Abstract
Cronobacter spp. are opportunistic food-borne pathogens that are responsible for rare but highly fatal cases of meningitis and necrotizing enterocolitis in neonates. While the operon responsible for yellow pigmentation in Cronobacter sakazakii strain ES5 was described recently, the involvement of additional genes in pigment expression and the influence of pigmentation on the fitness of Cronobacter spp. have not been investigated. Thus, the aim of this study was to identify further genes involved in pigment expression in Cronobacter sakazakii ES5 and to assess the influence of pigmentation on growth and persistence under conditions of environmental stress. A knockout library was created using random transposon mutagenesis. The screening of 9,500 mutants for decreased pigment production identified 30 colorless mutants. The mapping of transposon insertion sites revealed insertions in not only the carotenoid operon but also in various other genes involved in signal transduction, inorganic ions, and energy metabolism. To determine the effect of pigmentation on fitness, colorless mutants (ΔcrtE, ΔcrtX, and ΔcrtY) were compared to the yellow wild type using growth and inactivation experiments, a macrophage assay, and a phenotype array. Among other findings, the colorless mutants grew at significantly increased rates under osmotic stress compared to that of the yellow wild type while showing increased susceptibility to desiccation. Moreover, ΔcrtE and ΔcrtY exhibited increased sensitivity to UVB irradiation.
Cronobacter spp. (formerly Enterobacter sakazakii) are opportunistic food-borne pathogens that cause rare but life-threatening cases of meningitis, necrotizing enterocolitis, and septicemia in neonates (7, 30, 39, 40). While the pathogen appears to be ubiquitous, powdered infant formula (PIF) has been implicated as the main source of Cronobacter infection, necessitating effective means of both detecting this organism and preventing contamination in the PIF production environment (14, 26, 40).
Although white strains have been observed occasionally, the production of yellow pigment on tryptic soy agar (TSA) is still one of the key discriminative criteria in the identification of presumptive Cronobacter spp. isolates via the ISO/TS 22964 standard protocol (3, 6, 11, 25). Studies of which colorless or cream-white strains of Cronobacter spp. (formerly Enterobacter sakazakii) were identified have reported prevalence rates of 8, 13, and 21.4% (6, 11, 24).
The pigment's carotenogenic nature recently was identified in Cronobacter strain ES5 on a molecular and chemical level (31). Carotenoids are known to stabilize cellular membranes and influence membrane fluidity (13, 22, 48). Functioning as antioxidants, carotenoids scavenge reactive oxygen species (37, 54, 55). Moreover, pigments play a role in the survival of bacteria in harmful environments and have been found to increase the virulence of pathogens such as Staphylococcus aureus and Erwinia chrysanthemi (32, 33, 44, 55). In Cronobacter strain ES5, a gene cluster comprised of seven genes (crtE-idi- crtXYIBZ) was found to be responsible for carotenoid biosynthesis (31). While the study mentioned above identified the operon responsible for carotenoid production, the involvement of other genes in pigment expression has not been investigated.
Because no research exists on the influence of pigmentation on the fitness and persistence of Cronobacter spp., the potential implications of failing to detect colorless strains of this organism in the PIF production environment are difficult to assess. Thus, the aim of this study was to further describe the genetic basis of the pigmented phenotype of Cronobacter strain ES5 by isolating and characterizing isogenic white mutants via random transposon mutagenesis and subsequent sequencing, and to identify the impact of pigmentation on persistence and growth under conditions of environmental stress by comparing white mutants to the yellow wild type in a variety of growth and inactivation experiments, a macrophage assay, and a phenotype array.
MATERIALS AND METHODS
Bacterial strains and plasmids.
Wild-type and mutant strains as well as plasmids used in this study are listed in Table 1.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Genotype/characteristic(s) | Source or reference |
|---|---|---|
| Strains | ||
| Cronobacter sakazakii ES5 | Human isolate | Institute of Medical Microbiology, University of Zurich (21, 31) |
| Escherichia coli XL1-Blue | recA1 endA1 gyrA96 thi-1 hsdR17 supE44 relA1 lac [F′ proAB lacIqZΔM15 Tn10 (Tetr)] | Stratagene |
| Plasmids and transposons | ||
| pCR2.1 | Kmr, Apr; TA cloning vector, lacZΔ | Invitrogen |
| pUC19 | Apr; cloning vector | Epicentre |
| EZ-Tn5 <KAN-2> | Kmr, mini-Tn5 transposon | Epicentre |
| pSTn2 | Containing SonTn-f1/SonTnr1 amplified probe fragment | This study |
Media and growth conditions.
Medium ingredients were obtained from Difco Laboratories (Detroit, MI), Fluka (Buchs, Switzerland), Merck (Darmstadt, Germany), and Sigma (Buchs, Switzerland). Unless stated otherwise, stationary-phase inocula (109 CFU/ml) of Cronobacter sakazakii and Escherichia coli were produced by transferring single colonies into 5 ml of modified Luria-Bertani (LB) broth (2) and incubating them overnight (16 to 18 h) at 37°C under agitation (220 rpm). For experiments under nutrient-deficient conditions, M9 minimal medium containing 2% glucose was used (21). Medium was solidified by the addition of 15 g/liter agar. If required, antibiotics were added at the following final concentrations: ampicillin, 100 μg/ml; and kanamycin, 50 μg/ml. Growth in liquid cultures was monitored by the measurement of the optical density at 600 nm (OD600) with an Ultrospec II spectrophotometer (Biochrom Ltd., Cambridge, United Kingdom) or by the viable cell count of serial dilutions in 0.85% NaCl on plate count agar (Oxoid, Cambridge, United Kingdom).
DNA extraction, manipulations, and sequencing.
All kits for DNA isolation and purification were obtained from Qiagen (Hilden, Germany) and handled by following the manufacturer's instructions. Unless otherwise stated, chromosomal DNA was isolated using the DNeasy Blood and Tissue kit. Plasmids were extracted with the QIAprep Spin Miniprep or Plasmid Midi kits. DNA fragments from PCRs, restriction digests, and agarose gels were purified using the MinElute PCR Cleanup kit and the MinElute Gel Purification kit, respectively. The concentration of nucleic acids was determined using a Nanodrop ND-1000 UV/Vis spectrophotometer (NanoDrop Technologies, Wilmington, DE). The cloning, restriction enzyme analysis, and transformation of C. sakazakii and E. coli were performed using standard techniques (46). Enzymes and respective buffers were obtained from Roche (Basel, Switzerland) or New England Biolabs (Ipswich, MA). All sequencing was outsourced (Microsynth, Balgach, Switzerland).
Transposon mutagenesis and screening.
A transposon mutagenesis library was constructed using the EZ-Tn5 <KAN-2>Tnp Transposome kit (Epicentre, Madison, WI) in accordance with protocols provided by the supplier. Briefly, 1 ml of an overnight culture of ES5 (in LB) was used to inoculate 1 liter of LB. The strain was allowed to grow at 37°C to an OD600 of 0.6, and cultures were cooled on ice for 30 min. Cells were harvested by centrifugation (5,500 rpm, 4°C, 15 min), and the supernatant was discarded. Subsequently, cells were washed and centrifuged (5,000 rpm, 4°C, 15 min) three times: first in 1 liter and second in 500 ml ice-cold distilled water, followed by a third wash in 10 ml 10% ice-cold glycerol. Finally, the competent cells were resuspended in 1 ml 10% glycerol and stored in 50-μl aliquots at −80°C.
