Abstract
The persistence of naturally occurring campylobacteria in aerobic compost constructed of manure from beef cattle that were administered chlortetracycline and sulfamethazine (AS700) or from cattle not administered antibiotics (control) was examined. Although there were no differences in population sizes of heterotrophic bacteria, the temperature of AS700 compost was more variable and did not become as high as that of control compost. There were significant differences in water content, total carbon (C), total nitrogen (N), and electrical conductivity but not in the C/N ratio or pH between the two compost treatments. Campylobacteria were readily isolated from pen manure, for up to day 15 from control compost, and throughout the active phase of AS700 compost. Campylobacter DNA (including Campylobacter coli, Campylobacter fetus, Campylobacter hyointestinalis, and Campylobacter jejuni) was detected over the ca. 10-month composting period, and no reductions in quantities of C. jejuni DNA were observed over the duration of the active phase. The utilization of centrifugation in combination with ethidium monoazide (EMA) significantly reduced (>90%) the amplification of C. jejuni DNA that did not originate from cells with intact cell membranes. No differences were observed in the frequency of Campylobacter DNA detection between EMA- and non-EMA-treated samples, suggesting that Campylobacter DNA amplified from compost was extracted from cells with intact cell membranes (i.e., from viable cells). The findings of this study indicate that campylobacteria excreted in cattle feces persist for long periods in compost and call into question the common belief that these bacteria do not persist in manure.
Campylobacter jejuni and, to a lesser extent, Campylobacter coli incite serious acute and chronic afflictions. Enteritis caused by C. jejuni (i.e., campylobacteriosis) is the most common cause of bacterial enteritis in Canada (http://dsol-smed.phac-aspc.gc.ca/dsol-smed/ndis/index-eng.php). Although the epidemiology of campylobacteriosis is poorly understood, sporadic outbreaks of campylobacteriosis involving contaminated water have occurred when water treatment has failed. The most serious outbreak in Canada occurred in Walkerton Ontario in 2000; more than 2,300 people became infected with waterborne Escherichia coli O157:H7 and/or C. jejuni originating from cattle feces (3). Alberta, Canada, possesses a very large beef cattle population (≈6 million animals) primarily concentrated in the southern region of the province, and ≈2 million of these animals are in finishing feedlots (1). Large quantities of manure are produced by feedlot cattle. For example, in the Chinook Health Region of Southwestern Alberta in which Lethbridge is situated, there are ≈700,000 cattle in feedlots at any given time, producing ≈12 million kg of manure (fresh weight) per day. Several Campylobacter species, including C. jejuni and C. coli, are frequently shed in beef cattle feces in large numbers (15, 16). Although the impact of cattle-borne campylobacters on human health has not been definitely determined, the southern region of Alberta possesses one of the highest rates of campylobacteriosis in Canada among its human inhabitants, concomitant with the very high density of cattle in this region.
Large-scale windrow composting of bovine manure from intensive cattle operations is practiced by some Alberta feedlots. Composting is an aerobic process in which organic matter in manure is stabilized into a humus-like product (30). The process results in water loss and mass reduction, nutrient transformation (22), alteration of physical structure (23), elimination of weed seeds (21), and the inactivation of coliform bacteria (25), protozoan cysts and oocysts (34), and viruses (39). Limited research has investigated the impact of manure management systems, such as aerobic composting, on deactivation of campylobacters. Furthermore, the impact of antimicrobial agents excreted into the manure on the efficacy of the composting process on Campylobacter deactivation has not been investigated. Most studies conducted to date have indicated that campylobacters do not persist well in solid manure once excreted (7, 11, 12, 26, 32, 39). Although it is difficult to isolate or enumerate Campylobacter species within microbiologically complex substrates, molecular detection and/or quantification methods have not been extensively applied to study the persistence of campylobacteria. Furthermore, the persistence of naturally shed campylobacteria has largely been overlooked. Thus, the overall objective of the current study was to measure the ability of campylobacteria naturally shed in bovine feces to persist in manure compost using a combination of culture- and culture-independent methods. Specific objectives were (i) to develop and utilize a centrifugation method to facilitate isolation and detection of DNA from Campylobacter species in bovine manure compost, (ii) to apply qualitative and quantitative PCR methods to evaluate persistence of campylobacteria in compost, (iii) to validate the molecular methods used to amplify DNA from viable cells, and (iv) to contrast the persistence of Campylobacter species in composted manure obtained from beef cattle maintained on a diet supplemented with chlortetracycline and sulfamethazine (AS700) with composted manure from animals not administered antimicrobial agents.
MATERIALS AND METHODS
Centrifugation method.
To facilitate isolation and detection of DNA from campylobacteria in manure and compost, a two-step centrifugation method to remove large substrate particles and concentrate Campylobacter cells was developed and evaluated. Beef cattle manure determined to contain small amounts of C. jejuni DNA by direct PCR (13) was collected and frozen at −20°C until used. C. jejuni 81-176 was grown in Columbia broth (Becton, Dickinson and Co., Sparks, MD) for 16 h at 40°C under microaerobic conditions (3% H2, 5% O2, 10% CO2, and 82% N2). The turbidity of cells (optical density at 625 nm [OD625]) was measured, and cell density was adjusted to 1 × 109 cells ml−1 in Columbia broth. The suspension was diluted in a 10-fold dilution series in Columbia broth, and 0.5 ml of each dilution was thoroughly mixed into 5 g of thawed feces with a pipette tip. Target densities of viable C. jejuni cells in feces were 108, 107, 106, 105, 104, 103, and 0 (i.e., uninoculated) cells g−1.
Manure (5 g) was added to 45 ml of Columbia broth in a Stomacher 80 bag (BA6040; Seward Ltd., Worthing, United Kingdom) and homogenized for 120 s at the high setting using a Stomacher 80 homogenizer (Seward Ltd.). The homogenate was then removed from the bag and centrifuged at 4,000 × g for 10 min to remove particulate matter, and the supernatant containing C. jejuni cells was collected. To concentrate Campylobacter cells, the supernatant was centrifuged at 14,900 × g for 10 min, and the supernatant was removed and discarded; culture enumeration and PCR revealed that only small numbers C. jejuni cells or none at all were present in the supernatant (data not presented). The pellet was resuspended in 2 ml of Columbia broth, 200-μl aliquots were placed in 2-ml tubes, and an internal amplification control (IAC; 10 μl containing 700 copies μl−1) was added to each tube (13). DNA was extracted using a QIAamp DNA Stool Mini Kit according to the manufacturer's protocol (Qiagen Inc., Mississauga, ON, Canada), and the copy number of C. jejuni (mapA gene) was measured (in duplicate) in each fecal sample (14). C. jejuni cells in the suspended pellet were also enumerated by dilution plating (in duplicate) on Karmali agar amended with Selective Supplement ([KS agar] SR167; Oxoid Ltd., Nepean, ON, Canada). Cultures were maintained in a microaerobic environment at 40°C for 72 h, and colonies were enumerated in duplicate at the dilution yielding 30 to 300 colonies per plate. The experiment was conducted three times on separate occasions (i.e., replicates in time), and the linear relationship between log10 copy number and log10 CFU g−1 was determined.
Detection of viable campylobacterial cells.
