Abstract
Mannheimia (Pasteurella) haemolytica is the only pathogen that consistently causes severe bronchopneumonia and rapid death of bighorn sheep (BHS; Ovis canadensis) under experimental conditions. Paradoxically, Bibersteinia (Pasteurella) trehalosi and Pasteurella multocida have been isolated from BHS pneumonic lungs much more frequently than M. haemolytica. These observations suggest that there may be an interaction between these bacteria, and we hypothesized that B. trehalosi overgrows or otherwise inhibits the growth of M. haemolytica. Growth curves (monoculture) demonstrated that B. trehalosi has a shorter doubling time (∼10 min versus ∼27 min) and consistently achieves 3-log higher cell density (CFU/ml) compared to M. haemolytica. During coculture M. haemolytica growth was inhibited when B. trehalosi entered stationary phase (6 h) resulting in a final cell density for M. haemolytica that was 6 to 9 logs lower than expected with growth in the absence of B. trehalosi. Coculture supernatant failed to inhibit M. haemolytica growth on agar or in broth, indicating no obvious involvement of lytic phages, bacteriocins, or quorum-sensing systems. This observation was confirmed by limited growth inhibition of M. haemolytica when both pathogens were cultured in the same media but separated by a filter (0.4-μm pore size) that limited contact between the two bacterial populations. There was significant growth inhibition of M. haemolytica when the populations were separated by membranes with a pore size of 8 μm that allowed free contact. These observations demonstrate that B. trehalosi can both outgrow and inhibit M. haemolytica growth with the latter related to a proximity- or contact-dependent mechanism.
The bighorn sheep (BHS; Ovis canadensis) population in North America has declined from an estimated two million at the beginning of the 19th century to fewer than 70,000 today (7, 30). The decline of BHS populations is presumably due to loss of habitat, competition for forage with domestic livestock, predation, and disease (9, 19). The most important disease that has limited the growth of BHS populations is pneumonia (13, 14, 19, 31). Bacteria associated with BHS pneumonia are members of the genera Mannheimia and Pasteurella, particularly, the species Mannheimia (Pasteurella) haemolytica, Bibersteinia (Pasteurella) trehalosi, and Pasteurella multocida (6-9, 15, 20, 25, 31). Several independent studies have revealed that M. haemolytica is a major cause of BHS pneumonia. In fact, M. haemolytica is the only pathogen that has been shown to consistently cause severe bronchopneumonia and rapid death of BHS under experimental conditions (10, 14, 23). B. trehalosi has been isolated more often than M. haemolytica from the upper respiratory tract of healthy BHS (10, 12-14, 26, 31). Large numbers of B. trehalosi have also been isolated from the pneumonic lungs of BHS experimentally inoculated with M. haemolytica alone (10). Furthermore, our recent studies with M. haemolytica wild type and leukotoxin deletion mutants in BHS have revealed that the leukotoxin deletion mutant does not cause the death of BHS but instead induces only mild lung lesions, confirming the finding in cattle that leukotoxin is the most important virulence factor of M. haemolytica (10, 24, 29). Our recently concluded BHS inoculation study revealed that only leukotoxin producing strains of B. trehalosi can cause pneumonia, indicating that leukotoxin is the most important virulence determinant in B. trehalosi as well. More than 85% of the B. trehalosi isolates obtained from BHS, however, do not produce leukotoxin (28, 32). Therefore, this observation, together with the results from the animal experiments, indicates that B. trehalosi is unlikely to be the major cause of pneumonia outbreaks in BHS.
These observations prompted us to hypothesize that B. trehalosi outgrows or otherwise inhibits the growth of M. haemolytica. The objectives of the present study were to (i) characterize in vitro growth kinetics of M. haemolytica and B. trehalosi; (ii) develop M. haemolytica-specific and B. trehalosi-specific PCR assays to detect either species in mixed cultures; and (iii) determine whether B. trehalosi inhibits the growth of M. haemolytica in vitro and, if it does, characterize the mechanism of inhibition.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
M. haemolytica and B. trehalosi cultures were maintained as frozen stocks (−80°C) in brain heart infusion broth (BHI) with 10% glycerol. From the stocks, M. haemolytica serotype A1 (89010807N [21]) and other BHS isolates, as well as B. trehalosi serotype T10 (ATCC 33374) and other BHS isolates, were individually cultured in BHI agar supplemented with 5% defibrinated sheep blood (Remel, Lenexa, KS) and 37°C overnight incubation. A loopful of bacteria was transferred into each one of several tubes containing 10 ml of BHI broth and cultured overnight at 37°C with constant shaking (200 rpm). The chromosomally encoded ampicillin-resistant M. haemolytica strain developed by Murphy et al. (21) was cultured in BHI containing 10 μg of ampicillin/ml. We developed a rifampin-resistant B. trehalosi strain by culturing the wild-type B. trehalosi on BHI agar plates containing 10 μg of rifampin/ml overnight. The resistant clones were isolated and maintained in medium with rifampin. Antibiotic-sensitive B. trehalosi and M. haemolytica, as well as ampicillin-resistant (Ampr) M. haemolytica and rifampin-resistant (Rifr) B. trehalosi were used in subsequent studies.