For electroporation, 1 μl of transposon DNA was added to 50 μl of competent cells on ice. A micropulser (Bio-Rad, Reinach, Switzerland) was used for transformation before the electroporated cells were transferred to 1 ml super optimal broth with catabolite repression (SOC) for recovery and incubated for 45 min at 37°C. Cells were diluted 1:100 in prewarmed SOC medium. To select for transposon insertion clones, aliquots of 100 μl were plated onto LB plates containing 50 μg/ml kanamycin, and plates were incubated overnight (37°C).
Single mutants were picked in 96-well polystyrene microtiter dishes (Nunc, Dietikon, Switzerland) containing 150 μl/well of LB supplemented with 7.5% glycerol and 50 μg/ml of kanamycin, grown overnight at 37°C, and subsequently stored at −80°C. The screening of 9,500 transposon mutants for a decrease in pigment production was performed by the replica plating of mutants from storage plates onto screening plates (LB agar supplemented with 50 μg/ml kanamycin) using a 96-pin replicator (Nalge Nunc International, Naperville, IL). Following incubation overnight at 37°C, plates were stored at room temperature for 3 days under constant light to enhance color formation. Mutant colonies were visually inspected for a decrease in pigmentation.
Determination of chromosomal transposon copy number by Southern blot analysis.
To generate a template for probe synthesis, a fragment of the kanamycin resistance cassette of EZ-Tn5 <KAN-2> (Table 1) was amplified with SonTn-f1 and SonTn-r1 primers (Table 2) using the FastStart PCR system (Roche, Mannheim, Germany) with the following reaction conditions: (i) 5 min at 95°C; (ii) 30 cycles of 30 s at 95°C, 30 s at 56°C, and 1 min at 72°C; and (iii) 8 min at 72°C. The product was cloned into pCR2.1 (Table 1) using the TA cloning kit (Invitrogen, Carlsbad, CA) by following the manufacturer's instructions to yield pSTn2 (Table 1). The insert identity was verified by sequencing with M13 and M13r primers (Table 2). For the synthesis of the digoxigenin (DIG)-labeled probe STn, the probe fragment was amplified from pSTn2 with the above-mentioned primers using the PCR DIG Probe Synthesis kit (Roche, Mannheim, Germany) as recommended by the kit supplier. Chromosomal DNA for Southern analysis was prepared as follows: bacteria grown in LB at 37°C were harvested by centrifugation and incubated in lysis buffer (100 mM NaCl, 10 mM Tris-HCl, pH 8.0, 50 mM EDTA, pH 8.0, 0.5% [wt/vol] sodium dodecyl sulfate, 0.1 mg/ml proteinase K) at 50°C for at least 6 h. DNA was extracted with phenol-chloroform (1:1), precipitated with isopropanol, and dissolved in Tris-EDTA (TE) buffer. Southern analysis was performed nearly entirely as described by Sambrook et al. (46). Briefly, 5 μg of SphI-digested DNA was separated on a 1% agarose gel and transferred to a nylon membrane (GE Osmonics, Minnetonka, MN). Hybridization was performed at 48°C, followed by stringency washing at 68°C. Signal was detected with anti-DIG-AP-Fab fragments (Roche, Mannheim, Germany) by following the manufacturer's recommendations.
TABLE 2.
Primers used in this study
| Name | Nucleotide sequence (5′→3′) | Source |
|---|---|---|
| KAN-2 FP1 | ACC TAC AAC AAA GCT CTC ATC AAC C | Epicentre |
| M13 | TGT AAA ACG ACG GCC AG | New England Biolabs |
| M13r | CAG GAA ACA GCT ATG ACC | New England Biolabs |
| SonTn-f1 | ATG TTA CAG ATG AGA TGG | This study |
| SonTn-r1 | AGC ATC AAA TGA AAC TGC | This study |
Mapping of transposon insertion sites.
Chromosomal DNA of the transposon mutants was digested with SphI. The fragments ligated into pUC19 (Table 1) were digested with the same enzyme. The ligation mixture then was electroporated into E. coli XL1-Blue (10). Transformants carrying a plasmid containing the transposon sequence were selected by incubating them on LB containing 50 μg/ml of kanamycin. Plasmids were extracted from the selected clones, and transposon-flanking regions were sequenced with primer KAN-2 FP1 (Table 2). Transposon insertion sites were determined by sequencing the junctions between the Tn5 transposon sites and ES5 chromosomal DNA. The sequence obtained from each mutant was aligned with the C. sakazakii BAC 9E10 (accession no. AM384990.1) sequence and the NCBI assembly ATCC BAA-894 complete genome (accession no. CP000783.1) of C. sakazakii. Similarity searches were performed using BLASTn and BLASTx at the NCBI website (http://www.ncbi.nlm.nih.gov/sites/entrez?db=genomeprj&cmd=Retrieve&dopt=Overview&list_uids=12720) (1). Clusters of orthologous groups of proteins (COGs) were used for the classification of all mutants (http://www.ncbi.nlm.nih.gov/COG/).
Determination of carotenoid content.
The wild-type strain as well as all transposon mutant strains were assayed for carotenoid content using a spectrophotometric approach (38). Briefly, all strains were cultured in LB for 24 h (37°C). Cells were harvested from 850 μl of the culture by centrifugation at 10,000 × g for 1 min and washed twice using double-distilled water. Resuspension in 200 μl of methanol was followed by being heated at 55°C for 3 min and the removal of cell debris using centrifugation (15,000 × g, 1 min). The extraction was repeated once, and extracts were combined. Methanol was added up to the final volume of 1 μl. Absorbance at the beta carotene peak (465 nm) was normalized, and relative absorbance was measured using an Ultrospec II spectrophotometer (Biochrom Ltd., Cambridge, United Kingdom).
Persistence and growth experiments.
Selected white mutants (ΔcrtE, ΔcrtX, and ΔcrtY) were further characterized using a macrophage assay, various growth and inactivation experiments, and a phenotype microarray.
Macrophage assay.
These assays were performed as described by Garner et al. (18, 19) using the murine-macrophage-like cell line J774A.1 (ATCC TIB-67). All culture media, additives, and phosphate-buffered saline (PBS) were obtained from Gibco (Invitrogen AG, Basel, Switzerland) unless stated otherwise. A gentamicin concentration of 0.01 mg/ml was used. Previous experiments had shown this antibiotic concentration to inactivate up to 106 CFU/ml extracellular bacteria within an hour without influencing the total number of macrophage-like cells.
Briefly, cells were seeded at a density of approximately 4.0 × 105 macrophage cells/ml per well in 24-well tissue culture plates (Techno Plastic Products AG) using Dulbecco's modified Eagle's medium (DMEM; Trasadingen, Switzerland) supplemented with 10% fetal bovine serum (PAA Laboratories, Pasching, Austria). Monolayers were produced by incubation in 5% CO2 at 37°C for 48 h. Medium was changed 30 min prior to infection. Monolayers were infected by inoculation with bacterial overnight cultures grown to stationary phase, resulting in a final infection dose of 107 CFU/well (multiplicity of infection [MOI], 10:1). Medium was aspirated 45 min postinfection, and cells were washed twice using 1 ml PBS to remove any extracellular bacteria. Prewarmed, fresh, antibiotic-free medium (1 ml) was added and replaced 60 min postinfection by medium containing 10 μg/ml of gentamicin to kill any remaining extracellular bacteria. At 2 and 8 h postinfection, macrophages were lysed using 1 ml of ice-cold sterile distilled water. Total surviving engulfed bacteria were quantified by bacterial enumeration.
