Abstract
The transfer range of phage genes was investigated at the single-cell level by using an in situ DNA amplification technique. After absorption of phages, a phage T4 gene was maintained in the genomes of non-plaque-forming bacteria at frequencies of 10−2 gene copies per cell. The gene transfer decreased the mutation frequencies in nonhost recipients.
Recently, whole-genome analyses have revealed that many bacterial genomes contain foreign genes, especially phage genes (9). The phage genes in bacterial genomes include genes for virulence or fitness factors such as extracellular toxins, superantigens, lipopolysaccharide-modifying enzymes, and proteins conferring serum resistance, etc. (1). These findings suggest that the horizontal transfer of phage genes has contributed significantly to the acquisition of new genetic traits and to the genetic diversity of bacteria (1, 9, 10). To truly appreciate the mechanisms behind phage-associated evolution, it is important to understand the frequency and range of transfer of phage genes.
Most phage genomes consist of many genes derived from different origins (5, 8). Some genes are similar to those of other phages with phylogenetically different hosts or are found in the genomes of bacteria that are not the phage hosts. The mosaic nature of phage genomes has been known for some time, and a body of molecular genetic studies of phages to explain the mechanisms that drive this feature have been attempted previously (1, 5). More importantly, the horizontal transfer of phage genes has emerged as a major factor in the evolution of the phage genome. Since recombination between phage and phage/prophage can occur when these elements coexist in the same cell, coinfection with multiple phage species may result in the production of hybrid phage genomes (5). The pathways by which phages exchange genetic material vary dramatically in concert with host ranges. However, conventional plaque assays have shown that the host ranges of the phages studied are narrow. We hypothesized that phage genes can be transferred to more diverse species than previously thought.
In order to accurately quantify DNA movement, gene targeting that does not require cultivation or gene expression is necessary (7). In situ DNA amplification methods allow the visualization of specific DNA sequences inside bacterial cells. In this study, we employed cycling primed in situ amplification-fluorescent in situ hybridization (CPRINS-FISH) to examine the possible range and frequency of the transfer of phage genes. CPRINS uses one primer and results in linear amplification of the target DNA inside cells, and multiply labeled fluorescent probe sets are applied for detection of the amplicons to improve the specificity and sensitivity of CPRINS (3). Previously, CPRINS-FISH did clarify the movement of DNA of a specific gene among Escherichia coli cells at the single-cell level (4).
Enterobacterial phages P1 and T4 infect E. coli and have been well studied. P1 can exist as circular DNA within the bacterial cell as if it were a plasmid. Phage T4 is capable of undergoing only a lytic life cycle and not the lysogenic life cycle. Conventional methods using plaque assays have shown that the host of P1 and T4 is E. coli, but orthologous phage genes have been found in bacteria other than E. coli (6, 8). In the present study, strains of Enterobacteriaceae were allowed to grow on agar medium after the phage was adsorbed, and the maintenance of the transferred phage gene in the bacterial genomes was examined at the community level by quantitative real-time PCR and at the single-cell level by CPRINS-FISH.
The following bacterial strains were used for maintenance experiments: Citrobacter freundii IFO 12681, Enterobacter aerogenes BM 2688, E. coli NBRC 12713, a Proteus mirabilis clinical isolate, Salmonella enterica serovar Enteritidis IID 640, and Yersinia enterocolitica IID 981. The bacterial strains were grown in Luria-Bertani (LB) medium (1% tryptone, 0.5% yeast extract, 0.5% NaCl; Nacalai Tesque Inc., Kyoto, Japan) at 37°C overnight.
