Abstract
During productive infection, herpes simplex virus type 1 (HSV-1) induces the formation of discrete nuclear foci containing cellular chaperone proteins, proteasomal components, and ubiquitinated proteins. These structures are known as VICE domains and are hypothesized to play an important role in protein turnover and nuclear remodeling in HSV-1-infected cells. Here we show that VICE domain formation in Vero and other cells requires the HSV-1 immediate-early protein ICP22. Since ICP22 null mutants replicate efficiently in Vero cells despite being unable to induce VICE domain formation, it can be concluded that VICE domain formation is not essential for HSV-1 productive infection. However, our findings do not exclude the possibility that VICE domain formation is required for viral replication in cells that are nonpermissive for ICP22 mutants. Our studies also show that ICP22 itself localizes to VICE domains, suggesting that it could play a role in forming these structures. Consistent with this, we found that ICP22 expression in transfected cells is sufficient to reorganize the VICE domain component Hsc70 into nuclear inclusion bodies that resemble VICE domains. An N-terminal segment of ICP22, corresponding to residues 1 to 146, is critical for VICE domain formation in infected cells and Hsc70 reorganization in transfected cells. We previously found that this portion of the protein is dispensable for ICP22's effects on RNA polymerase II phosphorylation. Thus, ICP22 mediates two distinct regulatory activities that both modify important components of the host cell nucleus.
Soon after herpes simplex virus type 1 (HSV-1) infects a susceptible cell, its genome enters the nucleus. There, the ∼80 viral genes are transcribed by the host cell RNA polymerase II (Pol II). Viral gene expression occurs in a tightly regulated cascade in which there are three phases of gene expression: the immediate-early (IE), delayed-early (DE), and late (L) phases (reviewed in reference 50). Four of the five IE genes encode proteins that are important regulators of viral gene expression: ICP0, ICP4, ICP22, and ICP27. Production of these proteins is critical for expression of both DE and L genes. Many of the DE genes encode proteins directly or indirectly involved in viral DNA synthesis, and soon after their expression, viral DNA replication commences. The process of DNA replication drives high-level expression of L genes, most of which encode viral structural proteins. Late in infection, capsid assembly and genome packaging occur in the nucleus, and genome-containing capsids are transported to the cytoplasm for maturation and secretion.
HSV-1 replication in the nucleus is accompanied by a dramatic physical reorganization of this host cell organelle. Incoming viral genomes interact with ICP4 and ICP27 (13) and nucleate the formation of nuclear domain 10 (ND10/PML)-like bodies (12), which are subsequently disrupted by ICP0 (11). The viral genomes then associate with several other viral and cellular proteins, and these complexes ultimately develop into large structures called viral replication compartments (RCs) (30, 31, 46). RCs are believed to be sites of viral DNA synthesis (47), transcription (41), and capsid assembly (26). Host cell chromatin is excluded from RCs and becomes marginated at the nuclear periphery (47, 52). In addition, cellular splicing speckles coalesce and are pushed to the nuclear margins (36), the nuclear lamina is disrupted (55), and the nucleolus is significantly altered in its morphology (2) and composition (34). Late in infection, nuclear actin filaments form (14), possibly to promote the transport of assembled capsids to the nuclear envelope (16). Accompanying these many alterations, the nucleus approximately doubles in volume during the course of HSV-1 infection (37).
The massive reorganization of the HSV-1-infected nucleus likely involves host cell protein quality control systems that regulate the folding, assembly/disassembly, and degradation of host and viral proteins and protein complexes. The major set of host factors involved in such protein quality control events are the molecular chaperones (4, 29). It is thus noteworthy that HSV-1 infection induces nuclear foci which are enriched for cellular chaperones (5, 6). These structures are termed virus-induced chaperone-enriched (VICE) domains and usually form adjacent to RCs. They contain several cellular chaperones (Hsc70, Hsp70, Hsp40, and Hsp90), proteasomal components, ubiquitinated proteins, and at least one viral protein, capsid portal protein UL6. Although the function of VICE domains is not entirely clear, they are hypothesized to play a role in protein remodeling and quality control (6, 31, 32) and may contribute to RC formation (31) and transcriptional regulation (28). The reorganization of the host chaperone machinery could help the virus to reduce potential toxic effects of misfolded proteins, including the premature induction of apoptosis, which could limit viral replication. VICE domains may also provide viral polypeptides access to protein remodeling machinery to allow for their correct folding and assembly into multimeric protein complexes.
We have been investigating the functions of the HSV-1 IE protein ICP22. This protein is nonessential for HSV-1 replication in some cultured cells, including Vero cells, but in certain other cell lines it is required for efficient expression of a subset of L genes and for viral growth (38, 43, 54). Such restrictive cells include many rodent lines, human embryonic lung (HEL) cells, RAB-9 rabbit skin cells, and primary African green monkey kidney cells. ICP22 also alters the forms of RNA Pol II that are present in the infected cell. Two effects of ICP22 have been noted in this regard. First, ICP22 induces the loss of Pol II forms having phosphorylation on the serine-2 residue of the Pol II C-terminal domain (17, 18). Second, it induces the accumulation of an unusual hyperphosphorylated form of RNA Pol II known as RNA Pol III (18, 49). The latter but not the former effect is dependent on the viral protein kinase UL13 (33), which is thought to phosphorylate ICP22 during infection (45). UL13 is also required for ICP22's cell-type-dependent effects on viral L gene expression and growth (44).
One property of ICP22 that is not well understood is its ability to localize to small nuclear bodies (NBs) during infection (21, 27). To date, the nature of these NBs is unknown. Here, we report that these structures correspond to VICE domains. Moreover, we demonstrate that ICP22 is required for VICE domain formation and that its expression in uninfected cells is sufficient to reorganize the VICE domain component Hsc70 into nuclear inclusion bodies that resemble VICE domains. Our work thus reveals a surprising connection between ICP22 and the host cell chaperone machinery.
MATERIALS AND METHODS
Cells and viruses.
Vero (African green monkey kidney) cells were used for most experiments. In some immunofluorescence experiments, HEL cells were also used, while HeLa (human cervical cancer) cells were used in the coimmunoprecipitation analysis. All cells were originally obtained from the American Type Culture Collection. Cells were grown in Dulbecco modified Eagle medium supplemented with 5% (Vero) or 10% (HEL and HeLa) heat-inactivated fetal bovine serum (FBS), 50 U of penicillin/ml, and 50 g of streptomycin/ml. All tissue culture reagents except FBS were purchased from Invitrogen (Carlsbad, CA). FBS was purchased from Atlas Biologicals (Ft. Collins, CO). KOS1.1 (20) or KOS were the wild-type (WT) HSV-1 strains used in this study. The HSV-1 ICP22 mutants TF22, TF1.5, TF22BA, TF22AP, and TF22PS have all been recently described (1). Other mutants used were d22lacZ (33), d13lacZ (33), n212 (7), d27lacZ (48), and d120 (10). KOS1.1 and d13lacZ were grown in Vero cells, while d22lacZ, n212, d27lacZ, and d120 were grown in V22 (39), U2OS (58), V27 (48), and E5 (10) cells, respectively.
Infections and transfections.
Cells were infected in phosphate-buffered saline containing 0.1% glucose and 0.1% heat-inactivated newborn calf serum (NCS). After 1 h of viral adsorption at 37°C, the inoculum was replaced with medium 199 containing 2% heat-inactivated NCS, 50 U penicillin/ml, and 50 g of streptomycin/ml. Transfections were carried out as previously described (17) using Lipofectamine 2000 (Invitrogen). The plasmids pcDNA22, pcDNAUS1.5, pcDNA22-BA, pcDNA22-AP, and pcDNA22-PS (1, 17) were used to express WT or mutant forms of ICP22. Plasmid pCMVβ-c (40) was used to express β-galactosidase. In the experiment shown in Fig. 2, below, a transfection/infection protocol was used. Vero cells were transfected with pcDNA22 for 16 h and then infected with KOS at a multiplicity of infection (MOI) of 10 for 6 h.