Growth experiments.
All growth experiments followed the same basic protocol. Stationary-phase cultures (109 CFU/ml) of wild-type (wt) and mutant strains were prepared in 10 ml LB and 10 ml nutrient-deficient M9 medium and then 10-fold serially diluted in 0.85% NaCl. Aliquots of 100 μl were transferred to 10 ml of LB/M9, resulting in starter cultures containing the desired CFU/ml. Growth was determined by colony count. For growth experiments in highly osmotic conditions, the NaCl concentration was adjusted to 7% for LB and 3% for M9. Growth (at 37°C without agitation) was monitored after 0, 4, 8, 24, 36, and 48 h. For growth experiments under cold stress, cultures were incubated at 10°C without agitation for 20 days, with measurements at day 0, 1, 2, 3, 6, 9, 13, 16, and 20. For growth under acidic conditions, LB broth was preheated to 37°C and adjusted to pH 4.5 directly before inoculation using HCl. Growth (at 37°C without agitation) was monitored after 0, 4, 8, 24, and 36 h.
Desiccation assays.
Stationary-phase cultures (10 ml) of wild-type and mutant strains in LB/M9 were adjusted to an OD600 of 0.005 and grown for 22 h at 37°C. Cells were harvested by centrifugation (10 min, 10,000 × g), resuspended in 1 ml LB, and then 10-fold serially diluted in LB to 10−2 CFU/ml, resulting in 12 dilution levels (from 109 to 10−2 CFU/ml). The two lowest levels served as negative controls. Three 96-well microtiter plates were loaded identically with eight aliquots of 10 μl of each of the 12 dilution levels and dried for 4 h in a sterile cabinet. Prior to rehydration, plates were stored in a dark, sealed box containing Drierite desiccant bags (Drierite Co. Ltd., Xenia, OH). The rehydration of the plates was performed immediately (time point zero [T0]) and after 8 and 16 weeks, respectively, using buffered peptone water (200 μl/well), followed by incubation overnight at 37°C. Using a 96-pin replicator (Nalge Nunc International, Naperville, IL), an inoculum of 1 μl was transferred to a 96-well microtiter plate containing mannit indicator (phenol red broth supplemented with 5 g/liter mannit; Difco). Following overnight incubation at 37°C, positive wells were identified by color change due to mannit fermentation. The most-probable-number (MPN) technique was applied using the Food and Drug Administration's Bacteriological Analytical Manual. The pattern of positive and negative wells allowed for the calculation of the MPN. Comparison of the MPN for T0, T8, and T16 resulted in a log kill due to 8 and 16 weeks of desiccation.
Photosensitivity.
Because the highest level of sensitivity to UV irradiation in bacteria is known to coincide with the phase of active growth, bacterial strains were grown to late exponential phase (108 CFU/ml) in LB for 8 h in the dark (20, 36). Aliquots of 1 ml were 10-fold serially diluted to 106 CFU/ml. Cells were harvested by centrifugation (10 min, 10,000 × g), and the sediment was resuspended in 10 ml of 0.85% NaCl. Aliquots of 100 μl (104 cells) were spotted onto plate count agar in quadruplicate (27). Plates were irradiated with UVB at 312 nm using a TFX-20.M transilluminator (Vilber Lourmat, Marne la Vallée, France) at an intensity of 8 mW/cm2 for 10, 15, 20, 25, 30, and 35 s, respectively. To avoid photo reactivation, plates were stored in the dark. Plates were incubated overnight at 37°C, and bacterial colonies were enumerated to determine survival.
Oxidative stress and acid sensitivity assays.
Aliquots of 1 ml LB overnight cultures were used to inoculate 9 ml of fresh LB supplemented with 12.5 mM cumene peroxide. Samples were taken after 10, 20, and 30 min incubation at room temperature. For acid killing, the pH of the LB cultures was adjusted to 2.5 using HCl. Samples were taken after 15, 30, and 45 min at room temperature. Aliquots were serially diluted in 0.85% NaCl, plated on plate count agar, and incubated overnight before colonies were counted.
Phenotype microarray analysis.
The phenotype microarray (Biolog Inc., Hayward, CA) was performed with the wild-type strain and ΔcrtE using the Omnilog V.1.5 Comparison Module. Panels included carbon, nitrogen, phosphorus, sulfur, nutrient, osmotic and growth controls, and chemical sensitivity panels. To chart correlation plots of the independent runs, reproducibility was determined using metabolic and sensitivity threshold values.
Statistics.
DMFit 2.0 was used to model the time variation of the logarithm of cell concentrations of the batch cultures used for the determination of growth under various stress conditions (4) and allowed for the determination of the potential maximum growth rate (μmax). Statistical analysis was performed using SPSS Statistics 17.0 (SPSS Inc., Chicago, IL). Bacterial colony counts were converted into log CFU/ml. The means as well as standard deviations were calculated. Differences in growth and survival between the wild type and mutants were identified using both univariate analysis of variance (ANOVA) at a significance level of α ≤ 0.05 and posthoc multiple comparisons applying the Tukey honestly significant difference (Tukey HSD) test. Statistically significant differences were acknowledged for P ≤ 0.050.
RESULTS
The screening of 9,500 knockout mutants identified 30 colorless mutants. One mutant was excluded from the study because Southern blotting revealed that it possessed more than one transposon insertion. As depicted in Table 3, clusters of orthologous groups of proteins (COGs) were used for the further classification of colorless mutants (http://www.ncbi.nlm.nih.gov/COG/). Thus, mutants could be assigned to COGs comprising genes responsible for the transport and metabolism of coenzymes, carbohydrates, amino acids, lipids, and inorganic ions, as well as genes with functions in biosynthesis, transport, and the catabolism of secondary metabolites, energy production and conversion, and signal transduction.
TABLE 3.
