Scholar]
- 3.Brumme, C., C. Chui, C. Woods, B. Wynhoven, R. Hogg, J. Montaner, P. Harrigan, and Z. Brumme. 2006. Specific HLA-B alleles known to influence untreated HIV-1 disease progression do not appear to affect response to highly active antiretroviral therapy. Can. J. Infect. Dis. Med. Microbiol. 17(Suppl. A):114.18418485 [Google Scholar]
- 4.Brumme, Z. L., C. J. Brumme, C. Chui, T. Mo, B. Wynhoven, C. K. Woods, B. M. Henrick, R. S. Hogg, J. S. Montaner, and P. R. Harrigan. 2007. Effects of human leukocyte antigen class I genetic parameters on clinical outcomes and survival after initiation of highly active antiretroviral therapy. J. Infect. Dis. 195:1694-1704. [DOI] [PubMed] [Google Scholar]
- 5.Burnham, K. P., and D. R. Anderson. 2002. Model selection and multimodel inference: a practical-theoretic approach, 2nd ed. Springer-Verlag, New York, NY.
- 6.Dorak, M. T., J. Tang, S. Tang, A. Penman-Aguilar, R. A. Coutinho, J. J. Goedert, R. Detels, and R. A. Kaslow. 2003. Influence of human leukocyte antigen-B22 alleles on the course of human immunodeficiency virus type 1 infection in 3 cohorts of white men. J. Infect. Dis. 188:856-863. [DOI] [PubMed] [Google Scholar]
- 7.Doytchinova, I. A., P. Guan, and D. R. Flower. 2004. Identifying human MHC supertypes using bioinformatic methods. J. Immunol. 172:4314-4323. [DOI] [PubMed] [Google Scholar]
- 8.Emu, B., E. Sinclair, H. Hatano, A. Ferre, B. Shacklett, J. N. Martin, J. M. McCune, and S. G. Deeks. 2008. HLA class I-restricted T-cell responses may contribute to the control of human immunodeficiency virus infection, but such responses are not always necessary for long-term virus control. J. Virol. 82:5398-5407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Feeney, M. E., Y. Tang, K. A. Roosevelt, A. J. Leslie, K. McIntosh, N. Karthas, B. D. Walker, and P. J. Goulder. 2004. Immune escape precedes breakthrough human immunodeficiency virus type 1 viremia and broadening of the cytotoxic T-lymphocyte response in an HLA-B27-positive long-term-nonprogressing child. J. Virol. 78:8927-8930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Flores-Villanueva, P. O., E. J. Yunis, J. C. Delgado, E. Vittinghoff, S. Buchbinder, J. Y. Leung, A. M. Uglialoro, O. P. Clavijo, E. S. Rosenberg, S. A. Kalams, J. D. Braun, S. L. Boswell, B. D. Walker, and A. E. Goldfeld. 2001. Control of HIV-1 viremia and protection from AIDS are associated with HLA-Bw4 homozygosity. Proc. Natl. Acad. Sci. USA 98:5140-5145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Frahm, N., S. Adams, P. Kiepiela, C. H. Linde, H. S. Hewitt, M. Lichterfeld, K. Sango, N. V. Brown, E. Pae, A. G. Wurcel, M. Altfeld, M. E. Feeney, T. M. Allen, T. Roach, M. A. St John, E. S. Daar, E. Rosenberg, B. Korber, F. Marincola, B. D. Walker, P. J. Goulder, and C. Brander. 2005. HLA-B63 presents HLA-B57/B58-restricted cytotoxic T-lymphocyte epitopes and is associated with low human immunodeficiency virus load. J. Virol. 79:10218-10225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gao, X., G. W. Nelson, P. Karacki, M. P. Martin, J. Phair, R. Kaslow, J. J. Goedert, S. Buchbinder, K. Hoots, D. Vlahov, S. J. O'Brien, and M. Carrington. 2001. Effect of a single amino acid change in MHC class I molecules on the rate of progression to AIDS. N. Engl. J. Med. 344:1668-1675. [DOI] [PubMed] [Google Scholar]
- 13.Geczy, A. F., H. Kuipers, M. Coolen, L. J. Ashton, C. Kennedy, G. Ng, R. Dodd, R. Wallace, T. Le, C. H. Raynes-Greenow, W. B. Dyer, J. C. Learmont, and J. S. Sullivan. 2000. HLA and other host factors in transfusion-acquired HIV-1 infection. Hum. Immunol. 61:172-176. [DOI] [PubMed] [Google Scholar]
- 14.Glynn, M. K., and P. Rhodes. 2005. Estimated HIV prevalence in the United States at the end of 2003. 2005 Natl. HIV Prev. Conf., Atlanta, GA.