FIG. 2.
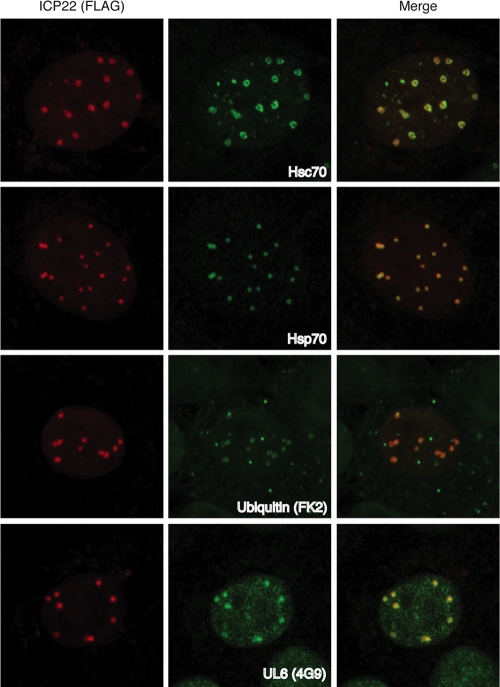
ICP22 NBs colocalize with markers of VICE domains. Vero cells transiently expressing FLAG-tagged ICP22 were infected with strain KOS at an MOI of 10 for 6 h. Cells were colabeled with antibodies recognizing the FLAG tag and one marker of VICE domains: Hsc70, Hsp70, mono/polyubiquitinated proteins (FK2 monoclonal antibody), or the HSV-1 capsid portal protein UL6 (4G9 monoclonal antibody). Imaging was performed using a 63× 1.4 objective lens and a Zeiss LSM 510 Meta confocal microscope. Images were collected at 2× digital zoom.
Immunofluorescence experiments.
For immunofluorescence experiments, cells were plated on glass coverslips and grown to ∼80% confluence prior to infection or transfection. For all immunofluorescence experiments except that shown in Fig. 2, analysis was done as follows. Cells were fixed in 3.7% formaldehyde for 10 min, followed by a 2-min permeabilization in cold acetone (46). To analyze Hsc70, a 1:100 dilution of a rat monoclonal antibody (catalog no. SPA-815; StressGen Corp.) was used as the primary antibody. When cells were costained for ICP22, a 1:10,000 dilution of anti-FLAG M2 mouse monoclonal antibody (Sigma) was included. When they were costained for ICP4, a 1:800 dilution of mouse monoclonal antibody H1114 (Rumbaugh-Goodwin Institute for Cancer Research, Plantation, FL) was included. Secondary staining was done with a 1:100 dilution of Cy2-conjugated donkey anti-rat IgG and a 1:2,000 dilution of Cy3-conjugated donkey anti-mouse IgG. When Escherichia coli β-galactosidase was detected in combination with Hsc70, the primary antibody included rabbit IgG specific for β-galactosidase (Rockland Immunochemicals, Gilbertsville, PA) diluted 1:200. In this case, secondary staining was done with a 1:100 dilution of Cy2-conjugated donkey anti-rat IgG and a 1:400 dilution of Cy3-conjugated goat anti-rabbit IgG. Samples were viewed and analyzed using an Olympus FluoView 1000 confocal laser scanning microscope.
For the experiment shown in Fig. 2, analysis was done as follows. Cells were fixed for 10 min in 4% paraformaldehyde, permeabilized for 10 min in 1% Triton X-100, and blocked overnight in 3% normal goat serum at 4°C. Cells were double labeled for 1 h at room temperature with antibodies recognizing the FLAG tag (polyclonal Sigma F7425; 1:100) and one of four markers of VICE domains: Hsc70 (monoclonal Stressgen SPA-815; 1:200), Hsp70 (monoclonal Stressgen SPA-810), ubiquitin (monoclonal FK2 Biomol PW-8810; 1:100), and the HSV-1 capsid portal protein UL6 (monoclonal 4G9 courtesy of Jay Brown, University of Virginia Health System, Charlottesville; 1:100). After extensive washing in 1× phosphate-buffered saline (PBS), cells were incubated with Alexa Fluor goat anti-rabbit 594 and goat anti-mouse or goat anti-rat 488 secondary antibodies (Molecular Probes) for 30 min followed by washing and mounting to slides. Imaging was performed using a 63× planapochromat objective lens (numerical aperture, 1.4) and a Zeiss LSM 510 Meta confocal microscope equipped with argon and helium lasers.
Estimation of relative particle-to-PFU ratios in HSV-1 stocks.
To estimate relative particle-to-PFU ratios in viral stocks, stocks were diluted in sterile water to give 107 PFU (as determined by plaque assay on the appropriate permissive cell line). An equal volume of 2× SDS-PAGE sample buffer (1× SDS-PAGE sample buffer is 62 mM Tris-HCl [pH 6.8], 2% SDS, 10% [vol/vol] glycerol, 0.002% bromophenol blue) was then added. The samples were boiled for 2 min and then analyzed by immunoblotting as described previously (1) to determine the levels of capsid protein VP5 and tegument protein VP16. To detect VP5, a 1:1,000 dilution of a mouse monoclonal antibody specific for VP5 (Abcam Inc.) was used. To detect VP16, a 1:10,000 dilution of a rabbit antiserum (57) was used.
Coimmunoprecipitations.
HeLa cells were grown to confluence in 150-cm2 tissue culture flasks and either mock infected or infected with HSV-1. At 6 h postinfection (hpi), the cells were rinsed twice with PBS and then lysed at room temperature for 30 min with shaking in 2 ml of lysis buffer (50 mM Tris-HCl [pH 7.4], 150 mM NaCl, 1 mM EDTA, 1% Triton X-100). A protease inhibitor cocktail (Roche Diagnostics, Indianapolis, IN) was added to the PBS and lysis buffer prior to use at the concentrations recommended by the manufacturer. After lysis, extracts were poured into tubes and frozen at −80°C. To immunoprecipitate FLAG-tagged proteins, M2 anti-FLAG affinity gel (product code A220; Sigma) was used, following the manufacturer's recommended protocol. Briefly, for each immunoprecipitation, 40 μl of washed affinity gel was combined in an Eppendorf tube with 1 ml of lysate and mixed overnight at 4°C. The resin was pelleted in a microcentrifuge and washed three times with 0.5 ml of Tris-buffered saline (50 mM Tris-HCl [pH 7.4], 150 mM NaCl). To elute bound protein, 20 μl of 2× SDS-PAGE sample buffer was added and the samples were boiled for 3 min. The resin was repelleted and the supernatant was analyzed by SDS-PAGE and immunoblotting. Blots were probed with rat anti-Hsc70 antibody, diluted 1:10,000, or with rabbit anti-ICP22 antisera (a gift from John Blaho) diluted 1:1,000.
RESULTS
ICP22 colocalizes with Hsc70 in VICE domains.
ICP22 localizes predominantly to the nucleus during infection, in part to small NBs of unknown identity (21, 27). These can be seen in the immunofluorescence micrograph shown in Fig. 1A (two are indicated by arrows), wherein FLAG-specific serum was used to stain ICP22 in cells infected with TF22 (1), a phenotypically WT HSV-1 strain that encodes a FLAG-tagged ICP22 (Fig. 1B). We first considered whether the ICP22 NBs might correspond to nuclear splicing speckles, which are reorganized in HSV-1-infected cells (36) and contain Ser-2-phosphorylated RNA Pol II (18). However, immunofluorescence analysis indicated that ICP22 NBs and splicing speckles are distinct structures (data not shown). We next tested whether ICP22 NBs could correspond to VICE domains. To determine this, TF22- or mock-infected Vero cells were fixed at 6 hpi and processed for immunofluorescence. The cells were stained for both FLAG and Hsc70, the latter being a marker for VICE domains (6). Consistent with our expectation, no FLAG staining was seen in mock-infected cells (Fig. 1C, panel a), while Hsc70 was detected in both the cytoplasm and nucleus (panel b), as previously reported (6). Also as expected, Hsc70 was dramatically rearranged in cells infected by TF22, with much of the Hsc70 being redistributed into nuclear VICE domains (panel f). Although much of the ICP22 in TF22-infected nuclei was diffuse, some of it localized to NBs (panel e), and these corresponded to VICE domains (panels g and h). These results suggest that the ICP22-containing NBs formed in WT HSV-1 infection are VICE domains.