The utility of using ethidium monoazide (EMA) (29) to selectively amplify DNA from Campylobacter cells possessing intact cell membranes in manure was investigated. Treatments consisted of (i) viable C. jejuni cells in bovine feces, (ii) heat-killed C. jejuni cells in feces, and (iii) uninoculated feces. C. jejuni 81-176 cells were grown on Karmali agar under microaerobic conditions at 40°C; cells at the late log stage of development were scraped from the surface of the medium and suspended in 5 ml of Columbia broth, and the suspension was divided into 1.0-ml aliquots in 2-ml tubes. Based on turbidity, the estimated density of cells in the inoculum was 2.3 × 109 CFU ml−1. To disrupt the cell membrane, one of the tubes was placed in boiling water for 30 min. The other treatment tubes were maintained at 4°C for 30 min. Viable cells in inocula were enumerated on Karmali agar in duplicate in a microaerobic environment at 40°C for 48 h; the mean densities of viable cells in the inocula were 5.4 × 109 and 0 CFU ml−1 for the viable and heat-treated cells, respectively. Equal volumes (200 μl) of viable or heat-killed cells were added to 5 g of bovine feces known to contain small amounts of C. jejuni DNA, and cells were thoroughly mixed in the feces using a sterile pipette tip. For uninoculated feces, 200 μl of Columbia broth alone was added to feces. Following inoculation, C. jejuni cells were recovered by centrifugation as described previously, except that the pellet was resuspended in 500 μl of Columbia broth. Densities of C. jejuni cells in feces were determined by dilution plating on KS agar in duplicate as described previously. Aliquots (200 μl) of the pellet were placed in four 2-ml tubes. Ethidium monoazide (Invitrogen Canada Inc., Burlington, ON, Canada) was added to two tubes (4 μl; final concentration of 100 μg ml−1), and water alone (4 μl) was added to the other two tubes under low-light conditions. Tubes were placed in the dark for 5 min, lids were opened, and all tubes were exposed to light emitted from a 500-W halogen light bulb for 1 min on ice; the light source was situated 10 cm from the samples. The IAC was then added to each tube. DNA was extracted using a QIAamp DNA Stool Mini Kit and subjected to conventional PCR for the Campylobacter genus (13) and to quantitative PCR (QPCR) in duplicate for C. jejuni (14). The experiment was conducted three times on separate occasions. Data were analyzed as a randomized complete block design using the mixed procedure of SAS (SAS Institute Inc., Cary, NC). In conjunction with a significant F test, the least square means statement (lsmeans) with the probability of difference option (pdiff) of SAS was used to conduct the least significant difference test.
Bovine manure compost.
Manure from Angus-cross steers maintained in an experimental feedlot at the Lethbridge Research Centre (LRC) from 29 November 2004 to 17 July 17 2005 was utilized. All steers were maintained on a base diet of barley (Hordeum vulgare) silage and rolled barley according to standard practices. One half of the cattle were administered 350 mg of chlortetracycline animal−1 day−1 and 350 mg of sulfamethazine (Aureo S-700 G; Alpharma Inc., NJ) animal−1 day−1 in their feed (i.e., AS700 treatment). The remaining cattle were not administered antimicrobial agents (i.e., control treatment). For each treatment, cattle were housed in five pens with 10 animals per pen. No animals were administered antimicrobial agents therapeutically during the experiment. Cattle were bedded using barley straw as needed. On 4 October 2005, 79 days after animals departed the feedlot, manure and bedding were scraped from the pens with a loader and transported to an adjacent experimental composting facility by truck. To prevent cross-contamination, manure from control treatment pens was removed first, and the loader bucket and truck were thoroughly washed before transfer of manure from AS700 treatment pens. Manure was combined across all pens per treatment (i.e., control and AS700), mixed with a manure spreader, divided into two equal volumes, and allocated into two replicate windrows per treatment that were ≈12 m long by 2.5 m wide and 2 m high. Windrows were not protected from precipitation. Industry standard manure windrow composting practices were used. The “active” composting period commenced on 4 October and lasted 126 days (Fig. 1). During the active composting phase, the compost was turned nine times with a tractor-pulled EarthSaver windrow turner (Fuel Harvesters Equipment, Midland, TX). After the active composting phase (turning), the compost entered a “curing” phase (no turning) for a further 105 days. The curing phase commenced on 7 February 2006 and ended 23 May 2006.
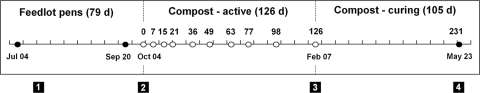
FIG. 1.
Compost experiment time course (2005 to 2006). Manure was maintained in feedlot pens for 79 days (d) before construction of compost windrows. Cattle were removed from the feedlot on 17 July 2005 (box 1), compost windrows were established on 4 October 2005 (box 2), and the active compost period lasted 126 days; the compost curing phase commenced on 7 February 2006 (box 3) and lasted 91 days, and the experiment was terminated on 23 May 2006 (box 4). Filled circles indicate sampling periods without compost turning, whereas open circles indicate sampling periods followed by compost aeration. The experimental duration was 310 days (ca. 10 months).
Manure and compost sampling.
Manure samples were collected from the pen floors of all feedlot pens ca. 2 weeks before removal of the animals on 4 July 2005 and again on 20 September 2005 before manure was composted. To obtain pen samples, subsamples of manure (≈200 g) were collected from five arbitrarily selected locations from the pen floor within each pen, and the samples were combined into a composite sample. During compost establishment, five subsamples (≈200 g) were obtained per treatment and were pooled. In addition, samples were obtained during the active compost phase immediately before compost turning. To obtain compost samples, two 2.0-m-wide cuts were made with a front-end loader (i.e., a Bobcat) perpendicular to the windrow's length at two locations (approximately one-third and two-thirds down the windrow); the front-end loader blade was washed thoroughly between compost treatments. Exposed faces from each cut location were sampled at the center and top of the face (approximately one-third from the top of the compost on the vertical axis). Subsamples of compost (≈2 kg) were removed from each side of the cut per location, placed on ice in a plastic bag free of Campylobacter, and transported to the laboratory within 15 min of collection. Samples were obtained on 4 October 2005 (day 0), 11 October 2005 (day 7), 19 October 2005 (day 15), 25 October 2005 (day 21), 9 November 2005 (day 36), 22 November 2005 (day 49), 6 December 2005 (day 63), 20 December 2005 (day 77), 10 January 2006 (day 98), 7 February 2006 (day 126), and 23 May 2006 (day 231) (Fig. 1).
Biological and physical parameters of compost.
To enumerate heterotrophic bacterial populations, a 10-g subsample of compost was placed in a stomacher bag with 90 ml of sodium phosphate buffer (0.05 M; pH 6.5), and the sample was homogenized for 2 min using a Stomacher blender set on high (Model 400; Seward Medical). The resultant homogenate was diluted in phosphate buffer in a 10-fold dilution series, and 100 μl from each dilution was spread in duplicate onto nutrient agar (Difco, Becton-Dickinson, Sparks, MD). Cultures were maintained aerobically at 37°C for 48 h, colonies were enumerated at the dilution yielding 30 to 300 colonies, and the number of CFU g−1 was determined.
Temperature was determined with thermocouples placed at the center and top (≈10 cm from the surface) of each windrow (two locations per windrow corresponding to sampling sites) at the time that windrows were established and after each turning; compost temperatures were logged every 20 min using a data logger (Sciemetric, Nepean, ON, Canada). Degree days (55°C base temperature) were calculated as Σ(daily mean temperature − 55°C). Ambient temperatures were recorded at a weather station located adjacent to the compost site. At each sample collection time, compost samples were analyzed for moisture, dry matter (DM), organic matter (OM), pH, electrical conductivity (EC), total carbon (C), and total nitrogen (N) (24). Moisture content was determined after oven drying (60°C) to a constant weight for 6 days. Oven-dried compost samples were subsequently ground to pass through a 0.25-mm-pore-size screen for further analysis of OM, C, and N. Ash content was determined after samples were burned in a muffle furnace at 600°C for 2 h (AOAC official method 942.05). The pH and EC of compost (10 g) were determined after the sample was soaked in 90 ml of distilled water for 30 min, and measurements were taken with a pH/conductivity meter (Accumet pH meter 50; Fisher Scientific, Hampton, NH). Total C and N were determined using a Carlo Erba NA 1500 Carbon-Nitrogen elemental analyzer (Carlo Erba Strumentazione, Rodano, Milan, Italy). Total mass (dry matter plus water) loss was calculated using initial (day 0) and final (day 231) weights of materials. The mass losses of DM, OM, C, and N were estimated using compost chemical properties and total mass values. Statistical analyses were performed using SAS (SAS Institute Inc.). Temperature data and compost properties (water content, C, N, C/N ratio, pH, and EC) were analyzed using the mixed procedure, with time treated as a repeated measure; in the case of temperature, weekly averages were analyzed. Appropriate covariance structures were utilized according to the lowest Akaike's information criterion (AIC).
Isolation and identification of campylobacteria from manure and compost.