Mapping of transposon insertions sites that result in white phenotype in C. sakazakii ES5
| COG functional categorya | COG functional class | Annotationb,c |
|
|---|---|---|---|
| Homologue | Gene product | ||
| Mutation in pigment operon | |||
| H: coenzyme transport and metabolism | Geranylgeranyl pyrophosphate synthase | crtE | Geranylgeranyl pyrophosphate synthase |
| GC: carbohydrate transport and metabolism/signal transduction mechanisms | Glucosyl transferases, related to UDP-glucosyltransferase | crtX | Zeaxanthin glucosyl transferase |
| R/E: general function prediction only/amino acid transport and metabolism | Acetyltransferase/choline dehydrogenase and related flavoproteins | crtY | Lycopene cyclase |
| Q: secondary metabolites biosynthesis, transport and catabolism | Phytoene dehydrogenase and related proteins | crtI | Phytoene dehydrogenase |
| I: lipid transport and metabolism | Phytoene/squalene synthase | crtB | Phytoene synthase |
| Mutation outside pigment operon | |||
| C: energy production and | FoF1-type ATP synthase, subunit alpha | ESA_04012 | FoF1 ATP synthase subunit alpha |
| conversion | FoF1-type ATP synthase, subunit beta | ESA_04006 | FoF1 ATP synthase subunit beta |
| FoF1-type ATP synthase, subunit gamma | ESA_04007 | FoF1 ATP synthase subunit gamma | |
| FoF1-type ATP synthase, subunit epsilon (mitochondrial delta subunit) | ESA_04005 | FoF1 ATP synthase subunit epsilon | |
| Pyruvate/2-oxoglutarate dehydrogenase complex, dihydrolipoamide acetyltransferase (E1) component, and related enzymes | ESA_02622 | sucA 2-oxoglutarate dehydrogenase E1 component | |
| Pyruvate/2-oxoglutarate dehydrogenase complex, dihydrolipoamide acetyltransferase (E2) component, and related enzymes | ESA_02621 | Dihydrolipoamide acetyltransferase | |
| Pyruvate/2-oxoglutarate dehydrogenase complex, dihydrolipoamide acetyltransferase (E3) component, and related enzymes | ESA_03222 | aceF dihydrolipoamide acetyltransferase | |
| Malate/lactate dehydrogenases | ESA_03622 | Malate dehydrogenase | |
| Succinate dehydrogenase/fumarate reductase, flavoprotein subunit | ESA_02624 | Succinate dehydrogenase flavoprotein subunit | |
| P: inorganic ion transport and metabolism | Na+/H+ antiporter | ESA_03316 | pH-dependent sodium/proton antiporter |
| T: signal transduction mechanisms | cAMP-binding proteins, catabolite gene activator, and regulatory subunit of cAMP-dependent protein kinases | ESA_04376 | cAMP regulatory protein |
| DnaK suppressor protein | ESA_03194 | DnaK transcriptional regulator DksA | |
| S: function unknown | Uncharacterized conserved protein | ESA_04343 (Ent638_3811)d | Hypothetical protein (intracellular growth attenuator IgA, Enterobacter sp. 638)d |
| ESA_03563 (ETA_03450)d | Hypothetical protein (YhbC-like protein, Erwinia tasmaniensis Et1/99)d | ||
| ESA_00549 (AAG53883)d | Hypothetical protein (sigma factor RpoS, Escherichia coli)d | ||
NCBI clusters of orthologous groups (COG) of proteins.
Cronobacter sakazakii ES5 BAC 9E10 for mutations within the pigment operon (accession no. AM384990.1).
NCBI assembly ATCC BAA-894 C. sakazakii complete genome for mutations outside pigment operon (accession no. CP000783.1).
Closest annotated homolog.
The wild type and all transposon mutants were assayed for carotenoid content. The means of two independent absorbance measurements at the beta carotene peak (465 nm) resulted in an OD465 of 0.12 for the yellow wild type and an OD465 of <0.01 for all mutant strains (data not shown).
Many of the identified transposition sites were located within genes outside of the pigment operon. In addition to playing a role in carotenoid formation, these genes presumably also have other effects on growth and inactivation patterns. As the aim of this study was to determine the effect of pigmentation on fitness, only strains with insertions in the pigment operon were selected for stress response experiments, in order to prevent factors other than pigmentation from influencing results. Specifically mutants ΔcrtE, ΔcrtX, and ΔcrtY were chosen, for which the loss of transcript expression was verified by reverse transcription-PCR (data not shown).
Growth experiments.
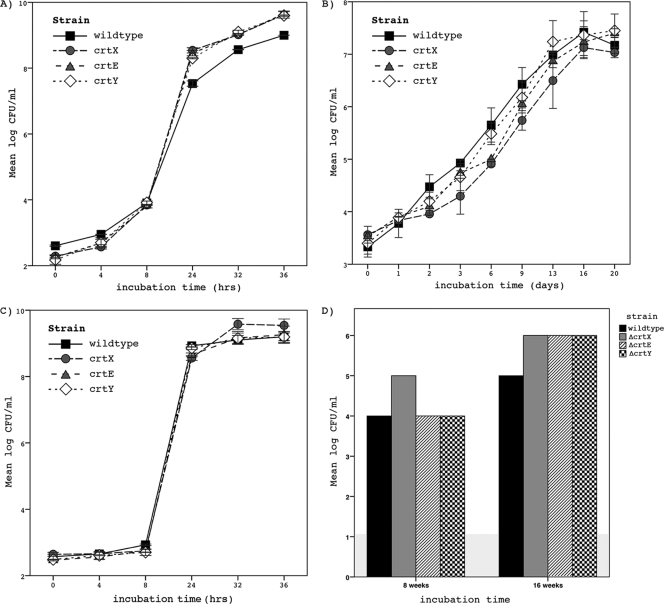
To examine the effect of salt stress, the growth of ΔcrtE, ΔcrtX, and ΔcrtY was compared to that of the wild type in LB adjusted to 7% NaCl and in M9 adjusted to 3% NaCl at 37°C. These represented the highest NaCl concentrations at which all strains grew to stationary phase within 48 h. In LB, as depicted in Fig. 1A, a significantly increased maximum specific rate of growth (μmaxwt, 0.23; μmaxΔcrtX, 0.32; μmaxΔcrtE, 0.31; and μmaxΔcrtY, 0.29) of all mutant strains was observed compared to that of the wild type (P = 0.000). In M9 (data not shown), consistently with the results for LB, all mutant strains showed significantly increased maximum rates of growth (μmaxwt, 0.14; μmaxΔcrtX, 0.22; μmaxΔcrtE, 0.24; and μmaxΔcrtY, 0.19; P = 0.000).
FIG. 1.
Evaluation of behavior of wild type and isogenic unpigmented mutants ΔcrtE, ΔcrtX, and ΔcrtY under a variety of stress conditions. (A) Growth experiments in LB adjusted to 7% NaCl. All mutants grew at significantly increased rates compared to the wild type (P = 0.000). (B) Growth kinetics in M9 at cold stress temperatures (10°C). All strains grew at similar maximum specific growth rates. (C) Growth under acidic conditions (pH 4.5) showed no significant differences for the wild type and the isogenic white mutants. The results represent means (± standard deviations) from duplicates of two independent experimental runs. (D) Inactivation by desiccation for 8 and 16 weeks in M9. Results were determined by the most probable number technique and depicted as log reduction in CFU/ml. While only ΔcrtX showed higher sensitivity to drying than the wild type after 8 weeks, all mutant strains showed higher inactivation numbers after 16 weeks of desiccation.
Cold stress experiments were performed by growing ΔcrtE, ΔcrtX, and ΔcrtY in LB and M9 medium at 10°C (for results in M9, see Fig. 1B). Under these conditions, no significant differences in maximum specific growth rates were detected for ΔcrtY, ΔcrtE, and ΔcrtX in both LB and M9 compared to those of the wild type (in LB, μmaxwt = 0.04, μmaxΔcrtX = 0.03, μmaxΔcrtE = 0.03, and μmaxΔcrtY = 0.04; in M9, μmaxwt = 0.01, μmaxΔcrtX = 0.01, μmaxΔcrtE = 0.01, and μmaxΔcrtY = 0.01).
To evaluate growth under acidic conditions, wild-type and mutant strains were incubated in LB adjusted to pH 4.5 at 37°C (Fig. 1C). The wild type and all mutant strains grew at comparable maximum specific growth rates (μmaxwt = 0.48, μmaxΔcrtX = 0.48, μmaxΔcrtE = 0.45, and μmaxΔcrtY = 0.50).
Inactivation experiments.
