Abstract
Inactivation of TK1761, the reporter gene established for Thermococcus kodakarensis, revealed the presence of a second β-glycosidase that we have identified as the product of TK1827. This enzyme (pTK1827) has been purified and shown to hydrolyze glucopyranoside but not mannopyranoside, have optimal activity at 95°C and from pH 8 to 9.5, and have a functional half-life of ∼7 min at 100°C. To generate a strain with both TK1761 and TK1827 deleted, a new selection/counterselection protocol has been developed, and the levels of β-glycosidase activity in T. kodakarensis strains with TK1761 and/or TK1827 deleted and with these genes expressed from heterologous promoters are described. Genetic tools and strains have been developed that extend the use of this selection/counterselection procedure to delete any nonessential gene from the T. kodakarensis chromosome. Using this technology, TK0149 was deleted to obtain an agmatine auxotroph that grows on nutrient-rich medium only when agmatine is added. Transformants can therefore be selected rapidly, and replicating plasmids can be maintained in this strain growing in rich medium by complementation of the TK0149 deletion.
Members of the Thermococcales, hyperthermophilic Euryarchaea, grow readily under laboratory conditions and are the focus of many basic and applied research projects (2, 8). Their investigation has, however, been limited by the lack of genetics, and it was a seminal advance therefore when Thermococcus kodakarensis (formerly Thermococcus kodakaraensis [1]) was shown to be naturally competent and to recombine added DNA into its genome (21, 23). T. kodakarensis is attractive as an experimental system as a fermentative heterotroph that grows rapidly on a variety of different substrates (1), optimally at 85°C, including substrates from which it generates substantial levels of hydrogen (11). The T. kodakarensis genome sequence and genome microarray assays are established (7, 12, 18) and, since the discovery of transformation (21), inactivation and manipulation of chromosomal genes has revealed novel biochemical pathways, facilitated in vivo evaluations of the archaeal gene expression machinery, and simplified enzyme purifications (3, 5, 6, 9, 10, 12, 15-20, 22-24, 28). For complementation assays and to facilitate heterologous gene expression in T. kodakarensis, shuttle plasmids have been constructed that replicate and confer selectable phenotypes in both T. kodakarensis and Escherichia coli (19), and TK1761 expression has been established as a reporter system that can be used to identify and quantify regulatory elements in T. kodakarensis (18, 20).
During the development of the TK1761 reporter system, a T. kodakarensis strain, designated TS416, was constructed with a nonsense mutation in TK1761 (18). This mutation had no discernible effects on growth, confirming that TK1761 was not an essential gene, but lysates of T. kodakarensis TS416 retained a low level β-glycosidase activity. T. kodakarensis apparently, therefore, had a second β-glycosidase, but since this activity was very low and remained constant with changes in TK1761 expression, its presence did not compromise the use of TK1761 expression as a reporter system in the laboratory media used. The existence of a second β-glycosidase did, nevertheless, raise a potential concern for TK1761 assays in cells grown under different conditions in which expression of the gene encoding this second β-glycosidase might not be constant. To address this, we purified and characterized the second β-glycosidase and, to eliminate the concern for the reporter assay, we constructed a T. kodakarensis strain with both TK1761 and the gene (TK1827) that encodes the second β-glycosidase deleted.
To construct the double-deletion strain, we developed a new selection/counterselection protocol and have extended this into a procedure that can be used to delete any nonessential gene from the T. kodakarensis genome. A two-gene cassette has been constructed that can be integrated into the T. kodakarensis chromosome at any desired locus by homologous recombination of flanking genes. Expression of the cassette provides a positive selection for transformants and confers sensitivity to 6-methyl purine (6MPs). Mutants then isolated that are spontaneously resistant to 6-methyl purine (6MPr) have both the cassette and the adjacent target gene(s) precisely deleted.
Sato et al. (21, 23) established the T. kodakarensis transformation protocol by selecting transformants of tryptophan (trpE) and uracil (pyrF) auxotrophs that grew on minimal medium without tryptophan or uracil. Overexpression of the hydroxy-methylglutaryl-coenzyme A reductase encoded by PF1848, cloned from Pyrococcus furiosus, was later found to confer to resistance to simvastatin (15) and mevinolin (19), allowing the selection of T. kodakarensis transformants on rich media that contain either antibiotic. Mutants spontaneously resistant to these antibiotics do, however, occur at experimentally significant frequencies, and these are very expensive reagents for routine use and prohibitively expensive for incorporation into large preparative cultures. With this in mind, to develop an alternative selection that might be used in nutrient-rich media, we used the 6MP cassette system to delete TK0149. As predicted (6), the T. kodakarensis ΔTK0149 strain generated was an agmatine auxotroph that only grows in nutrient-rich media when agmatine is added. When transformed with DNA expressing TK0149, transformants of this strain can be selected directly on standard nutrient-rich media, and complementation of the ΔTK0149 mutation can be used to maintain the presence of an expression plasmid in T. kodakarensis cells grown in large-volume, rich-medium cultures for enzyme purification.
MATERIALS AND METHODS
Reagents and enzymes.
Except where otherwise noted, all chemicals were purchased from Sigma Chemicals (St. Louis, MO), restriction enzymes from Invitrogen (Carlsbad, CA) and New England Biolabs (Waltham, MA), plasmid and PCR-product purification kits from Qiagen Inc (Valencia, CA) and Zymo Research (Orange, CA), and all chromatography columns from GE Healthcare (Piscataway, NJ).
Strains and plasmids.
The origins and genetic features of the T. kodakarensis strains and plasmids used in the present study are listed in Table 1 (all of the T. kodakarensis strains and plasmids listed in Table 1 are available from T. J. Santangelo on request). The numerical and letter designations of the T. kodakarensis genes, and their genome annotations (7; http://cmr.jcvi.org/) are listed in Table 2. Standard molecular biology procedures were used to PCR amplify DNA, construct plasmids, transform E. coli DH5α, select β-lactamase-resistant transformants, and isolate plasmid preparations. DNA molecules PCR amplified from T. kodakarensis genomic DNA (the primer sequences are available from T. J. Santangelo) were cloned into pUC118 (27) or pLC70 (19). In all constructs, trpE (TK0254) was positioned for constitutive transcription from PTK2279 (18-20). For TK1827 overexpression and for constitutive transcription of TK0664 in the trp-6MPs cassette, these genes were transcribed from PhmtB (18-20). In plasmid pTS535, the trp-6MPs cassette was positioned between two multiple cloning sites (MCS1 and MCS2; see Fig. S1 in the supplemental material). Plasmids pTS538, pTS541, pT559, and pTS561 (Table 1) were constructed by ligating PCR-amplified fragments of the T. kodakarensis genome into MCS1 and MCS2 on both sides of the trp-6MPs cassette in pTS535 (see Fig. S2A and B in the supplemental material).