Isolation of bacteria from pneumonic BHS lungs.
Dacron swabs taken from the lungs were inoculated onto Columbia blood agar (Becton Dickinson, Sparks, MD) supplemented with 5% (vol/vol) sheep blood and onto a Pasteurellaceae selective agar medium (16, 17). Each plate was streaked for colony isolation. The plates were incubated at 37°C in 10% CO2 and inspected at 24 and 48 h. All representative colonies were picked and grown in wedges on Columbia blood agar. Isolates were determined to be M. haemolytica or B. trehalosi if they were Gram stain negative; pleiomorphic; MacConkey, urea, and indole negative; oxidase, nitrate, glucose, and sucrose positive; and xylose or trehalose positive. Each isolate was biovariant-typed based on its ability to ferment various sugars in addition to other biochemical tests (a total of 21 phenotypic characteristics) according to the protocol of Jaworski et al. (16). P. multocida isolates fit the above criteria except that they were indole positive. P. multocida was further characterized according to a previously described protocol (5).
Bacterial growth curves.
Overnight cultures of B. trehalosi and M. haemolytica were diluted and inoculated (∼106 CFU) separately into two culture tubes, each containing 10 ml of BHI broth, followed by incubation (37°C, 200 rpm). At multiple time points up to 24 h, the optical density at 600 nm of the cultures was recorded, and a small aliquot of the culture was taken for CFU counts. Samples collected at each time point were serially diluted with BHI broth, placed on BHI agar plates, and incubated at 37°C. After overnight growth, bacterial colonies were counted and plotted against time.
Plaque assay.
Supernatant fluid was collected from single cultures of B. trehalosi or M. haemolytica or cocultures of B. trehalosi and M. haemolytica at different time points (1, 6, 12, and 24 h) and filter-sterilized (Acrodisc 0.2- or 0.45-μm-pore-size Supor membrane syringe filters; Pall Life Sciences, East Hills, NY). A total of 100 μl of M. haemolytica (106 CFU) and 300 μl of culture supernatant fluids were mixed and incubated at 37°C for 20 min to facilitate adsorption of bacteriophage (if any), after which 3 ml of BHI soft molten agar (0.7%, 45°C) was added into each tube. The tube was mixed gently and immediately poured onto preheated BHI agar plates with a swirling movement. The plates were kept at room temperature for 5 min for the soft agar to solidify and then incubated at 30, 37, and 42°C. Culture plates were examined for plaque (zone of lysis) formation for up to 24 h. M. haemolytica without culture supernatant fluids, but with soft molten agar, was used as negative control.
Bactericidal assay.
The presence of any bactericidal compounds in B. trehalosi and coculture supernatant fluids was evaluated by solid BHI agar and broth assays, described below. To minimize loss of bactericidal compounds (if any) during filtration, culture supernatants were filtered through low protein binding filters (Acrodisc 0.2- or 0.45-μm-pore-size Supor membrane syringe filters; Pall Life Sciences). BHI broth (100 μl) containing 106 CFU of M. haemolytica was gently mixed with 3 ml of BHI soft molten agar and poured onto preheated BHI agar plates with a swirling movement. Plates were kept at room temperature for 5 min. Discs soaked with filtered culture supernatant fluids were placed onto the solidified soft agar and incubated at 37°C. Plates were examined up to 24 h for signs of inhibition, as revealed by clear zones around the discs. Putative bacteriocin(s) can be precipitated from culture supernatant by using ammonium sulfate (4). Putative bacteriocins in M. haemolytica and B. trehalosi culture supernatants (400 ml) were precipitated with 40% ammonium sulfate and dialyzed against phosphate-buffered saline (pH 7.2). Portions (100 to 150 μg) of solubilized proteins were tested for bacteriocin activity on M. haemolytica plates by the spot-on-lawn assay. A bactericidal assay was also performed using broth culture. Filter-sterilized culture supernatant fluid (5 ml) was mixed with equal volumes of 2× BHI broth containing M. haemolytica (106 CFU) and cultured for 24 h at 37°C (200 rpm). M. haemolytica CFU counts were determined as described above. M. haemolytica without any culture supernatant fluids, but with BHI broth, served as the negative control. To determine whether the filter membrane has any undesirable effect on the putative bactericidal compounds, a direct inhibitory assay was performed as follows. M. haemolytica in soft agar was overlaid onto BHI agar as described earlier. Then, BHI containing 105 CFU (10 μl) of actively growing B. trehalosi was placed onto the soft agar, followed by incubation at 37°C for up to 24 h. The plates were examined for zones of inhibition, indicated by a clear halo around the colony.
Detection of M. haemolytica and B. trehalosi in cocultures by conventional culture techniques.
