Abstract
Viruses of the family Coronaviridae have recently emerged through zoonotic transmission to become serious human pathogens. The pathogenic agent responsible for severe acute respiratory syndrome (SARS), the SARS coronavirus (SARS-CoV), is a member of this large family of positive-strand RNA viruses that cause a spectrum of disease in humans, other mammals, and birds. Since the publicized outbreaks of SARS in China and Canada in 2002-2003, significant efforts successfully identified the causative agent, host cell receptor(s), and many of the pathogenic mechanisms underlying SARS. With this greater understanding of SARS-CoV biology, many researchers have sought to identify agents for the treatment of SARS. Here we report the utility of the potent antiviral protein griffithsin (GRFT) in the prevention of SARS-CoV infection both in vitro and in vivo. We also show that GRFT specifically binds to the SARS-CoV spike glycoprotein and inhibits viral entry. In addition, we report the activity of GRFT against a variety of additional coronaviruses that infect humans, other mammals, and birds. Finally, we show that GRFT treatment has a positive effect on morbidity and mortality in a lethal infection model using a mouse-adapted SARS-CoV and also specifically inhibits deleterious aspects of the host immunological response to SARS infection in mammals.
The Coronaviridae are a group of enveloped positive-strand RNA viruses of the group Nidovirales. This group of viruses was not, until recently, of major concern as a matter of public health, although they were long recognized as important agents of serious disease in domestic and companion animals. The recent evidence of zoonotic transfer of this family of viruses from bats to animals such as palm civet cats and then to humans during the 2002-2003 outbreak greatly increased scientific interest in the Coronaviridae (7, 14, 19). The best-known coronavirus (CoV) is the causative agent of severe acute respiratory syndrome (SARS), termed the SARS-related coronavirus (SARS-CoV) (7, 14, 19). The lethal SARS outbreaks in China and Canada in 2002-2003 first brought SARS-CoV to public attention. The subsequent identification of two new human coronaviruses associated with acute respiratory infections in humans further illuminated the continuing potential threat that coronaviruses present to public health (31, 36).
Infection with SARS-CoV results from the binding of SARS-CoV spike glycoprotein (S) to angiotensin-converting enzyme 2 (ACE2) on the surface of susceptible cells in the lung followed by viral fusion with host cell membranes and transfer of virion contents into the cell (12, 25, 27). The infection stimulates significant cytokine responses in lung tissue that, together with pathologies associated with rapidly replicating virus, cause damage to the airway epithelium and alveolar membranes resulting in edema, respiratory distress, and (in ∼10% of cases) death (5). Due to the proven threat from SARS-CoV infections and the possibility of future zoonotic transmission of coronaviruses, efforts have been initiated to identify agents that could either reduce infection or suppress the deleterious cytokine response to SARS-CoV infection (8, 29).
The molecular physiology of the SARS-CoV life cycle and the host response to infection have provided numerous potential targets for chemotherapeutic intervention. In addition to vaccine development strategies, various research groups have targeted the SARS-CoV-specific main protease or viral attachment, entry, and fusion for intervention. SARS-CoV protease inhibitors which inhibit the enzyme at concentrations from 0.5 to 7 μM have been reported (2). The SARS-CoV papain-like protease (PLP) has also been successfully developed as a target for small-molecule antivirals, some of which are active in the 100 nM range (22). Viral entry inhibitors include SARS-CoV S glycoprotein heptad repeat peptides identified as potential inhibitors of viral fusion (3). Another broad-spectrum antiviral approach involves targeting the high-mannose oligosaccharides that are commonly found on viral surface glycoproteins. For example, carbohydrate-binding lectins, including Urtica dioica agglutinin (UDA), have been reported to bind to the SARS-CoV S protein and inhibit viral fusion and entry (33).
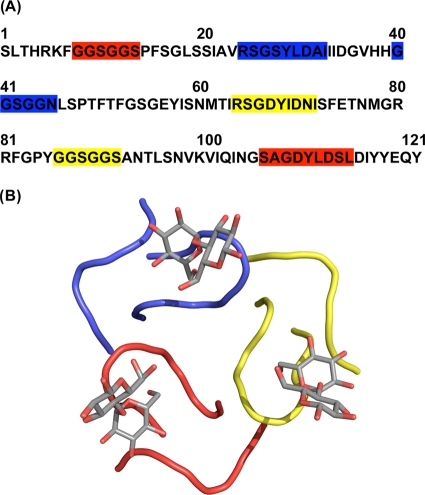
The antiviral protein griffithsin (GRFT) was originally isolated from the red alga Griffithsia sp. based upon its activity against the human immunodeficiency virus (HIV) (17). This unique 12.7-kDa protein was shown to bind specifically to oligosaccharides on the surface of the HIV envelope glycoprotein gp120. GRFT was shown to possess three largely identical carbohydrate-binding domains orientated as an equatorial triangle and affording multivalent binding and thereby increasing potency (37) (Fig. 1). Due to GRFT's ability to bind to specific oligosaccharides on envelope glycoproteins and block viral entry, it was hypothesized that GRFT might show broad-spectrum antiviral activity against other viruses, including SARS-CoV (38). Here we report the testing of GRFT for antiviral activity against a spectrum of coronaviruses, including SARS-CoV. In addition we present data on the specific binding interactions between GRFT and the SARS-CoV S protein. Finally, we evaluate the in vivo efficacy of intranasal administration of GRFT against infection with SARS-CoV in a lethal mouse model of pulmonary infection and explore the effects that GRFT treatment has on the induction of host cytokine response to SARS-CoV infection.
FIG. 1.
The amino acid sequence and carbohydrate binding domains of griffithsin. Griffithsin monomers contain three distinct, nonlinear, and uniform binding sites for monosaccharides such as mannose and glucose. The binding sites (red, blue, and yellow) are shown both in the amino acid sequence of griffithsin (A) and binding to the disaccharide maltose in a three-dimensional representation derived from the X-ray crystal structure (B).
MATERIALS AND METHODS
In vitro antiviral testing methods. (i) Compounds.
Ribavirin was obtained from ICN Pharmaceuticals (Costa Mesa, CA). Multiferon (multisubtype, human alpha interferon [IFN-α], consisting of a1, a2, a8, a10, a14, and a21 human IFN-α subtypes) was kindly provided by Douglas Lind (Viragen Inc., Plantation, FL), and the SARS-CoV protease inhibitor was provided by Sui Xiong Cai (Maxim Pharmaceuticals, San Diego, CA). GRFT was produced by recombinant production in Nicotiana benthamiana and purified for use as reported previously (18). SARS-CoV spike glycoprotein was obtained through the NIH Biodefense and Emerging Infections Research Resources Repository, NIAID, NIH (SARS-CoV spike [S] protein with histidine tag, recombinant from baculovirus, NR-686). ACE2 was purchased from R & D Systems (Minneapolis, MN). Recombinant HIV-1IIIB gp120, produced in a baculovirus expression system, was purchased from Immunodiagnostics Inc. (Woburn, MA).
(ii) Cells and virus.
Human ileocecal colorectal human adenocarcinoma cells (HRT-18G), mouse embryonic liver cells (BNL), and human diploid fibroblast cells (MRC-5) obtained from the American Type Culture Collection (ATCC, Manassas, VA) were grown in Dulbecco's modified essential medium (DMEM) supplemented with 10% fetal bovine serum (FBS; HyClone Laboratories, Thermo-Fisher Scientific, Logan, UT) and 0.37% NaHCO3. Chicken embryonic fibroblast cells (UMNSAH/DF-1, East Lansing line), pig testis fibroblast cells (ST), human ileocecal colorectal human adenocarcinoma cells (HCT-8), and African green monkey kidney cells (Vero 76), all from the ATCC, were grown in Eagle's formulation of minimal essential medium (MEM) supplemented with 10% FBS and 0.22% NaHCO3 and propagated at 37°C.