Inactivation assays included the lethal exposure of the wild type and ΔcrtE, ΔcrtX, and ΔcrtY to desiccation, UV irradiation, and oxidative and acidic stress.
Desiccation experiments in LB for 8 and 16 weeks led to similar inactivation rates (data not shown). A 4 log10 reduction was detected in all strains. In M9, desiccation for 8 weeks resulted in a 4 log10 reduction of the wild type, ΔcrtE, and ΔcrtY, while ΔcrtX was reduced by 5 log10. After 16 weeks, the wild type was reduced by 5 log10, while ΔcrtE, ΔcrtX, and ΔcrtY were reduced by 6 log10 (Fig. 1D).
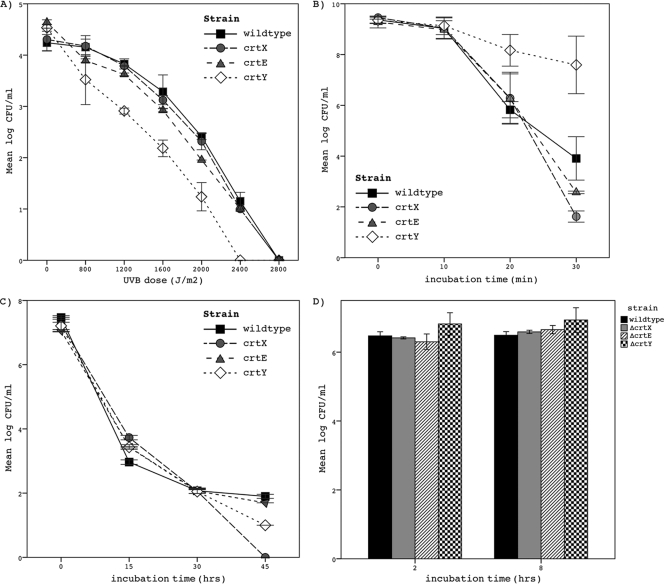
Upon UVB irradiation (Fig. 2A), ΔcrtX cells were inactivated, similarly to the wild type, by 2,000 J/m2 of UVB, whereas the same UVB dose resulted in the inactivation of significantly more cells of ΔcrtE and ΔcrtY (P = 0.009 for ΔcrtE and P = 0.000 for ΔcrtY). The following irradiation doses resulted in a 1 log10 and 4 log10 kill, respectively: 1,600 and 2,800 J/m2 for ΔcrtX and the wild-type strain, 1,200 and 2,800 J/m2 for ΔcrtE, and 800 and 2,400 J/m2 for ΔcrtY.
FIG. 2.
Inactivation of the wild type and white mutants ΔcrtE, ΔcrtX, and ΔcrtY of the same strain in view of various stressors. (A) Sensitivity to irradiation with 312 nm UVB at an intensity of 8 mW/cm2. Mutant strains crtE and crtY were inactivated more strongly than the wild type by 2,000 J/m2 (ΔcrtE, P = 0.009; ΔcrtY, P = 0.000). (B) Survival after 30 min of incubation in the presence of 12.5 mM cumene peroxide. Cell numbers of ΔcrtX and ΔcrtE declined more rapidly (P = 0.000 for ΔcrtX and P = 0.021 for ΔcrtE), whereas those of ΔcrtY declined more slowly than the cell numbers of the wild type (P = 0.000). (C) Inactivation by exposure to LB adjusted to pH 2.0. All mutant strains were inactivated in higher numbers than the wild type after 45 min (P = 0.000). (D) Intake and persistence of the wild type and mutants in macrophage-like cells. Cells of mutant strain crtY were engulfed at significantly higher numbers than those of the wild type (P = 0.000). All strains showed equal persistence within macrophage-like cells. Error bars represent the standard deviation from the mean for duplicates of two (B, C, and D) or three (A) independent experimental runs.
Strains were exposed to oxidative stress (Fig. 2B) by incubating them in LB supplemented with 12.5 mM cumene peroxide for a period of 30 min. The cell numbers of all strains were compared after 30 min of incubation and showed the higher resistance of ΔcrtY to oxidative stress and the impaired resistance of ΔcrtE and ΔcrtX compared to that of the wild type (P = 0.021 for ΔcrtE and P = 0.000 for ΔcrtX and ΔcrtY).
An acidic stress assay was performed by incubating the wild type and ΔcrtE, ΔcrtX, and ΔcrtY in LB at pH 2.0 for a period of 45 min. The comparison of surviving cell numbers of ΔcrtE, ΔcrtX, and ΔcrtY and the wild type after 45 min suggested the increased sensitivity of all white mutants to acidic inactivation (P = 0.000). The results of this assay are displayed in Fig. 2C.
Macrophage assay.
In the macrophage assay, both engulfment into and persistence within macrophage-like cells were assessed. The survival of all strains in macrophage-like cells did not differ 2 and 8 h postinfection (Fig. 2D). As for previous phagocytosis, insertion in the crtE and crtX genes did not alter intake into macrophage-like cells, whereas a significantly higher number of the ΔcrtY cells was engulfed compared to that of the wild-type strain (P = 0.000).
Phenotype microarray analysis.
The phenotype microarray was performed on ΔcrtE using the wild-type strain as a reference. The crtE mutant was selected for this experiment, as crtE is the first gene in the carotenoid pathway and, thus, is essential in the formation of all intermediates as well as the final pigment. A change of phenotype of ΔcrtE compared to that of the wild type was observed on all microarray panels. On the carbon panels, l-ornithine, maltitol, melibionic and succinic acid, and mono-methylsuccinate resulted in lost phenotypes. Nitrogen panels revealed lost phenotypes for l-aspartic acid, l-threonine, and several dipeptides. On phosphorus and sulfur panels, the phenotype was lost for adenosine-5′-monophosphate. On nutrient stimulation panels, the mutant strain crtE grew under all conditions tested, whereas the wild type grew only in l-homoserine-lactone, uridine, 2′-deoxyuridine, and Tween 20. Osmotic and growth control panels revealed the relative resistance of the ΔcrtE to sodium chloride, potassium chloride, sodium sulfate and lactate, and urea. On chemical sensitivity panels, ΔcrtE was resistant to tolyfluanid and aminoglycosides while exhibiting sensitivity to a variety of other chemicals, including myricetin, coumarin, and lomefloxacin (data not shown).
DISCUSSION
Characterization of nonpigmented transposon mutants.
While transposon mutagenesis caused insertions in most genes of the pigment operon, colorless mutants with an insertion in the gene idi or crtZ were not observed. Although the lack of colorless mutants with insertions in these genes may be coincidental, other explanations also seem plausible. In a study examining various environmental Enterobacteriaceae strains, a comparison of several gene clusters responsible for carotenoid synthesis demonstrated that idi is not obligatory for the expression of pigment. Rather, it appears to increase carotenoid titers when present (48). Similarly, crtZ also may not be obligatory for color formation, as it encodes beta carotene hydroxylase, which is responsible for the cyclization of beta carotene, resulting in the formation of zeaxanthin. As zeaxanthin's precursor beta carotene is also a yellow pigment, knocking out crtZ will not affect yellow pigmentation. Furthermore, at present it is unclear whether crtZ is even a functional gene in Cronobacter sakazakii. Bioinformatic analysis predicts that its stop codon lies 112 nucleotides downstream from the last gene of the crtE-idi-crtXYIB operon, thus overlapping with crtB (31). This indicates that crtZ is not transcribed at all, because while functionally related genes within an operon commonly are located near each other, each gene unit generally is defined as having a unique position within a chromosome (28). Alternatively, due to its overlap with crtB, crtZ may be transcribed from the opposite direction (31). Further studies are required to determine the role of crtZ in Cronobacter sakazakii.