TABLE 1.
T. kodakarensis strains and plasmids used and/or constructed in this study
| Strain or plasmid | Origin and/or genetic featuresa | Source or referencef |
|---|---|---|
| T. kodakarensis strains | ||
| KW128 | ΔpyrF; ΔtrpE::pyrF | 21 |
| TS372 | KW128 with trpE-PhmtB-TK1761 insertion | 18 |
| TS416 | TS372 with nonsense mutation at codon 3 of TK1761 | 18 |
| TS502 | KW128 with trpE-PhmtB-(His)10TK1827 insertion | This study |
| TS503 | KW128 with trpE-PhmtB-TK1827 insertion with TAG at codon 5 | This study |
| TS517 | KW128 with ΔTK0664b | This study |
| TS521 | KW128 with ΔTK0664::trpE | This study |
| TS538 | TS517 with ΔTK1761-TK1762-TK1763 | This study; Fig. S2 |
| TS541 | TS517 with ΔTK1827 | This study; Fig. S2 |
| TS1079 | TS541 with ΔTK1761-TK1762-TK1763 | This study |
| TS559 | TS517 with ΔTK0149c | This study |
| TS561 | TS517 with ΔTK0148-TK0149 | This study |
| PdaD | KU216 ΔpyrF with ΔTK0148-pdaD | 6 |
| Plasmids | ||
| pTS502 | pUC118::PhmtB-(His)10TK1827-trpE-TK1828 | This study |
| pTS503 | pUC118::PhmtB-TK1827-trpE-TK1828 with TAG at codon 5 of TK1827 | This study |
| pTS517 | pUC118::TK0662-TK0663-TK0664-trpE-TK0662-TK0663-TK0665 | This study |
| pTS521 | pUC118::TK0662-TK0663-trpE-TK0665 | This study |
| pTS535 | pUC118 with MCS1-(trp-6MPs)-MCS2d | This study; Fig. S1 |
| pTS538 | pTS535::TK1760-TK1764-(trp-6MPS)-TK1761 | This study; Fig. S2 |
| pTS541 | pTS535::TK1825-TK1826-TK1828-(trp-6MPs)-TK1827-TK1828 | This study; Fig. S2 |
| pTS559 | pTS535::TK0147-TK0148-TK0150-(trp-6MPs)-TK0150-TK0151 | This study |
| pTS561 | pTS535::TK0147-TK0150-(trp-6MPs)-TK0150-TK0151 | This study |
| pLC70 | trpE; Pgdh-PF1848e | 19 |
| pTS436 | pLC70::PhmtB-TK1761 | This study |
| pTS543 | pLC70::PhmtB-TK0149 | This study |
Loss of TK0664 results in resistance to 6-methyl purine (6MPr).
Loss of TK0149, or loss of TK0148 plus TK0149, results in agmatine auxotrophy.
The trp-6MPs cassette has P2279 and PhmtB directing the divergent transcription of trpE (TK0254) and TK0664, respectively. TK0664 expression results in sensitivity to 6-methyl purine (6MPs). Flanking the cassette are multiple cloning sites 1 and 2 (MCS1 and MCS2) that have single cleavage sites for HindIII, SphI, BglII, BclI, ClaI, NheI, SpeI, NotI, SacII, ApaI, AccI, AscI, EcoRV, and BstEII and for EcoRI, KpnI, Acc65I, BamHI, XbaI, respectively (see Fig. S1 in the supplemental material).
Figures S1 and S2 may be found in the supplemental material.
TABLE 2.
Correlation of numerical and symbol designations and genome annotations of the T. kodakarensis genes examined in this study
| Gene |
Genome annotationa | |
|---|---|---|
| TK no. | Name | |
| 0147 | speE | Spermidine synthase |
| 0148 | Hypothetical protein | |
| 0149 | pdaD | Arginine decarboxylase |
| 0150 | gcvH | Glycine cleavage system protein H |
| 0151 | Transcription regulator | |
| 0254 | trpE | Anthranilate synthase |
| 0661 | ABC-type ion transport system | |
| 0662 | Hypothetical protein | |
| 0663 | Hypothetical protein | |
| 0664 | Hypoxanthine/guanine phosphoribosyltransferase | |
| 0665 | acdA | Acetyl-coenzyme A synthetase I (α-subunit) |
| 1760 | ABC-type peptide transport system | |
| 1761 | β-Glycosidase | |
| 1762 | Hypothetical protein | |
| 1763 | Nucleic acid-binding protein | |
| 1764 | Tk-Dac | N-Acetylchitobiose deacetylase |
| 1825 | Hypothetical protein | |
| 1826 | Transcription regulator | |
| 1827 | bglA | β-Glycosidase |
| 1828 | galT | Galactose-1-phosphate uridylyltransferase |
| 1829 | Permease | |
| 2276 | pyrF | Orotidine-5-phosphate decarboxylase |
| 2279 | Sugar nucleotidyltransferase | |
As presented elsewhere (7; http://cmr.jcvi.org/).
Media and growth conditions.
T. kodakarensis cultures were grown anaerobically at 85°C in nutrient-rich ASW-YT-S° medium or minimal ASW-AA-S° medium that contained 2 g of sulfur/liter (1, 21, 23). Tryptophan (0.73 mM) and/or agmatine (6.7 mM) were added as required. Cells competent for DNA uptake were prepared as described previously (21). T. kodakarensis ΔtrpE transformants capable of growth without tryptophan were selected on ASW-AA plates, T. kodakarensis ΔTK0149 transformants capable of growth without agmatine were selected on ASW-YT plates, and T. kodakarensis transformants resistant to mevinolin were selected on ASW-YT plates containing 15 μM mevinolin (19). The media were solidified by incorporation of 1% Gelrite.
Isolation and identification of the β-glycosidase (pTK1827) encoded by TK1827.