Calf diarrheal virus (bovine coronavirus [BCoV]) was obtained from the ATCC and passaged in HRT-18G cells. Infectious bronchitis virus strain Connecticut A5968 (IBV; ATCC) was routinely grown in DF-1 cells. Mouse hepatitis virus (MHV) strain JHM (ATCC) and porcine transmissible gastroenteritis virus strain Miller (PCoV; ATCC) were passaged in BNL cells and ST cells, respectively. Human coronavirus strain OC43 (HCoV-OC43; ATCC) was passaged in HCT-8 cells (ATCC), and human coronavirus strain 229E (HCoV-229E; ATCC) was passaged in MRC-5 cells. The following reagent was obtained through the NIH Biodefense and Emerging Infections Research Resources Repository, NIAID, NIH: human coronavirus, NL63 (Amsterdam 1), NR-470. NL63 was grown in rhesus monkey kidney cells (LLC-MK2 cells) in MEM supplemented with 5% FBS, later reduced to 2% FBS in antiviral assays Severe acute respiratory syndrome-associated coronavirus strain Urbani (SARS-CoV) was obtained from the Centers for Disease Control and Prevention (Atlanta, GA), the Tor-II strain was from Heinz Feldman (National Microbiology Laboratory, Winnipeg, Manitoba, Canada), the Frankfurt strain was from Jindrich Cinatl (Klinikum der J. W. Goethe Universität, Frankfurt am Mein, Germany), and the CuHK strain was from Paul K. S. Chan (Chinese University of Hong Kong, Hong Kong, People's Republic of China). They were all passaged in Vero 76 cells. When viruses were grown, the FBS in each medium used above was reduced to 2% and the NaHCO3 was reduced to 0.22% with the exception of BCoV. For BCoV, the growth and antiviral test media were MEM without serum, 0.18% NaHCO3, 20 IU trypsin/ml, 2.0 μg EDTA/ml. For all antiviral assays, 50 μg/ml gentamicin was also added to the antiviral test medium.
(iii) Preparation of compounds for testing.
Compounds in solution were diluted in test medium (MEM without serum) through a series of eight 1/2-log10 dilutions for evaluation. A positive-control drug was also included to ensure that an overwhelming amount of virus was not used in the assay. The latter compounds also were diluted in test medium as described above.
(iv) CPE inhibition assay.
Cells were seeded into 96-well flat-bottomed tissue culture plates (Corning Glass Works, Corning, NY), 0.2 ml/well, at the proper cell concentration, and incubated overnight at 37°C in order to establish a cell monolayer. When the monolayer was established, the growth medium was decanted and the various dilutions of test compound were added to each well (3 wells/dilution, 0.1 ml/well). Compound diluent medium was added to cell and virus control wells (0.1 ml/well). Virus (viral multiplicity of infection [MOI] = 0.01 to 0.001), diluted in test medium, was added to compound test wells (3 wells/dilution of compound) and to virus control wells at 0.1 ml/well. Virus was added approximately 5 min after compound. Test medium without virus was added to all toxicity control wells (2 wells/dilution of each test compound) and to cell control wells at 0.1 ml/well. The plates were incubated at 37°C or at 33°C (HCoV-OC43, HCoV-299E, and HCoV-NL63) in a humidified incubator with a 5% CO2-95% air atmosphere until virus control wells had adequate cytopathic effect (CPE) readings. This was achieved in 3 to 10 days after virus exposure to cells, depending on the virus. Cells were then examined microscopically for CPE, this being scored from 0 (normal cells) to 4 (maximal, 100% CPE). The cells in the toxicity control wells were observed microscopically for morphological changes attributed to cytotoxicity. This cytotoxicity was also graded as T (100% toxicity, complete cell sloughing from plate), PVH (80% cytotoxicity), PH (60% cytotoxicity), P (40% cytotoxicity), PSl (20% cytotoxicity), and 0 (normal cells). The 50% effective dose (EC50) and 50% cytotoxic dose (IC50) was calculated by regression analysis of the virus CPE data and the toxicity control data, respectively. The therapeutic index (SI) for each compound tested was calculated using the formula SI = IC50/EC50.
(v) NR uptake assay of CPE inhibition and compound cytotoxicity.
The neutral red (NR) uptake assay was done on the same CPE inhibition test plates as described above to verify the inhibitory activity and the cytotoxicity observed by visual observation. The NR assay was performed using a modified method of Cavanaugh et al. (4) as described by Barnard et al. (1). Medium was removed from each well of a plate, 0.034% NRF (0.34% neutral red in phosphate-buffered saline [PBS] supplemented with formalin at 10%) was added to each well of the plate, and the plate was incubated for 2 h at 37°C in the dark. The NR solution was then removed from the wells and rinsed, and the remaining dye was extracted using ethanol buffered with Sörenson's citrate buffer. Absorbances at 540 nm/405 nm were read with a microplate reader (Opsys MR; Dynex Technologies, Chantilly, VA). Absorbance values were expressed as percentages of untreated controls and EC50, IC50, and SI values were calculated as described above.
(vi) ELISA studies.
The binding of GRFT to SARS-CoV spike (S) glycoprotein was analyzed in enzyme-linked immunosorbent assay (ELISAs) with two different modalities. For standard ELISA, purified S protein was immobilized at 10 ng/well to a 96-well protein-binding plate (Nunc; Maxisorp) by incubation for 2 h at room temperature. After being rinsed twice with PBS containing 0.1% Tween (PBS-T) and three times with blocking buffer (Superblock buffer in PBS containing 0.05% Tween 20; Pierce, Rockford, IL), the plate was incubated with blocking buffer for 3 h at room temperature and rinsed again with PBS-T. The wells were then incubated with 0.5 log10 serial dilutions of GRFT, followed by anti-GRFT rabbit polyclonal antibodies for 1 h at room temperature. After washing with PBS-T, the amount of bound GRFT was determined by adding a 1:5,000 dilution of goat anti-rabbit-horseradish peroxidase (HRP; ImmunoPure). The plate was washed again, and horseradish peroxidase substrate (Kirkegaard & Perry Laboratories) was added. The reaction was stopped by the addition of 50 μl/well of 2 M H2SO4, and absorbance was measured at 450 nm. To measure the inhibition of binding of GRFT to S protein by mannose, increasing concentrations of mannose were added to 100-μl aliquots of GRFT (1 μM in PBS) or PBS alone and incubated at room temperature for 30 min on a rocker. The aliquots were then added to an S-protein-coated plate and incubated for 1 h at room temperature. Binding of GRFT was measured as described above.
In studies to measure the ability of GRFT to prevent the binding of S protein to the ACE2 cellular receptor, S protein was bound to the wells of a 96-well plate as above and then, after blocking and washing, was treated with increasing concentrations of GRFT (0.0003 to 100 pmol/well). Following incubation with GRFT (2 h), the plate was then incubated with 50 ng/well of recombinant ACE2 (R & D Systems) and incubated for 1 h prior to washing and visualization with primary polyclonal goat-anti-ACE2 antibodies followed by horseradish peroxidase-ligated mouse anti-goat secondary antibodies monitored by absorbance at 450 nm.
(vii) Isothermal titration calorimetry.
The calorimetric binding experiments were carried out on a VP-ITC microcalorimeter (MicroCal, Inc., Northampton, MA). In the experiments, 5-μl aliquots of a GRFT solution (289.3 μM) were injected from a 250-μl syringe into a rapidly mixing (300 rpm) solution of either SARS-CoV spike (1.8 μM) or HIV-1IIIB gp120 (2.5 μM) contained within the calorimetric cell (1.4426 ml). Both experiments were carried out at 25°C in 10 mM sodium phosphate buffer, 60 mM NaCl, 0.02% NaN3 (pH 7.0). The isotherms were corrected for dilution/buffer effects and fitted using the Origin ITC analysis software according to the manufacturer's protocols. Concentrations of all proteins were determined by amino acid analysis. A nonlinear least-square method was used to fit the titration data and to calculate the errors. From the binding curve, values for enthalpy, stoichiometry, and binding affinity were extracted. The other thermodynamic parameters (free energy of binding and entropy values) were calculated using ΔG = −RTlnKa, ΔG = ΔH − TΔS, and R = 1.985 cal/(mol·K), where R is the universal gas constant, T is temperature in Kelvin, and Ka is the reaction equilibrium constant.
In vivo mouse testing methods. (i) Animals.