Our study also identified a large number of genes outside of the carotenoid operon with an influence on pigmentation. The majority of these genes were found to be associated with energy production and conversion, as well as energy transport. Thus, several colorless mutants showed insertions in genes encoding FoF1 ATPase subunits. While not mandatory for the survival of the bacteria, carotenoid biosynthesis requires ATP. Therefore, impaired energy metabolism resulting from defective FoF1 ATPase subunits ultimately may result in the omission of pigmentation production in favor of other, more vital functions.
Another insertion resulting in the lack of pigmentation was identified in the gene encoding the cyclic AMP (cAMP) regulatory protein. A study of Erwinia herbicola has determined that the expression of yellow pigment is mediated by cAMP (41). In our study, incubating the colorless cAMP-defective mutant on LB agar plates supplemented with 100 mM cAMP resulted in the expression of yellow pigment within 3 days, suggesting that pigment production in Cronobacter sakazakii is dependent on cAMP (data not shown).
Two white mutants contained insertions in the gene encoding the dnaK transcriptional regulator dksA. A deletion in this gene was reported to result in the loss of yellow pigmentation in the Gram-negative bacterium Myxococcus xanthus (17). Knocking out dksA also has been found to block rpoS induction by the nutritional stress signal ppGpp (9). The gene rpoS functions as a general regulator that is involved in the regulation of at least 50 genes (9). It plays a role in the survival of famine conditions and near-UV irradiation as well as in protection against osmotic, acidic, oxidative, and heat stress (5, 34). The white ΔrpoS mutant in our study is consistent with a study of Erwinia herbicola and a transformed Escherichia coli strain that identified impaired carotenoid formation in ΔrpoS (47).
Altered growth and inactivation patterns under various stress conditions.
Past studies have suggested that Cronobacter spp. are capable of surviving long periods of osmotic and dry stress, resulting in a competitive advantage in the production environment of PIF factories (8, 45). As the influence of pigmentation on survival has not been investigated previously, osmotic stress and desiccation experiments were performed. Our results demonstrated increased maximum specific growth rates of ΔcrtE, ΔcrtX, and ΔcrtY under osmotic stress compared to the wild type. Phenotypic microarray analysis of ΔcrtE confirmed this finding.
In contrast, all colorless mutants were more sensitive to prolonged desiccation (16 weeks) than the wild type. While these findings may appear contradictory, studies have shown that the effect of desiccation on cells is fundamentally different from the effect of a highly osmotic environment (42, 43). This can be explained by the fact that desiccation exposes cell surfaces to an atmosphere as opposed to an aqueous phase that exhibits decreased water activity (42).
Colorless mutants may be more vulnerable to desiccation because they lack photoprotective pigments that are thought to confer resistance to drying. One proposed mechanism is based on alterations in membrane structure (42). The presence of carotenoids in cell membranes and their role in membrane structure supports the hypothesis that these pigments confer protection against desiccation (22). Furthermore, extreme desiccation leads to an increase in reactive oxygen species, thereby causing severe oxidative damage to bacterial cells (29). Carotenoids reduce the extent of oxidative damage by quenching oxygen radicals (50, 55).
Similarly, carotenoids are known to act as important photoprotectants in the face of near-UV rays that inflict damage on DNA and cell membranes (5, 50, 51, 54). As the production environment of Cronobacter sakazakii allows only dry cleaning, UV irradiation is one potential strategy to improve hygienic measures in the future. Near-UV irradiation, especially at wavelengths below 320 nm, is highly absorbed by the DNA molecule, resulting in direct and indirect damage by reactive oxygen species (15). Accordingly, in our study, the resistance of white mutants ΔcrtE and ΔcrtY to near-UV irradiation (312 nm) was reduced compared to that of the wild type. However, ΔcrtX (zeaxanthin glycosylase) and the wild type show equally high resistance to UVB. This might be due to the formation of zeaxanthin, an intermediate of the beta carotene pathway that has been observed to confer protection against photosensitized lipid peroxidation (52).
Because near-UV irradiation causes cell damage by producing reactive oxygen species, its effect on cells is similar to the oxidative stress caused by peroxide. Therefore, it is not surprising that carotenoids are known to play a protective role against this stressor as well (5, 32, 33, 50, 54). Carotenoids rigidify the fluid phase of membranes and limit oxygen penetration to the hydrophobic membrane core, which is susceptible to oxidative degradation (22). In our study, we found ΔcrtY to be more resistant, and ΔcrtE and ΔcrtX to be less resistant, to oxidative stress than the wild type after 30 min of incubation in LB supplemented with 12.5 mM cumene peroxide. Although this was an unexpected result, the increased resistance of white ΔcrtY (lycopene cyclase) to peroxidase has been reported before. Possibly due to the retarded formation of degradation products of the pigment, lycopene itself has been observed to act as a prooxidant while exhibiting an antioxidant effect in combination with y-tocopherol (23).
Carotenoids were reported to play a protective role against neutrophil oxidative bursts in Staphylococcus aureus, thus increasing neutrophil survival (33). In the macrophage experiments in this study, all strains were equally persistent. We hypothesized that the supposed protective effect of carotenoids against oxidative burst can be counterbalanced by other nonoxidative bactericidal factors. Alternatively, recognizing differences in persistence also may require experiments of longer duration.
Past studies have suggested that carotenoids function as global regulators in response to cell stress from cold shock, thus contributing to membrane stabilization (12, 16, 22). This hypothesis is supported by the observation that the lowering of the cultivation temperature led to an increase in carotenoid production in the psychrotrophic bacterium Anthrobacter agilis, possibly indicating a protective role of these pigments at low temperatures (16). The results in this study showed that all white mutants grew at comparable maximum specific growth rates under cold stress compared to that of the wild type. A protective role of carotenoids therefore could not be validated for Cronobacter sakazakii ES5 under the tested conditions.
To date, the role of carotenoids in protection against acidic stress is poorly elucidated, although pigment formation has been observed to confer resistance to oleic acid in Staphylococcus aureus (53). In our study, the selected white mutants exhibited resistance to acidic stress equal to that of the wild type during growth and increased sensitivity to acidic inactivation after incubation at pH 2 for 45 min. All strains not only survived under conditions of low pH but also were able to grow to approximately 109 CFU/ml within 24 h at pH 4.5. This is of particular interest in terms of the survival of Cronobacter sakazakii in the stomach after ingestion, since neonates tend to show higher gastric pH levels than adults, frequently reaching values above pH 4.0 (35).
In the phenotype microarray, osmotic panels confirmed an increase in resistance to salt stress in the white phenotype. Moreover, white mutants were more responsive to nutrient stimulation. This might be due to alterations in membrane structure in the absence of carotenoids (12, 22, 49), facilitating the intake of a variety of molecules. To ensure that the changes in phenotype are direct effects of the transposon insertion in the crt genes, complementation data would be required.