- 15.Goulder, P. J., C. Brander, K. Annamalai, N. Mngqundaniso, U. Govender, Y. Tang, S. He, K. E. Hartman, C. A. O'Callaghan, G. S. Ogg, M. A. Altfeld, E. S. Rosenberg, H. Cao, S. A. Kalams, M. Hammond, M. Bunce, S. I. Pelton, S. A. Burchett, K. McIntosh, H. M. Coovadia, and B. D. Walker. 2000. Differential narrow focusing of immunodominant human immunodeficiency virus gag-specific cytotoxic T-lymphocyte responses in infected African and caucasoid adults and children. J. Virol. 74:5679-5690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Goulder, P. J., R. E. Phillips, R. A. Colbert, S. McAdam, G. Ogg, M. A. Nowak, P. Giangrande, G. Luzzi, B. Morgan, A. Edwards, A. J. McMichael, and S. Rowland-Jones. 1997. Late escape from an immunodominant cytotoxic T-lymphocyte response associated with progression to AIDS. Nat. Med. 3:212-217. [DOI] [PubMed] [Google Scholar]
- 17.Hall, H. I., R. Song, P. Rhodes, J. Prejean, Q. An, L. M. Lee, J. Karon, R. Brookmeyer, E. H. Kaplan, M. T. McKenna, and R. S. Janssen. 2008. Estimation of HIV incidence in the United States. JAMA 300:520-529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Harrer, E. G., S. Bergmann, K. Eismann, M. Rittmaier, A. Goldwich, S. M. Muller, B. M. Spriewald, and T. Harrer. 2005. A conserved HLA B13-restricted cytotoxic T lymphocyte epitope in Nef is a dominant epitope in HLA B13-positive HIV-1-infected patients. AIDS 19:734-735. [DOI] [PubMed] [Google Scholar]
- 19.Hendel, H., S. Caillat-Zucman, H. Lebuanec, M. Carrington, S. O'Brien, J. M. Andrieu, F. Schachter, D. Zagury, J. Rappaport, C. Winkler, G. W. Nelson, and J. F. Zagury. 1999. New class I and II HLA alleles strongly associated with opposite patterns of progression to AIDS. J. Immunol. 162:6942-6946. [PubMed] [Google Scholar]
- 20.Kaslow, R. A., M. Carrington, R. Apple, L. Park, A. Munoz, A. J. Saah, J. J. Goedert, C. Winkler, S. J. O'Brien, C. Rinaldo, R. Detels, W. Blattner, J. Phair, H. Erlich, and D. L. Mann. 1996. Influence of combinations of human major histocompatibility complex genes on the course of HIV-1 infection. Nat. Med. 2:405-411. [DOI] [PubMed] [Google Scholar]
- 21.Keet, I. P., J. Tang, M. R. Klein, S. LeBlanc, C. Enger, C. Rivers, R. J. Apple, D. Mann, J. J. Goedert, F. Miedema, and R. A. Kaslow. 1999. Consistent associations of HLA class I and II and transporter gene products with progression of human immunodeficiency virus type 1 infection in homosexual men. J. Infect. Dis. 180:299-309. [DOI] [PubMed] [Google Scholar]
- 22.Kiepiela, P., A. J. Leslie, I. Honeyborne, D. Ramduth, C. Thobakgale, S. Chetty, P. Rathnavalu, C. Moore, K. J. Pfafferott, L. Hilton, P. Zimbwa, S. Moore, T. Allen, C. Brander, M. M. Addo, M. Altfeld, I. James, S. Mallal, M. Bunce, L. D. Barber, J. Szinger, C. Day, P. Klenerman, J. Mullins, B. Korber, H. M. Coovadia, B. D. Walker, and P. J. Goulder. 2004. Dominant influence of HLA-B in mediating the potential co-evolution of HIV and HLA. Nature 432:769-775. [DOI] [PubMed] [Google Scholar]