Overnight cultures of B. trehalosi and M. haemolytica were diluted in BHI broth and added into 10 ml of BHI broth (∼10:1 inhibitor/target ratio; that is, B. trehalosi [∼107 CFU/ml] to M. haemolytica [∼106 CFU/ml]). The same numbers of B. trehalosi and M. haemolytica were individually inoculated into 10 ml of BHI broth as well. All three cultures were incubated at 37°C for 24 h (200 rpm), and samples were collected at different time points (1, 3, 6, 12, and 24 h). One set of samples was submitted to the Washington Animal Disease Diagnostic Laboratory at Washington State University to analyze independently where standard bacteriological methods are used to identify B. trehalosi and M. haemolytica. The other set of samples were serially diluted and plated on BHI agar plates, followed by colony PCR (described below). In the same coculture experiments, antibiotic sensitive bacteria were replaced with Ampr M. haemolytica and Rifr B. trehalosi strains. Serially diluted bacteria from the individual (mono-) and cocultures collected at different time points were placed either on ampicillin plates (for M. haemolytica) or rifampin plates (for B. trehalosi) and cultured at 37°C overnight. Colonies in each of the plates were counted and expressed as CFU/ml.
Multiplex PCR assay.
To differentiate M. haemolytica from B. trehalosi, O-sialoglycoprotein endopeptidase (gcp; GenBank accession numbers AY839677 and AY839681) and manganese-dependent superoxide dismutase (sodA; GenBank accession numbers AY702551 and AY702549) sequences were aligned by using CLUSTAL W program (http://www.ebi.ac.uk/Tools/clustalw). The regions exhibiting the least degree of sequence identity were selected, and the following species-specific primers were designed for multiplex PCR assay: M. haemolytica-specific gcp forward primer MhgcpF (5′-AGA GGC CAA TCT GCA AAC CTC G-3′) and reverse primer MhgcpR (5′-GTT CGT ATT GCC CAA CGC CG-3′), and B. trehalosi-specific sodA forward primer BtsodAF (5′-GCC TGC GGA CAA ACG TGT TG-3′) and reverse primer BtsodAR (5′-TTT CAA CAG AAC CAA AAT CAC GAA TG-3′). The multiplex PCR was carried out in a final volume of 50 μl with GoTaq PCR SuperMix (Promega, Inc., Madison, WI) with 0.2 μM concentrations of each primer and 2 μl of bacterial culture (colony or broth). The PCR cycling conditions consisted of an initial denaturation at 95°C for 5 min, followed by 35 cycles of denaturation at 95°C for 30 s, annealing at 55°C for 30 s, and extension at 72°C for 40 s, and a final elongation at 72°C for 5 min. The PCR products were visualized (gcp, 267 bp; sodA, 144 bp) after electrophoresis through 1% agarose gels run at 7.0 V/cm and stained with ethidium bromide. The specificity of the multiplex PCR primers for M. haemolytica and B. trehalosi was evaluated by using multiple field and reference strains. At least 30 individual colonies at each time point were assessed by the multiplex PCR.
Proximity-dependent inhibition assay.
To determine whether B. trehalosi-mediated growth inhibition of M. haemolytica was due to contact-dependent inhibition, we used cell culture inserts with two different pore sizes in the competition assay. Generally, bacteria do not pass through 0.4-μm pores, but they can readily pass through 8.0-μm pores, whereas macromolecules such as bacteriocin and other molecules can pass through 0.4-μm pores by simple diffusion. Therefore, we used 0.4- and 8.0-μm-pore-size polyethylene terephthalate (PET) track-etched membrane cell culture inserts (BD Falcon; BD Biosciences, Franklin Lakes, NJ) to further characterize the observed growth inhibition. Placing these inserts in six-well cell culture plates creates upper and lower chambers. Approximately 108 CFU of Rifr B. trehalosi in 10 μl of BHI were added into the upper chambers containing 2.5 ml of antibiotic-free BHI, and ∼107 CFU of Ampr M. haemolytica (at a 10:1 inhibitor/target ratio) in 10 μl of BHI were added into the lower chambers containing the same volume of antibiotic-free BHI. The culture plates were covered with the lids and wrapped with parafilm to minimize evaporation of the culture medium. These plates were incubated at 37°C with constant shaking at 100 rpm. Samples from both upper and lower chambers were collected at 24 h postincubation and serially diluted with BHI broth and placed on BHI agar plates containing either ampicillin or rifampin. As controls, Rifr B. trehalosi and Ampr M. haemolytica were individually cultured in six-well culture plates with the same culture volume, and CFU assays were performed.
Statistical analysis.
B. trehalosi and M. haemolytica numbers at different time points were expressed as mean CFU/ml with their corresponding standard deviations. The data were statistically analyzed by the Student t test, and P values were determined by using NCSS 2004 (Number Cruncher Statistical System, Kaysville, UT). The term “significant” indicates P < 0.05 with corrections for experimental error made using Bonferroni intervals.
RESULTS AND DISCUSSION
Prevalence of B. trehalosi, M. haemolytica, and P. multocida in pneumonic BHS lungs.