T. kodakarensis KW128 cells (∼320 g [wet weight]) were resuspended, under aerobic conditions, in ∼1 liter of lysis buffer (25 mM Tris-HCl [pH 8], 100 mM NaCl, 10% [vol/vol] glycerol, 10 mM β-mercaptoethanol) and lysed by sonication. Particulates were removed by centrifugation (JA25-50 rotor; 25,000 rpm for 20 min at 4°C), and aliquots of the clarified lysate were loaded onto a 15-ml HiTrap heparin column equilibrated with lysis buffer. Proteins were eluted by using a linear 0.1 to 1 M NaCl gradient in lysis buffer. An aliquot of each fraction was assayed for β-glycosidase activity by incubation with 2.8 mM ortho-nitrophenyl-β-d-glucopyranoside (ONP-gluco) at 85°C in 50 mM sodium phosphate buffer (pH 6.5). Increases in the A405 revealed two peaks of β-glycosidase activity that were separated by the heparin chromatography. Comparison with the activity in a lysate of T. kodakarensis TS416, the strain with a nonsense mutation in TK1761 (Table 1) (18), identified the fractions that contained the β-glycosidase not encoded by TK1761. These were pooled, dialyzed into 25 mM Tris-HCl (pH 8.8)-50 mM NaCl-10% (vol/vol) glycerol, clarified by centrifugation (JA16-250 rotor; 16,000 rpm for 40 min at 4°C), and loaded onto a 5-ml HiTrapQ-FF column. Fractions were eluted by using a linear 0.1 to 1 M NaCl gradient. The fractions that contained β-glycosidase activity were pooled and diluted with 25 mM Tris-HCl (pH 8.8)-10% (vol/vol) glycerol to a conductivity below that of 100 mM NaCl. This solution was loaded onto a 1-ml MonoQ column and eluted with a linear 0.1 to 1 M NaCl gradient in 20 mM Tris-HCl (pH 8.8). The fractions containing β-glycosidase were pooled, and the polypeptides present were visualized by Coomassie blue staining, and identified by liquid chromatography-tandem mass spectrometry (LC-MS/MS) after separation by denaturing polyacrylamide gel electrophoresis (SDS-PAGE).
Purification of recombinant His10-tagged pTK1827 from T. kodakarensis TS502.
The purification was carried out at room temperature, with equipment in a Coy anaerobic chamber in an atmosphere of 5% H2 and 95% N2. T. kodakarensis TS502 cells (∼6 g [wet weight]) were harvested near the end of exponential growth from cultures incubated at 85°C in ASW-YT-S° medium. The cells were resuspended (200 mg/ml) in 20 mM Tris-HCl (pH 8.8)-250 mM NaCl and frozen and thawed three times by immersion of the tube in liquid N2, and lysis was completed by repetitive passage through a small-gauge needle. Particulates were removed by centrifugation (JA25-50 rotor, 25,000 rpm for 20 min at 4°C), and the clarified lysate was loaded onto a 5-ml HiTrap chelating column charged with NiSO4 and equilibrated with 250 mM NaCl dissolved in 20 mM Tris-HCl (pH 8.8). Proteins were eluted by using a linear 0 to 250 mM imidazole gradient in 20 mM Tris-HCl (pH 8.8)-250 mM NaCl. An aliquot of each fraction was assayed for β-glycosidase activity by measuring ONP-gluco hydrolysis. Fractions that contained this activity were pooled and diluted with 20 mM Tris-HCl (pH 8.8) until the conductivity was below that of 0.1 M NaCl. The solution was loaded on a 1-ml MonoQ column, and fractions were eluted by using a linear 0.1 to 1 M NaCl gradient in 20 mM Tris-HCl (pH 8.8). The fractions containing β-glycosidase activity were pooled and dialyzed against 1 M NaCl-20 mM Tris-HCl (pH 8.8), and this solution was loaded onto a 5-ml HiTrap chelating column charged with NiSO4 and equilibrated with 1 M NaCl-20 mM Tris-HCl (pH 8.8). Fractions were eluted with a linear gradient from 0 to 250 mM imidazole in 250 mM NaCl-20 mM Tris-HCl (pH 8.8). The fractions that contained β-glycosidase activity were pooled, concentrated, and the buffer exchanged by using 30-kDa cutoff centrifugal concentrators (Millipore, Danvers, MA). The purified activity (∼1-mg yield) was dialyzed into 25 mM sodium phosphate (pH 7), 100 mM NaCl, and 50% (vol/vol) glycerol. The polypeptides present in an aliquot were visualized by Coomassie blue staining after separation by SDS-PAGE.
Characterization and optimization of the TK1827-encoded β-glycosidase.
Purified pTK1827 (1 μg) was added to reaction mixtures (500 μl) that contained 0 to 10 mM ONP-gluco, ortho-nitrophenyl-β-d-mannopyranoside (ONP-manno), or ortho-nitrophenyl-β-d-galactopyranoside (ONP-galacto) in 25 mM sodium phosphate (pH 6.5). After incubation at 85°C for 10 min, the increase in A405 was measured. A small change in A405 resulted from chemical hydrolysis of these substrates in the absence of pTK1827 that was subtracted from the experimental values.
To establish the optimum temperature for catalysis, reaction mixtures containing 1 μg of pTK1827 and 2.8 mM ONP-gluco were incubated for 10 min at different temperatures. The changes in A405 were measured, and the activity at each temperature was calculated as a percentage of the maximum activity observed. To establish the optimum pH for catalysis, reaction mixtures containing 1 μg of pTK1827 and 2.8 mM ONP-gluco dissolved in 25 mM sodium citrate (pH 4 to 5.5), sodium phosphate (pH 6 to 7.5), sodium borate (pH 8 to 9), or glycine-NaOH (pH 9.5 to 10) were incubated for 10 min at 85°C. The changes in A405 were measured, and the activity at each pH was calculated as a percentage of the maximum activity observed. To determine the rate of inactivation of pTK1827 at 100°C, aliquots (250 μl) that contained 1 μg of pTK1827 in 25 mM sodium phosphate (pH 6.5) were incubated at 100°C. Over time, 250 μl of 5.6 mM ONP-gluco in 25 mM sodium phosphate (pH 6.5) was added to each aliquot, and the reaction mixture was placed at 85°C for 10 min. Changes in A405 were measured, and the activity present was calculated as a percentage of the activity present in a reaction mixture that was not incubated at 100°C.
RESULTS AND DISCUSSION
Identification and properties of the β-glycosidase encoded by TK1827.