The complementation of the white transposon mutants was attempted but was not successful. Plasmid cloning vector pUC19 resulted in alterations in the phenotype for the wild type and therefore was considered unsuitable. In an alternative approach to knock back the transposon in the disrupted genes to restore the wild-type allele by homologous recombination, ES5 seemed to be refractory to the integration of sequences from the suicide vector. Therefore, no complementation data could be included and the direct effects of the transposon insertions into the crt genes could not be validated.
Acknowledgments
This work was supported by grant 55170102 from the University of Zurich.
Footnotes
Published ahead of print on 28 December 2009.
REFERENCES
- 1.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 2.Andersen, J. B., C. Sternberg, L. Kongsbak Poulsen, S. Petersen Bjørn, M. Givskov, and S. Molin. 1998. New unstable variants of green fluorescent protein for studies of transient gene expression in bacteria. Appl. Environ. Microbiol. 64:2240-2246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Anonymous. 2006. Milk and milk products—detection of Enterobacter sakazakii. Technical specification ISO/TS 22964. ISO/TS 22964:2006(E) and IDF/RM 210:2006(E), 1st ed. International Organization for Standardization, Geneva, Switzerland.
- 4.Baranyi, J., and T. A. Roberts. 1994. A dynamic approach to predicting bacterial growth in food. Int. J. Food Microbiol. 23:277-294. [DOI] [PubMed] [Google Scholar]
- 5.Becker-Hapak, M., E. Troxtel, J. Hoerter, and A. Eisenstark. 1997. RpoS dependent overexpression of carotenoids from Erwinia herbicola in OXYR deficient Escherichia coli. Biochem. Biophys. Res. Commun. 239:305-309. [DOI] [PubMed] [Google Scholar]
- 6.Besse, N. G., A. Leclercq, V. Maladen, C. Tyburski, and B. Lombard. 2006. Evaluation of the International Organization for Standardization-International Dairy Federation (ISO-IDF) draft standard method for detection of Enterobacter sakazakii in powdered infant food formulas. J. AOAC Int. 89:1309-1316. [PubMed] [Google Scholar]
- 7.Bowen, A. B., and C. R. Braden. 2006. Invasive Enterobacter sakazakii disease in infants. Emerg. Infect. Dis. 12:1185-1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Breeuwer, P., A. Lardeau, M. Peterz, and H. M. Joosten. 2003. Desiccation and heat tolerance of Enterobacter sakazakii. J. Appl. Microbiol. 95:967-973. [DOI] [PubMed] [Google Scholar]
- 9.Brown, L., D. Gentry, T. Elliott, and M. Cashel. 2002. DksA affects ppGpp induction of RpoS at a translational level. J. Bacteriol. 184:4455-4465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bullock, W. O., J. M. Fernandez, and J. M. Short. 1987. XL1-blue: a high efficiency plasmid transforming recA Escherichia coli strain with B-galactosidase selection. Biotechniques 5:376-379. [Google Scholar]
- 11.Cawthorn, D.-M., S. Botha, and R. C. Witthuhn. 2008. Evaluation of different methods for the detection and identification of Enterobacter sakazakii isolated from South African infant formula milks and the processing environment. Int. J. Food Microbiol. 127:129-138. [DOI] [PubMed] [Google Scholar]
- 12.Chattopadhyay, M. K. 2006. Mechanism of bacterial adaptation to low temperature. J. Biosci. 31:157-165. [DOI] [PubMed] [Google Scholar]
- 13.Chattopadhyay, M. K., M. V. Jagannadham, M. Vairamani, and S. Shivaji. 1997. Carotenoid pigments of an antarctic psychrotrophic bacterium Micrococcus roseus: temperature dependent biosynthesis, structure, and interaction with synthetic membranes. Biochem. Biophys. Res. Commun. 239:85-90. [DOI] [PubMed] [Google Scholar]
- 14.Drudy, D., N. R. Mullane, T. Quinn, P. G. Wall, and S. Fanning. 2006. Enterobacter sakazakii: an emerging pathogen in powdered infant formula. Clin. Infect. Dis. 42:996-1002. [DOI] [PubMed] [Google Scholar]
- 15.Fernández Zenoff, V., F. Siñeriz, and M. E. Farías. 2006. Diverse responses to UV-B radiation and repair mechanisms of bacteria isolated from high-altitude aquatic environments. Appl. Environ. Microbiol. 72:7857-7863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fong, N. J. C., M. L. Burgess, K. D. Barrow, and D. R. Glenn. 2001. Carotenoid accumulation in the psychrotrophic bacterium Arthrobacter agilis in response to thermal and salt stress. Appl. Microbiol. Biotechnol. 56:750-756. [DOI] [PubMed] [Google Scholar]
- 17.García-Moreno, D., M. C. Polanco, G. Navarro-Avilés, F. J. Murillo, S. Padmanabhan, and M. Elías-Arnanz. 2009. A vitamin B12-based system for conditional expression reveals dksA to be an essential gene in Myxococcus xanthus. J. Bacteriol. 191:3108-3119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Garner, M. R., B. L. Njaa, M. Wiedmann, and K. J. Boor. 2006. Sigma B contributes to Listeria monocytogenes gastrointestinal infection but not to systemic spread in the guinea pig infection model. Infect. Immun. 74:876-886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Garner, M. R., K. E. James, M. C. Callahan, M. Wiedmann, and K. J. Boor. 2006. Exposure to salt and organic acids increases the ability of Listeria monocytogenes to invade Caco-2 cells but decreases its ability to survive gastric stress. Appl. Environ. Microbiol. 72:5384-5395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gascón, J., A. Oubiña, A. Pérez-Lezaun, and J. Urmeneta. 1995. Sensitivity of selected bacterial species to UV radiation. Curr. Microbiol. 30:177-182. [DOI] [PubMed] [Google Scholar]
- 21.Grimm, M., R. Stephan, C. Iversen, G. G. G. Manzardo, T. Rattei, K. Riedel, A. Ruepp, D. Frishman, and A. Lehner. 2008. Cellulose as an extracellular matrix component present in Enterobacter sakazakii biofilms. J. Food Prot. 71:13-18. [DOI] [PubMed] [Google Scholar]
- 22.Gruszecki, W. I., and K. Strzalka. 2005. Carotenoids as modulators of lipid membrane physical properties. Biochim. Biophys. Acta 1740:108-115. [DOI] [PubMed] [Google Scholar]
- 23.Haila, K. M., S. M. Lievonen, and M. I. Heinonen. 1996. Effects of lutein, lycopene, annato, and y-tocopherol on autoxidation of triglycerides. J. Agric. Food Chem. 44:2096-2100. [Google Scholar]
- 24.Iversen, C., A. Lehner, N. Mullane, J. Marugg, S. Fanning, R. Stephan, and H. Joosten. 2007. Identification of “Cronobacter” spp. (Enterobacter sakazakii). J. Clin. Microbiol. 45:3814-3816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Iversen, C., P. Druggan, S. Schumacher, A. Lehner, C. Feer, K. Gschwend, H. Joosten, and R. Stephan. 2008. Development of a novel screening method for the isolation of “Cronobacter” spp. (Enterobacter sakazakii). Appl. Environ. Microbiol. 74:2550-2553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Iversen, C., and S. J. Forsythe. 2003. Risk profile of Enterobacter sakazakii, an emergent pathogen associated with infant milk formula. Trends Food Sci. Technol. 14:443-454. [Google Scholar]