Six- to eight-week-old BALB/c female mice were purchased from NCI. The animals were maintained in an approved animal care facility and transferred to a biosafety level 3 (BSL3) facility prior to infection. This study was approved by the University of Iowa Animal Care and Use Committee. The animals used for this study were part of a four-armed experiment including sham-treated, positive-control (SARS-CoV-infected), griffithsin-treated, and rhesus theta-defensin-treated animals. The results with rhesus theta-defensin-treated and some of the positive-control and sham-treated animals were previously published (34).
(ii) Griffithsin administration.
Recombinant GRFT protein was prepared as described previously (18). Mice were lightly anesthetized, and GRFT (5 mg/kg of body weight/day) was delivered intranasally 4 h prior to administration of MA15 SARS-CoV, followed by 2 doses daily (2.5 mg/kg/dose) for 4 days following infection. This dose was adopted from a previous study of a similar viral entry inhibitor, cyanovirin-N, with influenza virus (28).
(iii) MA15 Urbani SARS-CoV infection.
Mice were anesthetized and inoculated intranasally with 3 × 105 PFU of mouse-adapted Urbani SARS-CoV (MA15) (23) in 30 μl sterile 1× PBS in an approved BSL3 facility. Mice were weighed and examined daily. Animals were euthanized if they lost >25% of their body weight. To obtain specimens for lung histology, virus titers, lung homogenates, and serum collection, mice were euthanized at baseline and at day 2, day 4, and day 10 postinfection (p.i.). The left lung was fixed in zinc formalin, and the right lung was homogenized and stored at −80°C.
(iv) Plaque assay.
The right lung was excised aseptically, placed in sterile 1× PBS, and homogenized manually with a tissue homogenizer. The homogenates were centrifuged at 12,000 rpm for 10 min, and supernatant was removed. Lung homogenate supernatants were diluted and applied to Vero cells and incubated for 1 h, and an overlay medium containing agarose was added to cell surface. Three days postinfection, cells were fixed in 10% formaldehyde and stained with 0.1% crystal violet. Formed plaques were counted.
(v) Immunohistochemistry.
To detect SARS-CoV antigen, zinc formalin-fixed paraffin sections were incubated with a monoclonal antibody to the SARS-CoV N protein (gift of John Nicholls, University of Hong Kong, Hong Kong) and expression was detected using a biotin-avidin system. Sections were examined and photographed under light microscopy.
(vi) Histopathology.
Fixed tissues were processed, paraffin embedded, sectioned (4 μm), and hematoxylin and eosin stained. All slides were screened by a veterinary pathologist to first identify the histopathologic parameters that were appreciably altered. Subsequently, the slides were scored in a blinded fashion for lesion severity. Edema scores included the following: 1, absent to rare; 2, mild, detectable foci of eosinophilic seroproteinaceous fluid within alveoli and airways; and 3, moderate to severe foci of eosinophilic seroproteinaceous fluid pooling in alveoli and airways. Perivascular cellular inflammation scores included the following: 1, absent to rare; 2, mild perivascular cellular infiltrates starting to form small cellular aggregates; and 3, moderate to severe perivascular cellular infiltrates often forming marked aggregates with variable disruption of adjacent tissue architecture. Necrotizing bronchiolitis scores included the following: 1, absent to rare; 2, scattered foci of airway epithelium necrosis and sloughing; and 3, multifocal and coalescing to circumferential airway epithelium necrosis and sloughing. Data from the mice used in the negative-control (sham-treated) and positive-control (SARS-CoV without GRFT) groups were included in a previously published study (35).
(vii) Multiplex cytokine assay.
To identify changes in cytokine protein expression, we performed a multiplex bead-based immunoassay using the Bio-Plex cytokine assay (Bio-Rad Laboratories) as previously reported (35). Lung tissue homogenates were studied at baseline and day 2, day 4, and day 10 postinfection. The Bio-Plex cytokine 23-Plex kit measured mouse interleukin-1α (IL-1α), IL-1β, IL-2, IL-3, IL-4, IL-5, IL-6, IL-9, IL-10, IL-1(p40), IL-12(p70), IL-13, IL-17, eotaxin, granulocyte colony-stimulating factor (G-CSF), granulocyte/macrophage CSF (GM-CSF), IFN-α, KC, monocyte chemoattractant protein 1 (MCP-1), MIP-1α, MIP-1β, RANTES, and tumor necrosis factor alpha (TNF-α) expression patterns and was used following the manufacturer's instructions. Fifty microliters of each lung tissue homogenate specimen was transferred to appropriate microtiter wells containing diluted antibody-coated bead complexes and incubation buffer. Samples were incubated for 30 min at room temperature and 18 h (both in the dark) at 4°C, with continuous shaking. Following a rinse with wash buffer, 25 μl detection antibody was added to each well, incubated for 30 min at room temperature (in the dark), and washed as described above. Next 50 μl of streptavidin-phycoerythrin was added to all wells and incubated for 10 min at room temperature (in the dark). All microtiter wells received a final wash, and beads were resuspended in 125 μl assay buffer and analyzed on the Bio-Plex suspension array system using Bio-Plex Manager software. Cytokine concentrations were automatically calculated based on standard curve data. Data from the mice used in the negative-control (sham-treated) and positive-control (SARS-CoV without GRFT) groups were included in a previously published study (35).
(viii) Statistical analysis.
The indicated statistical tests were used in experimental analyses. Student's t test was applied at the 95% confidence level (P < 0.05) to each paired comparison at each of their respective time points using Excel. Analysis of variance (ANOVA) was applied with Bonferroni correction for multiple comparisons.
RESULTS
GRFT inhibits in vitro infection of Coronaviridae.
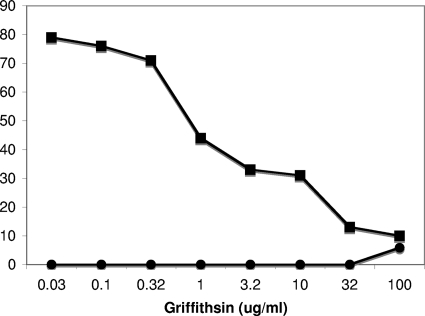
In initial studies of the effect of GRFT against SARS-CoV (Urbani strain), GRFT was shown to potently inhibit the cytopathic effect of SARS-CoV on Vero 76 cells (38). As can be seen in Fig. 2, GRFT reduced the percentage of cells killed by SARS-CoV (Urbani) in a concentration-dependent manner (EC50 = 48 nM) while showing minimal toxicity to the control cells. The antiviral activity of GRFT against SARS-CoV was not limited to the Urbani strain of the virus, as three additional strains of SARS-CoV were also sensitive to GRFT at similarly low concentrations (Table 1). In addition, GRFT demonstrated activity against a broad spectrum of other coronaviruses that infect mammals and birds in testing using a variety of cell lines. Of the coronaviruses tested several were susceptible to inhibition by GRFT below the lowest dose utilized (∼1 nM) (Table 2). It is interesting that GRFT mutants, in which the carbohydrate-binding domains were altered to prevent carbohydrate binding, were largely inactive against coronaviruses (EC50 of >50 μg/ml) (data not shown). Though all of the human coronaviruses were sensitive to GRFT at nanomolar concentrations, two coronaviruses, infectious bronchitis virus (IBV) and HCoV (NL63), were the most sensitive to inhibition (EC50 of <2.5 nM and <0.25 nM, respectively). It is interesting that these viruses belong to the group 1 and group 3 phylogenetic groups of the Coronaviridae, respectively. NL63 uses the ACE2 receptor for binding (21), but IBV uses a receptor possessing α-2,3-linked sialic acid residues (34). Thus, at the receptor level, the two viruses appear to have nothing in common, other than their susceptibility to inhibition by carbohydrate-binding agents such as GRFT (13, 32). In addition, GRFT was shown to be active against both group 1 and 2 Coronaviridae which utilize different mechanisms for proteolytic cleavage of the spike glycoprotein (9).
FIG. 2.