Purification by sequential column chromatography generated β-glycosidase-containing fractions that, based on Coomassie blue staining after SDS-PAGE, contained only two predominant polypeptides. Gel elution, trypsin digestion, and LC-MS/MS revealed that both polypeptides were the full-length product of TK1827. The migration of this hyperthermophilic protein as two bands during SDS-PAGE likely resulted from incomplete denaturation, and the same phenomenon, resolution by SDS-PAGE into two distinct bands, was also reported for purified recombinant pTK1761 (4). The identification of the purified protein as pTK1827 was consistent with the annotated function of the TK1827 gene product (7) (Table 2). The pTK1827 sequence is ∼79% identical to that of a β-glycosidase, designated BglA, encoded by PF0442 in P. furiosus, a Thermococcales relative of T. kodakarensis (13, 14, 26).
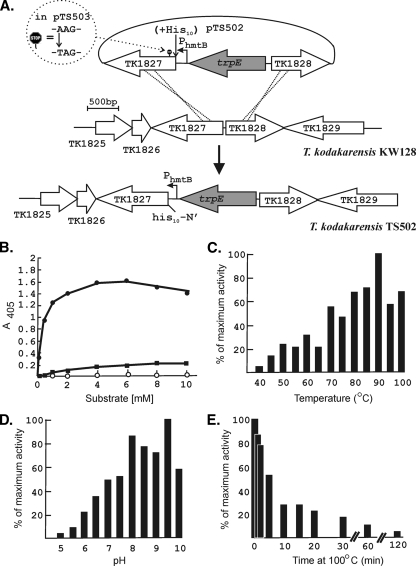
The very low abundance of pTK1827 in T. kodakarensis KW128 made its purification from this strain, in quantities sufficient for detailed characterization, impractical. To avoid potential concerns for misfolding or lack of posttranslation modifications if pTK1827 were generated in a mesophile or in a nonarchaeal expression system, we chose to overexpress TK1827 in T. kodakarensis. To do so, we constructed T. kodakarensis TS502 in which the native TK1827 promoter was replaced by the strong constitutive promoter PhmtB (19, 20) and, to simplify the purification, 10 histidine codons were added to the 5′ terminus (N-terminal His10-tag) of TK1827 (Fig. 1A). As a control, T. kodakarensis TS503 was constructed with TK1827 identically transcribed from PhmtB but with a nonsense mutation introduced at codon 5 of the TK1827 reading frame (Fig. 1A). PhmtB-directed expression of TK1827 resulted in a higher level of β-glycosidase activity in T. kodakarensis TS502 than in T. kodakarensis KW128, and this increase did not occur in T. kodakarensis TS503 (see below and Fig. 3). As observed for native pTK1827, aliquots of the His10-tagged pTK1827 purified from T. kodakarensis TS502 by Ni2+-affinity chromatography migrated to form two bands during SDS-PAGE.
FIG. 1.
Construction of T. kodakarensis TS502 and TS503 and properties of pTK1827. (A) Plasmids pTS502 and pTS503 were constructed by insertion of TK1827-trpE-TK1828 into pUC118 (27). As illustrated, in pTS502, 10 histidine codons were inserted in-frame at the 5′ terminus of TK1827 and, in pTS503, the TK1827 reading frame was terminated at codon 5 by a nonsense mutation. Aliquots of pTS502 and pTS503 DNA were used to transform T. kodakarensis KW128 and transformants, designated T. kodakarensis TS502 and TS503, were selected by growth on plates lacking tryptophan. PCR amplification and sequencing confirmed that T. kodakarensis TS502 had the genome structure shown, and that T. kodakarensis TS503 similarly had the predicted genome structure and retained the nonsense mutation in TK1827. (B) Hydrolysis of ONP-gluco (•), ONP-galacto (▪), and ONP-manno (○) during incubation with pTK1827 for 10 min at 85°C. (C) Effect of temperature on ONP-gluco hydrolysis expressed as a percentage of the maximum hydrolysis observed at 90°C. (D) Effect of pH on ONP-gluco hydrolysis. The results shown are average values of the percentages of the maximum hydrolysis observed at pH 9.5 measured in six replicates of this experiment. (E) Inactivation of pTK1827 by incubation at 100°C. The pTK1827 activity present after incubation at 100°C is shown as a percentage of the activity present before incubation at 100°C. The results shown are average values from two replicates of the experiment.
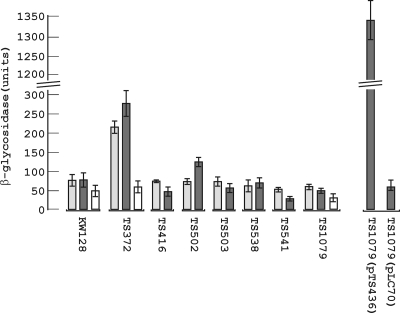
FIG. 3.
Assays of β-glycosidase activity in T. kodakarensis cell lysates. Cells were harvested from cultures of the different T. kodakarensis strains (see Table 1) grown in ASW-YT-S° medium to an OD600 of ∼0.6. The cells were lysed, and aliquots of the cleared lysates were incubated with ONP-manno (light gray bars) or ONP-gluco (dark gray bars) for 10 min at 85°C. The release of ONP was measured by the increase in A405. The results shown above each strain designation are the averages (with error bars) of triplicate assays of lysates from three separate cultures of that strain. The open bars show the results of assays with aliquots of T. kodakarensis KW128, TS372, and TS1079 lysates that were autoclaved for 20 min at 121°C before incubation with ONP-gluco.
Purified His10-tagged pTK1827 hydrolyzed ONP-gluco and had lower but detectable activity with ONP-galacto but did not hydrolyze ONP-manno (Fig. 1B). In contrast, pTK1761 was shown to have high activity with both ONP-gluco and ONP-manno and much lower activity with ONP-galacto (4). In common with pTK1761, pTK1827 was active in vitro at temperatures ranging from 45 to 100°C, optimally at ∼90°C (Fig. 1C), and hydrolyzed ONP-gluco from pH 5 to 10 and optimally at pH 8 and 9.5 (Fig. 1D). By extrapolation from Pyrococcus woesii (25), a related Thermococcales, the cytoplasm of T. kodakarensis is predicted to contain ∼800 mM K+ and the activity of pTK1827 was stimulated by 400 to 650 mM KCl addition, but no specific salt or salt concentration was required for activity. Under optimized conditions, the Km of pTK1827 for ONP-gluco was ∼0.5 mM, similar to the Km of 1.79 mM reported for this substrate for pTK1761 (4). Although pTK1827 was very active at 100°C (Fig. 1C), the activity was lost, with a half-life of ∼7 min during incubation at 100°C (Fig. 1E). Recombinant pTK1761 was similarly very active but also rapidly inactivated by incubation at 100°C (4).
