Abstract
Wetlands are sources of denitrification-derived nitrous oxide (N2O). Thus, the denitrifier community of an N2O-emitting fen (pH 4.7 to 5.2) was investigated. N2O was produced and consumed to subatmospheric concentrations in unsupplemented anoxic soil microcosms. Total cell counts and most probable numbers of denitrifiers approximated 1011 cells·gDW−1 (where DW is dry weight) and 108 cells·gDW−1, respectively, in both 0- to 10-cm and 30- to 40-cm depths. Despite this uniformity, depth-related maximum reaction rate (vmax) values for denitrification in anoxic microcosms ranged from 1 to 24 and −19 to −105 nmol N2O h−1· gDW−1, with maximal values occurring in the upper soil layers. Denitrification was enhanced by substrates that might be formed via fermentation in anoxic microzones of soil. N2O approximated 40% of total nitrogenous gases produced at in situ pH, which was likewise the optimal pH for denitrification. Gene libraries of narG and nosZ (encoding nitrate reductase and nitrous oxide reductase, respectively) from fen soil DNA yielded 15 and 18 species-level operational taxonomic units, respectively, many of which displayed phylogenetic novelty and were not closely related to cultured organisms. Although statistical analyses of narG and nosZ sequences indicated that the upper 20 cm of soil contained the highest denitrifier diversity and species richness, terminal restriction fragment length polymorphism analyses of narG and nosZ revealed only minor differences in denitrifier community composition from a soil depth of 0 to 40 cm. The collective data indicate that the regional fen harbors novel, highly diverse, acid-tolerant denitrifier communities capable of complete denitrification and consumption of atmospheric N2O at in situ pH.
Nitrous oxide (N2O) is a potent greenhouse gas with a global warming potential that is 300-fold higher than that of CO2, and its concentration increased from 270 ppb in 1750 to 319 ppb in 2005 (17). N2O can be produced in soils during denitrification, nitrification, the dissimilatory reduction of nitrate to nitrite and/or ammonium (hereafter referred to as dissimilatory nitrate reduction), or the chemical transformation of nitrite or hydroxylamine (5, 7, 49). The percentage of N2O produced in any of these processes is variable, depending mainly on the redox potential, pH, and C/N ratio (49). In anoxic ecosystems such as waterlogged soils, most of the N2O is considered to be denitrification derived (7, 9). Complete denitrification is the sequential reduction of nitrate to dinitrogen (N2) via nitrite, nitric oxide (NO), and N2O (75). The main product of denitrification varies with the organism and in situ conditions and is usually either N2O or N2 (68). N2O can occur as a by-product during dissimilatory nitrate reduction when accumulated nitrite interacts with nitrate reductase to form N2O (59). The production of N2O by dissimilatory nitrate reducers is favored in environments with large amounts of readily available organic carbon (65). Thus, their contribution to nitrate-dependent production of N2O in soils is likely insignificant compared to that of denitrifiers.
The oxidoreductases involved in denitrification are termed dissimilatory nitrate reductase (Nar, encoded by narGHJI, or Nap, encoded by napEDABC), nitrite reductase (Nir, encoded by nirK and nirS), NO reductase (cNor and qNor, encoded by norBC and norB, respectively), and N2O reductase (Nos, encoded by nosZ) (75). Nitrate reductase is also found in dissimilatory nitrate reducers (60). narG can therefore be used as a molecular marker to assess both denitrifiers and dissimilatory nitrate reducers, whereas nosZ is specific for the assessment of denitrifiers (25, 43, 48).
Denitrification in soils is regulated by temperature, pH, substrate (i.e., carbon) availability, and water content (10, 24, 66). Although denitrification increases with increasing temperature, it can still occur at temperatures below 0°C (10, 24). Low temperatures appear to limit the activity of N2O reductase more severely than other enzymes involved in denitrification and thus yield higher relative amounts of denitrification-derived N2O (24). Although denitrification activity usually decreases under acidic conditions, the relative percentage of N2O to total denitrification-derived nitrogenous gases increases with increasing acidity, a result attributed to the sensitivity of N2O reductase to low pH (27, 70). However, denitrifier communities can be adapted to the in situ pH of the system (40, 58, 73).
Wetlands are ecosystems in which denitrification is likely a dominant source of emitted N2O (7, 44, 45). The identification and analysis of main drivers for N2O production (i.e., the microbiota catalyzing N2O production and consumption) is thus of major concern in such environments. Fens are specialized wetlands characterized by soil acidity (67). However, information on acid-tolerant denitrifier communities of such wetlands is scarce. It is hypothesized that fens harbor a diverse, hitherto unknown, denitrifier community that is adapted to in situ conditions and associated with N2O fluxes (i.e., fen denitrifiers are acid tolerant and have a high affinity for nitrate and N2O). Thus, the main objectives of the present study were to evaluate the capacities of denitrifier communities of an N2O-emitting fen (20) to produce or consume N2O and to determine if a novel and diverse denitrifier community was associated with these capacities.
MATERIALS AND METHODS
Sampling site.
The minerotrophic fen Schlöppnerbrunnen is located in the Lehstenbach catchment in the Fichtelgebirge, Bavaria, Germany (50°07′53"N, 11°52′51"E), at approximately 700 m above sea level. The fen has a mean annual air temperature of 5.3°C and a mean annual precipitation of 1,160 mm (16). The fen soil is a fibric histosol on granite bedrock and was described previously (42, 72). Vegetation consists mainly of Molinia caerulea, Eriophorum vaginatum, Carex canescens, and Juncus effusus (42). Total carbon is 140 to 410 mg of C gDW−1 of soil (where DW is dry weight), and total nitrogen is 4 to 24 mg of N gDW−1 (42, 72). Two independent soil samples per sampling were taken from soil layers at 0 to 40 cm at 10-cm intervals with a soil corer. Soil was transported on ice in air-tight plastic bags and stored at 2°C for no longer than 4 days before processing. Soil for DNA extraction was stored immediately at −80°C.
Soil parameters.
Nitrate and nitrite concentrations as well as soil pH were determined in 2 M KCl extracts (3 g of soil in 7 ml KCl; extraction for 16 h at 2°C). Nitrate was measured by flow injection analysis (FIA-LAB; MLE, Dresden, Germany); nitrite was determined colorimetrically (23). pH was determined with a pH electrode (InLab 422; Mettler Toledo GmbH, Gießen, Germany) as a water-soil solution (1:4 dilution of fen soil with double-distilled water). Moisture content was determined by weighing the soil before and after drying at 40°C for 1 week.
Assessment of denitrification in fen soil microcosms.
Approximately 16 g of homogenized soil from four depths and two soil cores was diluted with 3 volumes of sterile water and placed into 125-ml infusion flasks that were then sealed with gas-tight rubber stoppers. The gas phase was sterile argon. Experiments were done in triplicate. Preliminary tests with unsupplemented microcosms (i.e., fen soil slurries) assessed denitrification potentials from in situ nitrate and nitrite. Microcosms were preincubated for 14 to 18 h at 15°C before being supplemented with nitrate (provided as NaNO3) or N2O; this preincubation was designed to consume traces of nitrate and nitrite by denitrification prior to the addition of nitrate or N2O.
Nitrate concentrations of 0 to 100 μM were used for apparent Michaelis-Menten kinetics, as up to 130 μM in situ nitrate has been observed (42, 55). N2O ranged from 0 to 56 μM (based on the volume of the aqueous phase), covering atmospheric N2O concentrations as well as concentrations where N2O consumption rates were maximal. The incubation time was 2 to 14 h, depending on the rate of N2O production or consumption. Acetylene blocks the reduction of N2O at the level of N2O reductase (74), and parallel nitrate-supplemented microcosms with and without acetylene (15% [vol/vol] in the headspace) were used to differentiate total denitrification and N2O production, respectively. Microcosms were incubated at 15°C at in situ pH in the dark unless otherwise indicated. Kinetic parameters (Km and the maximum reaction rate [vmax]) for nitrate-dependent denitrification were based on the production of N2O in microcosms supplemented with both nitrate and acetylene. Headspace concentrations of N2O were determined at three time points via gas chromatography for determining rates of N2O production or consumption. N2O was quantified with a Hewlett-Packard 5980 series II gas chromatograph equipped with an electron capture detector, 3396 series II integrator, and a Porapak Q-80/100 (Supelco, Bellefonte, Pa.) column (length, 4 m; inner diameter, 3.2 mm) with Ar-CH4 (95:5) as the carrier gas (40 or 20 ml per min); the injector temperature was 150°C, the column temperature was 60°C, and the detector temperature was 300°C (modified from reference 26).
