Abstract
Researchers agree that climate change factors such as rising atmospheric [CO2] and warming will likely interact to modify ecosystem properties and processes. However, the response of the microbial communities that regulate ecosystem processes is less predictable. We measured the direct and interactive effects of climatic change on soil fungal and bacterial communities (abundance and composition) in a multifactor climate change experiment that exposed a constructed old-field ecosystem to different atmospheric CO2 concentration (ambient, +300 ppm), temperature (ambient, +3°C), and precipitation (wet and dry) might interact to alter soil bacterial and fungal abundance and community structure in an old-field ecosystem. We found that (i) fungal abundance increased in warmed treatments; (ii) bacterial abundance increased in warmed plots with elevated atmospheric [CO2] but decreased in warmed plots under ambient atmospheric [CO2]; (iii) the phylogenetic distribution of bacterial and fungal clones and their relative abundance varied among treatments, as indicated by changes in 16S rRNA and 28S rRNA genes; (iv) changes in precipitation altered the relative abundance of Proteobacteria and Acidobacteria, where Acidobacteria decreased with a concomitant increase in the Proteobacteria in wet relative to dry treatments; and (v) changes in precipitation altered fungal community composition, primarily through lineage specific changes within a recently discovered group known as soil clone group I. Taken together, our results indicate that climate change drivers and their interactions may cause changes in bacterial and fungal overall abundance; however, changes in precipitation tended to have a much greater effect on the community composition. These results illustrate the potential for complex community changes in terrestrial ecosystems under climate change scenarios that alter multiple factors simultaneously.
Soil microbial communities are responsible for the cycling of carbon (C) and nutrients in ecosystems, and their activities are regulated by biotic and abiotic factors such as the quantity and quality of litter inputs, temperature, and moisture. Atmospheric and climatic changes will impact both abiotic and biotic drivers in ecosystems and the response of ecosystems to these changes. Feedbacks from ecosystem to the atmosphere may also be regulated by soil microbial communities (3). Although microbial communities regulate important ecosystem processes, it is often unclear how the abundance and composition of microbial communities correlate with climatic perturbations and interact to effect ecosystem processes. As such, much of the ecosystem climate change research conducted to date has focused on macroscale responses to climatic change such as changes in plant growth (43, 44), plant community composition (2, 37), and coarse scale soil processes (14, 18, 21, 26), many of which may also indirectly interact to effect microbial processes. Studies that have addressed the role of microbial communities and processes have most often targeted gross parameters, such as microbial biomass, enzymatic activity, or basic microbial community profiles in response to single climate change factors (22, 28, 29, 33, 61, 63).
Climate change factors such as atmospheric CO2 concentrations, warming, and altered precipitation regimes can potentially have both direct and indirect impacts on soil microbial communities. However, the direction and magnitude of these responses is uncertain. For example, the response of soil microbial communities to changes in atmospheric CO2 concentrations can be positive or negative, and consistent overall trends between sites and studies have not been observed (1, 28, 34-36). Further, depending on what limits ecosystem productivity, precipitation and soil moisture changes may increase or decrease the ratio of bacteria and fungi, as well as shift their community composition (8, 50, 58). Increasing temperatures can increase in microbial activity, processing, and turnover, causing the microbial community to shift in favor of representatives adapted to higher temperatures and faster growth rates (7, 46, 60, 64, 65). Atmospheric and climatic changes are happening in concert with one another so that ecosystems are experiencing higher levels of atmospheric CO2, warming, and changes in precipitation regimes simultaneously. Although the many single factor climate change studies described above have enabled a better understanding of how microbial communities may respond to any one factor, understanding how multiple climate change factors interact with each other to influence microbial community responses is poorly understood. For example, elevated atmospheric [CO2] and precipitation changes might increase soil moisture in an ecosystem, but this increase may be counteracted by warming (10). Similarly, warming may increase microbial activity in an ecosystem, but this increase may be eliminated if changes in precipitation lead to a drier soil condition or reduced litter quantity, quality, and turnover. Such interactive effects of climate factors in a multifactorial context have been less commonly studied even in plant communities (45), and detailed studies are rarer still in soil microbial communities (25). Clearly, understanding how microbial communities will respond to these atmospheric and climate change drivers is important to make accurate predications of how ecosystems may respond to future climate scenarios.