Inactivation of TK0664 confers resistance to 6-methyl purine (6MPr).
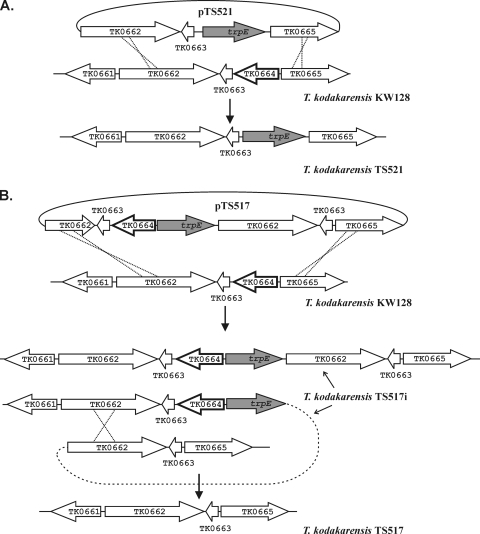
The nonsense mutations in TK1761 and TK1827, in strains T. kodakarensis TS416 (18) and T. kodakarensis TS503 (Fig. 1A), respectively, had no discernible effects on viability or growth. Construction of a T. kodakarensis strain with both genes deleted therefore seemed an attractive possibility to optimize the TK1761 reporter system. We tried to delete these genes by complementation of the uracil auxotrophy in T. kodakarensis KU216 to obtain targeted integration (pop-in), followed by selection of 5-fluoroorotic acid resistant mutants (pop-out/counterselection), as described by Sato et al. (23), but were unsuccessful. We therefore chose to use trpE complementation to direct integration in T. kodakarensis KW128 (Table 1) and screened additional nucleotide analogues to develop a different counterselection. The growth of T. kodakarensis KW128 was inhibited on plates containing 10 μM 6MP, 1 mM 6-thioguanine, or 1 mM 8-azaguanine and, based on colony formation, spontaneous resistance to these inhibitors occurred at a frequency of ∼1 per 108 cells. This frequency was consistent with mutations that resulted in the loss of a single enzyme involved in purine scavenging that conferred resistance by preventing the lethal incorporation of these toxic purine analogues. Based on the T. kodakarensis genome annotation (7), genes predicted to encode such enzymes were PCR amplified and sequenced from many analogue-resistant mutants. Resistance to 6MP correlated consistently with mutations in TK0664, and different missense and nonsense mutations, single- and multiple-base-pair insertions and deletions were identified in the TK0664 sequences amplified from different 6MPr mutants. To confirm that the loss of TK0664, annotated as encoding a hypoxanthine-guanine phosphoribosyl-transferase (7) (Table 2), resulted in 6MPr, TK0664 was replaced in the T. kodakarensis KW128 genome by trpE (Fig. 2A). The resulting strain, T. kodakarensis TS521, grew normally without added tryptophan and in the presence of up to ∼5 mM 6MP.
FIG. 2.
Construction of T. kodakarensis TS521 and TS517. In this figure, as well as in Fig. S2 in the supplemental material and Fig. 4, the gene(s) targeted for deletion are identified by a bold outline. (A) Plasmid pTS521 was constructed by cloning the genes, as shown, into pUC118 and used to transform T. kodakarensis KW128. A representative transformant, designated T. kodakarensis TS521, selected by growth on plates lacking tryptophan, had the genome structure shown. (B) Plasmid pTS517 was constructed by cloning the genes, as shown, into pUC118 and used to transform T. kodakarensis KW128. A representative transformant, designated T. kodakarensis TS517i, was selected by growth on plates lacking tryptophan and had the genome structure shown. Dilutions of a culture of T. kodakarensis TS517i were spread on plates containing 6MP, and a 6MPr tryptophan auxotroph, designated T. kodakarensis TS517, had the genome structure shown.
Use of 6MPr as a counterselection to isolate mutants with chromosomal deletions.
To determine whether 6MPr could be used to select T. kodakarensis mutants with targeted deletions and to obtain a strain with TK0664 precisely deleted and not replaced by trpE, plasmid pTS517 was constructed (Fig. 2B) and used to transform T. kodakarensis KW128. Transformants were selected by growth on minimal medium plates lacking tryptophan. PCR amplification and sequencing of genomic DNA confirmed that trpE (TK0254) was integrated into the genomes of several transformants adjacent to TK0664 and flanked by directly repeated copies of TK0662 and TK0663, as illustrated in Fig. 2B. Dilutions of a representative transformant, designated T. kodakarensis TS517i (strains intermediate in each construction are denoted by an “i” in the designation), were spread on plates that contained 6MP and tryptophan. Colonies grew at a frequency of ∼1 per 103 plated cells, and all of the spontaneous 6MPr mutants screened were also tryptophan auxotrophs. PCR amplification and sequencing of genomic DNA from several of these mutants confirmed that the trpE-TK0664 sequence was deleted, that only one copy of TK0662 and TK0663 remained, and that the intergenic sequence between TK0663 and TK0665 was identical to that in pTS517 (Fig. 2B). One isolate was retained for future studies and designated T. kodakarensis TS517 (Table 1).
Construction of the trp-6MPs cassette and 6MPr selection of TK1761 and TK1827 deletions.