Apparent Michaelis-Menten kinetics were fitted to the data points using the program SigmaPlot 2000 (SPSS Science Software GmbH, Erkrath, Germany) for calculation of Km and vmax according to the following equation (56): v = (vmax·[S])/(Km + [S]).
The influence of temperature, pH, and electron donors was tested at initial nitrate or N2O concentrations of 100 μM or 0.1 μM, respectively. Sodium salts of acetate, formate, succinate, or butyrate were supplied at 300 μM each, whereas ethanol was supplied at 500 μM. Consumption of electron donors was assessed by high-performance liquid chromatography (8, 31, 34).
Temperature and pH optima were calculated with SigmaPlot, version 10.0 (Systat Software Inc., San Jose, CA), based on the observed denitrification rates at different incubation temperatures (i.e., at 0.5, 5, 15, 27, 38, 48, 59, and 69°C) or incubation pH values. Temperature optima were approximated by the maximum of the following Ratkowsky equation (46): 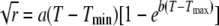 , where r is the rate of N2O production or consumption at a given temperature T (in K); Tmin and Tmax are the minimal and maximal temperatures (in K), respectively, at which N2O production or consumption approximates zero; and a and b are fit parameters. pH optima were approximated with a Gaussian model:
, where r is the rate of N2O production or consumption at a given temperature T (in K); Tmin and Tmax are the minimal and maximal temperatures (in K), respectively, at which N2O production or consumption approximates zero; and a and b are fit parameters. pH optima were approximated with a Gaussian model: 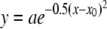 , where y is the observed N2O production rate at a given pH x, x0 is the optimum pH, and a and b are fit parameters.
, where y is the observed N2O production rate at a given pH x, x0 is the optimum pH, and a and b are fit parameters.
Total cell counts and enumeration of denitrifiers.
One milliliter of homogenized samples of 10−3 or 10−4 soil dilutions was incubated with 0.5 ml of phosphate-buffered saline (PBS; 10 mM sodium phosphate buffer [pH 7.4] containing 130 mM NaCl) containing 1 μg of 4′,6-diamidino-2-phenylindoldihydrochloride (DAPI) for 20 min on ice. Cells were fixed on polycarbonate filters (pore size, 0.2 μm; GTTP 4700; Millipore, Eschborn, Germany) via vacuum filtration and washed with PBS and then with 96% ethanol to reduce background fluorescence. Cells were quantified using a fluorescence microscope (Zeiss Axioscope 2; Carl Zeiss AG, Oberkochen, Germany) equipped with mercury vapor lamp HBO100. Cells were counted in 11 randomly chosen squares (square size was 15.25 mm2).
Most probable numbers (MPN) of denitrifiers were determined in triplicate (11). The mineral salts medium contained the following (in mg/liter) (modified from references 1 and 28): (NH4)2 SO4, 12.6; Na2SO4, 13.5; CaCl2·2 H2O, 10.0; MgCl2·2 H2O, 10.0; KH2PO4, 0.4; FeCl2·4 H2O, 10; MnSO4·1 H2O, 5; FeSO4·7 H2O, 1; CoCl2·6 H2O, 1; CaCl2·2 H2O, 1; ZnSO4·7 H2O, 1; CuSO4·5 H2O, 0.1; AlK(SO4)2·12 H2O, 0.2; H3BO3, 0.1; Na2MoO4·2 H2O, 0.1; and nitrilotriacetic acid, 15; the medium also contained vitamins ([μg/liter] biotin, 2; folic acid, 2; pyridoxine-HCl 10; thiamine-HCl, 5; riboflavin, 5; niacin, 5; dl-Ca-pantothenic acid, 5; vitamin B12, 0.1; p-aminobenzoic acid, 5; lipoic acid, 5). This medium was supplemented with nutrient broth (0.27 g/liter), NaNO3 (final concentration of 5 mM), and glutamate, succinate, butyrate, and ethanol (final concentration of 1.5 mM each). Anoxic medium were prepared using modified Hungate techniques (72); the pH was 5, and the headspace was sterile argon. Tubes were incubated in the dark at 15°C for 3 months. Denitrifiers were scored positive for growth when the optical density (at 660 nm) of culture tubes was greater than 0.04 and either N2 (at values 5-fold greater than that in uninoculated control tubes) or N2O (at values 5-fold greater than that of air) was produced.
Extraction of nucleic acids.
Nucleic acids were extracted using a bead-beating protocol (21), followed by separation of DNA and RNA using a Qiagen RNA/DNA Mini Kit (Qiagen GmbH, Hilden, Germany).
Amplification of narG and nosZ.
narG and nosZ were amplified using the primer pair narG1960f (TAY GTS GGS CAR GAR AA) and narG2650r (TTY TCR TAC CAB GTB GC) (43) and the pair nosZF (CGC TGT TCI TCG ACA GYC AG) and nosZR (ATG TGC AKI GCR TGG CAG AA), respectively (48). Each PCR was preceded by an initial denaturation (95°C for 5 min). Denaturation and elongation were at 95°C and 72°C, respectively. Seven precycles with annealing at 55°C were followed by 26 cycles with annealing at 51°C for narG; denaturation, annealing, and elongation were for 1, 1, and 2 min, respectively. Annealing was lowered stepwise from 58°C to 52°C in 10 precycles for nosZ, followed by 30 cycles with annealing at 52°C; denaturation, annealing, and elongation were for 0.5, 1, and 1 min, respectively. Final elongation was for 10 min at 72°C.
Cloning, screening, and sequencing.
PCR products of narG and nosZ were purified using a MinElute gel extraction kit (Qiagen GmbH, Hilden, Germany). Purified PCR products were ligated into vector pGEM-T (Promega, Mannheim, Germany) and transformed into competent Escherichia coli JM109 cells (Promega) according to the manufacturer's protocol. Clones were screened for insert-positive vectors via amplification with primers M13uni/M13rev (35). Gene libraries were screened by restriction fragment length polymorphism (RFLP) analysis. Products from PCRs using the M13 primers (M13-PCR) were digested with HaeIII and CfoI (2 U at 4 h at 37°C). Restriction fragments were separated on a 3% agarose gel (1 h at 100 V). One to two representative clones of each RFLP pattern were sequenced. M13-PCR products were purified prior to sequencing with a Millipore Multiscreen 96-well filtration system (Millipore Corp., Bedford, MA). Sequencing was done commercially by Macrogen (Seoul, South Korea).
Sequence analyses.
Sequence analyses were done with MEGA, version 4.0 (http://www.megasoftware.net/) (29). Sequences were edited, translated in silico, and aligned with reference sequences using the ClustalW algorithm implemented in MEGA, version 4.0. Sequences that had the same RFLP patterns with sequenced gene fragments were represented by the sequenced fragments. The abundance of each sequence in the alignment was adjusted according to the number of observations of the corresponding RFLP pattern in the gene library. The alignments were refined manually. Phylogenetic trees were constructed from in silico translated sequences. Each neighbor-joining (52) tree was constructed with 1,000 bootstrap replicates. Uncorrected distance matrices with sequences from different soil layers were created from the translated amino acid alignment and used for diversity analyses in DOTUR (54). Grouping of sequences into operational taxonomic units (OTUs), as well as estimations of species richness and species diversity (expressed by the Shannon diversity index), was conducted at sequence differences of 0 and 41% for narG and 0 and 14% for nosZ to assess maximal diversity as well as the species-level diversity of denitrifiers. The thresholds for estimating species-level diversity were obtained from comparisons of 16S rRNA similarities and structural gene similarities of cultured denitrifiers (39). Coverage (C) is the number of the detected genotypes relative to their expected total number in a gene library, and was calculated as follows: C = (1 − n × N−1) × 100, where n is the number of genotypes that occurred only once, and N is the number of clones screened (54).