FIG. 1.
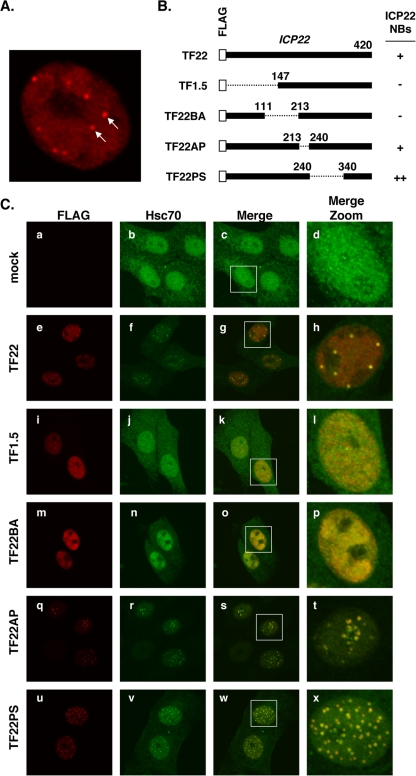
Colocalization of ICP22 and VICE domains. (A) Immunofluorescent staining of ICP22 in HSV-1-infected cells. Vero cells were infected at an MOI of 3 PFU/cell with HSV-1 strain TF22, encoding a FLAG-tagged ICP22. Cells were fixed at 6 hpi and stained with a FLAG-specific antiserum. The arrows point to two of the several NB structures that stained positively for ICP22. (B) Diagram of FLAG-tagged ICP22 polypeptides encoded by TF22 and related mutants. The propensity of the viruses to localize ICP22 to NBs (from reference 1) is indicated. (C) Localization of ICP22 and Hsc70 during infection. Vero cells were mock infected or infected with the indicated viruses at an MOI of 3 PFU/cell. Cells were fixed at 6 hpi and processed for immunofluorescence using antibodies specific for FLAG and Hsc70. The right-hand column shows enlargements of the images contained within the white squares.
To more closely examine the relationship between ICP22 NBs and VICE domains, we compared the localization of ICP22 with that of other known markers of VICE domains. To do this, Vero cells were transfected with plasmid pcDNA22, encoding FLAG-tagged ICP22, and infected 16 h later with WT HSV-1. Cells were fixed at 6 hpi and examined by immunofluorescence (Fig. 2). As seen above, ICP22 NBs colocalized with nuclear foci of Hsc70. ICP22 NBs also colocalized with two other VICE domain markers, the cellular chaperone Hsp70 and the HSV-1 capsid portal protein UL6 (6). Staining with monoclonal antibody FK2 (19) demonstrated that ICP22 also partially colocalized with mono- and polyubiquitinated proteins, which are present in VICE domains (6). The results of this experiment demonstrate that ICP22 NBs correspond to VICE domains. Thus, we have identified ICP22 as a VICE domain component.
In the experiment shown in Fig. 1, we also examined ICP22 and Hsc70 localization in cells infected with a panel of ICP22 deletion mutants that encode FLAG-tagged polypeptides (1) (Fig. 1B). We found that two of these mutants, TF1.5 and TF22BA, express ICP22 polypeptides that localize to the nucleus but not to NBs (Fig. 1C, panels i and m, respectively), while another mutant, TF22PS, expresses an ICP22 molecule that localizes to an unusually large number of NBs (panel u). Interestingly, the TF1.5 and TF22BA infections did not show evidence of VICE domain formation (panels j to l and n to p, respectively). However, in this and other experiments, we observed that TF22BA does have an effect on Hsc70, in that Hsc70 cytoplasmic staining is consistently diminished by infection with this mutant (panel n and data not shown). In contrast, VICE domains were formed in the TF22AP and TF22PS infections (panels r to t and v to x, respectively). Moreover, as in the WT infection, the ICP22 NBs largely colocalized with VICE domains, although a few Hsc70 foci in the TF22AP and TF22PS infections did not appear to stain for ICP22 (panels t and x, respectively, and data not shown). TF22AP also had an unusual phenotype, in that in some cells the ICP22 NBs/VICE domains appeared to coalesce in a small area of the nucleus. TF22PS, on the other hand, displayed an elevated number of VICE domains (panel v), consistent with its more numerous ICP22 NBs. This analysis demonstrates that ICP22 can dramatically influence the induction, location, and number of VICE domains, suggesting that it might play a role in the formation of these structures.
ICP22 is required for VICE domain formation.
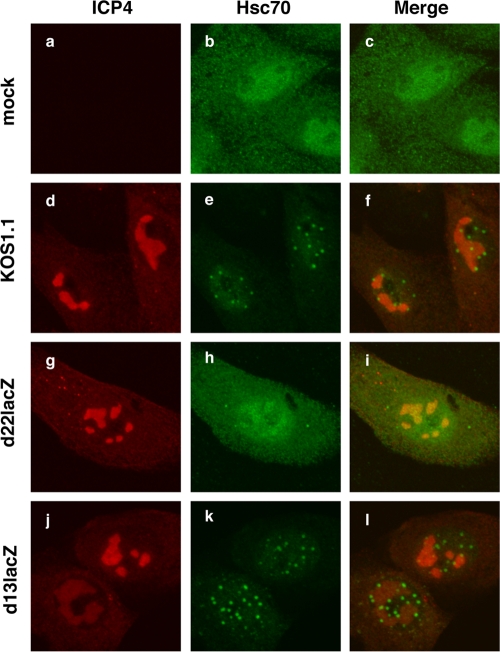
To determine whether ICP22 is involved in VICE domain formation, we examined the induction of VICE domains in cells infected with an ICP22 null mutant, d22lacZ (33). Since many of ICP22's functions are dependent on UL13, we also analyzed a UL13 mutant, d13lacZ (33). Mock- or HSV-1-infected Vero cells were fixed at 6 hpi and processed for immunofluorescence. To monitor VICE domains, the cells were stained for both Hsc70 and the IE protein ICP4, the latter being a marker for viral RCs (22). As mentioned previously, VICE domains are often found at the periphery of RCs (6, 28). As expected, Hsc70 was found in VICE domains in WT HSV-1-infected cells (Fig. 3d to f), and the level of cytoplasmic Hsc70 was reduced compared to mock-infected cells. Similar results were seen in d13lacZ-infected cells (Fig. 3j to l). In contrast, VICE domains did not form in the d22lacZ infection, despite well-developed RCs (Fig. 3g to i). In addition, the level of cytoplasmic Hsc70 remained high, similar to the mock-infected cells. We conclude that ICP22 but not UL13 is required for VICE domain formation in Vero cells, at least at 6 hpi.
FIG. 3.
ICP22 is required for formation of VICE domains. Vero cells were mock infected or infected at an MOI of 3 with WT HSV-1 or strains with mutations in ICP22 (d22lacZ) or UL13 (d13lacZ). At 6 hpi, the cells were fixed and VICE domain formation was assessed by immunofluorescence using antibodies directed against ICP4 and Hsc70.