Individual manure and compost samples were thoroughly mixed by hand, and a 5-g subsample from each was removed and homogenized in 45 ml of Columbia broth. Campylobacter cells were concentrated by centrifugation as described above and resuspended in 2 ml of Columbia broth. The suspension (25 μl) was spread onto KS agar in duplicate, and cultures were maintained in a microaerobic environment at 40°C and examined at 48 h and at 24 h thereafter for an additional 72 h. Cells from presumptive Campylobacter colonies (i.e., based on colony morphology, and/or microscopic examination of cell size, shape, and motility) were streaked onto Karmali agar and incubated in a microaerobic environment at 40°C, and cell biomass was stored at −80°C. In addition to direct plating, enrichment isolation was conducted. The suspension (150 μl) was added to 2 ml of Bolton broth (CM983; Oxoid Ltd.) containing Bolton broth selective supplement (SR048C), and cultures were maintained in a microaerobic environment at 175 rpm at 30°C for 3 h, at 37°C for 2 h, and at 40°C for 24 h. The enrichment broth (25 μl) was then spread in duplicate onto KS agar, cultures were incubated, and biomass was collected and stored as described above. Genomic DNA was extracted from stored biomass using an AutoGen 740 robot (AutoGen, Farmingham, MA), and DNA was subjected to genus- and taxon-specific PCR (13). In addition, PCR for the Arcobacter genus was conducted (10). The 16S rRNA gene was sequenced for Arcobacter-positive isolates using a 3130 Genetic Analyzer (Applied Biosystems Canada, Streetsville, ON, Canada), and sequences were compared directly with the NCBI GenBank nonredundant nucleotide database using BLASTN analysis.
Direct PCR detection and quantification of campylobacteria in compost.
Four 200-μl aliquots of the resuspended pellet from individual samples were placed in 2-ml tubes. EMA (4 μl) was added to two tubes, water (4 μl) was added to two tubes, and all tubes were exposed to light on ice as described previously. Samples were stored at −20°C until processed. An IAC was added to all tubes, and DNA was extracted using a QIAamp DNA Stool Mini Kit (Qiagen) (13). In total, DNA was extracted from 352 samples. Extracted DNA was subjected to taxon-specific PCR for the Campylobacter genus, C. jejuni, C. coli, C. hyointestinalis, C. fetus, and C. lanienae (13); only Campylobacter-negative samples in which the IAC was amplified were considered true negatives. Potentially false-negative samples (i.e., for which neither a Campylobacter nor an IAC amplicon occurred) were subsequently subjected to PCR using PrimeSTAR HS (Premix) DNA polymerase (Takara Bio Inc./Clontech Laboratories, Inc., Mountain View, CA) as this high-fidelity polymerase is less susceptible to PCR inhibition. The Mastermix (1×) consisted of 5 μl of PrimeSTAR premix, 0.25 μl of each primer (10 μM), 3.5 μl of Optima water, and 1 μl of template (10-μl total volume) on ice. Thermocycler conditions consisted of 35 cycles at 98°C for 10 s and 68°C for 1 min, a termination cycle at 72°C for 3 min, and a 4°C hold. Samples that were positive for C. jejuni using conventional PCR were subjected to QPCR targeting the mapA gene (14). Log copy number was converted to log CFU g−1.
Persistence of C. jejuni DNA in compost.
Cells of C. jejuni strain L93 (originally isolated from a dairy cow at LRC) were grown in Columbia broth under microaerobic conditions, harvested by centrifugation, and enumerated in duplicate on Karmali agar as described above for strain 81-176. Treatments consisted of (i) C. jejuni DNA, (ii) heat-killed C. jejuni cells, and (iii) viable C. jejuni cells. DNA was extracted using a DNAeasy Tissue Kit (Qiagen) according to the manufacturer's recommendations, and quantified by QPCR (14). Heat-killed cells were boiled for 30 min, and viable C. jejuni cells were maintained on ice for 30 min. The concentration of viable cells was determined by diluting the cell suspension in a 10-fold dilution series in Columbia broth, spreading 100-μl aliquots onto Karmali agar in duplicate, maintaining the cultures at 40°C in a microaerobic environment, and enumerating the colonies at the dilution yielding 30 to 300 colonies per dish. No viable cells were detected following boiling.
Cells and DNA were thoroughly distributed in 200 ± 10 mg of bovine feces known to contain low densities of C. jejuni DNA, and inoculated feces were placed within sterile bags (≈1 by 3 cm) constructed of nylon possessing 51-μm pores (B & SH Thompson, Scarborough, ON, Canada). It was determined in a preliminary study that populations of heterotrophic bacteria in manure within bags were equivalent to those in manure outside of bags during composting (data not presented). Approximately equivalent densities of cells from treatments two and three (determined by turbidity) and the equivalent concentration of DNA (i.e., treatment one) were added to fecal samples. Bags were heat sealed, attached to a nylon thread, and situated in a small-scale bovine manure compost windrow (under cover to protect from precipitation) at a depth of ≈20 cm on the day it was turned. The compost pile was ≈4.5 m long, 2.5 m wide, and 2 m high. Replicate bags (n = 2) were sampled immediately before placement (i.e., time zero), and at 2, 4, 8, 16, 32, 64, 128, and 168 h after placement in the compost. Ambient temperatures and compost temperature at a depth of 20 cm were recorded (hourly average) over the experimental period using a HOBO micrologger (HOBO H8 Pro; Onset Computer Corp., Cape Cod, MA).
After removal of samples from the compost, the small amounts of residual manure on the surfaces of the bags was carefully removed with a sterile spatula, bags were placed on ice, and within 15 min of collection they were frozen at −20°C. To extract DNA, bags were thawed, and DNA from the entire contents of each bag was extracted using a QIAamp DNA Stool Mini Kit (Qiagen) with an IAC, as described previously. To further remove PCR inhibitors (e.g., polyphenolic compounds), a post-clean-up protocol using Sepharose columns was applied using the basic method of Reuter et al. (28) except that the process was miniaturized. Briefly, Sepharose columns were prepared in 1-ml Centriprep spin column tubes (Amicon Plastics, Houston, TX), DNA (100 μl) was slowly pipetted onto center of the column, the column was centrifuged for 1 min at 100 × g, and the eluate containing DNA was collected and stored at −20°C. This method was found to adequately remove PCR inhibitors with negligible loss of DNA (data not presented). Samples were qualitatively assessed for Campylobacter genus DNA and for the IAC (13); C. jejuni DNA in duplicate samples was quantified by QPCR (14), and the mean of the two observations per replicate was calculated. Percent reductions in DNA concentration relative to time zero were calculated for each treatment by sample time. The experiment was arranged as a randomized complete block design with three levels of treatment, eight levels of time, and two levels of replicate. Given that the same compost windrows were sampled over time, the data were analyzed using the mixed model of SAS (SAS Institute Inc.), with time treated as a repeated measure. Appropriate covariance structures were selected according to the lowest AIC. In conjunction with a significant F test, the least significance difference test was applied.
RESULTS
Centrifugation method.
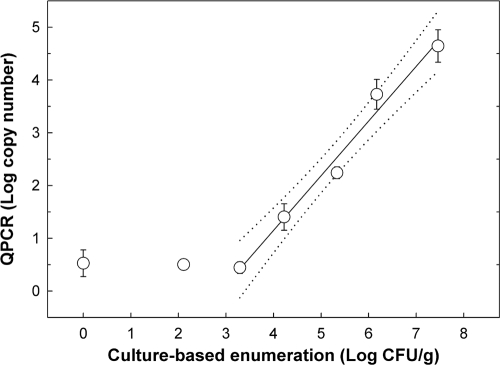
The majority of Campylobacter cells were recovered from the supernatant following centrifugation of the manure suspension at low speed. Densities of C. jejuni cells in the pellet after the fast spin were 7.46 ± 0.17, 6.17 ± 0.27, 5.33 ± 0.32, 4.22 ± 0.28, 3.29 ± 0.0, 2.11 ± 0.38, and 0 log10 CFU g−1 for the target density treatments of 108, 107, 106, 105, 104, 103, and 0 (i.e., uninoculated) cells g−1, respectively. The minimum detection limit of C. jejuni cells by QPCR was ≈103 cells g−1. A strong linear relationship (r2 = 0.98) was observed between the number of CFU and PCR target copy number, and the relationship can be described by the following equation: y = −2.997 + 1.036x (Fig. 2).