TRFLP analyses.
narG and nosZ were amplified with fluorescently labeled primers (narG1960f-DY681/narG2650r-DY781 and nosZF-DY681/nosZR-DY781) for terminal RFLP (TRFLP) analysis. Fluorescently labeled DNA was digested with mung bean nuclease (New England Biolabs, Frankfurt am Main, Germany) to remove single-stranded DNA and reduce the probability of pseudo-terminal restriction fragments (14). Digested DNA was purified using a QIAquick PCR Purification Kit (Qiagen GmbH, Hilden, Germany). PCR products of narG were digested with the restriction enzymes CfoI, HaeIII, or XhoI (New England Biolabs, Frankfurt am Main, Germany), and PCR products of nosZ were digested with BtgI, NlaIV, or PvuI plus SacI (New England Biolabs, Frankfurt am Main, Germany) (double digest). Gel electrophoresis was performed with an NEN model 4300 DNA analyzer (Licor, Lincoln, NE). The polyacrylamide gel consisted of 15 g of urea, 3.75 ml of 40% acrylamide-bisacrylamide solution (37.5:1; Bio-Rad, Hercules, CA), 6 ml of 5× Tris-borate-EDTA buffer (AppliChem GmbH, Darmstadt, Germany), and 9.25 ml of double-distilled H2O. A bind-silane solution (1:1 bind-silane [PlusOne; GE Healthcare, Piscataway, NJ] and 10% acetic acid) was applied to the glass plates for stabilizing the comb region of the gel. The gel was poured according to the manufacturer's protocol (Licor, Lincoln, NE). Electrophoresis was performed for 3 h at 1,500 V and 45°C. Gels were analyzed with GelQuest (Sequentix, Klein Raden, Germany). Terminal restriction fragments (TRFs) were assigned to narG or nosZ sequences via in silico TRF analysis in MEGA, version 4.0. Principal component analyses of the combined TRFLP profiles were conducted using RapidMiner (http://rapid-i.com/) for narG and nosZ.
Nucleotide sequence accession numbers.
Sequences are deposited in EMBL under accession numbers FN430426 to FN430490 (narG) and FN430491 to 430566 (nosZ).
RESULTS
Soil parameters.
Soil moisture content ranged from 37 to 90%, was highest in the 0- to 10-cm-depth soil, and decreased with increasing soil depth. Soil pH varied between 3.4 and 3.7 in KCl extracts and between 4.7 and 5.2 in water extracts (sampling dates, 2 July 2007 and 28 February 2008). Nitrate and nitrite concentrations were below the detection limit of 0.4 μM and 5 μM, respectively (sampling date, 2 July 2007).
Depth-related denitrification in acidic fen soil.
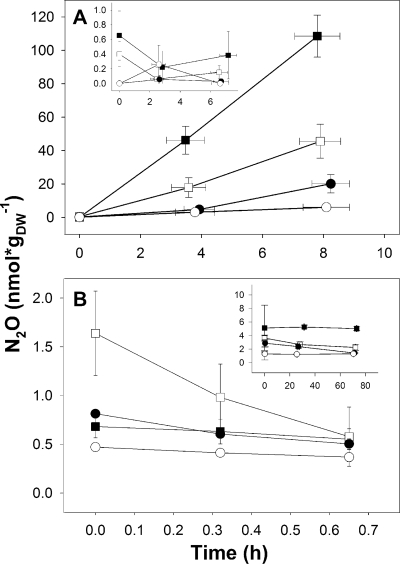
Unsupplemented anoxic fen soil produced only minor amounts of N2O (up to 40 nmol·gDW−1 after 3 days), which was in agreement with the low nitrate and nitrite concentrations in the soil. The N2O that was produced initially was completely consumed after 3 days in unsupplemented microcosms, demonstrating the potential of the acidic fen soil to consume N2O. Supplemental nitrate stimulated the production of N2O in all soil layers without apparent delay (Fig. 1A). The amount of N2O that accumulated in nitrate-supplemented (20 μM nitrate or less) microcosms leveled off or started to decrease after 2 h of incubation in microcosms without acetylene (data not shown), suggesting that N2O was subject to consumption under these conditions. The production of N2O was linear at all supplemental nitrate concentrations in microcosms with acetylene. The ratios of N2O to total N gases (i.e., N2 plus N2O) of 0- to 20-cm-depth soils were lower than those of 20- to 40-cm-depth soils; increased nitrate concentrations yielded increasingly higher ratios with 0- to 20-cm-depth soils (see Fig. S1 in the supplemental material).
FIG. 1.
Production (A) or consumption (B) of N2O by fen soil after the addition of 40 μM nitrate (A) or 0.02 μM N2O (B). Microcosms used for the experiments shown in panel A were supplemented with acetylene. Mean values and standard errors of three replicates are shown (sampling dates were 10 July 2007 for panel A and 17 July 2007 for panel B). The inset in panel A shows the control without supplemental nitrate; the inset in panel B shows the control treated with acetylene. Closed squares represent the soil layer at 0 to 10 cm, open squares represent the soil layer at 10 to 20 cm, closed circles represent the soil layer at 20 to 30 cm, and open circles represent the soil layer at 30 to 40 cm.
The consumption of supplemental N2O by fen soil was linear and without apparent delay and was blocked by acetylene (Fig. 1B). N2O was consumed to less than 100 ppb, a concentration less than that of atmospheric N2O, which approximates 319 ppb (18), indicating that acid-tolerant fen denitrifiers in all fen soil layers were capable of consuming atmospheric N2O. Initial nitrate-dependent N2O production rates and N2O-dependent N2O consumption rates followed apparent Michaelis-Menten kinetics (Table 1; see also Fig. S2 in the supplemental material). vmax values were highest in upper soil layers and decreased with soil depth, indicating that denitrification potentials in upper soil layers were greater than those of lower soil layers.
TABLE 1.
Kinetics, temperature optima, and pH optima of denitrification by denitrifiers in fen soil microcosms
| Depth of soil layer (cm) | In situ pH | Value in nitrate-amended soil |
Value in N2O-amended soil |
|||||
|---|---|---|---|---|---|---|---|---|
| vmax (nmol·h−1 gDW−1) | Km (μM) | Optimum temp (°C) | Optimum pH | vmax (nmol·h−1 gDW−1) | Km (μM) | Optimum temp (°C) | ||
| 0-10 | 5.2 | 24 ± 2 | 19 ± 3 | 34 | 5.5 | 83 ± 13 | 47 ± 13 | 20 |
| 10-20 | 4.8 | 9 ± 1 | 15 ± 4 | 34 | 4.5 | 105 ± 5 | 10 ± 2 | 20 |
| 20-30 | 4.7 | 3 ± 1 | 9 ± 6 | 46 | 4.2 | 40 ± 4 | 15 ± 4 | 30 |
| 30-40 | 4.8 | 1 ± 0 | 6 ± 0 | 38 | 4.3 | 19 ± 4 | 24 ± 9 | 6 |
Effects of temperature, pH, and electron donors on fen denitrifiers.
Denitrification, including the capacity to consume N2O, occurred at temperatures ranging from 0.5°C to 70°C. The optimal temperatures for nitrate-dependent N2O production and the consumption of N2O ranged from 34°C to 46°C and 6°C to 30°C, respectively (Table 1). Denitrification rates at temperatures above 60°C were minimal, a trend similar to that observed with other soils (33).
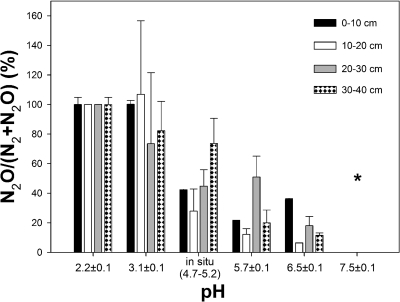
Denitrification occurred at pH 2 to 6.6 in all soil layers. Denitrification was observed at pH 7.5 only with 0- to 10-cm-depth soil. Highest denitrification rates were observed at in situ pH (i.e., 4.7 to 5.2). The ratio of N2O to total N gases tended to decrease with increasing pH (Fig. 2). At in situ pH, N2O approximated 40% of total N gasses in 0- to 30-cm-depth soils and around 80% in 30- to 40-cm-depth soils. N2O constituted nearly 100% of total N gases produced at pHs 3.1 and 2.2.
FIG. 2.
Effect of pH on the relative amount of N2O produced by fen soil. The asterisk indicates that N2O was detected in the presence but not in the absence of acetylene in 0- to 10-cm-depth soil and was not detected in 10- to 40-cm-depth soil microcosms. Mean values and standard errors of three replicates are shown (sampling date, 28 February 2008).