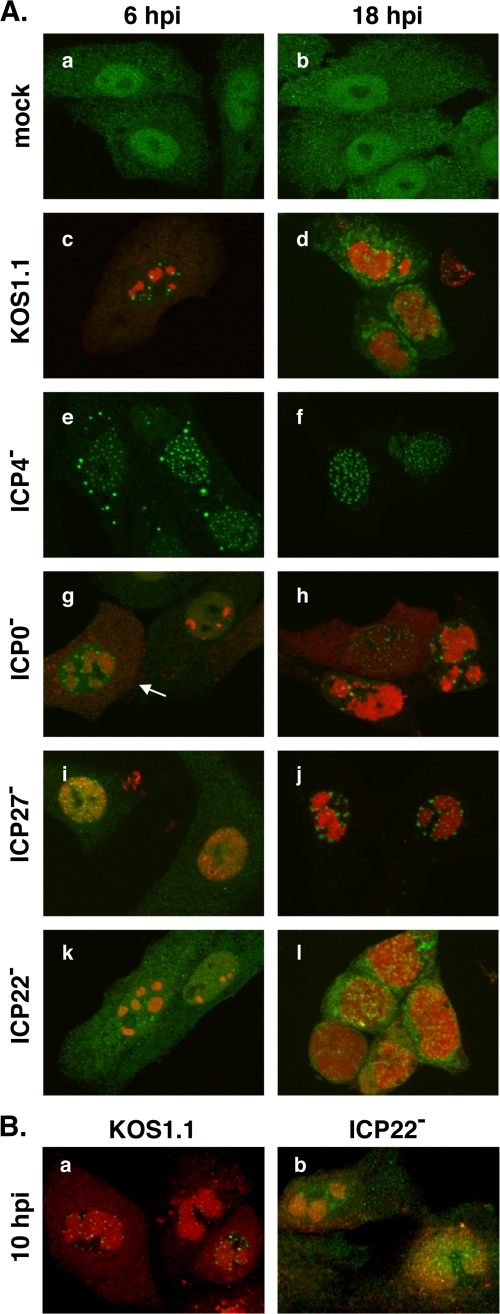
Two other IE proteins, ICP0 and ICP27, have previously been implicated in VICE domain formation (6, 28). In the case of ICP0, the requirement is not absolute, as it was shown that VICE domains form in ICP0 mutant-infected cells at late times after infection. To directly compare the roles of various IE proteins in VICE domain formation, we utilized HSV-1 null mutants in ICP4, ICP0, ICP27, and ICP22. Mock- or virus-infected cells were examined for VICE domains at both an early (6 hpi) and a late (18 hpi) time point by ICP4/Hsc70 costaining. At 6 hpi, WT HSV-1-infected cells formed VICE domains as expected (Fig. 4A, panel c). Nuclear inclusions of Hsc70 were also clearly visible in cells infected with the ICP4 mutant (panel e), as were some cytoplasmic inclusions. These nuclear inclusions resembled VICE domains, but they were more regularly spaced throughout the nucleus and more numerous. Their more regular distribution may be because ICP4 null mutants are not expected to form RCs due to their profound defect in viral DE/L gene expression (10). In contrast to the ICP4 mutant infection, Hsc70 foci/VICE domains were only observed in a fraction of the cells infected by the ICP0 mutant (Fig. 4A, panel g; for example, in the cell indicated by the arrow). VICE domain formation was also deficient in cells infected with an ICP27 null mutant (panel i). Moreover, and as previously reported (28), RCs formed poorly at this time point in ICP27 mutant-infected cells. Finally, as we observed in our earlier experiment, VICE domains did not form at 6 hpi in d22lacZ-infected cells (panel k), and the level of cytoplasmic Hsc70 remained high, similar to the mock infection. Thus, at 6 hpi, VICE domain formation is deficient in cells infected with ICP0, ICP27, and ICP22 mutants.
FIG. 4.
VICE domain formation in cells infected with IE mutants. (A) Vero cells were mock infected or infected at an MOI of 3 with WT strain KOS1.1 or null mutants in ICP4 (d120), ICP0 (n212), ICP27 (d27lacZ), or ICP22 (d22lacZ). The cells were fixed at 6 and 18 hpi, and VICE domain formation was analyzed by immunofluorescence as in Fig. 3. The arrow in panel g indicates an n212-infected cell nucleus that is positive for VICE domains. (B) Vero cells were infected as for panel A, with KOS1.1 or d22lacZ. The cells were fixed at 10 hpi and analyzed by immunofluorescence as for panel A.
We next examined VICE domain formation at 18 hpi (Fig. 4, right-hand panels). The results were quite different from those at the 6-h time point in that VICE domains were clearly seen in most cells infected with the ICP0 and ICP27 mutants (panels h and j, respectively). Hsc70-containing nuclear inclusions were also clearly present in ICP4 mutant-infected cells (panel f). In contrast, VICE domains were still not apparent in the ICP22 mutant-infected cells (panel l), although the rounding of the cells due to the cytopathic effects of infection made this somewhat difficult to assess. In another experiment, we compared Hsc70 staining in WT- and d22lacZ-infected Vero cells at 10 hpi (Fig. 4B). Hsc70 nuclear foci were lacking in most of the cells infected with the ICP22 mutant (Fig. 4B, panel b), although some cells had minute nuclear foci that showed no clear spatial relationship to RCs. In contrast, VICE domains adjacent to RCs were evident in the WT-infected cells (Fig. 4B, panel a). We also examined the role of ICP22 in VICE domain formation in HEL cells, which are nonpermissive for ICP22 mutants. Very similar results were obtained, i.e., WT- but not d22lacZ-infected cells formed VICE domains (data not shown). Together, the results of these experiments demonstrate that ICP22 is strictly required for the formation of VICE domains during HSV-1 infection. In contrast, ICP27 and ICP0 are not absolutely required for VICE domain formation, but they do help these structures to form during the early stages of infection.
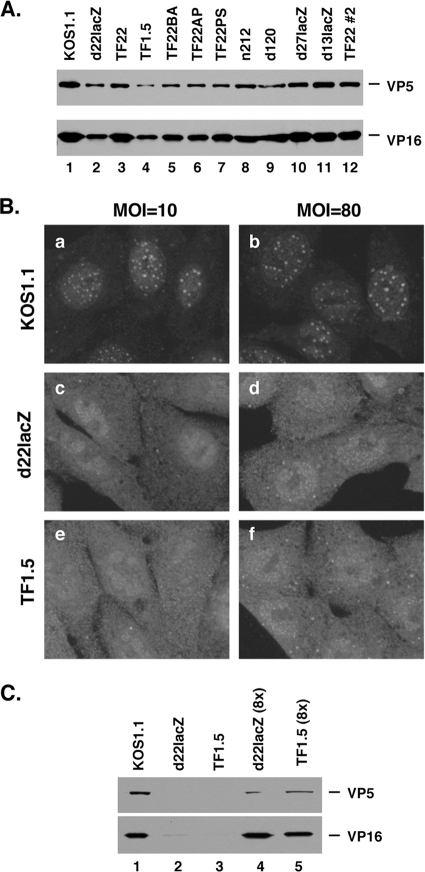
The inability of ICP22 mutants to form VICE domains is not due to altered particle-to-PFU ratios.
It has been observed that there is a correlation between the particle-to-PFU ratio of an HSV-1 stock and its ability to induce VICE domain formation, i.e., stocks with higher ratios tend to be more efficient at forming VICE domains (32). As ICP22 can affect virion composition and infectivity (39), we considered the possibility that ICP22 mutant stocks might be defective at inducing VICE domains because they have abnormal particle-to-PFU ratios. To investigate this, we used immunoblotting to estimate the relative particle-to-PFU ratio of the various viral stocks used in our experiments. Specifically, we assessed the relative levels of capsid protein VP5 and tegument protein VP16 in a defined amount of infectious virus (107 PFU; determined by plaque assay on the appropriate permissive cell line). This analysis, shown in Fig. 5A, indicated that the d22lacZ and TF1.5 stocks have somewhat lower particle-to-PFU ratios than the WT KOS1.1 stock and most of the other mutant stocks. Thus, it is conceivable that these two mutants are defective at inducing VICE domains (Fig. 1 and 3) because fewer than the WT number of viral particles are delivered to the host cell upon infection. To test this hypothesis, we asked whether d22lacZ or TF1.5 can trigger the formation of VICE domains if the MOI is raised so that significantly more viral particles are introduced into the cell. Thus, Vero cells were infected by WT HSV-1 or the ICP22 mutants at MOIs of 10 or 80, and VICE domain formation at 6 hpi was assessed by Hsc70 staining. VICE domains were clearly evident in WT HSV-1-infected cells at both MOIs (Fig. 5B, panels a and b). However, they were not observed at either MOI in the d22lacZ or TF1.5 infections (panels c and d and panels e and f, respectively). Control immunoblotting analysis showed that increasing the amount of d22lacZ or TF1.5 viral stocks by 8-fold increased the VP5/VP22 levels to approximately that seen in the WT stock (Fig. 5C). Thus, the inability of ICP22 mutants to induce VICE domain formation cannot be explained by altered particle/PFU ratios.