FIG. 2.
Linear relationship (r2 = 0.98) between log10 copy number and culture-based enumeration (log10 CFU g−1) using the centrifugation recovery method. The following equation describes this relationship: y = −2.997 + 1.036x (r2 = 0.983). Dotted lines represent 95% confidence intervals.
Detection of viable campylobacterial cells.
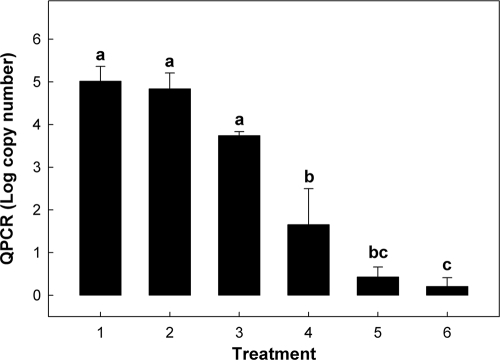
Densities in the inoculum were determined by dilution plating to be 8.73 ± 0.21 log10 CFU ml−1. The intensities of Campylobacter genus amplicons were similar between viable C. jejuni cell treatments regardless of the presence of EMA (Fig. 3, lanes 2 and 3). In contrast, a conspicuous difference in amplicon intensities was observed between the heat-killed C. jejuni treatments (Fig. 3, lanes 4 and 5). The amplicon for the EMA-treated heat-killed cells was noticeably lighter than the amplicon for cells not treated with EMA. A weak Campylobacter genus amplicon (equivalent intensity to that of lane 5) was observed for one of the two control samples (Fig. 3, lane 7) indicating that the feces utilized was not completely free of Campylobacter DNA. Similar results were observed for QPCR of C. jejuni DNA (Fig. 4). There were no significant differences (P = 0.79) between the two viable C. jejuni treatments. Furthermore, the quantity of heat-killed C. jejuni DNA that was not treated with EMA was similar (P ≥ 0.08) to the quantity in the viable C. jejuni treatments. Although greater variation was observed for EMA-treated feces, concentrations of C. jejuni DNA amplified from heat-killed cells treated with EMA (i.e., treatment 4) were significantly reduced (P = 0.009) relative to heat-killed cells not treated with EMA. This represented a decrease in amplification of 90.9 to 100%.
FIG. 3.
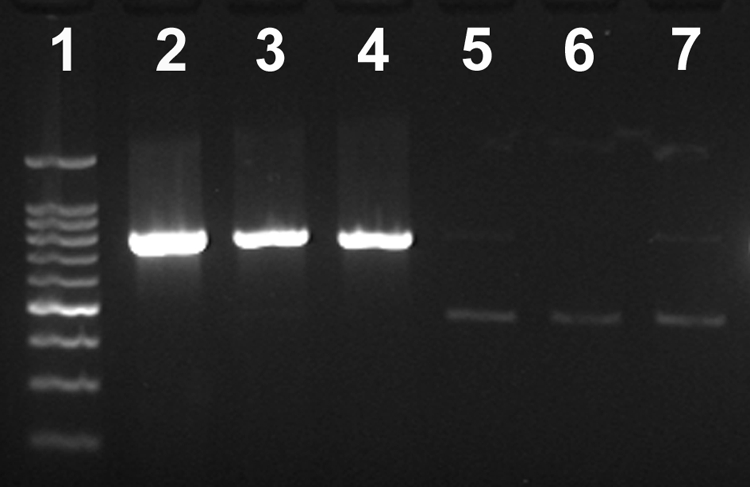
Effect of EMA on amplification of C. jejuni DNA from bovine feces using a Campylobacter genus primer set. The Campylobacter genus amplicon is 816 bp in size, and the IAC is 465 bp in size. Lane 1, 100-bp molecular weight marker; lane 2, feces inoculated with viable C. jejuni cells; lane 3, feces inoculated with viable cells treated with EMA; lane 4, feces inoculated with heat-killed cells; lane 5, feces inoculated with heat-killed cells treated with EMA (note the IAC control amplicon); lane 6, uninoculated feces; lane 7, uninoculated feces treated with EMA. No PCR products were observed in the negative control reaction (not shown).
FIG. 4.
Effect of EMA on quantities of amplifiable C. jejuni DNA in bovine feces. Data are for the following treatments: lane 1, feces inoculated with viable C. jejuni cells; lane 2, feces inoculated with viable cells and treated with EMA; lane 3, feces inoculated with heat-killed cells; lane 4, feces inoculated with heat-killed cells and treated with EMA; lane 5, uninoculated feces; and lane 6, uninoculated feces treated with EMA. Vertical bars associated with histograms represent standard errors of the mean, and histograms not indicated by the same letter are significantly different (P ≤ 0.05).
Bovine manure compost. (i) Biological and physical parameters.
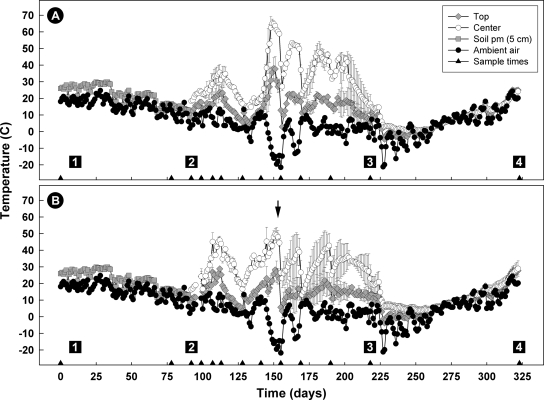
Large populations of heterotrophic bacteria (>8.0 log10 CFU g−1) were recovered for both the control and AS700 treatments upon compost establishment, and population sizes were sustained throughout the active composting period (data not presented). Nevertheless, compost temperatures following windrow establishment until day 49 were relatively moderate for both treatments, not exceeding 55°C (Fig. 5). Thereafter, temperatures at the center of the control but not the AS700 treatment compost exceeded 55°C; there was considerable variability between the two replicate windrows for the AS700 treatment during the later half of the active composting period (Fig. 5, arrow). The number of degree days (base of 55°C) attained at the center of the compost pile were 62 and 0 for the control and AS700 treatments, respectively. For both treatments, no degree days exceeding 55°C were attained at the top of the compost. During the curing phase, compost temperatures were similar to those of ambient air. Although water content, pH, EC, C, and N changed significantly (P < 0.001) over the composting period, the interaction between treatment and time was not significant (P > 0.05) for these variables. Averaged over time however, water content, EC, C, and N differed (P ≤ 0.05) between the AS700 and control treatments. The control treatment was 5.8% drier (0.425 versus 0.451 kg kg−1), EC was reduced by 11.8% (4.5 versus 5.1 dS m−1), and there was 11.2% (111 versus 125 kg tonne−1) and 10.1% (8.9 versus 9.9 kg tonne−1) less carbon and nitrogen, respectively. There was no difference averaged over time in pH values (8.2) and C/N ratios (12.5 to 12.6) between the two treatments. The initial pH of the compost was 8.4; it dropped to 7.9 by day 98 and remained there until the end of the active composting period.
FIG. 5.
Daily average ambient, soil (5-cm depth), and compost temperatures for the control (A) and AS700 (B) treatments; soil temperatures were used to estimate manure temperatures in feedlot pens. Triangles on the x axis indicate sample times. Compost temperatures (i.e., top and center) represent the mean of two replicates, and vertical bars indicate the standard error of the mean. Box 1 indicates the time at which cattle were removed from the feedlot (17 July 2005), box 2 indicates the time at which compost windrows were established (4 October 2005), box 3 indicates the end of the active composting period and the commencement of the curing compost phase, and box 4 indicates the termination of the composting period. Triangles located on the x axis indicate compost turning times. The arrow indicates a period of variable temperature between the replicate compost piles for AS700 compost.
(ii) Isolation of campylobacters from manure and compost.
A total of 345 presumptive campylobacteria isolates were recovered from pen manure and compost during the active composting period and subjected to taxon-specific PCR. Of these isolates, 144 were identified as either C. jejuni, C. fetus, C. hyointestinalis, or Arcobacter butlzeri (Table 1). Fourteen isolates were identified as campylobacters but were not identified to the species level. Although the majority of Campylobacter isolates were recovered at earlier stages of the compost period, isolates were recovered from AS700 treatment compost throughout the active period (i.e., up to 126 days). Fast-growing and spreading colonies of various ubiquitous bacteria (e.g., Bacillus, Paenibacillus, and Brevibacillus) were frequently encountered on KS agar at later sampling times, obscuring detection of campylobacteria on the medium.