Denitrification rates of formate-supplemented microcosms were up to 2.5-fold higher than those of unsupplemented microcosms (see Fig. S5 in the supplemental material). Denitrification rates of acetate- and ethanol-supplemented microcosms were up to 1.5 times higher than those of unsupplemented microcosms. These enhancements of denitrification were concomitant to the net consumption of up to 150 μM formate, 50 μM acetate, and 600 μM ethanol. Succinate and butyrate did not significantly augment denitrification (Fig. S5).
Enumeration of fen microbes and denitrifiers.
Total cell numbers (DAPI counts) approximated 3.2 × 1011 ± 0.5 × 1011 cells·gDW−1 and 1.0 × 1011 ± 0.5 × 1011 cells·gDW−1 in 0- to 10-cm and 30- to 40-cm-depth soils, respectively. Denitrifier counts were similar at different soil depths, approximating 7.5 × 107 (3.2 × 107 to 1.7 × 108) cells·gDW−1 and 5.9 × 107 (2.5 × 107 to 1.4 × 108) cells·gDW−1 in 0- to 10-cm and 30- to 40-cm-depth soils, respectively.
Phylogenetic analysis of fen denitrifiers.
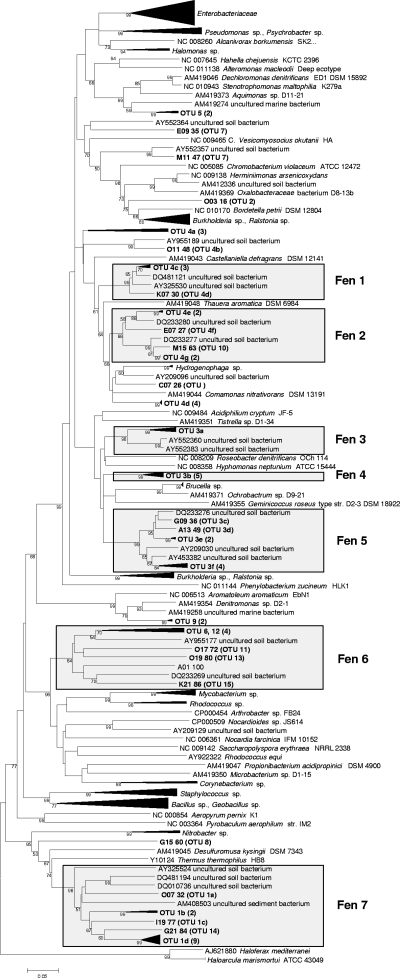
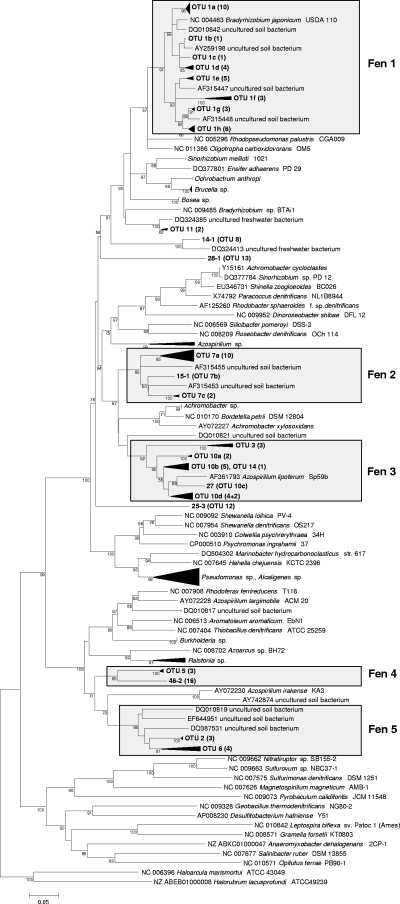
A total of 191 narG and 196 nosZ clones were analyzed by RFLP, and sequences were obtained from 64 and 75 clones, respectively. Translated amino acid sequences of representative narG and nosZ sequences were assigned to 15 and 18 OTUs, respectively (Table 2). The library coverages were 87 to 99% for narG at a sequence dissimilarity of 41% and 87 to 97% for nosZ at a sequence dissimilarity of 14% (39) (Table 2), indicating that the numbers of clones sampled were sufficient. narG and nosZ sequences from the acidic fen formed seven and five distinct clusters in the phylogenetic trees, respectively (Fig. 3 and 4). Most narG sequences were related to uncultured soil bacteria. Cultured relatives of fen sequences included Comamonas nitrativorans and species of Brucella and Hydrogenophaga (Fig. 3). nosZ sequences of clusters 1, 3, and 4 were closely related to Bradyrhizobium japonicum, Azospirillum lipoferum, and Azospirillum irakense, respectively, while nosZ sequences of cluster 2 were not closely related to cultured denitrifiers (Fig. 4).
TABLE 2.
Analyses of translated amino acid sequences of narG and nosZ derived from fen soil
| Depth of soil layer (cm) |
narG |
nosZ |
||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| No. of clonesa | % Dissimilarityb | % Library coverage | No. of OTUs observed | No. of OTUs estimated by:c |
Hd | No. of clonesa | % Dissimilarityb | % Library coverage | No. of OTUs observed | No. of OTUs estimated by:c |
Hd | |||||||
| ACE | Bootstrap | Chao1 | Jacknife | ACE | Bootstrap | Chao1 | Jacknife | |||||||||||
| 0-10 | 51 | 0 | 53 | 36 | 68 | 46 | 64 | 63 | 3.5 | 61 | 0 | 77 | 26 | 45 | 32 | 22 | 43 | 3.0 |
| 41 | 90 | 12 | 18 | 14 | 14 | 17 | 1.8 | 14 | 95 | 10 | 13 | 11 | 12 | 13 | 1.8 | |||
| 10-20 | 30 | 0 | 40 | 24 | 60 | 31 | 46 | 44 | 3.1 | 52 | 0 | 77 | 22 | 41 | 27 | 35 | 35 | 2.8 |
| 41 | 87 | 8 | 14 | 10 | 10 | 12 | 1.4 | 14 | 95 | 8 | 13 | 9 | 10 | 11 | 1.6 | |||
| 20-30 | 80 | 0 | 81 | 33 | 56 | 40 | 41 | 48 | 3.0 | 47 | 0 | 60 | 27 | 82 | 34 | 61 | 62 | 3.0 |
| 41 | 99 | 8 | 9 | 9 | 8 | 9 | 1.6 | 14 | 87 | 12 | 21 | 14 | 20 | 19 | 2.0 | |||
| 30-40 | 22 | 0 | 59 | 14 | 30 | 18 | 21 | 23 | 2.5 | 38 | 0 | 76 | 16 | 31 | 20 | 25 | 25 | 2.4 |
| 41 | 95 | 5 | 6 | 5 | 5 | 6 | 1.4 | 14 | 97 | 4 | 5 | 4 | 4 | 5 | 0.8 | |||
| Total | 191 | 0 | 94 | 62 | 66 | 70 | 65 | 73 | 3.9 | 196 | 0 | 87 | 51 | 96 | 62 | 84 | 88 | 3.3 |
| 41 | 98 | 15 | 19 | 19 | 18 | 20 | 2.0 | 14 | 97 | 18 | 25 | 21 | 22 | 24 | 2.1 | |||
Number in the library.
Percent sequence dissimilarity needed for OTU definition.
Number of genotypes estimated using the ACE, bootstrap, Chao1, and Jackknife richness estimators.
Shannon-Weaver diversity index.
FIG. 3.
Phylogenetic tree of narG sequences retrieved from the Schlöppnerbrunnen fen. The tree is based on translated amino acid sequences. The percentages of replicate trees in which the associated taxa clustered together in the bootstrap test (1,000 replicates) are shown next to the branches.
FIG. 4.
Phylogenetic tree of nosZ sequences retrieved from the Schlöppnerbrunnen fen. The tree is based on translated amino acid sequences. The percentages of replicate trees in which the associated taxa clustered together in the bootstrap test (1,000 replicates) are shown next to the branches.
The Shannon-Weaver diversity index for translated narG and nosZ as well as their estimated species richness values was highest in the upper soil layers and tended to decrease with increasing soil depth (Table 2).
Depth-resolved TRFLP fingerprints of fen denitrifiers.