FIG. 5.
The inability of ICP22 mutants to induce VICE domains is not due to altered particle-to-PFU ratios. (A) Determination of relative particle-to-PFU content in various viral stocks. Aliquots of 1 × 107 PFU of viral stocks, determined by plaque assay on the appropriate permissive cell line, were analyzed by SDS-PAGE and immunoblotting for the major capsid protein VP5 and tegument protein VP16. Note that two different stocks of TF22 were analyzed. (B) Vero cells were infected with the viruses shown, at an MOI of 10 or 80. At 6 hpi, cells were fixed and analyzed for VICE domains by Hsc70 staining. (C) Determination of VP5 and VP16 content in 1 × 107 PFU of KOS1.1 (lane 1), d22lacZ (lane 2), or TF1.5 (lane 3) or in 8 × 107 PFU of d22lacZ (lane 4) or TF1.5 (lane 5).
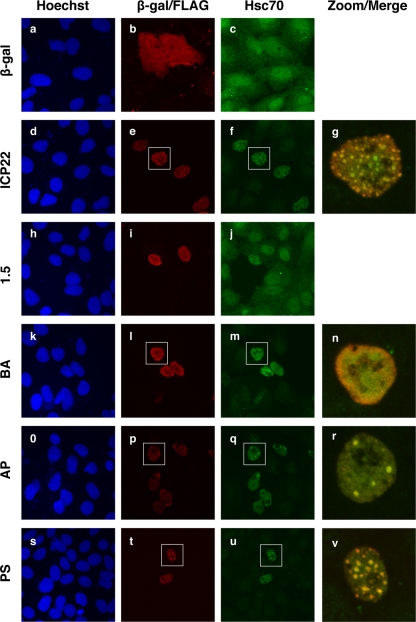
ICP22 is sufficient to reorganize Hsc70 in uninfected cells.
The experiments described above demonstrated that ICP22 can have striking effects on the localization of Hsc70 in infected cells. We therefore asked whether ICP22 can affect Hsc70 localization in uninfected cells. To address this, we utilized a set of expression plasmids encoding either FLAG-tagged ICP22 or the various mutant forms of it shown in Fig. 1B (1). These plasmids, or a control plasmid encoding E. coli β-galactosidase, were transfected into Vero cells. One day later, the cells were fixed and stained for Hsc70 and FLAG (or β-galactosidase). In the control transfection, the pattern of Hsc70 staining in the β-galactosidase-expressing cells could not be distinguished from the nonexpressing cells (Fig. 6a to c). This indicates that Hsc70 localization is not affected by β-galactosidase expression from the transfected plasmid. However, in cells transfected with the ICP22 construct (Fig. 6d to f), Hsc70 staining was dramatically altered, with significantly increased staining of Hsc70 in the nuclei of ICP22-expressing cells. Similar results were seen in cells transfected with the BA, AP, and PS mutant constructs (Fig. 6m, q, and u, respectively). (Note that the images shown in panels f, m, q, and u were captured so as to optimize Hsc70 staining in the most intensely staining cells, those being the cells expressing ICP22 polypeptides. Hsc70 staining is thus difficult to see in the nonexpressing cells.) The results with the TF1.5-encoded form of ICP22 (designated as 1.5 in Fig. 6) were quite different, however, as this mutant did not appear to affect Hsc70 localization Fig. 6h to j).
FIG. 6.
ICP22 can reorganize Hsc70 in transfected cells. Vero cells were transfected with expression plasmids encoding β-galactosidase, ICP22, or mutant forms of ICP22 as illustrated in Fig. 1B. One day later, the cells were fixed and processed for immunofluorescence to detect the expressed proteins and Hsc70. Note that expression of WT ICP22 as well as the BA, AP, and PS mutant forms of ICP22 increased the nuclear staining of Hsc70. For these constructs, the far right panels show merged enlargements of the images contained within the white squares.
We and others have previously shown that transiently expressed ICP22 partially localizes to nuclear foci that resemble the ICP22 NBs seen during viral infection (1, 8, 51, 56). These foci can be seen in Fig. 6e in the cells expressing ICP22. Interestingly, these ICP22 foci also contain Hsc70 and physically resemble VICE domains (Fig. 6f and g). Similar results were seen in cells expressing the AP and PS ICP22 mutant constructs (Fig. 6q and r and panels u and v, respectively). However, no Hsc70 foci were seen in cells transfected with the BA mutant. This is consistent with the inability of this mutant polypeptide to form ICP22 NBs in either transfected or infected cells (1). In summary, these transfection results indicate that expression of ICP22 or certain mutant forms of it is sufficient to both increase the overall nuclear staining of Hsc70 as well to induce its localization to ICP22-containing nuclear inclusions that resemble VICE domains.
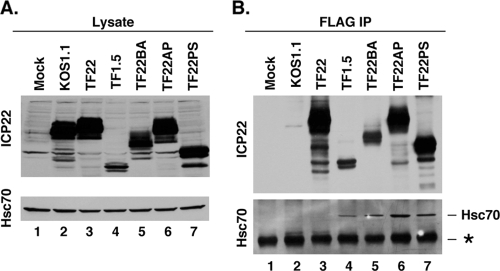
Interaction of Hsc70 with mutant ICP22 polypeptides.
Given the striking effects of ICP22 on the localization of Hsc70, we considered whether ICP22 and Hsc70 might physically interact. To test this, we carried out a coimmunoprecipitation experiment. HeLa cells were mock infected or infected with WT HSV-1, TF22, or the various FLAG-tagged mutants. At 6 hpi, total cell lysates were prepared. Analysis of the lysates by immunoblotting indicated that all of the infected cells expressed the expected forms of ICP22 (1) (Fig. 7A). In addition, Hsc70 levels were comparable in all lysates, consistent with previous analyses that indicated HSV-1 infection does not affect the abundance of Hsc70 (6, 28). We next used anti-FLAG monoclonal antibody affinity resin to immunoprecipitate the epitope-tagged proteins. Immunoblotting analysis of the precipitates with ICP22-specific antisera indicated that the immunoprecipitation was successful, as all of the FLAG-tagged ICP22 proteins were successfully pulled down (Fig. 7B, lanes 3 to 7). In contrast, only background levels of untagged ICP22 from KOS1.1 were precipitated (lane 2). Analysis of the immunoprecipitates with an Hsc70 antibody revealed that Hsc70 was present in the samples from all of the ICP22 mutants (lanes 4 to 7). However, Hsc70 was not detected in the TF22 immunoprecipitate (lane 3), suggesting that the full-length protein does not interact with Hsc70. It is formally possible that the FLAG tag interferes with the folding of full-length ICP22, or its ability to interact with Hsc70, thus explaining the negative binding results in the coimmunoprecipitation. However, this seems unlikely because the FLAG tag clearly does not interfere with the ability of mutant ICP22s to interact with Hsc70. Additionally, our prior work showed that full-length FLAG-tagged ICP22 is similar to untagged WT ICP22 in its ability to promote viral growth and gene expression in a restrictive cell line and to mediate the modification of RNA Pol II (1). Moreover, our present work shows that FLAG-ICP22 is capable of mediating striking effects on Hsc70 in both infected (Fig. 1) and transfected (Fig. 6) cells. Thus, the preponderance of evidence suggests that full-length, FLAG-tagged ICP22 is fully functional and correctly folded. Therefore, the most straightforward interpretation of our coimmunoprecipitation results is that the mutant forms of ICP22 stably interact with Hsc70, whereas the WT protein does not. If this is correct, then the data indicate that the ability of the various mutant and WT ICP22 polypeptides to induce VICE domain formation and reorganize Hsc70 does not correlate with their Hsc70 binding. This suggests that ICP22's effect on VICE domains and Hsc70 does not involve a physical interaction with Hsc70.
