Abstract
Conversion of forests to farmland permanently lowers atmospheric methane consumption due to unresolved reasons. Alphaproteobacterial methanotrophs were predominant in forested soils and gammaproteobacterial species were predominant in farmland soils of subtropical ferralsols in Brazil. The capability of atmospheric methane consumption was obliterated in farmland soils, suggesting a shift from oligotrophic to copiotrophic species.
Aerobic methanotrophic communities in aerated soils are the largest biological sink for atmospheric methane (CH4) (3, 4, 10, 17). Forest soil communities exhibit the highest consumption rates on a global scale (4, 26). The conversion to farmland lowers the sink capacity (26, 28, 29), but it can be restored by afforestation (27). Sink reconstitution is faster in tropical regions than in temperate regions (12, 15). The impact of this land use change on atmospheric CH4-consuming communities has scarcely been addressed (21, 25). The objective of the present study was to gain insight into CH4 oxidation kinetics and the composition of methanotrophic communities in soils from a naturally forested site, an afforested site, and two farmland sites from a subtropical region in South America (Mata Atlântica, Brazil).
Soils from an area in Brazil to the west of São Paulo and close to Caucaia do Alto (46°55′W to 47°06′W, 23°39′S to 23°47′S) were sampled in April 2005. The soil type at any site was a ferralsol (8a). The natural forest site was not managed for over 100 years (pH 3.6), whereas the reforested site was restored 20 years ago (pH 3.8). The humus layer was removed before sampling the forest soils. A nearby located conventionally farmed acre (pH 6.1) and an organically farmed acre (pH 6.2) were also sampled. Both sites were forests 50 to 100 years ago, and the forest sites were part of a formerly continuously forested region. Soils were sampled in three spatial replicates from the top 10 cm. Samples were pooled, transported within 2 weeks to Germany, and manually homogenized, and aliquots were stored at −20°C for molecular analyses and at 2°C for activity analyses.
Atmospheric CH4 consumption and kinetic parameters.
Ten grams of sieved soil samples was incubated in 150-ml gastight vials in triplicate at 22°C. The headspace contained air with 1.75 parts per million by volume (ppmv) CH4 or a specific mixing ratio adjusted by adding pure CH4 (99.000%; Riessner-Gase, Germany). Headspace mixing ratios were measured over a period of 6 h using gas chromatography with a flame ionization detector (GC-FID) (6). Soil samples from the forest sites had the potential to oxidize atmospheric CH4, whereas neither of the farmland soil samples did (Table 1). Measured atmospheric CH4 consumption rates were lower than those obtained from other Brazilian forest soil samples (20 to 33 pmol g [dry weight] of soil−1) (24), probably due to the long transport. Michaelis-Menten kinetics was determined according to a previous study (6). Apparent half-saturation constants [Km(app)] (Table 1) of the two forest soils were similar to those of other Brazilian forest soils (9). Thus, soils from these two forest sites might have been sinks for atmospheric CH4. Both of the farmland soil samples had higher maximal flux rates [νmax(app)] than the forest soil samples and lacked atmospheric CH4 consumption (Table 1), suggesting a soil indigenous CH4 source that provided in situ mixing ratios above atmospheric levels.
TABLE 1.
Methane oxidation kinetic parameters of soil samples
| Soil sample | Methane oxidation kinetic parametersa |
||
|---|---|---|---|
| vatm (pmol g [dry wt] of soil−1 h−1)b | Km(app) (ppm) | vmax(app) (pmol g [dry wt] of soil−1 h−1) | |
| Forest sites | |||
| Natural forest | 0.03 ± 0.5 | 89.5 ± 17.8 | 1.8 ± 0.01 |
| Afforested | 0.03 ± 0.3 | 260.0 ± 42.9 | 5.9 ± 0.04 |
| Farmland sites | |||
| Organic farming | ND | — | 81.0 ± 0.52 |
| Conventional farming | ND | — | 53.0 ± 0.31 |
Errors shown are standard errors of the means. ND, not detectable, —, not measured.
vatm, velocity at 1.75 ppmv methane mixing ratio.
Analyses of methanotrophic community structure.