Bacteria associated with BHS pneumonia are members of the genera Mannheimia and Pasteurella, in particular, the species M. haemolytica, B. trehalosi, and P. multocida (6-10, 12-15, 20). Bacteria from the lungs of sick and dead BHS are routinely isolated, characterized, and archived at the Caine Veterinary Teaching Center at Caldwell, ID. Analysis of these isolates over several years revealed that B. trehalosi has been isolated from pneumonic lungs of BHS at a much higher frequency than M. haemolytica (Table 1). However, >85% of B. trehalosi isolated from BHS do not produce the Lkt (28, 32). Furthermore, M. haemolytica has been shown to consistently cause severe bronchopneumonia and death of BHS under experimental conditions (10, 14, 23). These observations prompted us to investigate whether B. trehalosi out-competes or otherwise inhibits the growth of M. haemolytica.
TABLE 1.
Bacteria isolated from pneumonic lungs of BHS
| Region | Bacterium isolated | No. of BHSa | % Total |
|---|---|---|---|
| Hells Canyon | M. haemolytica | 10 | 17 |
| B. trehalosi | 27 | 46 | |
| P. multocida | 35 | 59 | |
| Total | 59 | ||
| Other regions | M. haemolytica | 41 | 50 |
| B. trehalosi | 54 | 66 | |
| P. multocida | 25 | 30 | |
| Total | 82 |
Some BHS were positive for different combinations of bacterial species.
Growth patterns of B. trehalosi and M. haemolytica in liquid cultures.
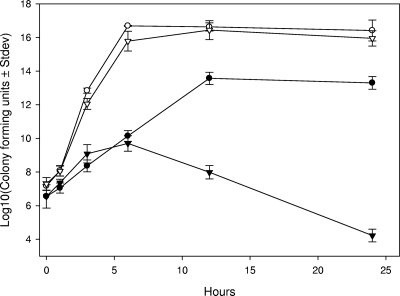
In vitro growth curves show very clearly that B. trehalosi grows faster than M. haemolytica and achieves a higher final cell density when grown in monocultures (Fig. 1) The estimated doubling time for B. trehalosi was ∼10 min compared to ∼27 min for M. haemolytica. The final CFU count at 24 h was 3 logs lower for M. haemolytica compared to B. trehalosi (Fig. 1). Incubation of either species beyond 24 h in the same culture media resulted in rapid decline in the number of live bacteria (not shown). The CFU of M. haemolytica/ml was significantly lower than that of B. trehalosi at all time points (P < 0.0002), except at the time of initial culture inoculation.
FIG. 1.
B. trehalosi exhibits a higher growth rate than M. haemolytica. The CFU for B. trehalosi (○) and M. haemolytica (•)/ml when grown in BHI broth as monocultures and the CFU of Rifr B. trehalosi (▵) and Ampr M. haemolytica (▴)/ml when grown in BHI broth as cocultures were determined. The results are the mean CFU from three independent experiments (± the standard deviation). The starting culture (P > 0.05) and 1-h (P > 0.02) CFU counts were equivalent for these experiments. All other comparisons between B. trehalosi and M. haemolytica were statistically different (P < 0.0002).
Differential detection of B. trehalosi from M. haemolytica in cocultures.
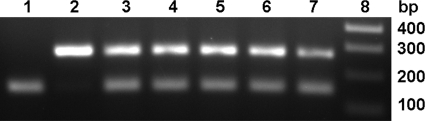
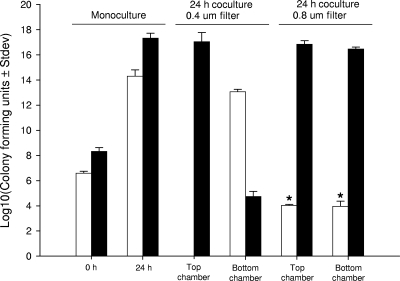
One of the intriguing questions in BHS pneumonia concerns which of the bacterial pathogens isolated from BHS is responsible for disease progression and death. The inability to consistently identify a single pathogen from pneumonic BHS lungs complicates the identification of the bona fide etiological agent or agents of pneumonia. The results of experimental inoculation studies reveal that, unlike many other bacterial pathogens, M. haemolytica alone can cause fatal pneumonia in BHS (10, 14, 23). As described in the previous section, when grown in vitro B. trehalosi has a faster growth rate and a higher final CFU count compared to M. haemolytica, and these findings are consistent with the idea that B. trehalosi overgrows M. haemolytica in pneumonic lungs. When grown as a coculture the dynamics of these populations change dramatically. By conventional methods based on colony morphology, the ability to ferment arabinose or trehalose, and oxidase and catalase activity, M. haemolytica was detected during early hours of coculture but not beyond 6 h (data not shown). In contrast, B. trehalosi was consistently isolated at all of the time points (1 to 24 h) in very high numbers. Serially diluted bacterial samples at each time point were grown on BHI agar and at least 30 colonies were screened by multiplex PCR. Consistent with the results from morphological and biochemical characterization, we could detect M. haemolytica only during early hours of cocultures. As expected, B. trehalosi was positive at all of the time points. We have examined 50 M. haemolytica and 50 B. trehalosi isolates of BHS and domestic sheep origin to evaluate the specificity and sensitivity of the multiplex PCR. Except one isolate (which was identified as M. haemolytica by biochemical tests), there were no discrepancy between culture identification methods and PCR, indicating that the multiplex PCR we developed in the present study is specific and can differentiate the two species. When multiplex PCR assay was performed with direct culture broth (rather than with individual colonies), M. haemolytica could be detected after 24 h of coculture (Fig. 2). This observation indicated the presence of viable M. haemolytica or DNA in cocultures at 24 h but, if viable, there were too few cells available for detection by conventional microbiological methods and colony PCR assay. To circumvent this problem we cocultured Rifr B. trehalosi and Ampr M. haemolytica and serially diluted samples collected