The effect of griffithsin against SARS-CoV (Urbani strain) in cell culture. Griffithsin inhibited the cytopathic effects of SARS-CoV on Vero cells in a concentration-dependent manner (▪) while displaying minimal toxicity to the same cells in the absence of viral challenge (•).
TABLE 1.
Antiviral activity of griffithsin against distinct strains of SARS-CoV
| Strain | EC50 (μg/ml) | IC50 (μg/ml) | SI |
|---|---|---|---|
| Urbani | 0.61 | >100 | >164 |
| Tor-II | 0.61 | >100 | >164 |
| CuHK | 0.78 | >100 | >128 |
| Frank | 1.19 | >100 | >83 |
TABLE 2.
Activity of GRFT on the cytopathic effects of selected coronavirusesa
| Virus, host, and treatment compound | Cytopathic effect assay |
Neutral red uptake assay |
||||
|---|---|---|---|---|---|---|
| EC50 | IC50 | SI | EC50 | IC50 | SI | |
| BCoV in human ileocecal colorectal human adenocarcinoma cells (HRT-18G) | ||||||
| GRFT | 0.057 | 32 | 560 | <0.032 | >100 | >3,100 |
| Ribavirin | 0.32 | 50 | 313 | <0.32 | 30 | 94 |
| IBV (Connecticut A5968) in chicken embryonic fibroblast cells (UMNSAH/DF-1) | ||||||
| GRFT | <0.032 | 68 | >2,100 | <0.032 | >100 | >3,100 |
| Multiferon | 0.0,012 | >0.03 | >25 | 0.0,029 | >0.03 | >10 |
| MHV (JHM) in mouse embryonic liver cells (BNL) | ||||||
| GRFT | 0.23 | 0.44 | 1.9 | 0.13 | 2.5 | 19 |
| Multiferon | 0.001 | >0.3 | >290 | 0.0,012 | >0.3 | >260 |
| PCoV (Miller) in pig testis fibroblast cells (ST) | ||||||
| GRFT | 0.57 | 57 | 100 | 0.77 | 46 | 59 |
| Multiferon | 0.000,095 | >0.03 | >320 | 0.00,017 | >0.03 | >180 |
| HCoV (OC43) in human ileocecal colorectal human adenocarcinoma cells (HCT-8) | ||||||
| GRFT | 0.16 | 52 | 320 | 0.048 | >100 | >2,100 |
| Multiferon | 0.0,006 | >0.03 | >50 | 0.0,008 | >0.03 | >37 |
| HCoV (229E) in human diploid fibroblast cells (MRC-5) | ||||||
| GRFT | 0.18 | >10 | >56 | 0.33 | >10 | >30 |
| Ribavirin | 18 | >1,000 | >56 | 26 | >1,000 | >38 |
| HCoV (NL63) in rhesus monkey kidney cells (LLC-MK2) | ||||||
| GRFT | <0.0,032 | 10 | >3,100 | <0.0,032 | >10 | >3,100 |
| Ribavirin | 0.63 | 320 | 500 | 1.6 | 120 | 77 |
EC50s and IC50s are shown in micrograms per milliliter for GRFT and ribavirin and in international units per milliliter for multiferon.
GRFT binds directly to the SARS-CoV spike glycoprotein.
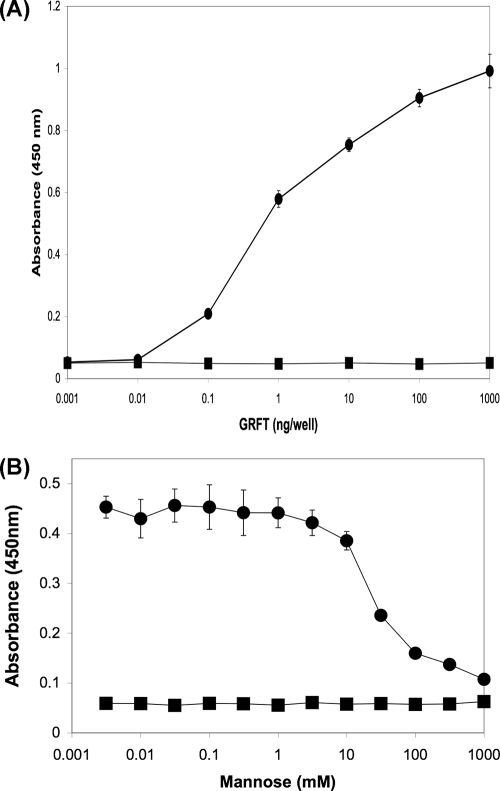
To determine if the activity of GRFT against SARS-CoV was due to specific interactions with the spike glycoprotein, ELISA studies utilizing recombinant SARS-CoV spike glycoprotein were performed which showed that GRFT binds to this protein in a concentration-dependent manner (Fig. 3A). Additional experiments showed that this binding interaction could be inhibited by the addition of excess mannose, thereby indicating that the association between GRFT and SARS-CoV spike glycoprotein is carbohydrate dependent (Fig. 3B). It should be noted, however, that the expression of the SARS-CoV spike glycoprotein in a baculovirus expression system could alter its glycosylation. Finally, ELISA studies were performed to determine if the binding of GRFT to SARS-CoV S glycoprotein was sufficient to inhibit the subsequent binding of the host cell receptor human ACE2 to S. These studies indicated that GRFT was unable to significantly inhibit the binding of the SARS-CoV S glycoprotein to ACE2 (data not shown). The inability of GRFT to inhibit S binding to ACE2 is similar to previous results with HIV-1 in which GRFT did not significantly inhibit the binding of gp120 to CD4 (17).
FIG. 3.
Griffithsin binds directly to SARS-CoV spike glycoprotein in a carbohydrate-dependent manner. Griffithsin bound directly to recombinant SARS-CoV spike protein (produced in a baculovirus expression system) (•) (A), and this binding was shown to be inhibited by increasing concentrations of mannose (•) (B) compared to the bovine serum albumin controls (▪) (A and B).
GRFT binds to SARS-CoV spike glycoprotein at multiple sites with high affinity.
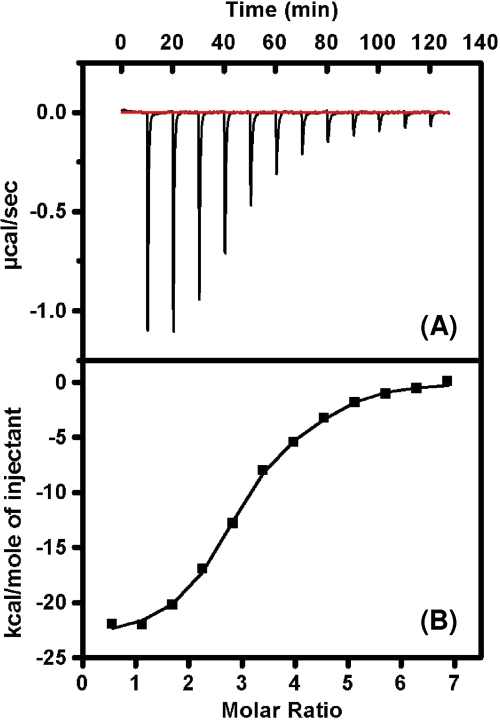
Since previous studies with carbohydrate-binding proteins and HIV gp120 have shown the importance of multiple binding interactions between these proteins and specific oligosaccharides on the envelope glycoprotein (26), we undertook isothermal titration calorimetric studies with recombinant SARS-CoV spike and GRFT. The results of this experiment provided the binding stoichiometry between these two proteins and also showed that GRFT interactions with spike are enthalpically driven (Fig. 4), resulting in a dissociation constant (Kd) of 24.9 nM (Table 3). The binding interaction of GRFT with SARS-CoV S was compared to that of GRFT with HIV-1 gp120. The results show that GRFT has an ∼10:1 binding stoichiometry with gp120 compared to a 3:1 binding stoichiometry with S, a finding which suggests a greater presence of high-mannose oligosaccharides on gp120 than on S. In line with the greater binding enthalpy (ΔH) for GRFT with gp120, the dissociation constant for gp120 was tighter (8.2 nM) than that seen with S.
FIG. 4.