From every pooled soil sample, a pmoA gene (encoding the hydroxylase of the particulate CH4 monooxygenase [pMMO]) library was set up. Phospholipid fatty acid (PLFA) stable isotope probing (SIP) at 0.5% CH4 was performed with previously unfrozen soil samples (forest sites) and frozen soil samples (farmland sites). DNA was extracted in four replicates by using a bead beating protocol (Fast Spin kit; Bio 101). pmoA genes were amplified from pooled DNA extracts using primers A189 and A682 (11), according to Ricke and coworkers (22). pmoA PCR products were cloned using the pGEM-T vector system II (Promega, Germany). Gene inserts were sequenced with the services of Macrogen Inc. (South Korea). Resulting pmoA sequences were phylogenetically affiliated using ARB (18) and implemented algorithms (neighbor joining; Tree-Puzzle) (see methods in File S2 of the supplemental material) (8, 19). Every library exhibited coverage of more than 90%. Thus, the analyzed number of sequences (Table 2) was sufficient to assess the expected genotypes. Sequences may be found in the EMBL database (accession numbers FN394081 to FN394214). The gene mmoX (encoding the hydroxylase subunit of the soluble MMO) could not be amplified (primers mmoXf945 and mmoXB1401) (1, 15), indicating that Methylocella species were not detectable, as well as type I or type II methanotrophs harboring this enzyme.
TABLE 2.
Frequency of genotypes in pmoA gene libraries
| pmoA gene library | Frequency of genotypes (%)a |
|||
|---|---|---|---|---|
| Forests |
Farmland |
|||
| Natural (n = 37) | Afforested (n = 35) | Organic (n = 31) | Conventional (n = 30) | |
| Methylococcaceae (type I/X) | ||||
| Methylococcus | ND | ND | ND | 36.7 |
| Crenothrix polyspora-like | ND | ND | 71.0 | ND |
| Methylocystaceae (type II) | ||||
| Methylocystis | ND | ND | ND | 63.3 |
| Beijerinckiaceae related (type II) | ||||
| USCα | 89.2 | 94.3 | 6.5 | ND |
| Cluster 5 | 5.4 | 2.8 | ND | ND |
| Ambiguous affiliation to pmoA or amoA | ||||
| Cluster 2 | ND | ND | 6.5 | ND |
| Cluster WC306-54 | 5.4 | 2.8 | ND | ND |
| Ammonia-oxidizing bacteria (Betaproteobacteria) | ND | ND | 12.9 | ND |
n, total number of phylogenetically analyzed gene inserts per library; ND, not detectable.
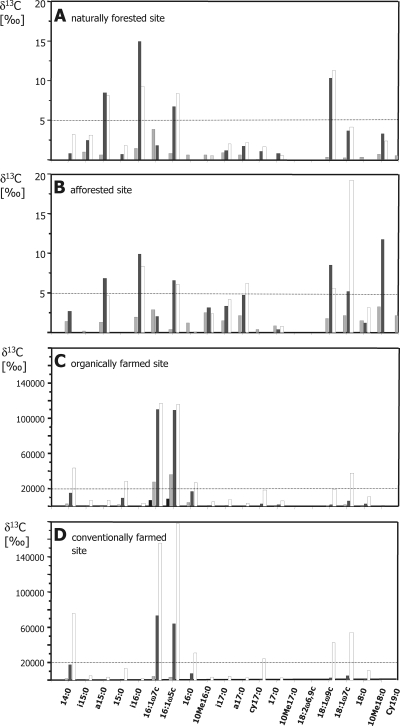
Four aliquots (10 g) of sieved fresh soil were mixed with sterile quartz sand to improve labeling efficiency, used to fill gastight 150-ml vials, and incubated with 0.5% 13CH4 at 30°C. The gravimetric water content was adjusted to 30 to 40% by addition of sterile water. Per site and 13C treatment, four incubations were run that were individually stopped at 2, 6, 13, or 30 days. CH4 concentrations were monitored by a GC-FID. Retrieved soil samples were immediately stored at −80°C until PLFAs were extracted and subjected to GC-combustion-isotope ratio mass spectrometry (GC-C-IRMS) (see methods in File S2 of the supplemental material). Absolute 13C incorporation, referred to as ng PLFA (see Table S1 and the methods in File S2 of the supplemental material), was converted to δ13C values for a better comparison of the samples (see Fig. 2).
FIG. 2.
δ13C values of detected PLFA at 2 (black), 6 (light gray), 13 (dark gray), and 30 (white) days from the natural forest (A), reforested (B), organically farmed (C), and conventionally farmed (D) soils. PLFAs are ordered according to their retention times.