TABLE 1.
Isolation of campylobacteria from bovine manure in pens and from manure compost for 231 days
| Samplea | No. of isolates by taxon in the indicated treatment groupb |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Control treatment |
AS700 treatment |
|||||||||||
| Cjej | Cfet | Chyo | Abut | Cspp | Subtotal | Cjej | Cfet | Chyo | Abut | Cspp | Subtotal | |
| Manure | 11 | 1 | 2 | 8 | 0 | 22 | 20 | 0 | 0 | 7 | 0 | 27 |
| MC-0 | 0 | 0 | 9 | 1 | 3 | 13 | 0 | 0 | 0 | 3 | 1 | 4 |
| MC-7 | 14 | 3 | 14 | 1 | 5 | 37 | 0 | 1 | 0 | 1 | 4 | 6 |
| MC-15 | 6 | 1 | 6 | 0 | 0 | 13 | 1 | 0 | 0 | 0 | 0 | 1 |
| MC-21 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 1 |
| MC-36 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 1 | 2 |
| MC-49 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 1 |
| MC-63 | 0 | 0 | 0 | 0 | 0 | 0 | 5 | 0 | 0 | 0 | 0 | 5 |
| MC-77 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 2 |
| MC-98 | 0 | 0 | 0 | 0 | 0 | 0 | 5 | 3 | 0 | 0 | 0 | 8 |
| MC-126 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 2 |
| MC-231 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Totalc | 31 | 5 | 31 | 10 | 8 | 85 | 37 | 5 | 0 | 11 | 6 | 59 |
Manure compost (MC) samples are identified by the day of sampling, e.g., MC-0 indicates manure compost at day zero.
Cjej, C. jejuni; Cfet, C. fetus; Chyo, C. hyointestinalis; Abut, A. butzleri; and Cspp, unidentified Campylobacter species. No C. coli or C. lanienae isolates were recovered.
The total number of campylobacteria isolates recovered and identified from manure in pens and compost was 144 (49 isolates from pen manure and 95 isolates from compost).
PCR detection and quantification of campylobacters.
Campylobacter DNA was frequently detected in pen manure samples and in control and AS700 compost throughout the active and curing periods (Tables 2 and 3). There were no conspicuous differences in detection rates between sample sites at the top or center of the compost piles for either control or AS700 compost. For control compost, rates of DNA detection were slightly lower for EMA-treated than for the non-EMA-treated samples (Table 2). Mean amplification rates (i.e., across sample times) for Campylobacter DNA were 90.5% ± 4.8% and 88.7% ± 2.3% at the top and center of the piles for non-EMA-treated control compost, respectively. For EMA-treated compost, amplification rates at the top and bottom of the piles were 58.4% ± 8.3% and 79.5% ± 2.3%, respectively. For AS700 compost, a reduction in the amplification rate was not observed for EMA-treated samples (Table 3); mean amplification rates for EMA-treated samples ranged from 94.1 to 100%, and for non-EMA-treated samples amplification rates ranged from 60.0 to 80.0%. Three (1.7%) inconclusive samples in which neither a genus nor an IAC amplicon was detected were encountered for the control compost, whereas 19 (10.8%) samples were inconclusive for AS700 compost.
TABLE 2.
Detection of Campylobacter DNA in control manure compost
| Day | PCR result for non-EMA-treated compost by locationa |
PCR result for EMA-treated compost by locationa |
||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Replicate 1 |
Replicate 2 |
Replicate 1 |
Replicate 2 |
|||||||||||||
| Top | Center | Top | Center | Top | Center | Top | Center | |||||||||
| 0 | − | I | + | + | − | + | − | − | + | + | − | + | + | + | + | + |
| 7 | + | + | + | + | + | I | + | + | − | + | − | + | − | − | − | − |
| 15 | + | + | − | + | + | + | + | + | + | − | − | + | − | − | + | + |
| 21 | − | + | − | + | + | + | + | + | − | + | + | − | − | − | + | − |
| 36 | + | + | + | + | + | + | + | + | − | − | + | + | − | − | + | − |
| 49 | + | + | + | − | + | + | + | + | − | I | + | + | − | − | − | + |
| 63 | + | − | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| 77 | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| 98 | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| 126 | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| 231 | + | + | + | + | + | + | + | + | − | + | + | + | + | − | + | + |
| Total positive (%) | 85.7 | 86.4 | 95.2 | 90.9 | 66.7 | 81.8 | 50.0 | 77.3 | ||||||||
Extracted DNA was subjected to Campylobacter genus-specific PCR. Each replicate compost pile was oriented east to west. For each day and location, the first result represents the east sample location, and the second result represents the west sample location. Results were considered inconclusive (I) if neither a genus nor an IAC amplicon was detected.
TABLE 3.
Detection of Campylobacter DNA in AS700 manure compost
| Day | PCR result for non-EMA-treated compost by locationa |
PCR result for EMA-treated compost by locationa |
||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Replicate 1 |
Replicate 2 |
Replicate 1 |
Replicate 2 |
|||||||||||||
| Top | Center | Top | Center | Top | Center | Top | Center | |||||||||
| 0 | − | − | − | − | − | + | − | − | I | + | + | I | + | + | I | + |
| 7 | − | − | + | − | − | − | − | − | + | + | + | I | + | + | + | + |
| 15 | + | + | I | + | + | I | + | I | + | I | + | + | + | + | + | + |
| 21 | + | I | + | + | − | + | + | I | + | + | + | + | + | + | + | + |
| 36 | + | + | − | + | + | + | + | + | + | + | + | + | + | + | + | + |
| 49 | + | + | + | + | − | + | + | + | + | + | + | + | + | + | + | + |
| 63 | + | + | + | + | + | + | + | + | I | + | + | + | + | + | + | + |
| 77 | + | I | − | + | − | + | + | + | + | + | + | + | I | I | + | + |
| 98 | + | + | − | I | I | + | + | + | + | I | + | + | + | + | + | + |
| 126 | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | I |
| 231 | + | + | + | + | − | − | + | + | I | − | + | + | + | + | + | + |
| Total positive (%) | 80.0 | 70.0 | 60.0 | 80.0 | 94.1 | 100.0 | 100.0 | 100.0 | ||||||||
Extracted DNA was subjected to Campylobacter genus-specific PCR. Each replicate compost pile was oriented east to west. For each day and location, the first result represents the east sample location and the second result represents the west sample location. Results were considered inconclusive (I) if neither a genus nor an IAC amplicon was detected.
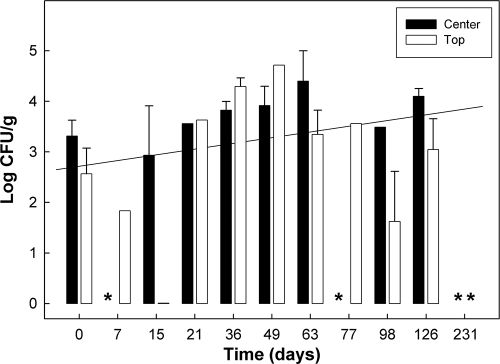
C. jejuni DNA was frequently detected (31.8 to 42.8%) in non-EMA-treated control compost and, to a lesser extent, in non-EMA-treated AS700 compost (0% to 15.0%). Quantities of C. jejuni DNA did not decrease substantively over time at either the top or center sample location (Fig. 6). DNA of C. coli and C. fetus were infrequently (<16%) detected in control and AS700 compost, and C. lanienae DNA was detected only in control compost (15.0 to 38.0%). C. hyointestinalis DNA was frequently (15.0 to 38.1%) detected in control compost but not in AS700 compost.
FIG. 6.
Temporal quantities of C. jejuni DNA in control treatment compost (center and top). Data obtained from EMA-treated and non-EMA-treated samples were combined. Vertical bars indicate standard errors of the mean, and asterisks indicate samples in which no C. jejuni DNA was detected. The diagonal line represents the linear relationship between time and C. jejuni DNA concentration (y = 2.72 + 0.113x).