Isothermal titration calorimetry measuring the binding interaction between GRFT and SARS-CoV spike glycoprotein. (A) Heat of interaction of multiple injections of GRFT into a solution of SARS-CoV spike shows a saturable binding interaction between GRFT and S. (B) Plotted thermogram of the saturable binding of GRFT and SARS-CoV spike from which affinity constants and stoichiometry are determined. The other thermodynamic parameters (free energy of binding and entropy values) were calculated using ΔG = −RTlnKa, ΔG = ΔH − TΔS, and R = 1.985 cal/(mol·K). (Table 3 has more data.)
TABLE 3.
Thermodynamic parameters of GRFT-spike and GRFT-gp120 binding interactions
| Envelope protein | Affinity, Kd (nM) | Enthalpy, ΔH (kcal/mol) | Entropy (TΔS) (kcal/mol) | Free energy (ΔGa) (kcal/mol) | Stoichiometry (GRFT:Env protein) |
|---|---|---|---|---|---|
| HIV-1 gp120 | 8.2 ± 4.3 | −30.40 ± 0.26 | −19.16 ± 0.28 | −11.20 ± 0.28 | 10.46 ± 0.18 |
| SARS CoV spike | 24.9 ± 2.0 | −23.10 ± 0.50 | −12.76 ± 0.50 | −10.40 ± 0.05 | 2.97 ± 0.12 |
ΔG = −RTlnKa.
GRFT pretreatment prevents lethal pulmonary infection in mice.
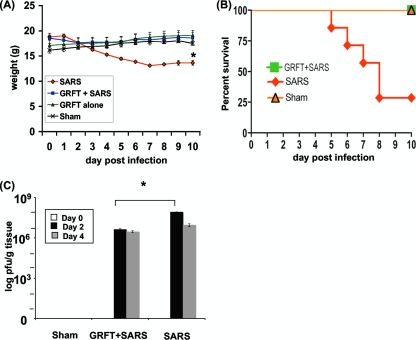
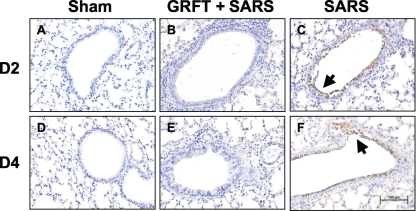
The mouse-adapted MA15 SARS-CoV causes a dose-dependent lung disease and significant morbidity and mortality in BALB/c mice (23). Groups of mice were inoculated with 3 × 105 PFU of virus, a dose previously shown to cause at least 75% mortality (35). One group of mice received concomitant treatment with intranasally administered GRFT peptide as described in Materials and Methods. As shown in Fig. 5A, MA15-infected mice began to lose weight within 2 to 3 days of inoculation, and this continued until they died from the infection or recovered. Figure 5B shows the survival curve for sham-treated, SARS-CoV-infected, and GRFT-treated mice. In contrast to the outcomes in untreated mice, animals that received GRFT 4 h before inoculation with MA15 followed by twice-daily treatment for 4 days did not lose weight and exhibited 100% survival. Animals receiving sham treatment or GRFT treatment alone survived and exhibited no weight loss. We evaluated MA15 titers in lung tissue 0, 2, and 4 days postinoculation. As shown in Fig. 5C, GRFT treatment significantly diminished the tissue virus titers on day 2 postinfection (mean virus titers [PFU/g tissue]: GRFT plus SARS-CoV, day 2, 4.2 × 106, and day 4, 2.9 × 106; SARS-CoV alone, day 2, 8.5 × 107, and day 4, 1.1 × 107). In agreement with this observation, GRFT-treated animals also had reduced pulmonary viral antigen load assessed by localization of the SARS-CoV N gene antigen by immunohistochemistry (Fig. 6).
FIG. 5.
GRFT treatment protects mice against morbidity from SARS-CoV infection. Mice were treated with sham control (no virus), GRFT alone, SARS-CoV alone, or GRFT followed with SARS-CoV infection as described in Materials and Methods. Animals were monitored daily for weight loss (A) and survival (B). GRFT-treated mice exhibited no weight loss. SARS-CoV-infected mice without GRFT treatment had a 30% survival rate and a ∼25% decrease in weight in those that survived. Lung tissue was harvested from mice, and viral titer levels were determined (C). Results in panels A and C are presented as means ± standard errors. n = 7 in panels A and B, and n = 3 in panel C. *, P ≤ 0.05 by t test or ANOVA.
FIG. 6.
SARS N gene product in lung tissue. Antigen labeling (brown stain) was detected primarily in lungs infected with SARS-CoV at day 2 and day 4 postinfection (C and F), mainly in airway epithelial cells and sloughed cells within the airway lumen (indicated by arrows) compared to lungs treated with vehicle (A and D). Lungs treated with GRFT and infected with SARS-CoV exhibited limited N antigen expression (B and E). n = 3 at each time point. Scale bar = 100 μM.
GRFT treatment decreases pulmonary pathology during SARS-CoV infection.
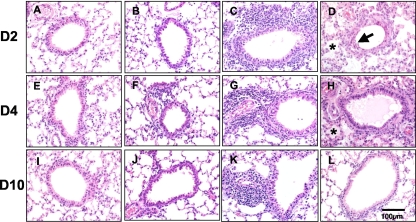
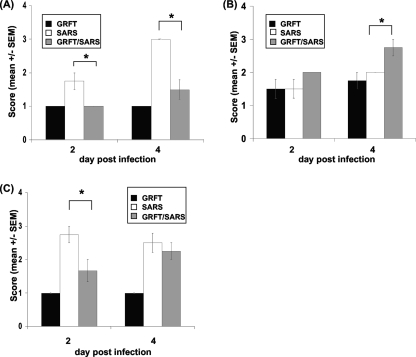
In humans and animal models SARS-CoV causes significant pathological changes in pulmonary tissues, characterized by necrotizing bronchiolitis, perivascular cellular inflammation, and alveolar edema (7, 14, 23). We assessed pulmonary histopathology at 2, 4, and 10 days postinfection in untreated, sham-treated (no virus), and GRFT-treated animals. Mice receiving GRFT alone developed a modest level of perivascular infiltrate that was largely resolved by 10 days (6 days following the last dose of GRFT) (Fig. 7). In contrast, mice treated with SARS-CoV alone manifested necrotizing bronchiolitis, perivascular infiltrates, and alveolar edema that were resolving by 10 days postinfection (in surviving animals). While mice receiving GRFT treatment and SARS-CoV exhibited robust perivascular infiltrates at levels greater than those with SARS-CoV alone, possibly due to the increased immunogenicity of GRFT-aggregated viral particles, the GRFT-treated mice had reduced levels of pulmonary edema at both 2 and 4 days postinfection and reduced severity of necrotizing bronchiolitis at 2 days postinfection. The histopathology scores are presented graphically in Fig. 8.
FIG. 7.
Lung tissue histopathology in SARS-CoV-infected mice with or without GRFT treatment. Hematoxylin-and-eosin-stained tissues were examined 2, 4, and 10 days postinfection. See Results for additional experimental details. Representative histopathology in sham-treated (A, E, and I), GRFT control-treated (B, F, and J), and SARS-CoV-infected mice with (C, G, and K) or without (D, H, and L) GRFT treatment. Arrows indicate necrotizing bronchiolitis; asterisks indicate alveolar edema. n = 4 at each time point. Scale bar = 100 μM.
FIG. 8.
Pulmonary histopathology scores in SARS-CoV-infected mice with or without GRFT treatment. Tissues were harvested 2 and 4 days postinfection and scored as described in Materials and Methods for evidence of alveolar edema (A), perivascular cellular infiltrates (B), and necrotizing bronchiolitis (C). Results are presented as means ± standard errors. n = 4; *, P ≤ 0.05, by two-way ANOVA with Bonferroni posttest correction.
GRFT treatment modifies cytokine responses in infected lung tissue.