FIG. 7.
Interaction of Hsc70 with mutant forms of ICP22. HeLa cells were mock infected or infected at an MOI of 10 with the mutants shown. At 6 hpi, total cell lysates were made and analyzed (A). (B) FLAG-tagged proteins were immunoprecipitated by incubation with affinity resin bearing anti-FLAG monoclonal antibody M2. After washing the immunoprecipitates, bound proteins were released and analyzed by immunoblotting with ICP22- or Hsc70-specific antisera. The asterisk in panel B denotes a nonspecific band corresponding to the mouse IgG heavy chain.
DISCUSSION
VICE domain formation is induced by ICP22 and is not required for productive HSV-1 infection.
At present, the function of VICE domains during HSV-1 productive infection is unknown. These structures have been suggested to serve a protein quality control role in the infected cell nucleus, perhaps facilitating the major nuclear alterations that accompany viral replication (5, 6). The present study has resulted in two significant findings with regard to the formation and function of VICE domains. First, we have discovered that the IE protein ICP22 is required for VICE domain formation. In contrast, the IE proteins ICP0 and ICP27 help VICE domains to form early in infection, consistent with the results of earlier studies (6, 28), but they are not absolutely required for the assembly of these structures. Second, since ICP22 mutants replicate efficiently in Vero cells despite being unable to induce VICE domain formation, it can be concluded that VICE domains are not essential for the completion of a productive HSV-1 infection. Consistent with this, it has been shown that varicella-zoster virus (VZV), another human alphaherpesvirus, can replicate in human cells without forming VICE domains (24, 25).
It is important to point out that our results do not exclude the possibility that VICE domains could play an important role in HSV-1 replication under some conditions. Although HSV-1 ICP22 mutants grow productively in Vero and some other cells, they are significantly defective for growth in several other cell lines (38, 43, 54) and show striking replication defects in animal models of infection (39, 42, 54). Even in Vero cells, ICP22 mutants are significantly defective when infections are carried out in cells synchronized in the S phase (38). Thus, VICE domain formation may be critical for replication in certain cell types or under particular conditions of infection. Consistent with this possibility, we have found that the ICP22 mutant TF1.5, which is defective for VICE domain formation, replicates very poorly in HEL cells, similar to an ICP22 null mutant (1). Further work, including additional genetic studies, will now be required to determine if and how the VICE domain-inducing activity of ICP22 impacts the viral replication cycle.
Identification of ICP22 NBs as VICE domains.
Work from Roizman's laboratory originally showed that ICP22 localizes to small NBs during HSV-1 infection (21, 27). It was demonstrated that ICP22 preferentially localizes to these foci early in infection and then begins to accumulate in RCs as viral DNA synthesis begins. It was also shown that the ICP22 foci contain the HSV-1 late proteins UL3 and UL4 (21, 35). In this study, we have discovered that ICP22 NBs correspond to VICE domains, which were originally described by Burch and Weller and demonstrated to contain the HSV-1 portal protein UL6 (6). Thus, the available evidence indicates that VICE domains contain at least four viral proteins: ICP22, UL3, UL4, and UL6. As discussed below, it is likely that ICP22 plays a direct role in the formation of VICE domains. The roles of UL3, UL4, and UL6 in VICE domains and the features that cause these particular proteins to localize there are presently unknown.
Modulation of Hsc70 localization by ICP22.
Hsc70 is a constitutively and widely expressed member of the Hsp70 family of molecular chaperones (9). As it appears to be a major constituent of VICE domains (6), we used it in our studies as a marker for these structures. Interestingly, recent data indicate that Hsc70 activity is required for efficient HSV-1 replication (28; C. Livingston and S. Weller, unpublished results). It is therefore intriguing that ICP22 can induce striking changes to Hsc70 localization in both infected and uninfected cells. In both contexts, one of its major effects appears to be to redistribute Hsc70 from the cytoplasm to the nucleus. Such a redistribution could be an important biological function of ICP22, allowing HSV-1 to recruit cytoplasmic Hsc70 into the nucleus to support its replication. In addition, ICP22 induces the condensation of Hsc70 into nuclear foci. The fact that the TF22BA-encoded ICP22 can trigger Hsc70 nucleocytoplasmic distribution without inducing nuclear foci of Hsc70 is interesting. This suggests that ICP22 mediates two distinct effects on Hsc70, one affecting its nucleocytoplasmic distribution and the other influencing its condensation into nuclear bodies.
At present, the mechanism(s) by which ICP22 affects Hsc70 localization and VICE domain formation is unknown. Although we initially suspected that these effects involve a direct interaction between ICP22 and Hsc70, our coimmunoprecipitation experiments failed to provide evidence for this. We demonstrated that Hsc70 interacts with mutant forms of ICP22, but we did not see evidence of an interaction with WT ICP22. Since Hsc70 bound to all mutant forms of ICP22, it is possible that the interactions detected reflect the chaperone activity of Hsc70, which might bind to mutant proteins because they are partially misfolded. It is conceivable that a more subtle and/or transient physical interaction between ICP22 and Hsc70 is responsible for the VICE domain formation/Hsc70 relocalization, but this interaction was not detected in our coimmunoprecipitation assay. Alternatively, it is possible that ICP22 affects Hsc70 through a mechanism that does not involve a physical interaction. In this regard, it is interesting that cellular stresses such as heat shock can prevent the normal nucleocytoplasmic shuttling of Hsc70 and cause its accumulation in the nucleus and nucleolus (23). It is possible that ICP22 carries out an activity that leads to cellular stress and therefore has a similar effect on Hsc70. It is notable that another viral protein, the human papillomavirus late capsid protein 2, can also induce the nuclear accumulation of Hsc70, resulting in its appearance in ND10 bodies (15). In this case, the effect appears to involve a physical interaction between L2 and Hsc70.
The N-terminal region of ICP22 is critical for VICE domain formation and Hsc70 effects.
We used a panel of HSV-1 mutants to map the sequences in ICP22 that are required for its effects on VICE domains and Hsc70. The abilities of ICP22 viral mutants to induce VICE domain formation correlate with their ability to localize ICP22 to NBs (Fig. 1B and C). Thus, only TF1.5 and TF22BA were unable to induce VICE domain formation. However, the TF22BA mutant does have an effect on Hsc70, in that it decreases the level of cytoplasmic Hsc70 staining. The only viral mutant completely unable to affect Hsc70 is TF1.5, which encodes an ICP22 molecule lacking residues 1 to 146. Likewise, in our plasmid transfection experiments, this mutant was the only construct unable to affect Hsc70 localization. Thus, both the virus and plasmid analyses implicate the N-terminal 146 residues in ICP22's effects on VICE domains and Hsc70. It is noteworthy that a single amino acid alteration in this region of ICP22 at residue 34 determines the severity of HSV-1 ocular disease in mice (3), raising the possibility that ICP22's effects on VICE domains/Hsc70 can impact viral pathogenesis. It is also worth pointing out that the N-terminal 146 residues of ICP22 are outside the central portion of the polypeptide that is widely conserved among homologous proteins encoded by the alphaherpesviruses (53). This suggests that the ability of HSV-1 ICP22 to modulate Hsc70 localization may be a function of ICP22 that is not widely shared among alphaherpesviral ICP22 homologs. Consistent with this, we have found that transient expression of ORF63, the VZV homolog of ICP22, does not detectably alter Hsc70 cellular localization in Vero cells (T. Bastian and S. Rice, unpublished data).