Methanotrophic communities in forest and agricultural soils.
pmoA gene libraries from the naturally forested and afforested sites were dominated by genotypes next related to Beijerinckiaceae. In both gene libraries, the genotype USCα was predominant, and both clusters 5 and WC306-54 were detected. WC306-54-related genotypes could not be functionally assigned to the pmoA or amoA gene (hydroxylase of ammonia monooxygenase) due to their intermediate phylogenetic positions (Fig. 1; Table 2). PLFA 13C incorporation patterns were similar in soil samples from both forest soils (Fig. 2A and B). PLFAs a15:0, i16:0, 16:1ω5c, 18:1ω9c, and cy19:0 were significantly labeled in both soils (δ13C > 5‰) (Fig. 2A). 13C incorporation of PLFAs from forest soils was low compared with that from farmland soils, although the same CH4 concentrations were used (see Table S1 in the supplemental material). This indicates that primarily the same species that were in situ active and detectable by pmoA utilized 13CH4. It cannot be excluded that type I methanotrophs were also present, since a 16:1 PLFA was labeled. Type I methanotrophs were not present in gene libraries, suggesting that the 16:1 PLFA indicated cross-feeding by nonmethanotrophic soil microbes or type II methanotrophs of the genus Methylocystis (5).
FIG. 1.
Phylogenetic affiliation of PmoA/AmoA sequences retrieved from gene libraries using Tree-Puzzle. OA, organically farmed site; CA, conventionally farmed site; AF, afforested site; NF, natural forest. In parentheses, numbers of similar genotypes found in the same library that share ≥87% sequence similarity on amino acid level. Scale bar represents 0.1 change per amino acid. Rectangles, nodes that were confirmed by an alternative treeing method. Further details are given in the methods in File S2 of the supplemental material.
Communities in farmland soils differed from those in soils from both forest sites, as revealed by pmoA (Table 2). In the agricultural soil under conventional farming, genotypes of Methylocystis and Methylococcus spp. were prevalent. A Crenothrix polyspora-like genotype was frequently found in the organically farmed acre soil. Beijerinckiaceae spp. and cluster 2 were less frequently found (Table 2). 13CH4 incorporation in PLFAs was higher in farmland samples than in forest soil samples (Fig. 2), and consistently, maximal CH4 oxidation rates were higher in farmland soil samples (Table 1). These observations may indicate that methanotrophic species in farmland soils were adapted to mixing ratios above atmospheric levels. Farmland soil samples were frozen for transport before PLFA SIP. This did not affect the composition of methanotrophic community, since PLFA and pmoA data were basically congruent regarding the prevalence of type I or II methanotrophs. PLFAs known from Methylococcaceae (type I/X) (10) and Methylocystis (5) were significantly labeled; type II-specific ones at a later time point indicated that type II methanotrophs were less abundant or active (δ13C > 20,000‰) (Fig. 2C and D). At the end of the incubation period, PLFA 18:1ω7c was detected, with a δ13C value above 20,000‰, suggesting a background activity of the present Beijerinckiaceae species.
Conclusions.
The conversion of a natural subtropical forest to farmland led to a loss of atmospheric CH4 consumption and a shift from Beijerinckiaceae species to Methylococcaceae and Methylocystaceae species. The data also suggest a restoration of the original community and the atmospheric CH4 sink after afforestation. A similar phenomenon was described by a previous study of tropical soils in Thailand (15). In temperate New Zealand soils, type II methanotrophs were active under a pine forest, whereas type I methanotrophs were active in adjacent pastures. Afforested sites consumed more atmospheric CH4 than the nonforested ones (25, 27). In contrast to these results, afforestation of a boreal grassland led to reduced atmospheric CH4 uptake and a reduction of the methanotrophic biomass (20).
The current data suggest that soil capability of atmospheric CH4 consumption resulted from the dominance of few oligotrophic methanotrophic species (for examples, see reference 16). Methylocystis strains can utilize atmospheric CH4 (2, 7, 13). However, it is unlikely that the detected Methylocystis spp. in agricultural soils utilized atmospheric CH4, since the required pmoA2 gene was not detected (2). Consequently, detected Methylococcaceae and Methylocystaceae can be regarded as copiotrophic, whereas detected Beijerinckiaceae represented oligotrophs. Increased CH4 concentration and availability of nitrogen may alter methanotrophic communities (14, 22). Fertilization may also reduce the abundance of atmospheric CH4 consuming methanotrophs in soils (20). pH values shifted from approximately 3 to 6 after deforestation. Hence, the sink capacity might have been reduced after deforestation, since oligotrophic species were outcompeted by copiotrophic species due to fertilization, higher organic matter turnover in farmlands, and/or increased pH.