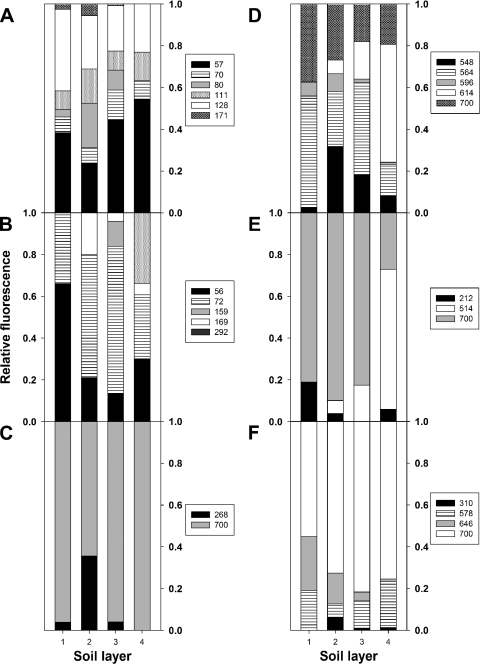
Different soil layers yielded dissimilar relative narG TRF intensities (Fig. 5). narG OTUs 1, 3, and 4 were consistently identified in the TRFs generated with the three restriction digests (see Table S1 in the supplemental material). A 57-bp TRF (TRF 57 bp) and TRF 128 bp (digestion with CfoI) were dominant in all soil layers. The relative abundance of TRF 57 bp increased from 40% in 0- to 10-cm-depth soil to 55% in 30- to 40-cm-depth soil, while the relative abundance of TRF 128 bp decreased from 40% in 0- to 10-cm-depth soil to 25% in 30- to 40-cm-depth soil, indicating a depth-related decrease of a portion of sequences belonging to narG OTUs 3 and 4 and a depth-related increase of another portion of sequences belonging to narG OTUs 3 and 4. TRFs 80 bp and 169 bp were not detected in 30- to 40-cm-depth soil, indicating the absence of a portion of sequences belonging to OTU 3 at this soil depth. The number of TRFs detected with HaeIII increased from two in 0- to 10-cm-depth soil to four in 30- to 40-cm-depth soil. The relative fluorescence of TRF 56 bp decreased significantly from 65% with 0- to 10-cm-depth soil to 15 to 30% with lower soil layers, indicating a decrease of a portion of sequences belonging to OTUs 1 and 3. TRF 159 bp was detected only in 20- to 30-cm-depth soil, while TRF 292 bp was detected only in 30- to 40-cm-depth soil, where it accounted for 30% of the relative fluorescence, indicating that a significant portion of sequences belonging to OTU 4 occurred only in those soil layers. Digestion with XhoI yielded only two TRFs (i.e., TRF 268 bp and TRF 700 bp). TRF 268 bp was detected at upper and middle soil depths and accounted for 35% relative fluorescence with 10- to 20-cm-depth soil, indicating the presence of a portion of sequences belonging to OTU 3 in this layer. Principal component analysis of the combined narG TRFLP profiles indicated that the nitrate reducer communities of 0- to 10-cm-, 20- to 30-cm-, and 30- to 40-cm-depth soils were similar and might differ from the community of 10- to 20-cm-depth soil (Fig. 5A to C; see also Fig. S3 in the supplemental material).
FIG. 5.
Comparative TRFLP analyses of narG (A to C) and nosZ (D to F) amplified from different soil layers of the acidic fen. PCR products were digested with CfoI (A), HaeIII (B), XhoI (C), BtgI (D), NlaIV (E), and PvuI and SacI (F). Mean values of three replicates are shown. Detected TRFs could be assigned to sequences from OTUs 1, 3, and 4 for narG and OTUs 1, 3, and 11 for nosZ. Soil layers 1, 2, 3, and 4 refer to soil depths of 0 to 10 cm, 10 to 20 cm, 20 to 30 cm, and 30 to 40 cm, respectively. Lengths of detected TRFs (in base pairs) are given in boxes next to the diagrams.
Different soil layers yielded dissimilar relative nosZ TRF intensities (Fig. 5). nosZ OTUs 1, 3, and 11 were consistently identified in the TRFs generated with the three restriction digests (see Table S1 in the supplemental material). BtgI yielded five TRFs, four of which occurred in all soil layers. TRF 614 bp had the highest relative fluorescence (65%) in 30- to 40-cm-depth soil, indicating a dominance of organisms associated with this TRF (belonging to a portion of sequences from OTU 1) in that soil layer. NlaIV yielded three TRFs, with TRF 700 bp being dominant (80 to 90% relative fluorescence) in 0- to 30-cm-depth soils, indicating that a portion of sequences from OTU 3 occurred more frequently in upper soil layers. TRF 514 bp was dominant (70% relative fluorescence) in 30- to 40-cm-depth soil, indicating that a portion of the OTU 1-affiliated sequences was dominant in deeper soils. Both PvuI and SacI yielded four TRFs. TRF 310 bp could not be assigned to any OTU. TRF 700 bp was dominant in all soil layers (55 to 80% relative fluorescence), indicating that a portion of sequences from OTUs 1 and 3 was dominant. TRF 646 bp decreased with increasing soil depth from approximately 25% to 2% relative fluorescence, indicating that a portion of OTU 11-affiliated sequences was less dominant in deeper soil layers. Principal component analysis of the combined nosZ TRFLP profiles indicated that minimal differences in the denitrifier community composition of the sampled soil layers might have occurred, with gradual changes between soil layers (Fig. 5D to F; see also Fig. S4 in the supplemental material).
DISCUSSION
Phylogenetically novel fen denitrifiers.
Cultured denitrifier numbers approximated 107 cells per gram of dry weight. Similar numbers of cultured fermenters and cultured aerobes occur in the Schlöppnerbrunnen fen (72), whereas the numbers of cultured Fe(III) reducers and cultured methanogens are lower, approximating 105 to 106 cells per gram of fresh weight and 104 to 105 cells per gram of dry weight, respectively (47, 72). Thus, denitrifiers appear to be a relatively abundant bacterial group capable of anaerobiosis in Schlöppnerbrunnen fen soil.
Novel narG and nosZ genotypes indicate that hitherto unknown denitrifiers occur in the Schlöppnerbrunnen fen, and statistical analyses verified a high phylogenetic diversity of the denitrifier community. Although novel genotypes were detected, some nosZ and narG sequences were related to sequences indicative of known soil genera (e.g., Azospirillum, Ralstonia, and Bradyrhizobium) (15, 25, 36, 43), indicating that part of the fen denitrifier community is similar to previously resolved genera. Communities of different soil layers were phylogenetically similar, indicating that fen denitrifiers were derived from the same pool of microorganisms. TRFLP analysis of nosZ revealed only minor differences between different soil layers. In contrast, narG-associated differences were more pronounced, suggesting that the detected dissimilatory nitrate reducers (which have narG but lack nosZ [60]) in the fen are more dissimilar between soil layers than are detected denitrifiers. The largest difference observed in the narG TRFLP profiles was between the profile of 10- to 20-cm-depth soil and profiles of the other soil layers, indicating that 10- to 20-cm-depth soil harbors a nitrate-reducing population that is not identical to the populations of the other soil layers.
In situ consequences of denitrifier activity.
Nitrate concentrations in the Schlöppnerbrunnen fen are generally low and often below the detection limit but can be as high as 0.13 mM in the upper 20 cm of soil (42, 55). Unsupplemented fen soil produced minor amounts of N2O, reflecting the low in situ nitrate concentrations in the fen (30, 42, 55). Supplemental nitrate caused a rapid increase in the production of N2O without apparent delay, indicating that (i) fen denitrifiers are poised to respond rapidly to nitrate and have a high potential to denitrify, and (ii) in situ denitrification is likely limited by nitrate availability. Increased concentrations of nitrate caused an increase in the relative proportion of N2O in total N gases, a phenomenon that has been observed with other soils (3, 18). The fen Schlöppnerbrunnen is a net source of N2O, and N2O concentrations of up to 100 ppm occur in the pore water (20).