at multiple time points. Samples were placed on both rifampin and ampicillin BHI agar plates. The antibiotic-resistant strains used in this experiment did not differ in their rate of growth or final CFU at 24 h when grown as monocultures (Fig. 3). When cocultured, M. haemolytica growth was not affected for up to 4 h, after which growth slowed and began to decline rapidly from 6 to 24 h (Fig. 1). Between 12 and 24 h there was an ∼9-log decline in the number of viable M. haemolytica compared to what would be expected in monoculture. The decline was observed initially as B. trehalosi was transitioning from log to stationary phase, which is consistent with B. trehalosi inhibiting the growth of M. haemolytica when cocultured. Collectively, these findings suggest that it is possible that the failure to consistently detect M. haemolytica in BHS pneumonic lungs is due to both overgrowth of B. trehalosi and simultaneous reduction in cell density of M. haemolytica. The failure of conventional culture methods to detect M. haemolytica in these samples could be due to the absence of this organism or its presence in insufficient numbers to be detected by the conventional methods. The problem of differential detection is compounded by inevitable delays between sample collection under field conditions and laboratory analysis. This delay may contribute to an opportunity for B. trehalosi to further outgrow and inhibit growth and viability of M. haemolytica. From a clinical diagnostic perspective, conventional culture methods are probably not sensitive enough to consistently detect M. haemolytica in field samples.
FIG. 2.
Multiplex PCR assays detect M. haemolytica in B. trehalosi/M. haemolytica cocultures, when colony PCR and bacteriological assays are negative. Lane 1, B. trehalosi monoculture (144 bp); lane 2, M. haemolytica monoculture (267 bp); lanes 3 to 7, B. trehalosi and M. haemolytica in cocultures collected after 0, 6, 12, 24, and 30 h of culture, respectively; lane 8, molecular weight markers. The results of one representative experiment out of three are shown.
FIG. 3.
B. trehalosi inhibits the growth of M. haemolytica by a proximity-dependent mechanism. Ampr M. haemolytica (□) and Rifr B. trehalosi (▪) were cultured in six-well culture plates to establish baseline CFU/ml counts at 0 and 24 h when grown as monocultures. Both strains were then cultivated in six-well plates with either a 0.4- or a 0.8-μm-pore-size filter to separate cells and with Rifr B. trehalosi placed in the top chamber and Ampr M. haemolytica placed in the bottom chamber. Samples were analyzed for CFU/ml after 24 h of culture on ampicillin and rifampin plates. Compared to the expected number of Ampr M. haemolytica grown in monoculture, there was a significant reduction in the number of Ampr M. haemolytica (P < 0.0001) when cultivated with 0.8-μm-pore-size filters where there was clear migration of both species between top and bottom wells. The results are the mean CFU counts from three independent experiments.
Evaluation of culture supernatants for bactericidal activity.
Because B. trehalosi inhibited the growth of M. haemolytica in cocultures, we next determined whether this inhibition was mediated by lytic bacteriophages, bactericidal compounds, or direct cell-to-cell contact. A diverse array of bacteriophages has been isolated from B. trehalosi, M. haemolytica, and P. multocida strains (1, 11), and some of the bacteriophage isolates were able to form plaques with indicator strains of the same species (11). We did not observe any plaque formation with either individual B. trehalosi, M. haemolytica, or coculture supernatants on B. trehalosi or M. haemolytica agar plates at 30, 37, and 42°C (Table 2). This observation suggested the absence of lytic bacteriophages in the culture supernatant fluids under our experimental conditions. No attempts were made to identify bacteriophages (if there were any in culture supernatant fluids) by electron microscopy due to the lack of lytic phage activity. Therefore, we could not rule out the presence of noninfectious prophages in the culture supernatant fluids. To identify any bactericidal compounds in the culture supernatant fluids, filter-sterilized B. trehalosi supernatant fluid-soaked discs or drops (100 to 150 μg of precipitated proteins in 10 μl) were placed in contact with M. haemolytica, but neither of them produced detectable growth inhibition (Table 2). To determine whether the association of both bacteria is needed for B. trehalosi to secrete inhibitory compounds, the same assays were performed with coculture supernatant fluids, but there was no inhibition of M. haemolytica growth on agar plates. Doubling the concentration of culture supernatant did not change this outcome, indicating that B. trehalosi does not produce any growth inhibitory compounds (data not shown). Direct plating of B. trehalosi on M. haemolytica plates was also performed to rule out the presence of inhibitory compounds with a very short half-life or low stability under culture conditions, but no inhibition was detected (data not shown). Taken together, these observations clearly indicate that the inhibition of M. haemolytica growth in cocultures was not due to lytic bacteriophage or secreted bactericidal compounds originating from B. trehalosi.