Supplementary Material
Acknowledgments
This study was financed by the German Ministry for Education and Research (grant BMBF 01 LB 0202), the University of Bayreuth (UBT), and the Deutsche Forschungsgemeinschaft (DFG; grants KO2912/2-1 and GL 327/4-4).
We thank R. Mertel for the construction of pmoA gene libraries, E. Bähr for soil sampling in Brazil, and the heads of the Department of Ecological Microbiology (H. L. Drake, UBT) and of the Institute of Soil Science and Soil Geography (W. Zech, UBT) for general support and advice.
Footnotes
Published ahead of print on 28 December 2009.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Reference deleted.
- 2.Baani, M., and W. Liesack. 2008. Two isozymes of particulate methane monooxygenase with different methane oxidation kinetics are found in Methylocystis sp. strain SC2. Proc. Natl. Acad. Sci. U. S. A. 105:10203-10208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Conrad, R. 1996. Soil microorganisms as controllers of atmospheric trace gases (H2, CO, CH4, OCS, N2O, and NO). Microbiol. Rev. 60:609-640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dalal, R. C., and D. E. Allen. 2008. Greenhouse gas fluxes from natural ecosystems. Aust. J. Bot. 56:369-407. [Google Scholar]
- 5.Dedysh, S. N., S. E. Belova, P. L. E. Bodelier, K. V. Smirnova, V. N. Khmelenina, A. Chidthaisong, Y. A. Trotsenko, W. Liesack, and P. F. Dunfield. 2007. Methylocystis heyeri sp. nov., a novel type II methanotrophic bacterium possessing ‘signature’ fatty acids of type I methanotrophs. Int. J. Syst. Evol. Microbiol. 57:472-479. [DOI] [PubMed] [Google Scholar]
- 6.Degelmann, D. M., S. Kolb, and W. Borken. 2009. Methane oxidation kinetics differ in European beech and Norway spruce soils. Eur. J. Soil Sci. 60:499-506. [Google Scholar]
- 7.Dunfield, P. F., and R. Conrad. 2000. Starvation alters the apparent half-saturation constant for methane in the type II methanotroph Methylcystis strain LR1. Appl. Environ. Microbiol. 66:4136-4138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Felsenstein, J. 1997. An alternating least squares approach to inferring phylogenies from pairwise distances. Syst. Biol. 46:101-111. [DOI] [PubMed] [Google Scholar]
- 8a.Food and Agriculture Organization of the United Nations. 1998. World reference base for soil resources. International Soil Reference and Information Centre, Rome, Italy.
- 9.Gulledge, J., A. P. Doyle, and J. P. Schimel. 1997. Different NH4+-inhibition patterns of soil CH4 consumption: a result of distinct CH4-oxidizer populations across sites? Soil Biol. Biochem. 29:13-21. [Google Scholar]
- 10.Hanson, R. S., and T. E. Hanson. 1996. Methanotrophic bacteria. Microbiol. Rev. 60:439-472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Holmes, A. J., A. Costello, M. E. Lidstrom, and J. C. Murrell. 1995. Evidence that particulate methane monooxygenase and ammonia monooxygenase may be evolutionarily related. FEMS Microbiol. Lett. 132:203-208. [DOI] [PubMed] [Google Scholar]
- 12.Keller, M., and W. A. Reiners. 1994. Soil atmosphere exchange of nitrous-oxide, nitric-oxide, and methane under secondary succession of pasture to forest in the Atlantic lowlands of Costa Rica. Global Biogeochem. Cycles 8:399-409. [Google Scholar]
- 13.Knief, C., and P. F. Dunfield. 2005. Response and adaptation of different methanotrophic bacteria to low methane mixing ratios. Environ. Microbiol. 7:1307-1317. [DOI] [PubMed] [Google Scholar]
- 14.Knief, C., S. Kolb, P. L. E. Bodelier, A. Lipski, and P. F. Dunfield. 2006. The active methanotrophic community in hydromorphic soils changes in response to changing methane concentration. Environ. Microbiol. 8:321-333. [DOI] [PubMed] [Google Scholar]