There is a large difference between nitrate input and detected nitrate concentrations in the fen soil as nearby oxic soils receive the same amount of nitrate input but have nearly 100-fold higher nitrate concentrations (42). This difference is suggestive of a high turnover of nitrate that is due in part to denitrification. The Km values (<20 μM) (Table 1) for nitrate are well below the maximum nitrate concentrations found in situ, indicating that fen denitrifiers have a high affinity for nitrate and can cope with low nitrate concentrations. Higher nitrate concentrations increased the relative amount of N2O formed by 0- to 20-cm-depth soils (see Fig. S1 in the supplemental material). Nitrate concentrations above 40 μM are rarely encountered in situ (30, 42, 55). Thus, the complete reduction of nitrate to N2 might occur under most in situ conditions, and the relative emissions of N2O versus N2 might increase when nitrate concentrations are periodically elevated. The rapid increase of N2O production in response to nitrate and the capacity of fen soil to consume supplemental N2O without apparent delay (Fig. 1) suggest that denitrifiers are active in situ.
High denitrification potentials in upper soil layers (0 to 20 cm) are coincident with higher concentrations of nitrate in those layers (42, 55). Rain events likely contribute to the larger amounts of nitrate in surface soils. The percentage of N2O in total N gases formed by fen soils increased with increasing soil depth, a trend that might be due to the limitation of readily available organic carbon in deeper layers in the Schlöppnerbrunnen fen (72). Electron donor limitation can enhance the percentage of N2O in total N gases produced by pure cultures of denitrifiers (53).
Nitrification versus denitrification as possible sources of N2O.
Isotope signatures of the N2O indicate that denitrification is the main source of the N2O emitted from the Schlöppnerbrunnen fen (20). Although denitrification tends to be the dominant source of N2O under water-saturated conditions (44), nitrification likely occurs in the acidic fen when oxic conditions are augmented during dryer periods. Nitrification during dryer and more oxic conditions would theoretically provide additional nitrate for denitrification in anoxic microzones or subsequent to a rain event. In this regard, N2O emissions from the fen increase after rewetting events following periods of drought (19).
Denitrification as an N2O sink.
Wetlands can consume N2O (4). The capacity of wetlands to consume N2O is influenced by environmental factors such as pH and temperature, as well as the composition of the microbial community (6). The capacity of Schlöppnerbrunnen fen soil to consume N2O to subatmospheric levels under anoxic conditions and the periodic occurrence of N2O at subatmospheric concentrations in fen pore water (20) are indirect evidence that N2O consumption occurs in situ. Isotope signatures of N2O from the fen indicate that the upward diffusion of the N2O produced in lower soil layers is subject to reduction to N2 in the upper soil layers (20). Indeed, N2O consumption rates were higher in upper soil layers than in lower soil layers. These collective findings suggest that the Schlöppnerbrunnen fen functions as not only an N2O source but also an N2O sink.
Ecophysiology of fen denitrifiers.
Km values for denitrification ranged from 6 to 19 μM nitrate, indicating that fen denitrifiers had a high affinity for nitrate. Km values were in the same range or lower than those of other soil types (32, 37, 62) and in the range of those of pure cultures that display a high affinity for nitrate (e.g., species of Alcaligenes, Pseudomonas, and Flavobacterium) (2, 41, 64).
Denitrification rates of different fen soil layers were optimal at 34 to 46°C, optimal temperatures that approximate those of many model soil denitrifiers (e.g., Pseudomonas denitrificans, which denitrifies optimally at 38°C) (69). In contrast, the highest denitrifier activity and highest numbers of cultured denitrifiers of different soils occur between 25 and 30°C (50, 51), indicating that denitrifiers in the Schlöppnerbrunnen fen have a temperature optimum that is slightly higher than the optima of denitrifiers from other soils. Enhanced denitrification capacities at temperatures that exceed most in situ conditions are a common phenomenon, and higher rates of denitrification in soils in summer can be attributed to increased soil temperatures (10, 24, 33).
The capacity of fen soil to consume N2O under anoxic conditions was blocked by acetylene (Fig. 1B) and is therefore assumed to be due to the reduction of N2O to N2 by N2O reductase (74). The consumption of N2O by different soil layers was highest between 6 and 30°C, temperatures lower than those for nitrate-dependent denitrification. These contrasting temperature optima suggest that different denitrifier subpopulations in the acidic fen have different temperature and electron acceptor (i.e., nitrate or N2O) preferences. The terminal reaction of the denitrification pathway appears to be more adapted to in situ temperatures than the preceding reactions. The degree to which N2O reductase of soil denitrifiers is inhibited by lower temperatures varies, with effects ranging from no inhibition to almost complete inhibition (12).
Denitrification rates were highest at in situ pH, indicating that Schlöppnerbrunnen fen denitrifiers are well adapted to the moderately acidic fen environment. Denitrification activities of acidic agricultural soils can be highest at in situ pH even though denitrification capacities might be higher in more pH-neutral soils (40), suggesting that soils of different pH values harbor distinct denitrifier communities adapted to in situ pH. Many pure cultures of denitrifiers (e.g., Pseudomonas sp.) have nearly neutral to slightly alkaline pH optima (63). Denitrification also occurred at very low pH by all fen soil layers, whereas only the upper 10-cm soil layer was capable of denitrification under slightly alkaline conditions. Therefore, alkaline conditions appear to be more limiting for fen denitrifiers than acidic conditions, which is consistent with the in situ conditions fen denitrifiers are subjected to. Acidic pH increases the percentage of N2O in total N gases (57, 70). Denitrification by Paracoccus denitrificans yields nitrite and nitrous oxide as transient intermediates at pH 5.5, whereas the amounts of these intermediates are low or not detectable at pH 8.5 (64). Up to 5 μM N2O occurs in the fen pore water (20), indicating that those intermediates occur in situ. The relative percentage of N2O in total N gases formed by Schlöppnerbrunnen fen soil was highest at pHs of 2 to 3 but was similar at in situ pH to values obtained at pH 7, indicating that the N2O reductases of fen denitrifiers are not inhibited by the moderately acidic in situ conditions.
Conclusions.
Schlöppnerbrunnen fen soil produces formate, ethanol, and acetate under anoxic conditions via fermentation (22, 72). Such substrates are utilized by pure cultures of denitrifiers such as Pseudomonas denitrificans, Pseudomonas stutzeri, and Paracoccus denitrificans (38, 61); formate and acetate are detectable in the fen pore water (22, 30, 72); and the augmentation of denitrification in fen soil microcosms by these substrates suggests that fen denitrifiers might form trophic links to fen fermenters. Denitrification optima by fen denitrifiers at moderately acidic pH are dissimilar to those of model denitrifiers (such as those listed above). That the temperature optima of fen denitrifiers are above temperatures usually occurring in situ indicates that the fen denitrifiers are prone to respond to global warming with increased activity. Thus, the source and sink functions of the fen for N2O might be enhanced. These physiological findings and the novel phylogeny of denitrifier community members indicate that the fen contains heretofore unknown denitrifiers that are adapted to in situ conditions and are integrated in the intermediary ecosystem metabolism (i.e., processes that link input and output) of the fen (13).
Supplementary Material
Acknowledgments
Support for this study was provided by the Deutsche Forschungsgemeinschaft (DFG DR 310/3-3 and DFG HO 4020/2-2) and the University of Bayreuth.