- 27.Jacobs, J. L., T. L. Carroll, and G. W. Sundin. 2005. Ultraviolet radiation (UVR) sensitivity analysis and UVR survival strategies of a bacterial community from the phyllosphere of field-grown peanut (Arachis hypogeae L.). Microb. Ecol. 49:104-113. [DOI] [PubMed] [Google Scholar]
- 28.Johnson, Z. I., and S. W. Chisholm. 2004. Properties of overlapping genes are conserved across microbial genomes. Genome Res. 14:2268-2272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kranner, I., W. J. Cram, M. Zorn, S. Wornik, I. Yoshimura, E. Stabentheiner, and H. W. Pfeifhofer. 2005. Antioxidants and photo protection in a lichen as compared with its isolated symbiotic partners. Proc. Natl. Acad. Sci. USA 102:3141-3146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lai, K. K. 2001. Enterobacter sakazakii infections among neonates, infants, children, and adults. Medicine 80:113-122. [DOI] [PubMed] [Google Scholar]
- 31.Lehner, A., M. Grimm, T. Rattei, A. Ruepp, D. Frishman, G. G. G. Manzardo, and R. Stephan. 2006. Cloning and characterization of Enterobacter sakazakii pigment genes and in situ spectroscopic analysis of the pigment. FEMS Microbiol. Lett. 265:244-248. [DOI] [PubMed] [Google Scholar]
- 32.Liu, G. Y., and V. Nizet. 2009. Color me bad: microbial pigments as virulence factors. Trends Microbiol. 17:406-413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu, G. Y., A. Essex, J. T. Buchanan, V. Datta, H. M. Hoffman, J. F. Bastian, J. Fierer, and V. Nizet. 2005. Staphylococcus aureus golden pigment impairs neutrophil killing and promotes virulence through its antioxidant activity. J. Exp. Med. 202:209-215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Loewen, P. C., and R. Hengge-Aronis. 1994. The role of sigma factor S (KatF) in bacterial global regulation. Annu. Rev. Microbiol. 48:53-80. [DOI] [PubMed] [Google Scholar]
- 35.Maffei, H. V. L., and F. J. Nóbrega. 1975. Gastric pH and microflora of normal and diarrhoeic infants. Gut 16:719-726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Martiny, H., T. Schubert, and H. Rüden. 1990. Use of UV radiation for the disinfection of water. V. Microbiological studies of the behaviour of bacterial cells from the logarithmic and from the stationary phase in cold and warm drinking water. Zentralbl. Hyg. Umweltmed. 190:380-394. [PubMed] [Google Scholar]
- 37.Mathews, M. M., and W. R. Sistrom. 1959. Function of carotenoid pigments in non-photosynthetic bacteria. Nature 184:1892-1893. [DOI] [PubMed] [Google Scholar]
- 38.Morikawa, K., A. Maruyama, Y. Inose, M. Higashide, H. Hayashi, and T. Otha. 2001. Overexpression of sigma factor, σ−B, urges Staphylococcus aureus to thicken the cell wall and to resist β-lactams. Biochem. Biophys. Res. Commun. 288:385-389. [DOI] [PubMed] [Google Scholar]
- 39.Muytjens, H. L., H. C. Zanen, H. J. Sonderkamp, L. A. Kollée, I. K. Wachsmuth, and J. J. Farmer III. 1983. Analysis of eight cases of neonatal meningitis and sepsis due to Enterobacter sakazakii. J. Clin. Microbiol. 18:115-120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nazarowec-White, M., and J. M. Farber. 1997. Enterobacter sakazakii: a review. Int. J. Food Microbiol. 34:103-113. [DOI] [PubMed] [Google Scholar]
- 41.Perry, K. L., T. A. Simonitch, K. J. Harrison-Lavoie, and S. T. Liu. 1986. Cloning and regulation of Erwinia herbicola pigment genes. J. Bacteriol. 168:607-612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Potts, M. 1994. Desiccation tolerance of prokaryotes. Microbiol. Rev. 58:755-805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Record, M. T., Jr., W. Zhang, and C. F. Anderson. 1998. Analysis of effects of salts and uncharged solutes on protein and nucleic acid equilibria and processes: a practical guide to recognizing and interpreting polyelectrolyte effects, Hofmeister effects, and osmotic effects of salts. Adv. Protein Chem. 51:281-353. [DOI] [PubMed] [Google Scholar]
- 44.Reverchon, S., C. Rouanet, D. Expert, and W. Nasser. 2002. Characterization of indigoidine biosynthetic genes in Erwinia chrysanthemi and role of this blue pigment in pathogenicity. J. Bacteriol. 184:654-665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Riedel, K., and A. Lehner. 2007. Identification of proteins involved in osmotic stress response in Enterobacter sakazakii by proteomics. Proteomics 7:1217-1231. [DOI] [PubMed] [Google Scholar]
- 46.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 47.Sandmann, G., W. S. Woods, and R. W. Tuveson. 1990. Identification of carotenoids in Erwinia herbicola and in a transformed Escherichia coli strain. FEMS Microbiol. Lett. 59:77-82. [DOI] [PubMed] [Google Scholar]
- 48.Sedkova, N., L. Tao, P. E. Rouvière, and Q. Cheng. 2005. Diversity of carotenoid synthesis clusters from environmental Enterobacteriaceae strains. Appl. Environ. Microbiol. 71:8141-8146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Subczynski, W. K., E. Markowska, W. I. Gruszecki, and J. Sielewiesiuk. 1992. Effects of polar carotenoids on dimyristoylphosphatidylcholine membranes: a spin-label study. Biochim. Biophys. Acta 1105:97-108. [DOI] [PubMed] [Google Scholar]
- 50.Tian, B., Z. Xu, Z. Sun, J. Lin, and Y. Hua. 2007. Evaluation of the antioxidant effects of carotenoids from Deinococcus radiodurans through targeted mutagenesis, chemiluminescence, and DNA damage analyses. Biochim. Biophys. Acta 1770:902-911. [DOI] [PubMed] [Google Scholar]
- 51.Tuveson, R. W., R. A. Larson, and J. Kagan. 1988. Role of cloned carotenoid genes expressed in Escherichia coli in protecting against inactivation by near-UV light and specific phototoxic molecules. J. Bacteriol. 170:4675-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wrona, M., W. Korytowski, M. Rózanowska, T. Sarna, and T. G. Truscott. 2003. Cooperation of antioxidants in protection against photosensitized oxidation. Free Radic. Biol. Med. 35:1319-1329. [DOI] [PubMed] [Google Scholar]
- 53.Xiong, Z., and F. A. Kapral. 1992. Carotenoid pigment levels in Staphylococcus aureus and sensitivity to oleic acid. J. Med. Microbiol. 37:192-194. [DOI] [PubMed] [Google Scholar]
- 54.Zhang, L., Q. Yang, X. Luo, C. Fang, Q. Zhang, and Y. Tang. 2007. Knockout of crtB or crtI gene blocks the carotenoid biosynthetic pathway in Deinococcus radiodurans R1 and its resistance to oxidative DNA-damaging agents due to change of free radicals scavenging ability. Arch. Microbiol. 188:411-419. [DOI] [PubMed] [Google Scholar]
- 55.Ziegelhoffer, E. C., and T. J. Donohue. 2009. Bacterial responses to photooxidative stress. Nat. Rev. Microbiol. 7:856-863. [DOI] [PMC free article] [PubMed] [Google Scholar]