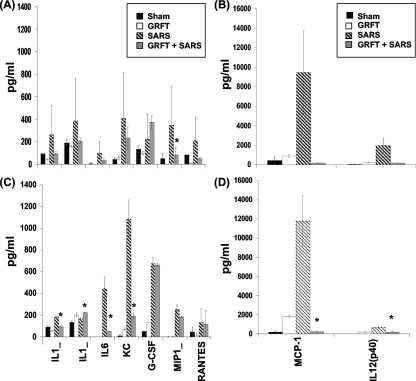
It has been hypothesized that the morbidity and mortality associated with SARS-CoV infection originate from maladaptive cytokine/chemokine responses or suppression of innate immune responses (6, 20). We measured cytokine levels in pulmonary tissue homogenates 2 and 4 days following MA15 inoculation. As shown in Fig. 9 (top panels), mice treated with GRFT and exposed to SARS-CoV showed significantly reduced levels of MIP-1α at day 2 postinfection compared to SARS-CoV alone. IL-1α and -β, RANTES, MCP-1, IL-12(p40), IL-6, and G-CSF also showed downward trends in GRFT-treated mice compared to SARS-CoV at day 2 p.i., but the differences were not statistically significant. At 4 days p.i. (Fig. 9, bottom panels), several cytokines were significantly reduced in GRFT-treated animals compared to those infected with SARS-CoV alone, including IL-1α and -β, IL-6, G-CSF, MCP-1, and IL-12(p40).
FIG. 9.
GRFT modifies pulmonary cytokine responses to SARS-CoV infection. Tissues were harvested at days 2 (A and B) and 4 (C and D) postinfection, and cytokine responses were assessed by multiplex analysis of tissue homogenates as described in Materials and Methods. Results are shown for sham (PBS alone), GRFT alone, SARS-CoV with GRFT treatment, and SARS-CoV alone. The x axis legend for day 4 applies to the corresponding panels above for day 2. Note differences in scale on left and right panels. Results are presented as means ± standard errors. n = 3/group; *, P ≤ 0.05 by t test.
DISCUSSION
The potent antiviral lectin GRFT displayed low nanomolar activity against SARS-CoV (Urbani strain) with an EC50 of 0.61 μg/ml (48 nM). Previously, we had speculated that GRFT would be likely to have activity against SARS-CoV due to its carbohydrate specificity and the known carbohydrate components of the SARS-CoV envelope glycoprotein S (17). Extending our previous work (38), here we detail the activity of GRFT against several strains of SARS-CoV and show consistent advantageous selectivity indices for all of the tested strains (Table 1). The carbohydrate-binding agent Urtica dioica agglutinin (UDA) has been reported by others to display anti-SARS-CoV activity (33), but the concentrations necessary for inhibition were significantly higher than those that we report here for GRFT. This difference in activity could be attributed to the multivalent interactions that the three independent carbohydrate-binding sites afford GRFT (Fig. 1). Other agents reported to show activity against SARS-CoV, including SARS-CoV protease inhibitors (2) and SARS-specific heptad repeat peptides (3), also show such activity only at significantly higher molar concentrations than GRFT. Though human recombinant alpha interferon (multiferon) and other host-targeted agents have demonstrated anti-SARS activity at low concentrations (Table 2), their potency does not significantly exceed that displayed by GRFT, which directly targets viral envelope glycoproteins.
In addition to GRFT's activity against SARS-CoV, GRFT demonstrated broad-spectrum activity against a variety of Coronaviridae, including those recently identified as human pathogens (Table 2). Both group 1 and group 2 coronaviruses were susceptible to GRFT with similarly low nanomolar sensitivities. GRFT was active against coronavirus strains that utilize protein-protein interactions for viral targeting (e.g., ACE2 as a cellular receptor, SARS-CoV, and HCoV-NL63) and those that utilize protein-carbohydrate interactions for viral attachment (i.e., α-2,3-linked sialic acid moieties, IBV-CoV, and HCoV-OC43). The broad range of Coronaviridae species sensitive to GRFT is a significant attribute for this antiviral protein, as this group of viruses appears to be capable of continuing zoonotic evolution and transfer to human hosts (24). GRFT was active against several coronaviruses at concentrations less than the lowest tested concentration, with HCoV-NL63, a strain recently identified as a human pathogen (31), displaying the greatest sensitivity (EC50 of <0.0032 μg/ml [0.25 nM]).
The molecular target through which GRFT appears to mediate its anticoronavirus activity is the surface envelope glycoprotein spike (S). GRFT binds directly to S as shown by ELISA studies showing the concentration-dependent binding to recombinant S (Fig. 3A). This mechanism is consistent with our previous studies of the mechanism of GRFT inhibition of HIV that revealed that GRFT binds to HIV-1 gp120 and prevents viral entry (18, 38). As with gp120, GRFT appears to bind to S via interaction with oligosaccharide moieties. Here we show that the binding of GRFT to S can indeed be inhibited by millimolar concentrations of mannose (Fig. 3B). GRFT is known to bind to select monosaccharides (mannose, glucose, and N-acetylglucosamine) in a multivalent manner via its three independent carbohydrate-binding domains (Fig. 1) (37). The unique equatorial triangular orientation of these three sites has been shown to be ideally situated so as to allow for engagement of multiple triantennary arms of specific high-mannose oligosaccharides such as oligomannose 9 (38). The oligosaccharide component of SARS-CoV S has been previously reported to contain multiple high-mannose oligosaccharides to which GRFT might bind (11). In isothermal titration calorimetry studies of the GRFT/S binding interactions, we determined that GRFT binds to S with a stoichiometry of 3:1, indicating that there are multiple binding sites for GRFT on S (Table 3). In this same study GRFT was shown to bind to S with a dissociation constant of 24.9 nM (Table 3). This affinity is weaker than that between GRFT and HIV gp120, an interaction that was shown to have a stoichiometry of ∼10:1. The difference in the affinity for GRFT between the two envelope glycoproteins is likely responsible for the difference seen in the antiviral activity of GRFT against HIV (0.048 to 0.63 nM) (17) and SARS-CoV (48 to 94 nM). Finally, we found that GRFT does not inhibit the interaction between the SARS-CoV S protein and the cellular receptor ACE2 (data not shown). Thus, the interaction between GRFT and S results in a complex that, though still able to bind to ACE2, may prevent the subsequent steps necessary for viral entry. The activity of GRFT against IBV suggests that this perturbation of viral entry by GRFT may be independent of the specific cellular receptor to which S binds. This mechanism would be similar to that seen with GRFT and HIV wherein GRFT binding to gp120 does not prevent the subsequent binding of gp120 to the cellular receptor CD4 and where GRFT activity is independent of the chemokine receptor tropism (CCR5 or CXCR4) of individual strains of HIV-1 (17, 34).
The potent in vitro activity of GRFT against SARS-CoV was confirmed using a mouse model system recently shown to recapitulate several aspects of the physiology of human disease, including a robust pulmonary disease component (23, 24). This mouse model was previously used to examine the impact of prior infection with SARS-CoV on immune responses and survival following subsequent reexposure to the virus (23, 24). Previously our group has shown that intranasal administration of the antiviral lectin cyanovirin-N (CV-N) was efficacious in the prevention of mortality in mice infected with a lethal strain of influenza virus H1N1 (28), and so, for the current study, GRFT was administered to mice via an intranasal route.
These studies showed that GRFT protected mice from a lethal inoculum of mouse-adapted SARS-CoV. Remarkably, 100% of the animals treated with 10 mg/kg/day GRFT survived viral challenge, in contrast to only 30% of control animals (Fig. 5B). The improved survival was further mirrored by prevention of weight loss and an improvement in lung histopathology scores and a reduction in lung tissue virus titers (Fig. 5A and C and Fig. 8), which indicated that, though GRFT-treated animals were infected by SARS-CoV, drug treatment significantly modified the disease course and outcome.