TABLE 2.
Bactericidal activity of M. haemolytica and B. trehalosi culture supernatant fluidsa
| Culture supernatant | Bactericidal activity | Plaque formation |
|---|---|---|
| M. haemolytica | - | - |
| B. trehalosi | - | - |
| Coculture | - | - |
Five different M. haemolytica and B. trehalosi strains or isolates were examined by using a lytic phage assay at 30, 37, and 42°C. -, lack of bactericidal activity or plaque formation.
B. trehalosi inhibits M. haemolytica growth by a proximity-dependent mechanism.
The failure of the culture supernatant fluid from B. trehalosi to inhibit M. haemolytica growth prompted us to investigate other mechanism(s) of growth inhibition. Aoki et al. (2) have described a novel phenomenon called “contact-dependent growth inhibition” whereupon some strains of Escherichia coli inhibit the growth of other strains E. coli (target). The inhibitory and target E. coli strains were genetically different from each other (2), and inhibition involved a specific ligand interaction between CdiAB from the inhibitory strains and BamA (YaeT), which is displayed on the surface membrane of target strains (3). To date, this phenomenon of contact-dependent inhibition has not been identified in any other bacterial species, although a potential BamA homologue has been annotated for the Pasteurella species (18). It is also possible that quorum-sensing systems may be triggered during coculture that causes M. haemolytica to alter growth phase (22, 27). To determine whether related mechanisms are relevant to our system, contact-dependent inhibition assays were performed using cell culture inserts with PET membranes as described in Materials and Methods. When B. trehalosi in the upper chamber was separated from M. haemolytica in the lower chamber by filters with a pore size of 0.4 μm, the number of B. trehalosi in the upper chamber was not significantly different from that in cultures containing B. trehalosi alone (Fig. 3). The number of M. haemolytica in the lower chamber was lower than that in cultures containing M. haemolytica alone, but this difference may be attributable to the presence of some B. trehalosi in the lower chamber (Fig. 3). The presence of B. trehalosi in the lower chamber was unexpected because most bacteria cannot pass through 0.4-μm pores. In order to ensure that there was no cross-contamination, the following experiments were performed. When M. haemolytica was placed in the upper chamber and B. trehalosi was placed in the lower chamber separated by 0.4-μm-pore-size membranes, a small number of M. haemolytica (∼102 CFU/ml) were detected in the lower chamber, while B. trehalosi was not detected in the upper chamber (data not shown). Similar results were observed in repeated experiments confirming the lack of cross-contamination. Thus, it appears that the chambers allow some passage of cells, presumably due to manufacturing defects. Nevertheless, this experiment was consistent with our earlier conclusion that M. haemolytica numbers decreased when cultured in close proximity to B. trehalosi. This result is consistent with either a contact-dependent or proximity-dependent mechanism where the latter would involve bacteriocins or quorum-sensing systems that are only functional at very high concentrations achieved by labile proteins that are in high concentration proximal to the interacting cells. When larger pore sizes (8.0 μm) were used, the concentrations of B. trehalosi were identical in both the upper and the lower chambers and were not different from that in cultures containing B. trehalosi alone (Fig. 3). This result indicated that B. trehalosi was able to easily cross the membrane boundary. The number of M. haemolytica in both chambers was significantly lower than the number in cultures containing M. haemolytica alone, a finding consistent with growth inhibition mediated by a contact-dependent or proximity-dependent mechanism.
In summary, we have clearly demonstrated here that B. trehalosi induces inhibition of M. haemolytica and that the inhibition is mediated by a mechanism that requires close proximity between the inhibitor (B. trehalosi) and the target (M. haemolytica). This mechanism could be explained by the presence of soluble signaling molecules (e.g., quorum sensing) or bacteriocins, although we found no evidence for such molecules with our assays, and these proteins would probably be quite labile to be undetected in the culture supernatant. Alternatively, such compounds might only be effective given very high concentrations achieved in close proximity but, presumably, such proximity-dependent concentrations would also be difficult to achieve while cultures are being shaken at 200 rpm. Thus, we submit that the more parsimonious explanation involves a contact-dependent mechanism. Regardless of the mechanisms involved, B. trehalosi can overgrow, while simultaneously inhibiting M. haemolytica growth, and if these patterns reflect in vivo conditions, then these results are consistent with the failure to routinely isolate M. haemolytica from pneumonic lungs from sick or dead BHS. Contact-dependent inhibition was first described among E. coli strains, but if a similar mechanism explains our observations it will be the first report of contact-dependent inhibition between two different bacterial species. The molecular basis underlying the inhibition of growth of M. haemolytica is currently under investigation in our laboratory.