Stationary-phase cultures of 500 μl were incubated with 500 μl of SM buffer (50 mmol liter−1 Tris-HCl [pH 7.5], 100 mmol liter−1 NaCl, 8 mmol liter−1 MgSO4, 0.01% gelatin) containing the phage P1kc NBRC 20008 (2) or T4GT7 (11) at 37°C for 10 min at a multiplicity of infection of 1:1 (ratio of PFU of the phage to CFU of the recipient bacterium). The concentration of bacterial cells was adjusted to 109 cells ml−1. After 10 min of incubation, the diluted cell suspension (105 cells) was filtered through a polycarbonate filter (Advantec, Tokyo, Japan) with a pore size of 0.2 μm and a diameter of 25 mm. Cells trapped on the filter were cultured on LB agar medium at 37°C for 24 h. The filter was transferred into a microtube, and cells on the filter were suspended in 1 ml of sterile deionized water. The numbers of cells in the suspension and cells remaining on the filter were determined by using an epifluorescence microscope (see below) after staining of the samples with 1 μg ml−1 of 4′,6-diamidino-2-phenylindole (DAPI; Sigma-Aldrich Japan, Tokyo). The level of recovery of cells from the filter into sterile deionized water was about 99%. The cultured cells were subjected to real-time PCR and CPRINS-FISH.
For real-time PCR, bacterial DNA was extracted using a QIAamp DNA isolation kit (Qiagen, Tokyo, Japan). The cell suspension was mixed with 10 mg ml−1 of lysozyme solution and incubated at 37°C for 1 h. DNA extraction was then performed according to the kit manufacturer's instructions. Table 1 lists the oligonucleotide primers for PCR and CPRINS and the polynucleotide probes used in the present study. Tail fiber genes from phages P1kc and T4GT7 were quantified by real-time PCR with a LightCycler system (Roche Diagnostics, Tokyo, Japan). LightCycler FastStart DNA master SYBR green I (Roche Diagnostics) was used with 5 mmol liter−1 Mg2+ and 0.5 μmol liter−1 (each) primers targeting the tail fiber genes of P1kc (P1-tail931f and P1-tail1148r) and T4GT7 (T4-tail2770f and T4-tail2983r). After a hot start for 10 min at 95°C, 40 cycles of PCR were run with denaturation at 94°C for 15 s, annealing at 60°C for 10 s, extension at 72°C for 10 s, and fluorescence detection at 83°C for 5 s. The known amounts of PCR products from the phage DNA (101 to 107 copies per reaction) were used for the standard curves to quantify the target DNA. To confirm the specificity of the reaction after real-time PCR, the PCR mixture was collected in a glass capillary and subjected to agarose gel electrophoresis in addition to a melting-curve analysis with the LightCycler system. The maintenance frequencies determined by real-time PCR were recorded as the copy number of the phage tail fiber gene per bacterial genome detected by staining with PicoGreen (Invitrogen, Tokyo, Japan) after cultivation of cells on LB agar medium for 24 h as described above. The frequencies were determined in triplicate for each sample. The increase in the phage gene copy number was determined by comparing the copy numbers in cells on the filter before and after cultivation. The phage gene copy number in cells on the filter was determined by the following formula: (total number of cells determined by DAPI staining) × (phage tail fiber gene copy number determined by real-time PCR)/(bacterial genome copy number determined by PicoGreen staining).
TABLE 1.