While studies of the prevention or treatment of SARS-CoV lung disease have been hampered by the lack of animal models that faithfully recapitulate the features of human disease, several agents have shown some efficacy in modifying disease outcomes. These include alpha interferon (10), small interfering RNA (siRNA) (15), and passive immunization (16, 30). Furthermore, in a previous study we showed that, during a simultaneous experiment, the cyclic antimicrobial peptide rhesus theta-defensin did not afford the complete protection from the morbidity of SARS-CoV infections seen with GRFT (34). Our studies with a robust model of SARS-CoV lung disease suggest that GRFT may modify disease outcome by more than one mechanism. First, by binding to the spike glycoprotein and interfering with productive infection, GRFT may reduce the overall virus burden during the first and subsequent rounds of infection. The reduced virus load in the lung at days 2 and 4 is consistent with this idea. In addition, enhanced peribronchial mononuclear cell infiltration and modification of cytokine responses suggest that GRFT also is immunomodulatory. Stimulation of leukocyte infiltration has been reported following high-dose topical application of GRFT to rabbit (but not human) cervical mucosa (18).
GRFT is currently being developed for potential use as an anti-HIV microbicide, and a recent report demonstrates the feasibility of large-scale production and purification from Nicotiana benthamia plants (18). This new production stream for GRFT greatly enables future development efforts based on GRFT's activity against enveloped viruses. Here we report that GRFT shows remarkable efficacy against lethal SARS-CoV infection and was potently active against a broad spectrum of human coronaviruses and other animal coronaviruses. Several questions remain, however, for the potential development of GRFT for use in the treatment of respiratory infections by coronaviruses. Though GRFT completely protected animals from SARS-CoV-induced death, the presence of perivascular infiltrates in GRFT-treated animals will need to be further characterized. Since GRFT-treated infected animals recover, this suggests that these cellular infiltrates may mediate protective immunity to SARS-CoV. Further, we used 10-mg/kg/day GRFT treatment in the in vivo studies reported here, but it is possible that lower doses of GRFT treatment would be equally efficacious. In summary, the antiviral protein GRFT shows noteworthy activity against Coronaviridae mediated via a novel mechanism of action. Its outstanding in vivo efficacy in SARS-CoV-infected mice suggests that this antiviral agent merits further investigation for the prophylaxis or treatment of respiratory infection by susceptible viruses.
Acknowledgments
We thank T. Moulaei and A. Wlodawer (Macromolecular Crystallography Laboratory, CCR, NCI) for assistance with the figures. We thank K. Gustafson and J. Beutler (MTDP, CCR, NCI), T. Gallagher (Loyola University), and S. Perlman (University of Iowa) for their reviews and helpful discussions of the manuscript.
This research was supported by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research (B.R.O., B.G., and J.B.M.). This project has been funded in whole or in part with federal funds from the National Cancer Institute, National Institutes of Health, under contract N01-CO-12400 and HHSN261200800001E (S.R.S.). P.B.M. is supported in part by NIH PO1 A1060699 and the Roy J. Carver Trust; K.E.P. is supported by a Pilot Project grant from the Executive Vice President of the University of Louisville.
The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government.
Footnotes
Published ahead of print on 23 December 2009.
REFERENCES
- 1.Barnard, D. L., C. W. Day, K. Bailey, M. Heiner, R. Montgomery, L. Lauridsen, P. K. Chan, and R. W. Sidwell. 2006. Evaluation of immunomodulators, interferons and known in vitro SARS-coV inhibitors for inhibition of SARS-coV replication in BALB/c mice. Antivir. Chem. Chemother. 17:275-284. [DOI] [PubMed] [Google Scholar]
- 2.Blanchard, J. E., N. H. Elowe, C. Huitema, P. D. Fortin, J. D. Cechetto, L. D. Eltis, and E. D. Brown. 2004. High-throughput screening identifies inhibitors of the SARS coronavirus main proteinase. Chem. Biol. 11:1445-1453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bosch, B. J., B. E. Martina, R. Van Der Zee, J. Lepault, B. J. Haijema, C. Versluis, A. J. Heck, R. De Groot, A. D. Osterhaus, and P. J. Rottier. 2004. Severe acute respiratory syndrome coronavirus (SARS-CoV) infection inhibition using spike protein heptad repeat-derived peptides. Proc. Natl. Acad. Sci. U. S. A. 101:8455-8460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cavanaugh, P. F., Jr., P. S. Moskwa, W. H. Donish, P. J. Pera, D. Richardson, and A. P. Andrese. 1990. A semi-automated neutral red based chemosensitivity assay for drug screening. Invest. New Drugs 8:347-354. [DOI] [PubMed] [Google Scholar]
- 5.Chen, J., and K. Subbarao. 2007. The immunobiology of SARS*. Annu. Rev. Immunol. 25:443-472. [DOI] [PubMed] [Google Scholar]
- 6.de Lang, A., T. Baas, T. Teal, L. M. Leijten, B. Rain, A. D. Osterhaus, B. L. Haagmans, and M. G. Katze. 2007. Functional genomics highlights differential induction of antiviral pathways in the lungs of SARS-CoV-infected macaques. PLoS Pathog. 3:e112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Drosten, C., S. Gunther, W. Preiser, S. van der Werf, H. R. Brodt, S. Becker, H. Rabenau, M. Panning, L. Kolesnikova, R. A. Fouchier, A. Berger, A. M. Burguiere, J. Cinatl, M. Eickmann, N. Escriou, K. Grywna, S. Kramme, J. C. Manuguerra, S. Muller, V. Rickerts, M. Sturmer, S. Vieth, H. D. Klenk, A. D. Osterhaus, H. Schmitz, and H. W. Doerr. 2003. Identification of a novel coronavirus in patients with severe acute respiratory syndrome. N. Engl. J. Med. 348:1967-1976. [DOI] [PubMed] [Google Scholar]
- 8.Golda, A., and K. Pyrc. 2008. Recent antiviral strategies against human coronavirus-related respiratory illnesses. Curr. Opin. Pulm. Med. 14:248-253. [DOI] [PubMed] [Google Scholar]
- 9.Gorbalenya, A. E., L. Enjuanes, J. Ziebuhr, and E. J. Snijder. 2006. Nidovirales: evolving the largest RNA virus genome. Virus Res. 117:17-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Haagmans, B. L., T. Kuiken, B. E. Martina, R. A. Fouchier, G. F. Rimmelzwaan, G. van Amerongen, D. van Riel, T. de Jong, S. Itamura, K. H. Chan, M. Tashiro, and A. D. Osterhaus. 2004. Pegylated interferon-alpha protects type 1 pneumocytes against SARS coronavirus infection in macaques. Nat. Med. 10:290-293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Han, D. P., M. Lohani, and M. W. Cho. 2007. Specific asparagine-linked glycosylation sites are critical for DC-SIGN- and L-SIGN-mediated severe acute respiratory syndrome coronavirus entry. J. Virol. 81:12029-12039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jia, H. P., D. C. Look, L. Shi, M. Hickey, L. Pewe, J. Netland, M. Farzan, C. Wohlford-Lenane, S. Perlman, and P. B. McCray, Jr. 2005. ACE2 receptor expression and severe acute respiratory syndrome coronavirus infection depend on differentiation of human airway epithelia. J. Virol. 79:14614-14621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Keyaerts, E., L. Vijgen, C. Pannecouque, E. Van Damme, W. Peumans, H. Egberink, J. Balzarini, and M. Van Ranst. 2007. Plant lectins are potent inhibitors of coronaviruses by interfering with two targets in the viral replication cycle. Antiviral Res. 75:179-187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ksiazek, T. G., D. Erdman, C. S. Goldsmith, S. R. Zaki, T. Peret, S. Emery, S. Tong, C. Urbani, J. A. Comer, W. Lim, P. E. Rollin, S. F. Dowell, A. E. Ling, C. D. Humphrey, W. J. Shieh, J. Guarner, C. D. Paddock, P. Rota, B. Fields, J. DeRisi, J. Y. Yang, N. Cox, J. M. Hughes, J. W. LeDuc, W. J. Bellini, and L. J. Anderson. 2003. A novel coronavirus associated with severe acute respiratory syndrome. N. Engl. J. Med. 348:1953-1966. [DOI] [PubMed] [Google Scholar]