- 15.Knief, C., S. Vanitchung, N. W. Harvey, R. Conrad, P. F. Dunfield, and A. Chidthaisong. 2005. Diversity of methanotrophic bacteria in tropical upland soils under different land uses. Appl. Environ. Microbiol. 71:3826-3831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kolb, S. 2009. The quest for atmospheric methane oxidizers in forest soils. Environ. Microbiol. Rep. 1(5):336-346. [DOI] [PubMed] [Google Scholar]
- 17.Kolb, S., C. Knief, P. F. Dunfield, and R. Conrad. 2005. Abundance and activity of uncultured methanotrophic bacteria involved in the consumption of atmospheric methane in two forest soils. Environ. Microbiol. 7:1150-1161. [DOI] [PubMed] [Google Scholar]
- 18.Ludwig, W., O. Strunk, R. Westram, L. Richter, H. Meier, Yadhukumar, A. Buchner, T. Lai, S. Steppi, G. Jobb, W. Forster, I. Brettske, S. Gerber, A. W. Ginhart, O. Gross, S. Grumann, S. Hermann, R. Jost, A. Konig, T. Liss, R. Lussmann, M. May, B. Nonhoff, B. Reichel, R. Strehlow, A. Stamatakis, N. Stuckmann, A. Vilbig, M. Lenke, T. Ludwig, A. Bode, and K. H. Schleifer. 2004. ARB: a software environment for sequence data. Nucleic Acids Res. 32:1363-1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Maxfield, P. J., E. R. C. Hornibrook, and R. P. Evershed. 2008. Acute impact of agriculture on high-affinity methanotrophic bacterial populations. Environ. Microbiol. 10:1917-1924. [DOI] [PubMed] [Google Scholar]
- 20.Menyailo, O. V., B. A. Hungate, W. R. Abraham, and R. Conrad. 2008. Changing land use reduces soil CH4 uptake by altering biomass and activity but not composition of high-affinity methanotrophs. Global Change Biol. 14:2405-2419. [Google Scholar]
- 21.Mohanty, S. R., P. L. E. Bodelier, V. Floris, and R. Conrad. 2006. Differential effects of nitrogenous fertilizers on methane-consuming microbes in rice field and forest soils. Appl. Environ. Microbiol. 72:1346-1354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ricke, P., S. Kolb, and G. Braker. 2005. Application of a newly developed ARB software-integrated tool for in silico terminal restriction fragment length polymorphism analysis reveals the dominance of a novel pmoA cluster in a forest soil. Appl. Environ. Microbiol. 71:1671-1673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Reference deleted.
- 24.Singh, B. K., K. R. Tate, G. Kolipaka, C. B. Hedley, C. A. Macdonald, P. Millard, and J. C. Murrell. 2007. Effect of afforestation and reforestation of pastures on the activity and population dynamics of methanotrophic bacteria. Appl. Environ. Microbiol. 73:5153-5161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Smith, K. A., K. E. Dobbie, B. C. Ball, L. R. Bakken, B. K. Sitaula, S. Hansen, R. Brumme, W. Borken, S. Christensen, A. Prieme, D. Fowler, J. A. Macdonald, U. Skiba, L. Klemedtsson, A. Kasimir-Klemedtsson, A. Degorska, and P. Orlanski. 2000. Oxidation of atmospheric methane in Northern European soils, comparison with other ecosystems, and uncertainties in the global terrestrial sink. Global Change Biol. 6:791-803. [Google Scholar]
- 26.Tate, K. R., D. J. Ross, S. Saggar, C. B. Hedley, J. Dando, B. K. Singh, and S. M. Lambie. 2007. Methane uptake in soils from Pinus radiata plantations, a reverting shrubland and adjacent pastures: effects of land-use change, and soil texture, water and mineral nitrogen. Soil Biol. Biochem. 39:1437-1449. [Google Scholar]
- 27.Verchot, L. V., E. A. Davidson, J. H. Cattanio, and I. L. Ackerman. 2000. Land-use change and biogeochemical controls of methane fluxes in soils of eastern Amazonia. Ecosystems 3:41-56. [Google Scholar]
- 28.Willison, T. W., K. W. T. Goulding, and D. S. Powlson. 1995. Effect of land-use change and methane mixing-ratio on methane uptake from United Kingdom soil. Global Change Biol. 1:209-212. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.