Acknowledgments
This research was supported by funds from the Foundation for North American Wild Sheep and its Eastern, Idaho, Oregon, and Washington chapters and by the Rocky Mountain Bighorn Society.
We thank Lisa Orfe for helpful discussions and technical support.
Footnotes
Published ahead of print on 28 December 2009.
REFERENCES
- 1.Ackermann, H. W., and L. Karaivanov. 1984. Morphology of Pasteurella multocida bacteriophages. Can. J. Microbiol. 30:1141-1148. [DOI] [PubMed] [Google Scholar]
- 2.Aoki, S. K., R. Pamma, A. D. Hernday, J. E. Bickham, B. A. Braaten, and D. A. Low. 2005. Contact-dependent inhibition of growth in Escherichia coli. Science 309:1245-1248. [DOI] [PubMed] [Google Scholar]
- 3.Aoki, S. K., J. C. Malinverni, K. Jacoby, R. Pamma, B. N. Trinh, S. Remers, J. Webb, B. A. Braaten, T. J. Silhavy, and D. A. Low. 2008. Contact-dependent growth inhibition requires the essential outer membrane protein BamA (YaeT) as the receptor and the inner membrane transport protein AcrB. Mol. Microbiol. 70:323-340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Avonts, L., E. V. Uytven, and L. D. Vuyst. 2004. Cell growth and bacteriocin production of probiotic Lactobacillus strains in different media. Int. Dairy J. 14:947-955. [Google Scholar]
- 5.Bisgaard, M., and R. Mutters. 1986. Re-investigations of selected bovine and ovine strains previously classified as Pasteurella haemolytica and description of some new taxa within the Pasteurella haemolytica-complex. Acta Pathol. Microbiol. Immunol. Scand. 94:185-193. [DOI] [PubMed] [Google Scholar]
- 6.Blackall, P. J., A. M. Bojesen, H. Christensen, and M. Bisgaard. 2007. Reclassification of [Pasteurella] trehalosi as Bibersteinia trehalosi gen. nov., comb. nov. Int. J. Syst. Evol. Microbiol. 57:666-674. [DOI] [PubMed] [Google Scholar]
- 7.Buechner, H. K. 1960. The bighorn sheep in the United States, its past, present, and future. Wildl. Monogr. 4:3-174. [Google Scholar]
- 8.Callan, R. J., T. D. Bunch, G. W. Workman, and R. E. Mock. 1991. Development of pneumonia in desert bighorn sheep after exposure to a flock of exotic wild and domestic sheep. J. Am. Vet. Med. Assoc. 198:1052-1056. [PubMed] [Google Scholar]
- 9.Coggins, V. L. 1988. The Lostine Rocky Mountain bighorn sheep die-offs and domestic sheep. Biennial Symp. Northern Sheep Goat Council 6:57-64. [Google Scholar]
- 10.Dassanayake, R. P., S. Shanthalingam, C. N. Herndon, P. K. Lawrence, E. F. Cassirer, K. A. Potter, W. J. Foreyt, K. D. Clinkenbeard, and S. Srikumaran. 2009. Mannheimia haemolytica serotype A1 exhibits differential pathogenicity in two related species, Ovis canadensis and Ovis aries. Vet. Microbiol. 133:366-371. [DOI] [PubMed] [Google Scholar]
- 11.Davies, R. L., and I. Lee. 2006. Diversity of temperate bacteriophages induced in bovine and ovine Mannheimia haemolytica isolates and identification of a new P2-like phage. FEMS Microbiol. Lett. 260:162-170. [DOI] [PubMed] [Google Scholar]
- 12.Dunbar, M. R., A. C. S. Ward, and G. Power. 1990. Isolation of Pasteurella haemolytica from tonsillar biopsies of Rocky Mountain bighorn sheep. J. Wildl. Dis. 26:210-213. [DOI] [PubMed] [Google Scholar]
- 13.Foreyt, W. J. 1989. Fatal Pasteurella haemolytica pneumonia in bighorn sheep after direct contact with clinically normal domestic sheep. Am. J. Vet. Res. 50:341-344. [PubMed] [Google Scholar]
- 14.Foreyt, W. J., K. P. Snipes, and R. W. Kasten. 1994. Fatal pneumonia following inoculation of healthy bighorn sheep with Pasteurella haemolytica from healthy domestic sheep. J. Wildl. Dis. 30:137-145. [DOI] [PubMed] [Google Scholar]
- 15.Hobbs, N. T., and M. W. Miller. 1992. Interactions between pathogens and hosts: simulation of pasteurellosis epidemics in bighorn sheep populations, p. 997-1007. In D. R. McCullough and R. H. Barrett (ed.), Wildlife 2001: population. Elsevier Applied Science, Essex, England.