- 15.Li, B. J., Q. Tang, D. Cheng, C. Qin, F. Y. Xie, Q. Wei, J. Xu, Y. Liu, B. J. Zheng, M. C. Woodle, N. Zhong, and P. Y. Lu. 2005. Using siRNA in prophylactic and therapeutic regimens against SARS coronavirus in Rhesus macaque. Nat. Med. 11:944-951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McCray, P. B., Jr., L. Pewe, C. Wohlford-Lenane, M. Hickey, L. Manzel, L. Shi, J. Netland, H. P. Jia, C. Halabi, C. D. Sigmund, D. K. Meyerholz, P. Kirby, D. C. Look, and S. Perlman. 2007. Lethal infection of K18-hACE2 mice infected with severe acute respiratory syndrome coronavirus. J. Virol. 81:813-821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mori, T., B. R. O'Keefe, R. C. Sowder II, S. Bringans, R. Gardella, S. Berg, P. Cochran, J. A. Turpin, R. W. Buckheit, Jr., J. B. McMahon, and M. R. Boyd. 2005. Isolation and characterization of griffithsin, a novel HIV-inactivating protein, from the red alga Griffithsia sp. J. Biol. Chem. 280:9345-9353. [DOI] [PubMed] [Google Scholar]
- 18.O'Keefe, B. R., F. Vojdani, V. Buffa, R. J. Shattock, D. C. Montefiori, J. Bakke, J. Mirsalis, A. L. d'Andrea, S. D. Hum., B. Bratcher, C. J. Saucedo, J. B. McMahon, G. P. Pogue, and K. E. Palmer. 2009. Scaleable manufacture of HIV-1 entry inhibitor griffithsin and validation of its safety and efficacy as a topical microbicide component. Proc. Natl. Acad. Sci. U. S. A. 106:6099-6104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Peiris, J. S., S. T. Lai, L. L. Poon, Y. Guan, L. Y. Yam, W. Lim, J. Nicholls, W. K. Yee, W. W. Yan, M. T. Cheung, V. C. Cheng, K. H. Chan, D. N. Tsang, R. W. Yung, T. K. Ng, and K. Y. Yuen. 2003. Coronavirus as a possible cause of severe acute respiratory syndrome. Lancet 361:1319-1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Perlman, S., and A. A. Dandekar. 2005. Immunopathogenesis of coronavirus infections: implications for SARS. Nat. Rev. Immunol. 5:917-927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pyrc, K., B. J. Bosch, B. Berkhout, M. F. Jebbink, R. Dijkman, P. Rottier, and L. van der Hoek. 2006. Inhibition of human coronavirus NL63 infection at early stages of the replication cycle. Antimicrob. Agents Chemother. 50:2000-2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ratia, K., S. Pegan, J. Takayama, K. Sleeman, M. Coughlin, S. Baliji, R. Chaudhuri, W. Fu, B. S. Prabhakar, M. E. Johnson, S. C. Baker, A. K. Ghosh, and A. D. Mesecar. 2008. A noncovalent class of papain-like protease/deubiquitinase inhibitors blocks SARS virus replication. Proc. Natl. Acad. Sci. U. S. A. 105:16119-16124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Roberts, A., D. Deming, C. D. Paddock, A. Cheng, B. Yount, L. Vogel, B. D. Herman, T. Sheahan, M. Heise, G. L. Genrich, S. R. Zaki, R. Baric, and K. Subbarao. 2007. A mouse-adapted SARS-coronavirus causes disease and mortality in BALB/c mice. PLoS Pathog. 3:e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rockx, B., T. Sheahan, E. Donaldson, J. Harkema, A. Sims, M. Heise, R. Pickles, M. Cameron, D. Kelvin, and R. Baric. 2007. Synthetic reconstruction of zoonotic and early human severe acute respiratory syndrome coronavirus isolates that produce fatal disease in aged mice. J. Virol. 81:7410-7423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sheahan, T., B. Rockx, E. Donaldson, A. Sims, R. Pickles, D. Corti, and R. Baric. 2008. Mechanisms of zoonotic severe acute respiratory syndrome coronavirus host range expansion in human airway epithelium. J. Virol. 82:2274-2285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shenoy, S. R., B. R. O'Keefe, A. J. Bolmstedt, L. K. Cartner, and M. R. Boyd. 2001. Selective interactions of the human immunodeficiency virus-inactivating protein cyanovirin-N with high-mannose oligosaccharides on gp120 and other glycoproteins. J. Pharmacol. Exp. Ther. 297:704-710. [PubMed] [Google Scholar]
- 27.Sims, A. C., R. S. Baric, B. Yount, S. E. Burkett, P. L. Collins, and R. J. Pickles. 2005. Severe acute respiratory syndrome coronavirus infection of human ciliated airway epithelia: role of ciliated cells in viral spread in the conducting airways of the lungs. J. Virol. 79:15511-15524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Smee, D. F., K. W. Bailey, M. H. Wong, B. R. O'Keefe, K. R. Gustafson, V. P. Mishin, and L. V. Gubareva. 2008. Treatment of influenza A (H1N1) virus infections in mice and ferrets with cyanovirin-N. Antiviral Res. 80:266-271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tong, T. R. 2006. SARS coronavirus anti-infectives. Recent Pat. Antiinfect. Drug Discov. 1:297-308. [DOI] [PubMed] [Google Scholar]
- 30.Tseng, C. T., C. Huang, P. Newman, N. Wang, K. Narayanan, D. M. Watts, S. Makino, M. M. Packard, S. R. Zaki, T. S. Chan, and C. J. Peters. 2007. Severe acute respiratory syndrome coronavirus infection of mice transgenic for the human angiotensin-converting enzyme 2 virus receptor. J. Virol. 81:1162-1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.van der Hoek, L., K. Pyrc, M. F. Jebbink, W. Vermeulen-Oost, R. J. Berkhout, K. C. Wolthers, P. M. Wertheim-van Dillen, J. Kaandorp, J. Spaargaren, and B. Berkhout. 2004. Identification of a new human coronavirus. Nat. Med. 10:368-373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.van der Meer, F. J., C. A. de Haan, N. M. Schuurman, B. J. Haijema, W. J. Peumans, E. J. Van Damme, P. L. Delputte, J. Balzarini, and H. F. Egberink. 2007. Antiviral activity of carbohydrate-binding agents against Nidovirales in cell culture. Antiviral Res. 76:21-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.van der Meer, F. J., C. A. de Haan, N. M. Schuurman, B. J. Haijema, M. H. Verheije, B. J. Bosch, J. Balzarini, and H. F. Egberink. 2007. The carbohydrate-binding plant lectins and the non-peptidic antibiotic pradimicin A target the glycans of the coronavirus envelope glycoproteins. J. Antimicrob. Chemother. 60:741-749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Winter, C., G. Herrler, and U. Neumann. 2008. Infection of the tracheal epithelium by infectious bronchitis virus is sialic acid dependent. Microbes Infect. 10:367-373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wohlford-Lenane, C. L., D. K. Meyerholz, S. Perlman, H. Zhou, D. Tran, M. E. Selsted, and P. B. McCray, Jr. 2009. Rhesus theta-defensin prevents death in a mouse model of severe acute respiratory syndrome coronavirus pulmonary disease. J. Virol. 83:11385-11390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Woo, P. C., S. K. Lau, C. M. Chu, K. H. Chan, H. W. Tsoi, Y. Huang, B. H. Wong, R. W. Poon, J. J. Cai, W. K. Luk, L. L. Poon, S. S. Wong, Y. Guan, J. S. Peiris, and K. Y. Yuen. 2005. Characterization and complete genome sequence of a novel coronavirus, coronavirus HKU1, from patients with pneumonia. J. Virol. 79:884-895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ziolkowska, N. E., B. R. O'Keefe, T. Mori, C. Zhu, B. Giomarelli, F. Vojdani, K. E. Palmer, J. B. McMahon, and A. Wlodawer. 2006. Domain-swapped structure of the potent antiviral protein griffithsin and its mode of carbohydrate binding. Structure 14:1127-1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ziolkowska, N. E., S. R. Shenoy, B. R. O'Keefe, J. B. McMahon, K. E. Palmer, R. A. Dwek, M. R. Wormald, and A. Wlodawer. 2007. Crystallographic, thermodynamic, and molecular modeling studies of the mode of binding of oligosaccharides to the potent antiviral protein griffithsin. Proteins 67:661-670. [DOI] [PMC free article] [PubMed] [Google Scholar]