Plasmid pTS535 was constructed with trpE (TK0254) and TK0664 adjacent but transcribed divergently from two strong constitutive promoters, PTK2279 and PhmtB (18-20), respectively, and flanked by multiple cloning sites designated MCS1 and MCS2 (see Fig. S1 in the supplemental material). When expressed in T. kodakarensis TS517 (ΔtrpE; ΔTK0664), these genes, designated the trp-6MPs cassette, conferred tryptophan-independent growth and sensitivity to 6MP. Plasmids pTS538 and pTS541 were generated from pTS535 by cloning PCR-amplified fragments of the T. kodakarensis genome into MCS1 and MCS2 and so flanking the trp-6MPs cassette (see Fig. S2A and B in the supplemental material; Table 1). The flanking sequences facilitated homologous recombination and directed the integration of the trp-6MPS cassette adjacent to TK1761 (see Fig. S2A) and TK1827 (see Fig. S2B) in pTS538 and pTS541 transformants of T. kodakarensis TS517, respectively. PCR amplification and sequencing of genomic DNA from several transformants confirmed the presence of the trp-6MPs cassette positioned between the designed duplications of chromosomal genes (see Fig. S2A and B). Dilutions of two representative transformants, designated T. kodakarensis TS538i and TS541i, were spread on plates containing 6MP and tryptophan, and sequencing of genomic DNA, PCR amplified from spontaneous 6MPr mutants, confirmed the absence of the trp-6MPs cassette. In the 6MPr mutants obtained from T. kodakarensis TS538i, the TK1761-TK1762-TK1763 operon was also deleted, and a representative was designated T. kodakarensis TS538 (see Fig. S2A in the supplemental material). In the 6MPr mutants obtained from T. kodakarensis TS541i, in addition to the loss of the trp-6MPs cassette, TK1827 was deleted, and a representative was designated T. kodakarensis TS541 (see Fig. S2B in the supplemental material). Consistent with deletion of the trp-6MPs cassette, all of the spontaneous 6MPr mutants isolated and screened from T. kodakarensis TS538i and TS541i were also tryptophan auxotrophs.
To construct a strain with both TK1761 and TK1872 deleted, T. kodakarensis TS541 (Fig. S2B; Table 1) was transformed with pTS538 DNA (Fig. S2A). Dilutions of a transformant selected by growth on plates lacking tryptophan were spread on plates that contained 6MP and tryptophan. PCR amplification and sequencing of genomic DNA from several spontaneous 6MPr mutants confirmed that both the trp-6MPs cassette and the TK1761-TK1762-TK1763 operon had been deleted, as illustrated for the construction of T. kodakarensis TS538 in Fig. S2A in the supplemental material. Screening confirmed that ∼80% of these 6MPr mutants were also tryptophan auxotrophs. PCR amplification and sequencing reconfirmed that TK1827 was also deleted in one such isolate that was retained for future studies and designated T. kodakarensis TS1079 (Table 1).
β-Glycosidase activity in T. kodakarensis deletion mutants.
Lysates of cells harvested at an optical density at 600 nm (OD600) of ∼0.6 from three independent exponentially growing cultures of T. kodakarensis KW128, TS372, TS416, TS502, TS503, TS538, TS541, and TS1079 (Table 1) were assayed for β-glycosidase activity. Expression of TK1761 from PhmtB in T. kodakarensis TS372 resulted in a three- to fivefold increase in ONP-gluco and ONP-manno hydrolyzing activity relative to the activity in T. kodakarensis KW128 (Fig. 3). This increase did not occur in T. kodakarensis TS416 that differs from T. kodakarensis TS372 only by the presence of a nonsense mutation in TK1761 (18). Transcription of TK1827 from PhmtB in T. kodakarensis TS502 (Fig. 1A) also resulted in an increase in ONP-gluco but not in ONP-manno hydrolyzing activity, and this increase similarly did not occur in T. kodakarensis TS503 that has a nonsense mutation in TK1827. Surprisingly, however, in contrast to these increases in activity that resulted from increased expression of TK1761 and TK1827, deletion of these genes did not significantly reduce the levels of ONP-gluco or ONP-manno hydrolyzing activity in T. kodakarensis TS538, TS541, and TS1079 below the levels in T. kodakarensis KW128, TS416, and TS503 (Fig. 3). Purification of pTK1827 provided no evidence for a third β-glycosidase, and it seemed possible therefore that this residual substrate hydrolysis was not enzyme catalyzed. To pursue this, knowing that both pTK1761 (4) and pTK1827 (Fig. 1E) were inactivated by incubation at 100°C, aliquots of T. kodakarensis KW128, TS372, and TS1079 lysates were autoclaved for 20 min at 121°C and then assayed for ONP-gluco hydrolyzing activity. Consistent with inactivation of pTK1761, autoclaving substantially reduced the β-glycosidase activity present in T. kodakarensis TS372 lysates but did not totally eliminate substrate hydrolysis and only marginally reduced the activity present in T. kodakarensis KW128 and TS1079 lysates (Fig. 3). Although this residual hydrolysis could still be catalyzed by one or more unidentified, very-heat-resistant β-glycosidases, it seems most likely that these colorimetric substrates undergo slow chemical hydrolysis when incubated at 85°C with components in a T. kodakarensis cell lysate. In either case, this very low level of background activity can be readily determined and subtracted, if necessary (see below), from experimental results.
To investigate the range of the TK1761 reporter signal, T. kodakarensis TS1079 was transformed with expression plasmids that replicate in the T. kodakarensis cytoplasm at a copy number of ∼3 per chromosome (19). A DNA molecule with TK1761 transcribed from PhmtB was PCR amplified from pTS372 (18) (Table 1) and cloned into pLC70 (19), generating pTS436 (Table 1). The ONP-gluco hydrolyzing activity in T. kodakarensis TS1079(pTS436) was ∼30-fold higher than in the control T. kodakarensis TS1079(pLC70) and ∼5-fold higher than in T. kodakarensis TS372 (18) that has a single copy of the PhmtB-TK1761 construct present in its genome (Fig. 3). In experiments with such large positive signals, the low background of substrate hydrolysis (see above) would not pose a serious experimental concern.
Deletion of TK0149, agmatine auxotrophy, and selection of transformants on rich media.