Persistence of C. jejuni DNA in compost.
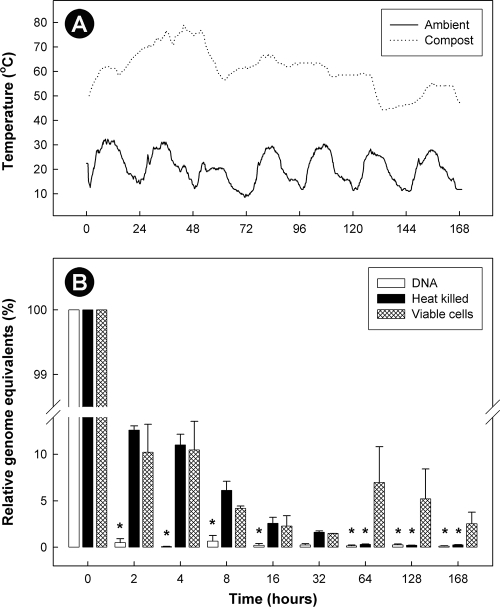
Temperatures (at a depth of ≈20 cm) of the small-scale compost exceeded 55°C for most (76%) of the experimental period (Fig. 7). Within 26 h, temperatures at this depth exceeded 70°C (maximum of 78.9°C) and remained above 70°C until the 54-h point, after which temperatures gradually decreased. Substantively less C. jejuni DNA was observed for all three treatments 2 h after placement in compost (Fig. 7); concentrations of DNA were reduced by 99.5%, 87.4%, and 89.7% for the naked DNA, heat-killed cell, and viable cell treatments, respectively. However, the persistence of C. jejuni DNA differed significantly (P < 0.001) for the three treatments as a function of time. DNA within heat-killed and viable C. jejuni cells persisted longer in compost than naked DNA, and DNA within viable cells persisted longer than in heat-killed cells.
FIG. 7.
Ambient and bovine manure compost temperatures (A) and quantities of C. jejuni DNA (relative to time zero) (B) over the 7-day duration of the ancillary experiment examining differential persistence of Campylobacter DNA in compost. Treatments consisted of naked DNA, heat-killed cells, and viable C. jejuni cells. Vertical bars indicate the standard error of the mean, and bars marked by an asterisk are significantly different (P ≤ 0.05) from the viable cell treatment.
DISCUSSION
Very large quantities of beef cattle manure are produced from confined feeding operations in Alberta, and composting has been proposed as a method of deactivating enteric pathogens present in the manure. In the current study, we examined the persistence of Campylobacter species in compost constructed from the manure of cattle administered chlortetracycline and sulfamethazine or no antibiotics. Composting is an aerobic technique used to manage solid manure and other organic waste. Effective composting relies on the action of complex microbial communities including the generation of metabolic heat and enzymatic decomposition processes. Salient benefits of manure composting include decreased water and bulk (≈50 to 70% reduction in volume) (31), destruction of weed seeds (21) and pathogens (34, 39), and stabilization of nutrients as organic compounds (22). Appropriate moisture (40 to 65%) and efficient aeration are critical components of effective composting. To facilitate aeration, we used the industry standard windrow method in which compost is periodically turned. Temperatures at or near 55°C are considered optimal (31), and we observed that a temperature of 55°C was reached or exceeded only late in the active composting phase of the control compost and never for AS700 compost. In addition, temperature is temporally and spatially variable within compost (39), consistent with the observations that temperatures were lower at the top than at the center of the compost and that a rapid rise in temperature followed compost turning. Nevertheless, heterotrophic bacterial populations typically exceeded 108 CFU g−1 (dry weight) regardless of treatment (i.e., control versus AS700) or location (i.e., top versus center).
Limited research has been conducted on the ability of Campylobacter species to persist in solid manure compost. In the current study we utilized both isolation and molecular methods to examine the ability of Campylobacter species naturally excreted in feces to persist in compost. A significant obstacle that we faced was the ability of fast-growing thermophilic bacteria (e.g., members of the Bacillaceae) to grow on the semiselective Campylobacter isolation medium that we employed, an observation made previously (39). The growth of these bacteria precluded effective isolation of campylobacters. Nevertheless, we did isolate 95 Campylobacter isolates from compost, including at later stages of the composting process (i.e., days 15 to 126). Using culture-based enumeration methods, previous studies concluded that campylobacters (primarily C. jejuni bacteria) do not persist well in solid manure (7, 11, 12, 26, 32, 39) or in thermophilic anaerobic manure digesters (35, 36) but can survive for prolonged periods in mesophilic anaerobic digesters (18, 19).
There are few reports in which persistence of Campylobacter in manure has been examined using molecular methods. Wéry et al. (38) utilized TaqMan QPCR to examine quantities of C. jejuni in various stages of urban wastewater treatment and determined that significant quantities of C. jejuni DNA were present in dewatered sludge (>104 gene copies g−1). In the current study, we utilized qualitative and quantitative PCR to investigate persistence of Campylobacter species. We observed that DNA of campylobacteria shed in bovine feces persisted in manure in pens for ca. 2.5 months and throughout the active and curing phases of composting (ca. 7.7 months). Furthermore, we observed no differences in the frequency of detection between the top and center sample locations, nor did we observe decreases in the concentration of C. jejuni DNA over the active composting period. Quantities of DNA equivalent to 104 C. jejuni CFU g−1 or higher were typically observed. The utilization of molecular methods provides the major advantages of eliminating reliance on isolation and its associated limitations, as well as the detection/quantification of viable but nonculturable cells. However, molecular methods also possess a number of disadvantages. The presence of PCR inhibitors and the sensitivity/specificity of primers are two important issues facing researchers. We utilized an IAC and encountered low to moderate inhibition (<10%). The primers we utilized have previously been determined to be very sensitive, yet the minimum detection threshold we observed for C. jejuni was ≈103 cells g−1. This is attributed to the necessity of extracting DNA from small volumes and the dilution effect of the DNA extraction process.
Arguably, the greatest problem with using molecular methods to study the fate of pathogens in chemically and microbiologically complex ecosystems is distinguishing dead from viable cells. In the current study, we evaluated an EMA method to amplify DNA only from cells possessing an intact cell membrane (29). This method relies on the ability of EMA to penetrate the cell membrane of dead cells and covalently bind to DNA following irradiation, thereby preventing the annealing of primers to DNA. Both EMA and propidium monoazide have been successfully used to quantify bacterial DNA from living cells (4), but the method has to be carefully evaluated. For example, the addition of EMA resulted in an underestimation of viable cells of C. jejuni and Staphylococcus species (6, 20) and is not effective for bacteria embedded in biofilms (27). We evaluated the EMA method in combination with differential centrifugation to remove large particles and to concentrate C. jejuni cells incorporated in bovine feces. The centrifugation method increased the accessibility of EMA to cells and their exposure to light irradiation and removed some PCR inhibitors such as insoluble humic acids (37). The application of the EMA method in combination with centrifugation did not affect amplification of DNA from viable cells in feces and significantly inhibited the amplification of DNA from heat-killed cells. However, DNA from ≈4 to 9% of nonviable cells was amplified in two of three replicates, whereas no DNA was amplified in the third replicate. Although not absolute, these results indicate that the vast majority of DNA amplified from compost samples treated with EMA originated from cells possessing intact cell membranes (i.e., viable cells).
To further corroborate that DNA was being detected from viable cells, the persistence of C. jejuni DNA (i.e., naked DNA, DNA in heat-killed cells, and DNA in viable cells) was determined in highly active compost. Almost no naked DNA was amplified 2 h after placement in active compost (i.e., ≥50°C). DNA in heat-killed cells lasted longer than naked DNA but for significantly less time than DNA in viable C. jejuni cells. Consistent with our observation, a transgene in corn placed in compost was rapidly degraded and was undetectable by day 14 (8). In the current study, we utilized a nylon bag possessing relatively large pores (i.e., 51 μm in diameter). It was essential to utilize a container to facilitate recovery of samples from compost, and the utilization of a bag possessing relatively large pores was deemed necessary to ensure that conditions within the bag (e.g., temperature and oxygen status) were representative of the compost ecosystem. However, the pore size of the bags was substantially larger than the size of Campylobacter cells, and we cannot definitively exclude the possibility that DNA and/or C. jejuni cells escaped the bag. However, the compost was relatively dry (≈40%), and we did not detect any reduction in mass of the manure put in the bags. It is unlikely that substantive loss of DNA from the bag occurred within the initial 2-h sample period, given the relatively low moisture content of the compost. Furthermore, it is very improbable that nonviable C. jejuni cells would have escaped containment at a greater rate than viable cells. Taken together, these results support our EMA findings that demonstrated that Campylobacter cells are capable of surviving for prolonged periods in a very inhospitable environment (e.g., compost).