ICP22 possesses multiple functional domains.
In this work, we have shown that an N-terminal region of ICP22 corresponding to residues 1 to 146 is critical for ICP22's ability to induce VICE domain formation during infection, as well as to modulate Hsc70 localization in uninfected cells. In contrast, we previously found that this N-terminal region is not required for ICP22's effects on RNA Pol II phosphorylation (1). Rather, the RNA Pol II-modifying function of ICP22 maps to the C-terminal half of the protein. Our studies thus demonstrate that ICP22 mediates two genetically separable activities that impact host cell systems involved in protein quality control and transcription, respectively. Therefore, similar to other HSV-1 IE proteins, ICP22 can be considered a multifunctional and multidomain regulatory protein.
Acknowledgments
We thank John Blaho for the gift of ICP22 antiserum and Tony Smith for technical assistance with the confocal microscopy. We also are grateful to the members of the Rice, Schiff, and Bresnahan laboratories for helpful discussions.
This research was supported by Public Health Service grants AI42737 (to S.A.R.) and AI21747 (to S.K.W.).
Footnotes
Published ahead of print on 23 December 2009.
REFERENCES
- 1.Bastian, T. W., and S. A. Rice. 2009. Identification of sequences in herpes simplex virus type 1 ICP22 that influence RNA polymerase II modification and viral late gene expression. J. Virol. 83:128-139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Besse, S., and F. Puvion-Dutilleul. 1996. Intranuclear retention of ribosomal RNAs in response to herpes simplex virus type 1 infection. J. Cell Sci. 109:119-129. [DOI] [PubMed] [Google Scholar]
- 3.Brandt, C. R., A. W. Kolb, D. D. Shah, A. M. Pumfery, R. L. Kintner, E. Jaehnig, and J. J. Van Gompel. 2003. Multiple determinants contribute to the virulence of HSV ocular and CNS infection and identification of serine 34 of the US1 gene as an ocular disease determinant. Invest. Ophthalmol. Vis. Sci. 44:2657-2668. [DOI] [PubMed] [Google Scholar]
- 4.Bukau, B., J. Weissman, and A. Horwich. 2006. Molecular chaperones and protein quality control. Cell 125:443-451. [DOI] [PubMed] [Google Scholar]
- 5.Burch, A. D., and S. K. Weller. 2005. Herpes simplex virus type 1 DNA polymerase requires the mammalian chaperone hsp90 for proper localization to the nucleus. J. Virol. 79:10740-10749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Burch, A. D., and S. K. Weller. 2004. Nuclear sequestration of cellular chaperone and proteasomal machinery during herpes simplex virus type 1 infection. J. Virol. 78:7175-7185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cai, W., T. L. Astor, L. M. Liptak, C. Cho, D. M. Coen, and P. A. Schaffer. 1993. The herpes simplex virus type 1 regulatory protein ICP0 enhances virus replication during acute infection and reactivation from latency. J. Virol. 67:7501-7512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cun, W., M. Hong, L. D. Liu, C. H. Dong, J. Luo, and Q. H. Li. 2006. Structural and functional characterization of herpes simplex virus 1 immediate-early protein infected-cell protein 22. J. Biochem. 140:67-73. [DOI] [PubMed] [Google Scholar]
- 9.Daugaard, M., M. Rohde, and M. Jaattela. 2007. The heat shock protein 70 family: highly homologous proteins with overlapping and distinct functions. FEBS Lett. 581:3702-3710. [DOI] [PubMed] [Google Scholar]
- 10.DeLuca, N. A., A. M. McCarthy, and P. A. Schaffer. 1985. Isolation and characterization of deletion mutants of herpes simplex virus type 1 in the gene encoding immediate-early regulatory protein ICP4. J. Virol. 56:558-570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Everett, R. D., and G. G. Maul. 1994. HSV-1 IE protein Vmw110 causes redistribution of PML. EMBO J. 13:5062-5069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Everett, R. D., and J. Murray. 2005. ND10 components relocate to sites associated with herpes simplex virus type 1 nucleoprotein complexes during virus infection. J. Virol. 79:5078-5089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Everett, R. D., G. Sourvinos, and A. Orr. 2003. Recruitment of herpes simplex virus type 1 transcriptional regulatory protein ICP4 into foci juxtaposed to ND10 in live, infected cells. J. Virol. 77:3680-3689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Feierbach, B., S. Piccinotti, M. Bisher, W. Denk, and L. W. Enquist. 2006. Alpha-herpesvirus infection induces the formation of nuclear actin filaments. PLoS Pathog. 2:e85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Florin, L., K. A. Becker, C. Sapp, C. Lambert, H. Sirma, M. Muller, R. E. Streeck, and M. Sapp. 2004. Nuclear translocation of papillomavirus minor capsid protein L2 requires Hsc70. J. Virol. 78:5546-5553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Forest, T., S. Barnard, and J. D. Baines. 2005. Active intranuclear movement of herpesvirus capsids. Nat. Cell Biol. 7:429-431. [DOI] [PubMed] [Google Scholar]
- 17.Fraser, K. A., and S. A. Rice. 2007. Herpes simplex virus immediate-early protein ICP22 triggers loss of serine 2-phosphorylated RNA polymerase II. J. Virol. 81:5091-5101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fraser, K. A., and S. A. Rice. 2005. Herpes simplex virus type 1 infection leads to loss of serine-2 phosphorylation on the carboxyl-terminal domain of RNA polymerase II. J. Virol. 79:11323-11334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fujimuro, M., H. Sawada, and H. Yokosawa. 1994. Production and characterization of monoclonal antibodies specific to multi-ubiquitin chains of polyubiquitinated proteins. FEBS Lett. 349:173-180. [DOI] [PubMed] [Google Scholar]
- 20.Hughes, R. G., Jr., and W. H. Munyon. 1975. Temperature-sensitive mutants of herpes simplex virus type 1 defective in lysis but not in transformation. J. Virol. 16:275-283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jahedi, S., N. S. Markovitz, F. Filatov, and B. Roizman. 1999. Colocalization of the herpes simplex virus 1 UL4 protein with infected cell protein 22 in small, dense nuclear structures formed prior to onset of DNA synthesis. J. Virol. 73:5132-5138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Knipe, D. M., D. Senechek, S. A. Rice, and J. L. Smith. 1987. Stages in the nuclear association of the herpes simplex virus transcriptional activator protein ICP4. J. Virol. 61:276-284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kodiha, M., A. Chu, O. Lazrak, and U. Stochaj. 2005. Stress inhibits nucleocytoplasmic shuttling of heat shock protein hsc70. Am. J. Physiol. Cell Physiol. 289:C1034-C1041. [DOI] [PubMed] [Google Scholar]
- 24.Kyratsous, C. A., and S. J. Silverstein. 2007. BAG3, a host cochaperone, facilitates varicella-zoster virus replication. J. Virol. 81:7491-7503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kyratsous, C. A., and S. J. Silverstein. 2008. The co-chaperone BAG3 regulates herpes simplex virus replication. Proc. Natl. Acad. Sci. U. S. A. 105:20912-20917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lamberti, C., and S. K. Weller. 1998. The herpes simplex virus type 1 cleavage/packaging protein, UL32, is involved in efficient localization of capsids to replication compartments. J. Virol. 72:2463-2473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Leopardi, R., P. L. Ward, W. O. Ogle, and B. Roizman. 1997. Association of herpes simplex virus regulatory protein ICP22 with transcriptional complexes containing EAP, ICP4, RNA polymerase II, and viral DNA requires posttranslational modification by the U(L)13 proteinkinase. J. Virol. 71:1133-1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li, L., L. A. Johnson, J. Q. Dai-Ju, and R. M. Sandri-Goldin. 2008. Hsc70 focus formation at the periphery of HSV-1 transcription sites requires ICP27. PLoS One 3:e1491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liberek, K., A. Lewandowska, and S. Zietkiewicz. 