Probes and primers designed in this study
| Name | Target | Type | Nucleotide sequence (5′-3′) |
|---|---|---|---|
| P1-tail931f | Tail fiber gene of phage P1 | Primer | AACGACCCGAATTACAGCAC |
| P1-tail1148r | Tail fiber gene of phage P1 | Primer | AGTGCTGCTGCAAGCTCATA |
| T4-tail2770f | Tail fiber gene of phage T4 | Primer | AGCACAAATGGTGAGCACAG |
| T4-tail2983r | Tail fiber gene of phage T4 | Primer | TTGCTACCGTGTGGGTATGA |
| T4-tail2664 | Tail fiber gene of phage T4 | Probe | GGCTTCAAGTACTGACTTAGGTACTAAAACCACATCAAGCTTTGACTATGGTACG |
| T4-tail2720 | Tail fiber gene of phage T4 | Probe | AAGGGAACTAACAGTACGGGTGGACACACTCACTCTGGTAGTGGTTCTA |
| T4-tail2769 | Tail fiber gene of phage T4 | Probe | TAGCACAAATGGTGAGCACAGCCACTACATCGAGGCATGGAATGG |
| T4-tail2818 | Tail fiber gene of phage T4 | Probe | GGTGTAGGTGGTAATAAGATGTCATCATATGCCATATCATACAGGGCGGG |
| T4-tail2869 | Tail fiber gene of phage T4 | Probe | GGGAGTAACACTAATGCAGCAGGGAACCACAGTCACACTTTCTCTTTTGGG |
| T4-tail2922 | Tail fiber gene of phage T4 | Probe | TAGCAGTGCTGGCGACCATTCCCACTCTGTAGGTATTGGTGCTCATA |
CPRINS-FISH targeting the tail fiber gene of phage T4GT7 was performed as described by Kenzaka et al. (3, 4), except for the probe/primer sequences and thermal conditions. After cell wall permeabilization by lysozyme treatment (3), the CPRINS reaction was performed under the following conditions: a hot start at 95°C for 9 min, denaturation at 94°C for 1 min, annealing at 60°C for 30 s, and extension at 72°C for 1.5 min for primer T4-tail2983r. Amplification was repeated for 30 cycles by using a thermal cycler (PTC-200; Bio-Rad Laboratories, Inc.). After amplification, filters were rinsed with 0.1% Nonidet P-40 and sterile deionized water, dehydrated in 99% ethanol, and vacuum dried. Hybridization with Alexa Fluor 546-labeled polynucleotide probes (T4-tail2664, T4-tail2720, T4-tail2769, T4-tail2818, T4-tail2869, and T4-tail2922), washing, and DAPI staining were performed as described in a previous study (4). In order to exclude the possibility of nonspecific probe binding to cell structures other than the target DNA in the target cells, FISH using laboratory strains without amplification of target DNA and CPRINS-FISH targeting the tail fiber genes in E. coli strains that did not carry the genes were performed.
In order to examine the infection ranges of phages, plaque assays and direct counting of phages were performed. Plaque assays were performed with LB soft agar (0.8% agar) as described by Kenzaka et al. (4). For the direct counting, phages were stained with 5× SYBR gold (Invitrogen, Tokyo, Japan) and trapped onto an Anodisc filter (Whatman Japan, Tokyo) with a pore size of 0.02 μm and a diameter of 25 mm.
The cells or phage particles on the filters were observed under an epifluorescence microscope (E-400; Nikon, Tokyo, Japan) with the Nikon filter sets UV-2A (EX300-350, DM400, and BA420) for DAPI, B-2A (EX450-490, DM505, and BA520) for SYBR gold, and HQ-CY3 (G535/50, FT565, and BP610/75) for Alexa Fluor 546. Images were acquired using a Retiga 2000R cooled charge-coupled device camera (QImaging, Surrey, BC, Canada), and at least 2,000 DAPI- or SYBR gold-stained objects per sample were counted. The maintenance frequencies determined by CPRINS-FISH were recorded as the number of CPRINS-FISH-positive cells divided by the total direct count of recipient cells after cultivation as described above. The frequencies were determined in triplicate for each sample.
After cultivation on LB agar medium for 24 h, the total number of cells on the filter as determined by DAPI staining increased by 8.7 × 102- to 1.1 × 104-fold (Table 2). Real-time PCR showed that the phage P1kc gene copy number increased only in plaque-forming strains (E. coli and E. aerogenes) and not in non-plaque-forming strains (Table 2). In contrast, the phage T4GT7 gene copy number increased in both plaque-forming and non-plaque-forming strains by 7.6 × 101- to 7.0 × 104-fold. The maintenance frequencies were more than 10−2 gene copies per bacterial genome (Table 2). Direct observation via epifluorescence microscopy showed that progeny phages were not produced in the non-plaque-forming strains (Table 2), and thus, fragments of phage genes were thought to integrate into the genomes of non-plaque-forming strains and replicate with the bacterial genomes.
TABLE 2.