- 23.Lazaryan, A., E. Lobashevsky, J. Mulenga, E. Karita, S. Allen, J. Tang, and R. A. Kaslow. 2006. Human leukocyte antigen B58 supertype and human immunodeficiency virus type 1 infection in native Africans. J. Virol. 80:6056-6060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Leslie, A., D. A. Price, P. Mkhize, K. Bishop, A. Rathod, C. Day, H. Crawford, I. Honeyborne, T. E. Asher, G. Luzzi, A. Edwards, C. M. Rousseau, J. I. Mullins, G. Tudor-Williams, V. Novelli, C. Brander, D. C. Douek, P. Kiepiela, B. D. Walker, and P. J. Goulder. 2006. Differential selection pressure exerted on HIV by CTL targeting identical epitopes but restricted by distinct HLA alleles from the same HLA supertype. J. Immunol. 177:4699-4708. [DOI] [PubMed] [Google Scholar]
- 25.Lund, O., M. Nielsen, C. Kesmir, A. G. Petersen, C. Lundegaard, P. Worning, C. Sylvester-Hvid, K. Lamberth, G. Roder, S. Justesen, S. Buus, and S. Brunak. 2004. Definition of supertypes for HLA molecules using clustering of specificity matrices. Immunogenetics 55:797-810. [DOI] [PubMed] [Google Scholar]
- 26.Magierowska, M., I. Theodorou, P. Debre, F. Sanson, B. Autran, Y. Riviere, D. Charron, and D. Costagliola. 1999. Combined genotypes of CCR5, CCR2, SDF1, and HLA genes can predict the long-term nonprogressor status in human immunodeficiency virus-1-infected individuals. Blood 93:936-941. [PubMed] [Google Scholar]
- 27.Ngumbela, K. C., C. L. Day, Z. Mncube, K. Nair, D. Ramduth, C. Thobakgale, E. Moodley, S. Reddy, C. de Pierres, N. Mkhwanazi, K. Bishop, M. van der Stok, N. Ismail, I. Honeyborne, H. Crawford, D. G. Kavanagh, C. Rousseau, D. Nickle, J. Mullins, D. Heckerman, B. Korber, H. Coovadia, P. Kiepiela, P. J. Goulder, and B. D. Walker. 2008. Targeting of a CD8 T cell env epitope presented by HLA-B*5802 is associated with markers of HIV disease progression and lack of selection pressure. AIDS Res. Hum. Retrovir. 24:72-82. [DOI] [PubMed] [Google Scholar]
- 28.Novitsky, V., P. Gilbert, T. Peter, M. F. McLane, S. Gaolekwe, N. Rybak, I. Thior, T. Ndung'u, R. Marlink, T. H. Lee, and M. Essex. 2003. Association between virus-specific T-cell responses and plasma viral load in human immunodeficiency virus type 1 subtype C infection. J. Virol. 77:882-890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Peter, J. B., and J. S. Sevall. 2004. Molecular-based methods for quantifying HIV viral load. AIDS Patient Care STDS 18:75-79. [DOI] [PubMed] [Google Scholar]
- 30.Qing, M., T. Li, Y. Han, Z. Qiu, and Y. Jiao. 2006. Accelerating effect of human leukocyte antigen-Bw6 homozygosity on disease progression in Chinese HIV-1-infected patients. J. Acquir. Immune Defic. Syndr. 41:137-139. [DOI] [PubMed] [Google Scholar]
- 31.Rogers, A. S., D. K. Futterman, A. B. Moscicki, C. M. Wilson, J. Ellenberg, and S. H. Vermund. 1998. The REACH Project of the Adolescent Medicine HIV/AIDS Research Network: design, methods, and selected characteristics of participants. J. Adolesc. Health 22:300-311. [DOI] [PubMed] [Google Scholar]
- 32.Rowland-Jones, S., J. Sutton, K. Ariyoshi, T. Dong, F. Gotch, S. McAdam, D. Whitby, S. Sabally, A. Gallimore, T. Corrah, et al. 1995. HIV-specific cytotoxic T-cells in HIV-exposed but uninfected Gambian women. Nat. Med. 1:59-64. [DOI] [PubMed] [Google Scholar]
- 33.Rowland-Jones, S. L., D. F. Nixon, M. C. Aldhous, F. Gotch, K. Ariyoshi, N. Hallam, J. S. Kroll, K. Froebel, and A. McMichael. 1993. HIV-specific cytotoxic T-cell activity in an HIV-exposed but uninfected infant. Lancet 341:860-861. [DOI] [PubMed] [Google Scholar]