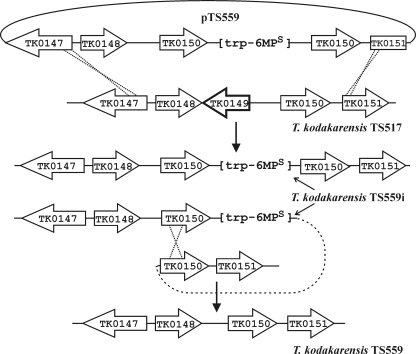
The observation that disruption of TK0149 (designated pdaD; Table 2) resulted in agmatine auxotrophy (6) suggested a positive selection that might be imposed on cells growing in nutrient rich media. We confirmed that the T. kodakarensis pdaD mutant grew in ASW-YT·S° medium only with agmatine added, but PCR amplification and sequencing revealed that both TK0149 and an adjacent 3′-region of TK0148 were deleted in this strain (6). To determine whether TK0149 expression was sufficient to complement this mutation, TK0149 was cloned downstream from PhmtB in pLC70, resulting in plasmid pTS543 (Table 1). Transformation of T. kodakarensis pdaD with pTS543 resulted in transformants that grew and formed colonies within 24 h on plates that contained ASW-YT medium without added agmatine. The deletion into TK0148 did not therefore play a critical role in conferring agmatine auxotrophy, but to confirm that deletion of TK0149 was sufficient and to obtain defined strains for further use, we constructed mutants with precise deletions of TK0149 alone and of TK0148 plus TK0149. T. kodakarensis TS517 was transformed with pTS559 (Fig. 4) or pTS561 (Table 1), and transformants were selected by growth on minimal medium plates without tryptophan but containing agmatine. PCR amplification and sequencing of genomic DNA from several transformants confirmed the integration of the trp-6MPs cassette, deletion of TK0149 (pTS559 transformants; Fig. 4) or of TK0148 and TK0149 (pTS561 transformants), and duplication of TK0150. Transformation with pTS559 or pTS561 resulted directly in the deletion of TK0149 or the deletion of TK0148 plus TK0149, respectively, with integration (pop-in) of the trp-6MPs cassette, rather than at the subsequent counterselection (pop-out) step as in the T. kodakarensis TS538 and TS541 constructions (see Fig. S2A and B in the supplemental material). Representative transformants were designated T. kodakarensis TS559i (Fig. 4) and TS561i (Table 1), and dilutions of these strains were spread on plates that contained 6MP, tryptophan, and agmatine. Spontaneous 6MPr mutants were screened for growth with or without tryptophan and with or without agmatine. Genomic DNA was PCR amplified from several 6MPr, tryptophan- and agmatine-requiring mutants, and sequencing confirmed the absence of the trp-6MPs cassette and the presence of only one copy of TK0150 directly adjacent to TK0148 in mutants from T. kodakarensis TS559i (Fig. 4) and directly adjacent to TK0147 in mutants from T. kodakarensis TS561i. A representative from each construction was confirmed to require agmatine addition for growth in rich media and designated T. kodakarensis TS559 (Fig. 4) and T. kodakarensis TS561 (Table 1), respectively. Transformation of these strains with plasmid pTS543 resulted in transformants that grew rapidly on plates containing ASW-YT-S° medium without agmatine. Expression of TK0149 from pTS543 also ensured the maintenance of this plasmid in the cytoplasm of transformants grown in nutrient rich media lacking agmatine.
FIG. 4.
Construction of T. kodakarensis TS559. Plasmid pTS559 was constructed by cloning the genes, as shown, into pTS535 (see Fig. S1 in the supplemental material) and was used to transform T. kodakarensis TS517 (Table 1). A transformant, designated T. kodakarensis TS559i, selected by growth on plates lacking tryptophan but containing agmatine, had the genome structure shown. Dilutions of a culture of T. kodakarensis TS559i were spread on plates containing 6MP, tryptophan, and agmatine, and a 6MPr, tryptophan, and agmatine auxotroph, designated T. kodakarensis TS559, had the genome shown. The same protocol was used to generate T. kodakarensis TS561 except that plasmid pTS561, which differed from pTS559 by the absence of TK0148 (Table 1), was used in the first step to transform T. kodakarensis TS517 to produce the intermediate strain, T. kodakarensis TS561i.
Conclusions.
T. kodakarensis provides an attractive experimental system for investigations of hyperthermophily, archaeal physiology, and molecular biology (2, 8). The results reported here increase and accelerate the technologies available to genetically manipulate and exploit T. kodakarensis for such research and for production-scale applications. We have identified, characterized, and deleted the β-glycosidase encoded by TK1827 that might otherwise have compromised the use of the TK1761 reporter system in T. kodakarensis under some growth conditions and established that the TK1761 reporter β-glycosidase can accumulate to very different levels without effecting growth. Two new positive selections (6MPr, agmatine complementation) have been added for transformants, and a simple procedure has been established, based on TK0664 expression (6MPs), that makes it possible to delete multiple genes sequentially from the T. kodakarensis chromosome. By routine cloning into plasmid pTS535 (see Fig. S1 in the supplemental material) and transformation of T. kodakarensis TS517 (Fig. 2B and Table 1), the 6MPs-based selection can be used to delete any nonessential T. kodakarensis gene. Deletion of TK0149 results in agmatine auxotrophy (6) and complementation of a ΔTK0149 mutation selects transformants on nutrient rich media, allowing much faster strain construction. Complementation of agmatine auxotrophy can also be used to maintain plasmids and so replace the need for antibiotics in large-volume preparative cultures.
Supplementary Material
Acknowledgments
This research was supported by grants from the DOE (DE-FG02-87ER13731) and the NIH (GM53185) to J.N.R. and by NIH fellowship (1F32-GM073336-01) support to T.J.S.
Footnotes
Published ahead of print on 18 December 2009.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Atomi, H., T. Fukui, T. Kanai, M. Morikawa, and T. Imanaka. 2004. Description of Thermococcus kodakaraensis sp. nov., a well studied hyperthermophilic archaeon previously reported as Pyrococcus sp. KOD1. Archaea 1:263-267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cavicchioli, R. 2007. Archaea: molecular and cellular biology. ASM Press, Washington, DC.
- 3.Dev., K., T. J. Santangelo, S. Rothenburg, D. Neculai, M. Dey, F. Sicheri, T. E. Dever, J. N. Reeve, and A. G. Hinnebusch. 2009. Archaeal aIF2B interacts with eukaryotic translation initiation factors eIF2α and eIF2Bα: implications for aIF2B function and eIF2B regulation. J. Mol. Biol. 392:701-722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ezaki, S., K. Miyaoku, K. Nishi, T. Tanaka, S. Fujiwara, M. Takagi, H. Atomi, and T. Imanaka. 1999. Gene analysis and enzymatic properties of thermostable β-glycosidase from Pyrococcus kodakaraensis KOD1. J. Biosci. Bioeng. 88:30-135. [DOI] [PubMed] [Google Scholar]
- 5.Fujiwara, S., R. Aki, M. Yoshida, H. Higashibata, T. Imanaka, and W. Fukuda. 2008. Expression profiles and physiological roles of two types of molecular chaperonins from the hyperthermophilic archaeon Thermococcus kodakaraensis. Appl. Environ. Microbiol. 74:7306-7312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fukuda, W., N. Morimoto, T. Imanaka, and S. Fujiwara. 2008. Agmatine is essential for the cell growth of Thermococcus kodakaraensis. FEMS Microbiol. Lett. 287:113-120. [DOI] [PubMed] [Google Scholar]
- 7.Fukui, T., H. Atomi, T. Kanai, R. Matsumi, S. Fujiwara, and T. Imanaka. 2005. Complete genome sequence of the hyperthermophilic archaeon Thermococcus kodakaraensis KOD1 and comparison with Pyrococcus genomes. Genome Res. 15:352-363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Garrett, R. A., and H.-P. Klenk. 2007. Archaea: evolution, physiology, and molecular biology. Blackwell Publishing, Oxford, United Kingdom.