Frequencies of maintenance of phage P1kc and T4GT7 genes in Enterobacteriaceae strains
| Phage | Recipient | Result for infection range indicator: |
Increase in total no. of cellsc | Increase in phage gene copy no. (SD)d | Maintenance frequency (SD) as determined bye: |
||
|---|---|---|---|---|---|---|---|
| Plaque formationa | Production of progenyb | Real-time PCR | CPRINS-FISH | ||||
| P1kc | C. freundii | − | − | 7.0 × 103 | None | <1.5 × 10−3 | ND |
| E. aerogenes | + | + | 1.7 × 103 | 7.7 × 103 (6.5 × 103) | 5.0 × 100 (4.2 × 100) | ND | |
| E. coli | + | + | 7.2 × 103 | 5.5 × 103 (2.7 × 103) | 9.1 × 10−1 (0.5 × 10−1) | ND | |
| P. mirabilis | − | − | 7.4 × 103 | None | <1.5 × 10−3 | ND | |
| S. Enteritidis | − | − | 8.4 × 103 | None | <1.7 × 10−4 | ND | |
| Y. enterocolitica | − | − | 4.6 × 103 | None | <1.8 × 10−4 | ND | |
| T4GT7 | C. freundii | − | − | 1.5 × 103 | 7.5 × 103 (4.0 × 103) | 8.3 × 10−1 (4.4 × 10−1) | 8.6 × 10−2 (3.4 × 10−2) |
| E. aerogenes | + | + | 8.7 × 102 | 1.2 × 103 (0.8 × 103) | 8.0 × 10−1 (5.0 × 10−1) | 4.0 × 10−1 (0.7 × 10−1) | |
| E. coli | + | + | 1.1 × 104 | 7.0 × 104 (2.7 × 104) | 8.0 × 101 (3.0 × 10) | 2.1 × 10−1 (0.4 × 10−1) | |
| P. mirabilis | − | − | 4.0 × 103 | 5.8 × 103 (4.2 × 103) | 3.3 × 10−1 (2.4 × 10−1) | 3.4 × 10−2 (2.2 × 10−2) | |
| S. Enteritidis | − | − | 1.0 × 104 | 7.6 × 101 (5.0 × 101) | 1.0 × 10−2 (0.7 × 10−2) | 8.8 × 10−2 (2.0 × 10−2) | |
| Y. enterocolitica | − | − | 3.6 × 103 | 1.6 × 104 (0.4 × 104) | 6.1 × 10−1 (1.6 × 10−1) | 2.2 × 10−2 (2.9 × 10−2) | |
Plaque formation on soft agar was tested.
The production of progeny phage particles was observed via epifluorescence microscopy.
The increase (n-fold) in the total number of cells during bacterial growth for 24 h was determined via epifluorescence microscopy.
The increase (n-fold) in the copy number of the phage tail fiber gene during bacterial growth for 24 h was determined by real-time PCR. Values in parentheses indicate standard deviations of results for triplicate samples.
Maintenance frequencies were determined by real-time PCR and CPRINS-FISH analyses targeting the phage tail fiber gene and are shown as the phage tail fiber gene copy numbers per bacterial genome and the numbers of gene-positive cells divided by the total numbers of cells, respectively. Values in parentheses indicate standard deviations of results for triplicate samples. ND, not determined.