- 34.Scherer, A., J. Frater, A. Oxenius, J. Agudelo, D. A. Price, H. F. Gunthard, M. Barnardo, L. Perrin, B. Hirschel, R. E. Phillips, and A. R. McLean. 2004. Quantifiable cytotoxic T lymphocyte responses and HLA-related risk of progression to AIDS. Proc. Natl. Acad. Sci. USA 101:12266-12270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schmitz, J. E., M. J. Kuroda, S. Santra, V. G. Sasseville, M. A. Simon, M. A. Lifton, P. Racz, K. Tenner-Racz, M. Dalesandro, B. J. Scallon, J. Ghrayeb, M. A. Forman, D. C. Montefiori, E. P. Rieber, N. L. Letvin, and K. A. Reimann. 1999. Control of viremia in simian immunodeficiency virus infection by CD8+ lymphocytes. Science 283:857-860. [DOI] [PubMed] [Google Scholar]
- 36.Schunk, D. 2008. A Markov chain Monte Carlo algorithm for multiple imputation in large surveys. Adv. Stat. Anal. 92:101-114. [Google Scholar]
- 37.Sette, A., and J. Sidney. 1998. HLA supertypes and supermotifs: a functional perspective on HLA polymorphism. Curr. Opin. Immunol. 10:478-482. [DOI] [PubMed] [Google Scholar]
- 38.Sette, A., and J. Sidney. 1999. Nine major HLA class I supertypes account for the vast preponderance of HLA-A and -B polymorphism. Immunogenetics 50:201-212. [DOI] [PubMed] [Google Scholar]
- 39.Sidney, J., B. Peters, N. Frahm, C. Brander, and A. Sette. 2008. HLA class I supertypes: a revised and updated classification. BMC Immunol. 9:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Singer, J. D. 1998. Using SAS PROC MIXED to fit multilevel models, hierarchical models, and individual growth models. J. Educ. Behav. Stat. 24:323-355. [Google Scholar]
- 41.Smith, D. K., D. L. Warren, D. Vlahov, P. Schuman, M. D. Stein, B. L. Greenberg, and S. D. Holmberg. 1997. Design and baseline participant characteristics of the Human Immunodeficiency Virus Epidemiology Research (HER) Study: a prospective cohort study of human immunodeficiency virus infection in US women. Am. J. Epidemiol. 146:459-469. [DOI] [PubMed] [Google Scholar]
- 42.Tang, J., S. Tang, E. Lobashevsky, A. D. Myracle, U. Fideli, G. Aldrovandi, S. Allen, R. Musonda, and R. A. Kaslow. 2002. Favorable and unfavorable HLA class I alleles and haplotypes in Zambians predominantly infected with clade C human immunodeficiency virus type 1. J. Virol. 76:8276-8284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tang, J., C. M. Wilson, S. Meleth, A. Myracle, E. Lobashevsky, M. J. Mulligan, S. D. Douglas, B. Korber, S. H. Vermund, and R. A. Kaslow. 2002. Host genetic profiles predict virological and immunological control of HIV-1 infection in adolescents. AIDS 16:2275-2284. [DOI] [PubMed] [Google Scholar]
- 44.Trachtenberg, E., B. Korber, C. Sollars, T. B. Kepler, P. T. Hraber, E. Hayes, R. Funkhouser, M. Fugate, J. Theiler, Y. S. Hsu, K. Kunstman, S. Wu, J. Phair, H. Erlich, and S. Wolinsky. 2003. Advantage of rare HLA supertype in HIV disease progression. Nat. Med. 9:928-935. [DOI] [PubMed] [Google Scholar]
- 45.Wilson, C. M., J. Houser, C. Partlow, B. J. Rudy, D. C. Futterman, and L. B. Friedman. 2001. The REACH (Reaching for Excellence in Adolescent Care and Health) project: study design, methods, and population profile. J. Adolesc. Health 29:8-18. [DOI] [PubMed] [Google Scholar]