- 9.Hirata, A., T. Kanai, T. J. Santangelo, M. Tajiri, K. Manabe, J. N. Reeve, T. Imanaka, and K. S. Murakami. 2008. Archaeal RNA polymerase subunits E and F are not required for transcription in vitro, but a Thermococcus kodakaraensis mutant lacking subunit F is temperature sensitive. Mol. Microbiol. 70:623-633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Imanaka, H., A. Yamatsu, T. Fukui, H. Atomi, and T. Imanaka. 2006. Phosphoenolpyruvate synthase plays an essential role for glycolysis in the modified Embden-Meyerhof pathway in Thermococcus kodakaraensis. Mol. Microbiol. 61:898-909. [DOI] [PubMed] [Google Scholar]
- 11.Kanai, T., H. Imanaka, A. Nakajima, K. Uwamori, Y. Omori, T. Fukui, H. Atomi, and T. Imanaka. 2005. Continuous hydrogen production by the hyperthermophilic archaeon, Thermococcus kodakaraensis KOD1. J. Biotechnol. 116:271-282. [DOI] [PubMed] [Google Scholar]
- 12.Kanai, T., J. Akerboom, S. Takedomi, H. J. van de Werken, F. Blombach, J. van der Oost, T. Murakami, H. Atomi, and T. Imanaka. 2007. A global transcriptional regulator in Thermococcus kodakaraensis controls the expression levels of both glycolytic and gluconeogenic enzyme-encoding genes. J. Biol. Chem. 282:33659-33670. [DOI] [PubMed] [Google Scholar]
- 13.Kaper, T., C. H. Verhees, J. H. G. Lebbink, J. F. T. van Lieshout, L. D. Kluskens, D. E. Ward, S. W. M. Kengen, M. M. Beerthuyzen, W. M. deVos, and J. van der Oost. 2001. Characterization of β-glycosylhydrolases from Pyrococcus furiosus. Methods Enzymol. 330:329-346. [DOI] [PubMed] [Google Scholar]
- 14.Kaper, T., H. H. van Heusden, B. van Loo, A. Vasella, J. van der Oost, and W. M. deVos. 2002. Substrate specificity engineering of β-mannosidase and β-glucosidase from Pyrococcus by exchange of unique active site residues. Biochemistry 41:4147-4155. [DOI] [PubMed] [Google Scholar]
- 15.Matsumi, R., K. Manabe, T. Fukui, H. Atomi, and T. Imanaka. 2007. Disruption of a sugar transporter gene cluster in a hyperthermophilic archaeon using a host-marker system based on antibiotic resistance. J. Bacteriol. 189:2683-2691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Orita, I., T. Sato, H. Yurimoto, N. Kato, H. Atomi, T. Imanaka, and Y. Sakai. 2006. The ribulose monophosphate pathway substitutes for the missing pentose phosphate pathway in the archaeon Thermococcus kodakaraensis. J. Bacteriol. 188:4698-4704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Santangelo, T. J., L. Čuboňová, C. L. James, and J. N. Reeve. 2007. TFB1 or TFB2 is sufficient for Thermococcus kodakaraensis viability and for basal transcription in vitro. J. Mol. Biol. 367:344-357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Santangelo, T. J., L. Čuboňová, R. Matsumi, H. Atomi, T. Imanaka, and J. N. Reeve. 2008. Polarity in archaeal operon transcription in Thermococcus kodakaraensis. J. Bacteriol. 190:2244-2248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Santangelo, T. J., L. Čuboňová, and J. N. Reeve. 2008. Shuttle vector expression in Thermococcus kodakaraensis: contributions of cis elements to protein synthesis in a hyperthermophilic Archaeon. Appl. Environ. Microbiol. 74:3099-3104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Santangelo, T. J., L. Čuboňová, K. M. Skinner, and J. N. Reeve. 2009. Archaeal intrinsic transcription termination in vivo. J. Bacteriol. 191:7102-7108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sato, T., T. Fukui, H. Atomi, and T. Imanaka. 2003. Targeted gene disruption by homologous recombination in the hyperthermophilic archaeon Thermococcus kodakaraensis KOD1. J. Bacteriol. 185:210-220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sato, T., H. Imanaka, N. Rashid, T. Fukui, H. Atomi, and T. Imanaka. 2004. Genetic evidence identifying the true gluconeogenic fructose-1,6-bisphosphatase in Thermococcus kodakaraensis and other hyperthermophiles. J. Bacteriol. 186:5799-5807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sato, T., T. Fukui, H. Atomi, and T. Imanaka. 2005. Improved and versatile transformation system allowing multiple genetic manipulations of the hyperthermophilic archaeon Thermococcus kodakaraensis. Appl. Environ. Microbiol. 71:3889-3899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sato, T., H. Atomi, and T. Imanaka. 2007. Archaeal type III RuBisCOs function in a pathway for AMP metabolism. Science 315:1003-1006. [DOI] [PubMed] [Google Scholar]
- 25.Scholz, S., J. Sonnenbichler, W. Schäfer, and R. Hensel. 1992. Di-myo-inositol-1,1′-phosphate: a new inositol phosphate isolated from Pyrococcus woesei. FEBS Lett. 306:239-342. [DOI] [PubMed] [Google Scholar]
- 26.VanFossen, A. L., D. L. Lewis, J. D. Nichols, and R. M. Kelly. 2008. Polysaccharide degradation and synthesis by extremely thermophilic anaerobes. Ann. N. Y. Acad. Sci. 1125:322-333. [DOI] [PubMed] [Google Scholar]
- 27.Vieira, J., and J. Messing. 1987. Production of single-stranded plasmid DNA. Methods Enzymol. 153:3-11. [DOI] [PubMed] [Google Scholar]
- 28.Yokooji, Y., H. Tomita, H. Atomi, and T. Imanaka. 2009. Pantoate kinase and phosphopantothenate synthetase, two novel enzymes necessary for CoA biosynthesis in the Archaea. J. Biol. Chem. 284:28137-28145. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.