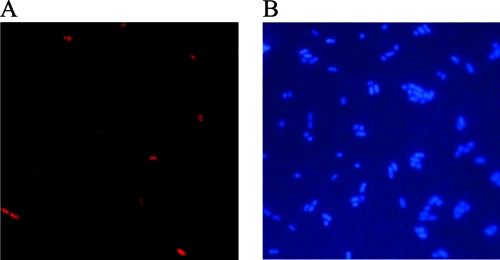
Real-time PCR provided a copy number for the target phage gene in the whole population, but the location of the target phage gene and the frequency of cells carrying the target gene were unclear. In addition, bacterial genomic DNA, which was measured using PicoGreen, included phage DNA, and thus the frequencies measured by dividing by the amount of bacterial genomic DNA were probably less accurate than those measured as described below. In order to confirm that the phage gene was located inside bacterial cells and determine a more accurate maintenance frequency for total cells, CPRINS-FISH targeting the tail fiber gene of phage T4GT7 was performed. CPRINS-FISH visualized the target phage gene in individual cells under an epifluorescence microscope (Fig. 1). It showed that the frequencies of maintenance of the tail fiber gene, expressed as the number of gene-positive cells divided by the total number of cells, were 2.1 × 10−1 to 4.0 × 10−1 for plaque-forming strains after growth on LB medium for 24 h (Table 2). Since phage T4GT7 is capable of undergoing only a lytic life cycle, CPRINS-FISH would detect cells in which the phage gene was replicating. For non-plaque-forming strains, the maintenance frequencies were 2.2 × 10−2 to 8.8 × 10−2 (Table 2). If the gene was amplified by the CPRINS reaction outside bacterial cells, the amplicon would not accumulate inside bacterial cells and they would not exhibit bright fluorescence. Therefore, CPRINS-FISH proved that a part of the phage T4GT7 gene was located inside cells of non-plaque-forming strains. The tail fiber gene is responsible for the phage tail structure. The DNA sequences of the phage genes responsible for phage morphology have been found in many bacterial genomes (1, 5).
FIG. 1.
Visualization of E. coli cells carrying the tail fiber gene transferred by phage T4GT7. (A) After being mixed with phages for 10 min, E. coli NBRC 12713 cells were cultured for 24 h and subjected to CPRINS-FISH targeting the phage gene. Only cells having amplified tail fiber gene products emitted the fluorescence of the Alexa Fluor 546-labeled probe under green excitation (exposure, 0.5 s). (B) All DAPI-stained bacterial cells were visualized under UV excitation (exposure time, 0.1 s).
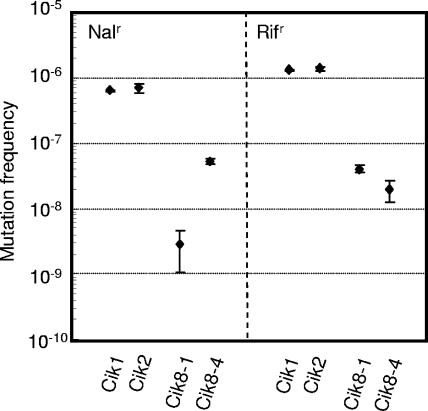
In order to explore the effect of integration of the phage gene into the bacterial genome on bacterial heredity, we determined the mutation frequency for a C. freundii strain that acquired the phage T4GT7 gene. Two colonies which acquired the phage T4GT7 gene were screened by colony PCR with T4-tail2770f and T4-tail2983r primers and designated Cik8-1 and Cik8-4. Mutation frequencies were determined with LB medium containing 150 μg ml−1of rifampin (rifampicin) or 10 μg ml−1of nalidixic acid. The mutation frequencies associated with nalidixic acid resistance decreased by 12- to 240-fold and the frequencies associated with rifampin resistance decreased by 40- to 83-fold compared to those for the parent strains (Fig. 2). Mutation increases genetic variation. The decreased mutation frequency would contribute to the genetic stability of the genome in individual cells but not to genetic variation in the population. Our results show that phage T4GT7 was capable of affecting the genomic properties of C. freundii, which was thought previously not to be the host, although the mechanism by which mutation frequencies decreased remains unknown. Further experiments are required to clarify the molecular mechanism by which mutation frequencies altered after gene transfer.
FIG. 2.
Mutation frequencies for T4GT7-infected C. freundii strains. Mutation frequencies were determined with LB agar medium containing nalidixic acid or rifampin. Cik8-1 and Cik8-4 were strains which acquired a phage gene transferred from phage T4GT7. Cik1 and Cik2 were the parent strains.