Footnotes
Published ahead of print on 18 December 2009.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Balch, W. E., G. E. Fox, L. J. Magrum, C. R. Woese, and R. S. Wolfe. 1979. Methanogens: reevaluation of a unique biological group. Microbiol. Rev. 43:260-296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Betlach, M. R., and J. M. Tiedje. 1981. Kinetic explanation for accumulation of nitrite, nitric oxide, and nitrous oxide during bacterial denitrification. Appl. Environ. Microbiol. 42:1074-1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blackmer, A. M., and J. M. Bremner. 1978. Inhibitory effect of nitrate on reduction of N2O to N2 by soil microorganisms. Soil Biol. Biochem. 10:187-191. [Google Scholar]
- 4.Blackmer, A. M., and J. M. Bremner. 1976. Potential of soil as a sink for atmospheric nitrous oxide. Geophys. Res. Lett. 3:739-742. [Google Scholar]
- 5.Bremner, J. M. 1997. Sources of nitrous oxide in soils. Nutr. Cycling Agroecosyst. 49:7-16. [Google Scholar]
- 6.Cavigelli, M. A., and G. P. Robertson. 2000. The functional significance of denitrifier community composition in a terrestrial ecosystem. Ecology 81:1402-1414. [Google Scholar]
- 7.Conrad, R. 1996. Soil microorganisms as controllers of atmospheric trace gases (H2, CO, CH4, OCS, N2O, and NO). Microbiol. Rev. 60:609-640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Daniel, S. L., T. Hsu, S. I. Dean, and H. L. Drake. 1990. Characterization of the H2- and CO-dependent chemolithotrophic potentials of the acetogens Clostridium thermoaceticum and Acetogenium kivui. J. Bacteriol. 172:4464-4471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Davidson, E. A. 1992. Sources of nitric oxide and nitrous oxide following wetting of dry soil. Soil Sci. Soc. Am. J. 56:95-102. [Google Scholar]
- 10.DeKlein, C. A. M., and R. S. P. VanLogtestijn. 1996. Denitrification in grassland soils in The Netherlands in relation to irrigation, N-application rate, soil water content and soil temperature. Soil Biol. Biochem. 28:231-237. [Google Scholar]
- 11.DeMan, J. C. 1975. The probability of most probable numbers. Eur. J. Appl. Microbiol. 1:67-78. [Google Scholar]
- 12.Dorsch, P., and L. R. Bakken. 2004. Low-temperature response of denitrification: comparison of soils. Eurasian Soil Sci. 37:S102-S106. [Google Scholar]
- 13.Drake, H. L., M. A. Horn, and P. K. Wüst. 2009. Intermediary ecosystem metabolism as a main driver of methanogenesis in acidic wetland soil. Environ. Microbiol. Rep. 1:307-318. [DOI] [PubMed] [Google Scholar]
- 14.Egert, M., and M. W. Friedrich. 2003. Formation of pseudo-terminal restriction fragments, a PCR-related bias affecting terminal restriction fragment length polymorphism analysis of microbial community structure. Appl. Environ. Microbiol. 69:2555-2562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Enwall, K., L. Philippot, and S. Hallin. 2005. Activity and composition of the denitrifying bacterial community respond differently to long-term fertilization. Appl. Environ. Microbiol. 71:8335-8343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Foken, T. 2003. Lufthygienische-bioklimatische Kennzeichnung des oberen Egertales Bayreuther Forum Ökologie 100:1-69. [Google Scholar]
- 17.Forster, P., V. Ramaswamy, P. Artaxo, T. Berntsen, R. Betts, D. W. Fahey, J. Haywood, J. Lean, D. C. Lowe, G. Myhre, J. Nganga, R. Prinn, G. Raga, M. Schulz, and R. VanDorland. 2007. Changes in atmospheric constituents and in radiative forcing, p 129-234. In S. Solomon, D. Qin, M. Manning, Z. Chen, M. Marquis, K. B. Averyt, M. Tignor, and H. L. Miller (ed.), Climate change 2007: the physical science basis. Contribution of working group I to the fourth assessment report of the intergovernmental panel on climate change. Cambridge University Press, Cambridge, United Kingdom.
- 18.Gaskell, J. F., A. M. Blackmer, and J. M. Bremner. 1981. Comparison of effects of nitrate, nitrite, and nitric oxide on reduction of nitrous oxide to dinitrogen by soil-microorganisms. Soil Sci. Soc. Am. J. 45:1124-1127. [Google Scholar]
- 19.Goldberg, S. D., K. H. Knorr, C. Blodau, G. Lischeid, and G. Gebauer. 2010. Impact of altering the water table height of an acidic fen on N2O and NO fluxes and soil concentrations. Glob. Change Biol. 16:220-233.
- 20.Goldberg, S. D., K. H. Knorr, and G. Gebauer. 2008. N2O concentration and isotope signature along profiles provide deeper insight into the fate of N2O in soils. Isotopes Environ. Health Stud. 44:377-391. [DOI] [PubMed] [Google Scholar]
- 21.Griffiths, R. I., A. S. Whiteley, A. G. O'Donnell, and M. J. Bailey. 2000. Rapid method for coextraction of DNA and RNA from natural environments for analysis of ribosomal DNA- and rRNA-based microbial community composition. Appl. Environ. Microbiol. 66:5488-5491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hamberger, A., M. A. Horn, M. G. Dumont, J. C. Murrell, and H. L. Drake. 2008. Anaerobic consumers of monosaccharides in a moderately acidic fen. Appl. Environ. Microbiol. 74:3112-3120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Harrigan, W. F., and M. E. Mc Cance. 1966. Laboratory methods in microbiology. Academic Press, London, United Kingdom.
- 24.Holtan-Hartwig, L., P. Dorsch, and L. R. Bakken. 2002. Low temperature control of soil denitrifying communities: Kinetics of N2O production and reduction. Soil Biol. Biochem. 34:1797-1806. [Google Scholar]
- 25.Horn, M. A., H. L. Drake, and A. Schramm. 2006. Nitrous oxide reductase genes (nosZ) of denitrifying microbial populations in soil and the earthworm gut are phylogenetically similar. Appl. Environ. Microbiol. 72:1019-1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Karsten, G., and H. L. Drake. 1997. Denitrifying bacteria in the earthworm gastrointestinal tract and in vivo emission of nitrous oxide (N2O) by earthworms and in vivo emission of dinitrogen by earthworms via denitrifying bacteria in the gut. Appl. Environ. Microbiol. 63:1878-1882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kesik, M., S. Blagodatsky, H. Papen, and K. Butterbach-Bahl. 2006. Effect of pH, temperature and substrate on N2O, NO and CO2 production by Alcaligenes faecalis p. J. Appl. Microbiol. 101:655-667. [DOI] [PubMed] [Google Scholar]
- 28.Kuhner, C. H., H. L. Drake, E. Alm, and L. Raskin. 1996. Methane production and oxidation by soils from acidic forest wetlands of east-central Germany, p. 304. Abstr. 96th Gen. Meet. Am. Soc. Microbiol. American Society for Microbiology, Washington, DC.
- 29.Kumar, S., M. Nei, J. Dudley, and K. Tamura. 2008. MEGA: a biologist-centric software for evolutionary analysis of DNA and protein sequences. Brief. Bioinform. 9:299-306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Küsel, K., and C. Alewell. 2004. Riparian zones in a forested catchment: hot spots for microbial reductive processes, p. 377-395. In E. Matzner (ed.), Biogeochemistry of forested catchments in a changing environment: a German case study. Springer Verlag, Berlin, Germany.