- 16.Jaworski, M. D., D. L. Hunter, and A. C. S. Ward. 1998. Biovariants of isolates of Pasteurella from domestic and wild ruminants. J. Vet. Diagn. Invest. 10:49-55. [DOI] [PubMed] [Google Scholar]
- 17.Jaworski, M. D., A. C. S. Ward, D. L. Hunter, and I. V. Wesley. 1993. DNA analysis of Pasteurella haemolytica biotype T isolates to monitor transmission in bighorn sheep (Ovis canadensis canadensis). J. Clin. Microbiol. 31:831-835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mahasreshti, P. J., G. L. Murphy, J. H. Wyckoff 3rd, S. Farmer, R. E. Hancock, and A. W. Confer. 1997. Purification and partial characterization of the OmpA family of proteins of Pasteurella haemolytica. Infect. Immun. 65:211-218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Miller, M. W. 2001. Pasteurellosis, p. 330-339. In E. S. Williams and I. K. Barker (ed.), Infectious diseases of wild mammals. Iowa State University Press, Ames.
- 20.Monello, R. J., D. L. Murray, and E. F. Cassirer. 2001. Ecological correlates of pneumonia epizootics in bighorn sheep herds. Can. J. Zool. 79:1423-1432. [Google Scholar]
- 21.Murphy, G. L., L. C. Whitworth, K. D. Clinkenbeard, and P. A. Clinkenbeard. 1995. Haemolytic activity of the Pasteurella haemolytica leukotoxin. Infect. Immun. 63:3209-3212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ng, W. L., and B. L. Bassler. 2009. Bacterial quorum-sensing network architectures. Annu. Rev. Genet. doi: 10.1146/annurev-genet-102108-134304. [DOI] [PMC free article] [PubMed]
- 23.Onderka, D. K., S. A. Rawluk, and W. D. Wishart. 1988. Susceptibility of Rocky Mountain bighorn sheep and domestic sheep to pneumonia induced by bighorn and domestic livestock strains of Pasteurella haemolytica. Can. J. Vet. Res. 52:439-444. [PMC free article] [PubMed] [Google Scholar]
- 24.Petras, S. F., M. Chidambaram, E. F. Illyes, S. Froshauer, G. M. Weinstock, and C. P. Reese. 1995. Antigenic and virulence properties of Pasteurella haemolytica leukotoxin mutants. Infect. Immun. 63:1033-1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rudolph, K. M., D. L. Hunter, R. B. Rimler, E. F. Cassirer, W. J. Foreyt, W. J. DeLong, G. C. Weiser, and A. C. Ward. 2007. Microorganisms associated with a pneumonic epizootic in Rocky Mountain bighorn sheep (Ovis canadensis canadensis). J. Zoo Wildl. Med. 38:548-558. [DOI] [PubMed] [Google Scholar]
- 26.Safaee, S., G. C. Weiser, E. F. Cassirer, R. R. Ramey, and S. T. Kelley. 2006. Microbial diversity in bighorn sheep revealed by culture-independent methods. J. Wildl. Dis. 42:545-555. [DOI] [PubMed] [Google Scholar]
- 27.Straight, P. D., and R. Kolter. 2009. Interspecies chemical communication in bacterial development. Annu. Rev. Microbiol. 63:99-118. [DOI] [PubMed] [Google Scholar]
- 28.Sweeney, S. J., R. M. Silflow, and W. J. Foreyt. 1994. Comparative leukotoxicities of Pasteurella haemolytica isolates from domestic sheep and free-ranging bighorn sheep (Ovis canadensis). J. Wildl. Dis. 30:523-528. [DOI] [PubMed] [Google Scholar]
- 29.Tatum, F. M., R. E. Briggs, S. S. Sreevatsan, E. S. Zehr, S. L. Hsuan, L. O. Whiteley, T. R. Ames, and S. K. Maheswaran. 1998. Construction of an isogenic leukotoxin deletion mutant of Pasteurella haemolytica serotype 1: characterization and virulence. Microb. Pathog. 24:37-46. [DOI] [PubMed] [Google Scholar]
- 30.Valdez, R., and P. R. Krausman. 1999. Mountain sheep of North America, p. 19. University of Arizona Press, Tucson.
- 31.Weiser, G. C., W. J. DeLong, J. L. Paz, B. Shafii, W. J. Price, and A. C. Ward. 2003. Characterization of Pasteurella multocida associated with pneumonia in bighorn sheep. J. Wildl. Dis. 39:536-544. [DOI] [PubMed] [Google Scholar]
- 32.Weiser, G. C., D. S. Miller, M. L. Drew, J. C. Rhyan, and A. C. Ward. 2009. Variation in Pasteurella (Bibersteinia) and Mannheimia spp. following transport and antibiotic treatment in free-ranging and captive Rocky Mountain bighorn sheep (Ovis canadensis canadensis). J. Zoo Wildl. Med. 40:117-125. [DOI] [PubMed] [Google Scholar]