In summary, during growth on agar medium after the phage was allowed to be adsorbed by strains of Enterobacteriaceae, the phage P1kc gene was not maintained in non-plaque-forming strains but the phage T4GT7 gene was maintained in more diverse species than previously expected. The transfer of foreign DNA molecules (DNA entry) into a bacterium is an important first step in genetic diversification through horizontal gene transfer. A previous study reported that phage P1kc is capable of injecting DNA into non-plaque-forming E. coli cells (4), but the phage P1kc gene was not maintained during bacterial growth in the present study. The results showing the difference in maintenance between phage P1kc and T4GT7 genes suggest that the maintenance of transferred phage genes depends on phage gene sequences or other phage factors. When maintained, the phage gene could alter the mutation frequency for bacteria that acquired the gene, affecting the genomic variability at the population level. Conventionally, phage-bacterium interaction has been studied with certain models consisting of a phage and a bacterium in which the phage can multiply (12, 13). Our results indicate the importance of the dynamic of phage genes among diverse bacteria that were previously thought not to be hosts and the hereditary impact of phage gene transfer on such bacteria.
Acknowledgments
This work was supported by a JSPS Grant-in-Aid for Young Scientists (B) (18780055).
Footnotes
Published ahead of print on 18 December 2009.
REFERENCES
- 1.Brüssow, H., C. Canchaya, and W. D. Hardt. 2004. Phages and the evolution of bacterial pathogens: from genomic rearrangements to lysogenic conversion. Microbiol. Mol. Biol. Rev. 68:560-602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Enomoto, M., and B. A. Stocker. 1974. Transduction by phage P1kc in Salmonella typhimurium. Virology 60:503-514. [DOI] [PubMed] [Google Scholar]
- 3.Kenzaka, T., S. Tamaki, N. Yamaguchi, K. Tani, and M. Nasu. 2005. Recognition of individual genes in diverse microorganisms by cycling primed in situ amplification. Appl. Environ. Microbiol. 71:7236-7244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kenzaka, T., K. Tani, A. Sakotani, N. Yamaguchi, and M. Nasu. 2007. High-frequency phage-mediated gene transfer among Escherichia coli cells, determined at the single-cell level. Appl. Environ. Microbiol. 73:3291-3299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lawrence, J. G., G. F. Hatful, and R. W. Hendrix. 2002. Imbroglios of viral taxonomy: genetic exchange and failings of phenetic approaches. J. Bacteriol. 184:4891-4905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Łobocka, M. B., D. J. Rose, G. Plunkett III, M. Rusin, A. Samojedny, H. Lehnherr, M. B. Yarmolinsky, and F. R. Blattner. 2004. Genome of bacteriophage P1. J. Bacteriol. 186:7032-7068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Maruyama, F., T. Kenzaka, N. Yamaguchi, K. Tani, and M. Nasu. 2005. Visualization and enumeration of bacteria carrying a specific gene sequence by in situ rolling circle amplification. Appl. Environ. Microbiol. 71:7933-7940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Miller, E. S., E. Kutter, G. Mosig, F. Arisaka, T. Kunisawa, and W. Rüger. 2003. Bacteriophage T4 genome. Microbiol. Mol. Biol. Rev. 67:86-156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ochman, H., J. G. Lawrence, and E. A. Groisman. 2000. Lateral gene transfer and the nature of bacterial innovation. Nature 405:299-304. [DOI] [PubMed] [Google Scholar]
- 10.Weinbauer, M. G., and F. Rassoulzadegan. 2004. Are viruses driving microbial diversification and diversity? Environ. Microbiol. 6:1-11. [DOI] [PubMed] [Google Scholar]
- 11.Wilson, G. G., K. Y. Young, G. J. Edlin, and W. Konigsberg. 1979. High-frequency generalised transduction by bacteriophage T4. Nature 280:80-82. [DOI] [PubMed] [Google Scholar]
- 12.Wommack, K. E., and R. R. Colwell. 2000. Virioplankton: viruses in aquatic ecosystems. Microbiol. Mol. Biol. Rev. 64:69-114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zinder, N. D., and J. Lederberg. 1952. Genetic exchange in Salmonella. J. Bacteriol. 64:679-699. [DOI] [PMC free article] [PubMed] [Google Scholar]