Of interest, the majority of DNA (>95%) from viable C. jejuni (strain L93) inoculated into active small-scale compost could not be amplified after only 7 days, contrasting sharply with the prolonged persistence of Campylobacter species that we observed in the large-scale compost study. Besides a reliance on nylon sample bags, two major differences between these two experiments were the use of a single laboratory strain of C. jejuni and the substantially higher compost temperatures achieved in the ancillary experiment; the experiment was commenced after the small-scale compost pile, in which compost conditions were optimal, was turned. The rapid death of C. jejuni strain L93 is consistent with previous studies that observed poor survival of inoculated C. jejuni in manure or compost (12, 26, 32, 39). A number of Campylobacter species are readily shed in beef cattle feces (15, 16), and the persistence of an array of naturally occurring campylobacters and strains was measured in the large-scale compost experiment. Natural populations of E. coli vary widely in their ability to persist in soils receiving swine manure (33). Whether variability exists in the survivability of specific genotypes of campylobacteria and/or whether Campylobacter cells shed in feces persist longer is currently uncertain and warrants investigation.
In North America, beef production relies heavily on the administration of antibiotic growth promoters (AGP) to cattle within confined feeding operations. Chlortetracycline in combination with sulfamethazine (AS700) is a commonly administered in-feed AGP, and it is typically used to improve weight gain and feed efficiency, to aid in the prevention of liver abscesses, reduce bacterial diarrhea, prevent foot rot, and/or reduce the incidence of bovine respiratory disease (17). In the current study, AS700 was administered to animals throughout the feeding period. Elmund et al. (5) showed that 75% of ingested chlortetracycline is excreted and subsequently recovered in cattle feces. In laboratory-scale composters, concentrations of chlortetracycline (and its epimer, epi-chlortetracycline) in nonsterile and sterilized bovine manure compost decreased by ≥98% (30 days), and the presence of the antibiotic had no observable effect on the composting process (2). Interestingly, these investigators concluded that degradation was not due to the presence of viable microorganisms in the compost and was directly or indirectly the result of elevated temperature. In our study, quantities of iso-chlortetracycline (783.5 μg kg−1 of manure at compost day 0) and sulfamethazine (149.7 μg kg−1 at day 0) decreased by 83.6% and 38.6% after ca. 30 days, respectively, and by 99.5% and 93.2% at the end of the 126-day active composting period, respectively (Allan Cessna, National Water Research Institute, Environment Canada, Saskatoon, SK, Canada; personal communication). No chlorotetracycline (i.e., unmetabolized) was detected, and only small quantities of 4-epi-iso-chlortetracycline were observed in the compost (Allan Cessna, personal communication). Although residual AS700 had no effect on persistence of campylobacteria or on heterotrophic bacterial populations, the chemical properties of compost (e.g., water content and nitrogen and carbon content) and temperature were different between the control and AS700 treatments.
In conclusion, we utilized culture-based and molecular methods to determine the degree to which Campylobacter species persist in bovine manure compost. Isolation was found to be ineffectual for assessing persistence, but the application of qualitative and quantitative PCR showed that campylobacteria persisted in manure and subsequently in compost for a prolonged period (ca. 10 months). Centrifugation was used in combination with EMA to ensure that DNA was amplified from Campylobacter cells possessing intact cell membranes. Furthermore, the results of an ancillary study that examined persistence of C. jejuni DNA placed in compost corroborated the EMA findings that amplified DNA originated from viable cells. Compost is a very inhospitable environment, and the ability of Campylobacter species to persist in this ecosystem for prolonged periods clearly challenges the common belief that campylobacters do not survive well outside of their hosts (9).
Acknowledgments
We acknowledge the excellent assistance provided by the following people at Agriculture and Agri-Food Canada (AAFC): Jenny Gusse for validating the centrifugation and EMA methods and for conducting PCR of Campylobacter DNA, Tara Shelton for carrying out taxon-specific PCR and QPCR, Kathaleen House for isolating and identifying campylobacteria, Grant Duke for conducting the ancillary DNA persistence experiment, Lorna Selinger and Ruth Barbieri for compost sample collection and for enumerating heterotrophic bacterial populations, Andrew Olson for collecting temperature data, Paul DeMaere and Dan Yagos for conducting chemical analyses of compost, and Brant Baker and Fred Van Herk for compost establishment. We also thank the two anonymous reviewers of the manuscript for their constructive suggestions.
This research was supported by the following sources: an AAFC GAPS grant entitled “Assessment and management of biological and chemical contaminants carried in livestock and poultry manure” to E.T., T.A.M., and F.J.L.; a Canada-Alberta Beef Industry Development Fund grant to G.D.I.; and an AAFC Peer Review Project grant entitled “Assessment and management of on-farm fate of microbial and chemical contaminants carried in human and animal wastes used as organic fertilizers” to G.D.I. and E.T.
Footnotes
Published ahead of print on 18 December 2009.
REFERENCES
- 1.Alberta Government. 2009. Cattle and calves on farms in Alberta. Agriculture and Rural Development, Government of Alberta, Edmonton, Alberta, Canada. http://www1.agric.gov.ab.ca/$department/deptdocs.nsf/all/sdd1492?.
- 2.Arikan, O. A., W. Mulbry, and C. Rice. 2009. Management of antibiotic residues from agricultural sources: use of composting to reduce chlortetracycline residues in beef manure from treated animals. J. Hazard. Mater. 164:483-489. [DOI] [PubMed] [Google Scholar]
- 3.Bruce-Grey-Owen Sound Health Unit (BGOSHU). 2000. The investigative report of the Walkerton outbreak of waterborne gastroenteritis, May-June 2000. Bruce-Grey-Owen Sound Health Unit, Walkerton, Ontario, Canada. http://water.sesep.drexel.edu/outbreaks/WalkertonReportOct2000/REPORT_Oct00.PDF.