2008. Chaperones in control of protein disaggregation. EMBO J. 27:328-335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liptak, L. M., S. L. Uprichard, and D. M. Knipe. 1996. Functional order of assembly of herpes simplex virus DNA replication proteins into prereplicative site structures. J. Virol. 70:1759-1767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Livingston, C. M., N. A. DeLuca, D. E. Wilkinson, and S. K. Weller. 2008. Oligomerization of ICP4 and rearrangement of heat shock proteins may be important for herpes simplex virus type 1 prereplicative site formation. J. Virol. 82:6324-6336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Livingston, C. M., M. F. Ifrim, A. E. Cowan, and S. K. Weller. 2009. Virus-induced chaperone-enriched (VICE) domains function as nuclear protein quality control centers during HSV-1 infection. PLoS Pathog. 5:e1000619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Long, M. C., V. Leong, P. A. Schaffer, C. A. Spencer, and S. A. Rice. 1999. ICP22 and the UL13 protein kinase are both required for herpes simplex virus-induced modification of the large subunit of RNA polymerase II. J. Virol. 73:5593-5604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lymberopoulos, M. H., and A. Pearson. 2007. Involvement of UL24 in herpes-simplex-virus-1-induced dispersal of nucleolin. Virology 363:397-409. [DOI] [PubMed] [Google Scholar]
- 35.Markovitz, N. S., and B. Roizman. 2000. Small dense nuclear bodies are the site of localization of herpes simplex virus 1 U(L)3 and U(L)4 proteins and of ICP22 only when the latter protein is present. J. Virol. 74:523-528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Martin, T. E., S. C. Barghusen, G. P. Leser, and P. G. Spear. 1987. Redistribution of nuclear ribonucleoprotein antigens during herpes simplex virus infection. J. Cell Biol. 105:2069-2082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Monier, K., J. C. Armas, S. Etteldorf, P. Ghazal, and K. F. Sullivan. 2000. Annexation of the interchromosomal space during viral infection. Nat. Cell Biol. 2:661-665. [DOI] [PubMed] [Google Scholar]
- 38.Orlando, J. S., T. L. Astor, S. A. Rundle, and P. A. Schaffer. 2006. The products of the herpes simplex virus type 1 immediate-early US1/US1.5 genes downregulate levels of S-phase-specific cyclins and facilitate virus replication in S-phase Vero cells. J. Virol. 80:4005-4016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Orlando, J. S., J. W. Balliet, A. S. Kushnir, T. L. Astor, M. Kosz-Vnenchak, S. A. Rice, D. M. Knipe, and P. A. Schaffer. 2006. ICP22 is required for wild-type composition and infectivity of herpes simplex virus type 1 virions. J. Virol. 80:9381-9390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Perkins, K. D., J. Gregonis, S. Borge, and S. A. Rice. 2003. Transactivation of a viral target gene by herpes simplex virus ICP27 is posttranscriptional and does not require the endogenous promoter or polyadenylation site. J. Virol. 77:9872-9884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Phelan, A., J. Dunlop, A. H. Patel, N. D. Stow, and J. B. Clements. 1997. Nuclear sites of herpes simplex virus type 1 DNA replication and transcription colocalize at early times postinfection and are largely distinct from RNA processing factors. J. Virol. 71:1124-1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Poffenberger, K. L., A. D. Idowu, E. B. Fraser-Smith, P. E. Raichlen, and R. C. Herman. 1994. A herpes simplex virus type 1 ICP22 deletion mutant is altered for virulence and latency in vivo. Arch. Virol. 139:111-119. [DOI] [PubMed] [Google Scholar]
- 43.Poffenberger, K. L., P. E. Raichlen, and R. C. Herman. 1993. In vitro characterization of a herpes simplex virus type 1 ICP22 deletion mutant. Virus Genes 7:171-186. [DOI] [PubMed] [Google Scholar]
- 44.Purves, F. C., W. O. Ogle, and B. Roizman. 1993. Processing of the herpes simplex virus regulatory protein alpha 22 mediated by the UL13 protein kinase determines the accumulation of a subset of alpha and gamma mRNAs and proteins in infected cells. Proc. Natl. Acad. Sci. U. S. A. 90:6701-6705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Purves, F. C., and B. Roizman. 1992. The UL13 gene of herpes simplex virus 1 encodes the functions for posttranslational processing associated with phosphorylation of the regulatory protein alpha 22. Proc. Natl. Acad. Sci. U. S. A. 89:7310-7314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Quinlan, M. P., L. B. Chen, and D. M. Knipe. 1984. The intranuclear location of a herpes simplex virus DNA-binding protein is determined by the status of viral DNA replication. Cell 36:857-868. [DOI] [PubMed] [Google Scholar]
- 47.Randall, R. E., and N. Dinwoodie. 1986. Intranuclear localization of herpes simplex virus immediate-early and delayed-early proteins: evidence that ICP 4 is associated with progeny virus DNA. J. Gen. Virol. 67:2163-2177. [DOI] [PubMed] [Google Scholar]
- 48.Rice, S. A., and D. M. Knipe. 1990. Genetic evidence for two distinct transactivation functions of the herpes simplex virus alpha protein ICP27. J. Virol. 64:1704-1715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rice, S. A., M. C. Long, V. Lam, P. A. Schaffer, and C. A. Spencer. 1995. Herpes simplex virus immediate-early protein ICP22 is required for viral modification of host RNA polymerase II and establishment of the normal viral transcription program. J. Virol. 69:5550-5559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Roizman, B., D. M. Knipe, and R. J. Whitley. 2007. Herpes simplex viruses, p. 2501-2601. In D. M. Knipe and P. M. Howley (ed.), Fields virology, 5th ed., vol. 2. Lippincott, Williams & Wilkins, Philadelphia, PA. [Google Scholar]
- 51.Salsman, J., N. Zimmerman, T. Chen, M. Domagala, and L. Frappier. 2008. Genome-wide screen of three herpesviruses for protein subcellular localization and alteration of PML nuclear bodies. PLoS Pathog. 4:e1000100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Schwartz, J., and B. Roizman. 1969. Concerning the egress of herpes simplex virus from infected cells: electron and light microscope observations. Virology 38:42-49. [DOI] [PubMed] [Google Scholar]
- 53.Schwyzer, M., U. V. Wirth, B. Vogt, and C. Fraefel. 1994. BICP22 of bovine herpesvirus 1 is encoded by a spliced 1.7 kb RNA which exhibits immediate early and late transcription kinetics. J. Gen. Virol. 75:1703-1711. [DOI] [PubMed] [Google Scholar]
- 54.Sears, A. E., I. W. Halliburton, B. Meignier, S. Silver, and B. Roizman. 1985. Herpes simplex virus 1 mutant deleted in the alpha 22 gene: growth and gene expression in permissive and restrictive cells and establishment of latency in mice. J. Virol. 55:338-346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Simpson-Holley, M., R. C. Colgrove, G. Nalepa, J. W. Harper, and D. M. Knipe. 2005. Identification and functional evaluation of cellular and viral factors involved in the alteration of nuclear architecture during herpes simplex virus 1 infection. J. Virol. 79:12840-12851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Stelz, G., E. Rucker, O. Rosorius, G. Meyer, R. H. Stauber, M. Spatz, M. M. Eibl, and J. Hauber. 2002. Identification of two nuclear import signals in the alpha-gene product ICP22 of herpes simplex virus 1. Virology 295:360-370. [DOI] [PubMed] [Google Scholar]
- 57.Weinheimer, S. P., B. A. Boyd, S. K. Durham, J. L. Resnick, and D. R. O'Boyle II. 1992. Deletion of the VP16 open reading frame of herpes simplex virus type 1. J. Virol. 66:258-269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yao, F., and P. A. Schaffer. 1995. An activity specified by the osteosarcoma line U2OS can substitute functionally for ICP0, a major regulatory protein of herpes simplex virus type 1. J. Virol. 69:6249-6258. [DOI] [PMC free article] [PubMed] [Google Scholar]