- 31.Küsel, K., and H. L. Drake. 1995. Effects of environmental parameters on the formation and turnover of acetate by forest soils. Appl. Environ. Microbiol. 61:3667-3675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Laverman, A. M., P. VanCappellen, D. VanRotterdam-Los, C. Pallud, and J. Abell. 2006. Potential rates and pathways of microbial nitrate reduction in coastal sediments. FEMS Microbiol. Ecol. 58:179-192. [DOI] [PubMed] [Google Scholar]
- 33.Malhi, S. S., W. B. Mcgill, and M. Nyborg. 1990. Nitrate losses in soils: effect of temperature, moisture and substrate concentration. Soil Biol. Biochem. 22:733-737. [Google Scholar]
- 34.Matthies, C., A. Freiberger, and H. L. Drake. 1993. Fumarate dissimilation and differential reductant flow by Clostridium formicoaceticum and Clostridium aceticum. Arch. Microbiol. 160:273-278. [Google Scholar]
- 35.Messing, J. 1983. New M13 vectors for cloning. Methods Enzymol. 101:20-78. [DOI] [PubMed] [Google Scholar]
- 36.Miller, M. N., B. J. Zebarth, C. E. Dandie, D. L. Burton, C. Goyer, and J. T. Trevors. 2008. Crop residue influence on denitrification, N2O emissions and denitrifier community abundance in soil. Soil Biol. Biochem. 40:2553-2562. [Google Scholar]
- 37.Murray, R. E., L. L. Parsons, and M. S. Smith. 1989. Kinetics of nitrate utilization by mixed populations of denitrifying bacteria. Appl. Environ. Microbiol. 55:717-721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nishimura, Y., T. Kamihara, and S. Fukui. 1979. Nitrite reduction with formate in Pseudomonas denitrificans ATCC 13867. Biochem. Biophys. Res. Commun. 87:140-145. [DOI] [PubMed] [Google Scholar]
- 39.Palmer, K., H. L. Drake, and M. A. Horn. 2009. Genome derived criteria for assigning environmental narG and nosZ sequences to operational taxonomic units of nitrate reducers. Appl. Environ. Microbiol. 75:5170-5174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Parkin, T. B., A. J. Sexstone, and J. M. Tiedje. 1985. Adaptation of denitrifying populations to low soil pH. Appl. Environ. Microbiol. 49:1053-1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Parsonage, D., A. J. Greenfield, and S. J. Ferguson. 1985. The high affinity of Paracoccus denitrificans cells for nitrate as an electron acceptor: analysis of possible mechanisms of nitrate and nitrite movement across the plasma membrane and the basis for inhibition by added nitrite of oxidase activity in permeabilized cells. Biochim. Biophys. Acta 807:81-95. [Google Scholar]
- 42.Paul, S., K. Küsel, and C. Alewell. 2006. Reduction processes in forest wetlands: tracking down heterogeneity of source/sink functions with a combination of methods. Soil Biol. Biochem. 38:1028-1039. [Google Scholar]
- 43.Philippot, L., S. Piutti, F. Martin-Laurent, S. Hallet, and J. C. Germon. 2002. Molecular analysis of the nitrate-reducing community from unplanted and maize-planted soils. Appl. Environ. Microbiol. 68:6121-6128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pihlatie, M., E. Syväsalo, A. Simojoki, M. Esala, and K. Regina. 2004. Contribution of nitrification and denitrification to N2O production in peat, clay and loamy sand soils under different soil moisture conditions. Nutr. Cycling Agroecosyst. 70:135-141. [Google Scholar]
- 45.Pinay, G., B. Gumiero, E. Tabacchi, O. Gimenez, A. M. Tabacchi-Planty, M. M. Hefting, T. P. Burt, V. A. Black, C. Nilsson, V. Iordache, F. Bureau, L. Vought, G. E. Petts, and H. Decamps. 2007. Patterns of denitrification rates in European alluvial soils under various hydrological regimes. Freshw. Biol. 52:252-266. [Google Scholar]
- 46.Ratkowsky, D. A., R. K. Lowry, T. A. McMeekin, A. N. Stokes, and R. E. Chandler. 1983. Model for bacterial culture growth rate throughout the entire biokinetic temperature range. J. Bacteriol. 154:1222-1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Reiche, M., G. Torburg, and K. Küsel. 2008. Competition of Fe(III) reduction and methanogenesis in an acidic fen. FEMS Microbiol. Ecol. 65:88-101. [DOI] [PubMed] [Google Scholar]
- 48.Rich, J. J., R. S. Heichen, P. J. Bottomley, K. Cromack, and D. D. Myrold. 2003. Community composition and functioning of denitrifying bacteria from adjacent meadow and forest soils. Appl. Environ. Microbiol. 69:5974-5982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Robertson, G. P., and J. M. Tiedje. 1987. Nitrous oxide sources in aerobic soils: nitrification, denitrification and other biological processes. Soil Biol. Biochem. 19:187-193. [Google Scholar]
- 50.Saad, O. A. L. O., and R. Conrad. 1993. Adaptation to temperature of nitric oxide-producing nitrate-reducing bacterial populations in soil. Syst. Appl. Microbiol. 16:120-125. [Google Scholar]
- 51.Saad, O. A. L. O., and R. Conrad. 1993. Temperature-dependence of nitrification, denitrification, and turnover of nitric-oxide in different soils. Biol. Fertil. Soils 15:21-27. [Google Scholar]
- 52.Saitou, N., and M. Nei. 1987. The neighbor-joining method—a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4:406-425. [DOI] [PubMed] [Google Scholar]
- 53.Schalk-Otte, S., R. J. Seviour, J. G. Kuenen, and M. S. M. Jetten. 2000. Nitrous oxide (N2O) production by Alcaligenes faecalis during feast and famine regimes. Water Res. 34:2080-2088. [Google Scholar]
- 54.Schloss, P. D., and J. Handelsman. 2005. Introducing DOTUR, a computer program for defining operational taxonomic units and estimating species richness. Appl. Environ. Microbiol. 71:1501-1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Schmalenberger, A., H. L. Drake, and K. Küsel. 2007. High unique diversity of sulfate-reducing prokaryotes characterized in a depth gradient in an acidic fen. Environ. Microbiol. 9:1317-1328. [DOI] [PubMed] [Google Scholar]
- 56.Segel, I. H. 1993. Enzyme kinetics: behavior and analysis of rapid equilibrium and steady-state enzyme systems. John Wiley & Sons, New York, NY.
- 57.Simek, M., and J. E. Cooper. 2002. The influence of soil pH on denitrification: progress towards the understanding of this interaction over the last 50 years. Eur. J. Soil Sci. 53:345-354. [Google Scholar]
- 58.Simek, M., L. Jisova, and D. W. Hopkins. 2002. What is the so-called optimum pH for denitrification in soil? Soil Biol. Biochem. 34:1227-1234. [Google Scholar]
- 59.Smith, M. S. 1983. Nitrous oxide production by Escherichia coli is correlated with nitrate reductase activity. Appl. Environ. Microbiol. 45:1545-1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Stolz, J. F., and P. Basu. 2002. Evolution of nitrate reductase: molecular and structural variations on a common function. Eur. J. Chem. Biol. 3:198-206. [DOI] [PubMed] [Google Scholar]
- 61.Strohm, T. O., B. Griffin, W. G. Zumft, and B. Schink. 2007. Growth yields in bacterial denitrification and nitrate ammonification. Appl. Environ. Microbiol. 73:1420-1424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Strong, D. T., and I. R. P. Fillery. 2002. Denitrification response to nitrate concentrations in sandy soils. Soil Biol. Biochem. 34:945-954. [Google Scholar]
- 63.Thomas, K. L., D. Lloyd, and L. Boddy. 1994. Effects of oxygen, pH and nitrate concentration on denitrification by Pseudomonas species. FEMS Microbiol. Lett. 118:181-186. [DOI] [PubMed] [Google Scholar]
- 64.Thomsen, J. K., T. Geest, and R. P. Cox. 1994. Mass spectrometric studies of the effect of pH on the accumulation of intermediates in denitrification by Paracoccus denitrificans. Appl. Environ. Microbiol. 60:536-541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Tiedje, J. M. 1988. Ecology of denitrification and dissimilatory nitrate reduction to ammonium, p. 179-244. In A. J. B. Zehnder (ed.), Biology of anaerobic microorganisms. John Wiley and Sons, New York, NY.
- 66.VanBeek, C. L., E. W. J. Hummelink, G. L. Velthof, and O. Oenema. 2004. Denitrification rates in relation to groundwater level in a peat soil under grassland. Biol. Fertil. Soils 39:329-336. [Google Scholar]
- 67.VanBremen, N. 1995. How Sphagnum bogs down other plants. Trends Ecol. Evol. 10:270-275. [DOI] [PubMed] [Google Scholar]
- 68.VanCleemput, O. 1998. Subsoils: chemo- and biological denitrification, N2O and N2 emissions. Nutr. Cycling Agroecosyst. 52:187-194. [Google Scholar]
- 69.Wang, J. H., B. C. Baltzis, and G. A. Lewandowski. 1995. Fundamental denitrification kinetic-studies with Pseudomonas denitrificans. Biotechnol. Bioeng. 47:26-41. [DOI] [PubMed] [Google Scholar]
- 70.Weier, K. L., and J. W. Gilliam. 1986. Effect of acidity on denitrification and nitrous oxide evolution from Atlantic coastal plain soils. Soil Sci. Soc. Am. J. 50:1202-1205. [Google Scholar]
- 71.Well, R., J. Augustin, J. Davis, S. Griffith, K. Meyer, and D. Myrold. 2001. Production and transport of denitrification gases in shallow ground water. Nutr. Cycling Agroecosyst. 60:65-75. [Google Scholar]
- 72.Wüst, P. K., M. A. Horn, and H. L. Drake. 2009. Trophic links between fermenters and methanogens in a moderately acidic fen soil. Environ. Microbiol. 11:1395-1409. [DOI] [PubMed] [Google Scholar]
- 73.Yamulki, S., R. M. Harrison, K. W. T. Goulding, and C. P. Webster. 1997. N2O, NO and NO2 fluxes from a grassland: effect of soil pH. Soil Biol. Biochem. 29:1199-1208. [Google Scholar]
- 74.Yoshinari, T., and R. Knowles. 1976. Acetylene inhibition of nitrous oxide reduction by denitrifying bacteria. Biochem. Biophys. Res. Commun. 69:705-710. [DOI] [PubMed] [Google Scholar]
- 75.Zumft, W. G. 1997. Cell biology and molecular basis of denitrification. Microbiol. Mol. Biol. Rev. 61:533-615. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.