- 4.Delgado-Viscogliosi, P., L. Solignac, and J. M. Delattre. 2009. Viability PCR, a culture-independent method for rapid and selective quantification of viable Legionella pneumophila cells in environmental water samples. Appl. Environ. Microbiol. 75:3502-3512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Elmund, G. K., S. M. Morrison, D. W. Grant, and S. M. Nevins. 1971. Role of excreted chlortetracycline in modifying the decomposition process in feedlot waste. Bull. Environ. Contam. Toxicol. 6:129-132. [DOI] [PubMed] [Google Scholar]
- 6.Flekna, G., P. Stefanic, M. Wagner, F. J. Smulders, S. S. Mozina, and I. Hein. 2007. Insufficient differentiation of live and dead Campylobacter jejuni and Listeria monocytogenes cells by ethidium monoazide (EMA) compromises EMA/real-time PCR. Res. Microbiol. 158:405-412. [DOI] [PubMed] [Google Scholar]
- 7.Gilpin, B. J., B. Robson, P. Scholes, F. Nourozi, and L. W. Sinton. 2009. Survival of Campylobacter spp. in bovine faeces on pasture. Lett. Appl. Microbiol. 48:162-166. [DOI] [PubMed] [Google Scholar]
- 8.Guan, J., J. L. Spencer, and B. L. Ma. 2005. The fate of the recombinant DNA in corn during composting. J. Environ. Sci. Health B 40:463-473. [DOI] [PubMed] [Google Scholar]
- 9.Guan, T. Y., and R. A. Holley. 2003. Pathogen survival in swine manure environments and transmission of human enteric illness—a review. J. Environ. Qual. 32:383-392. [DOI] [PubMed] [Google Scholar]
- 10.Harmon, K. M., and I. V. Wesley. 1997. Multiplex PCR for the identification of Arcobacter and differentiation of Arcobacter butzleri from other arcobacters. Vet. Microbiol. 58:215-227. [DOI] [PubMed] [Google Scholar]
- 11.Hoar, B. R., E. R. Atwill, C. Elmi, W. W. Utterback, and A. J. Edmondson. 1999. Comparison of fecal samples collected per rectum and off the ground for estimation of environmental contamination attributable to beef cattle. Am. J. Vet. Res. 60:1352-1356. [PubMed] [Google Scholar]
- 12.Hutchison, M. L., L. D. Walters, S. M. Avery, and A. Moore. 2005. Decline of zoonotic agents in livestock waste and bedding heaps. J. Appl. Microbiol. 99:354-362. [DOI] [PubMed] [Google Scholar]
- 13.Inglis, G. D., and L. D. Kalischuk. 2003. Direct detection of Campylobacter species in bovine feces using polymerase chain reaction. Appl. Environ. Microbiol. 69:3435-3447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Inglis, G. D., and L. D. Kalischuk. 2004. Direct quantification of Campylobacter jejuni and Campylobacter lanienae in feces of cattle by real-time polymerase chain reaction. Appl. Environ. Microbiol. 70:2296-2306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Inglis, G. D., L. D. Kalichuk, and H. W. Busz. 2003. A survey of Campylobacter species shed in faeces from beef cattle using polymerase chain reaction. Can. J. Microbiol. 49:655-661. [DOI] [PubMed] [Google Scholar]
- 16.Inglis, G. D., L. D. Kalichuk, and H. W. Busz. 2004. Chronic shedding of Campylobacter species in beef cattle. J. Appl. Microbiol. 97:410-420. [DOI] [PubMed] [Google Scholar]
- 17.Inglis, S. (ed.). 2005. Compendium of veterinary products, 8th ed. North American Compendiums, Ltd., Hensall, Ontario, Canada.
- 18.Kearney, T. E., M. J. Larkin, J. P. Frost, and P. N. Levett. 1993. Survival of pathogenic bacteria during mesophilic anaerobic digestion of animal waste. J. Appl. Bacteriol. 75:215-219. [DOI] [PubMed] [Google Scholar]
- 19.Kearney, T. E., M. J. Larkin, and P. N. Levett. 1993. The effect of slurry storage and anaerobic digestion on survival of pathogenic bacteria. J. Appl. Bacteriol. 74:86-93. [DOI] [PubMed] [Google Scholar]
- 20.Kobayashi, H., M. Oethinger, M. J. Tuohy, G. S. Hall, and T. W. Bauer. 2009. Unsuitable distinction between viable and dead Staphylococcus aureus and Staphylococcus epidermidis by ethidium bromide monoazide. Lett. Appl. Microbiol. 48:633-638. [DOI] [PubMed] [Google Scholar]
- 21.Larney, F. J., and R. E. Blackshaw. 2003. Weed seed viability in composted beef cattle feedlot manure. J. Environ. Qual. 32:1105-1113. [DOI] [PubMed] [Google Scholar]
- 22.Larney, F. J., B. H. Ellert, and A. F. Olson. 2005. Carbon, ash and organic matter relationships for feedlot manures and composts. Can. J. Soil Sci. 85:261-264. [Google Scholar]
- 23.Larney, F. J., A. F. Olson, A. A. Carcamo, and C. Chang. 2000. Physical changes during active and passive composting of beef feedlot manure in winter and summer. Bioresour. Technol. 75:139-148. [Google Scholar]
- 24.Larney, F. J., A. F. Olson, J. J. Miller, P. R. DeMaere, F. Zvomuya, and T. A. McAllister. 2008. Physical and chemical changes during composting of wood chip-bedded and straw-bedded beef cattle feedlot manure. J. Environ. Qual. 37:725-735. [DOI] [PubMed] [Google Scholar]
- 25.Larney, F. J., L. J. Yanke, J. J. Miller, and T. A. McAllister. 2003. Fate of coliform bacteria in composted beef cattle feedlot manure. J. Environ. Qual. 32:1508-1515. [DOI] [PubMed] [Google Scholar]
- 26.Nicholson, F. A., S. J. Groves, and B. J. Chambers. 2005. Pathogen survival during livestock manure storage and following land application. Bioresour. Technol. 96:135-143. [DOI] [PubMed] [Google Scholar]
- 27.Pisz, J. M., J. R. Lawrence, A. N. Schafer, and S. D. Siciliano. 2007. Differentiation of genes extracted from non-viable versus viable micro-organisms in environmental samples using ethidium monoazide bromide. J. Microbiol. Methods 71:312-318. [DOI] [PubMed] [Google Scholar]
- 28.Reuter, T., W. Xu, T. W. Alexander, K. Stanford, Y. Xu, and T. A. McAllister. 2009. Purification of polymerase chain reaction (PCR)-amplifiable DNA from compost piles containing bovine mortalities. Bioresour. Technol. 100:3343-3349. [DOI] [PubMed] [Google Scholar]
- 29.Rudi, K., B. Moen, S. M. Drømtorp, and A. L. Holck. 2005. Use of ethidium monoazide and PCR in combination for quantification of viable and dead cells in complex samples. Appl. Environ. Microbiol. 71:1018-1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rynk, R. 1992. On-farm composting handbook. Publication number NRAES-54. Natural Resource, Agriculture, and Engineering Service, Ithaca, NY.
- 31.Saskatchewan Ministry of Agriculture. 2008. Composting solid manure. Saskatchewan Ministry of Agriculture, Regina, Saskatchewan, Canada. http://www.agriculture.gov.sk.ca/adx/aspx/adxGetMedia.aspx?DocID=2880,2879,346,185,81,1,Documents&MediaID=6458&Filename=Composting+Solid+Manure+-+Printer+Friendly.pdf.
- 32.Sinton, L. W., R. R. Braithwaite, C. H. Hall, and M. L. Mackenzie. 2007. Survival of indicator and pathogenic bacteria in bovine feces on pasture. Appl. Environ. Microbiol. 73:7917-7925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Topp, E., M. Welsh, Y. C. Tien, A. Dang, G. Lazarovits, K. Conn, and H. Zhu. 2003. Strain-dependent variability in growth and survival of Escherichia coli in agricultural soil. FEMS Microbiol. Ecol. 44:3003-3308. [DOI] [PubMed] [Google Scholar]
- 34.Van Herk, F. H., T. A. McAllister, C. L. Cockwill, N. Gusselle, F. J. Larney, J. J. Miller, and M. E. Olson. 2004. Inactivation of Giardia cysts and Cryptosporidium oocysts in beef feedlot manure by thermophilic windrow composting. Compost Sci. Util. 12:235-241. [Google Scholar]
- 35.Wagner, A. O., G. Gstraunthaler, and P. Illmer. 2008. Survival of bacterial pathogens during the thermophilic anaerobic digestion of biowaste: laboratory experiments and in situ validation. Anaerobe 14:181-183. [DOI] [PubMed] [Google Scholar]
- 36.Wagner, A. O., C. Malin, G. Gstraunthaler, and P. Illmer. 2009. Survival of selected pathogens in diluted sludge of a thermophilic waste treatment plant and in NaCl-solution under aerobic and anaerobic conditions. Waste Manag. 29:425-429. [DOI] [PubMed] [Google Scholar]
- 37.Watson, R. J., and B. Blackwell. 2000. Purification and characterization of a common soil component which inhibits the polymerase chain reaction. Can. J. Microbiol. 46:633-642. [DOI] [PubMed] [Google Scholar]
- 38.Wéry, N., C. Lhoutellier, F. Ducray, J. P. Delgenès, and J. J. Godon. 2008. Behaviour of pathogenic and indicator bacteria during urban wastewater treatment and sludge composting, as revealed by quantitative PCR. Water Res. 42:53-62. [DOI] [PubMed] [Google Scholar]
- 39.Xu, W., T. Reuter, G. D. Inglis, F. J. Larney, T. W. Alexander, J. Guan, K. Stanford, Y. Xu, and T. A. McAllister. 2009. A biosecure composting system for disposal of cattle carcasses and manure following infectious disease outbreak. J. Environ. Qual. 38:437-450. [DOI] [PubMed] [Google Scholar]