To address how multiple climate change drivers will interact to shape soil microbial communities, we took advantage of a multifactor climatic change experiment that manipulated atmospheric CO2 (+300 ppm, ambient), warming (+3°C, ambient) and precipitation (wet and dry) in a constructed old-field ecosystem that had been ongoing for 3.5 years at the time of sampling. Previous work on this project has demonstrated direct and interactive effects of the treatments on plant community composition and biomass (15, 30), soil respiration (56), microbial activity (30), nitrogen fixation (21), and soil carbon stocks (20). These results led us to investigations of how the soil bacterial and fungal communities, important regulators of some of these processes, were responding using culture-independent molecular approaches. Our research addresses two overarching questions. (i) Do climatic change factors and their interactions alter bacterial and fungal abundance and diversity? (ii) Do climatic change factors and their interactions alter bacterial or fungal community composition?
MATERIALS AND METHODS
Site description and experimental design.
The Old-Field Community Climate and Atmospheric Manipulation (OCCAM) was established in 2002 on the National Ecological Research Park, Oak Ridge, TN (35°54′N, 84°20′W). The site was in agricultural use until 1943 (10, 15, 20, 21, 30, 45). The soils are of alluvial origin and classified as Captina silt loam (12). The mean January minimum temperature is −2.7°C, and the mean July maximum temperature is 31.2°C, with an annual mean precipitation of 1,322 mm, uniformly distributed throughout the year (56). The experimental setup was described previously in Wan et al. (56) and Garten et al. (21). Briefly, three blocks with five plots each were trenched to 75 cm and lined with polyvinyl chloride film and foam insulation to create 15 4-m-diameter plots. Existing vegetation and most of the seed bank was removed (15), and in July 2002 and April 2003 seven oldfield species that are common to the southeastern United States, were planted in a grid: Plantago lanceolata L., a herbaceous, annual C3 photosynthesis system dicot; Andropogon virginicus L., a cespitose C4 photosynthesis system bunchgrass; Festuca pratense L. syn F. elatior L., a C3 photosynthesis system bunchgrass; Dactylis glomerata L., a C3 photosynthesis system bunchgrass; Trifolium pratense L., a C3 photosynthesis system legume; Solidago canadensis, a perennial forb; and Lespedeza cuneata (Dum. Cours.) G. Don., a C3 photosynthesis system perennial, N2-fixing shrub. Open-top chambers were installed on each the plot in March 2003, and treatments were assigned randomly within each block and then maintained either elevated (+3°C) or ambient temperatures and elevated (+300 ppm) or ambient levels of CO2 (56). In addition, plots were trenched down the center to maintain a wet side and a dry side based in the annual precipitation of 25 mm week−1 at the site. To exclude incoming precipitation, a clear plastic canopy was placed over each open-top chamber, and rainwater was collected and added back to the plots weekly at 25 mm week−1, hereafter the wet (W) side of the plot, and 2 mm wk−1 to represent drought conditions, hereafter the dry (D) side of the plot (10, 20). The experiment was a completely randomized, split-plot block design with three blocks for each of four treatments—ambient CO2, ambient temperature (aCO2aTemp); elevated CO2, ambient temperature (eCO2aTemp); ambient CO2, elevated temperature (aCO2eTemp); and elevated CO2, elevated temperature (eCO2eTemp)—with their respective wet and dry sides.
Soil DNA extraction and bacterial and fungal ribosomal DNA quantitative PCR.
Soil samples (0 to 15 cm) were collected with a 5-cm-diameter hammer core in October 2006.
Three soil cores were collected from the dry and wet side of each plot. Samples from each plot were pooled and homogenized in the field. Subsamples were immediately frozen using liquid N2 and stored at −80°C until analysis. Soil DNA was extracted from 0.25 g of each samples by using the Power Soil DNA isolation kit (MoBio, Carlsbad, CA) according to the manufacturer's instructions. Quantitative PCR reactions were performed in 96-well plates on a iCycler iQ Multi-Color real-time PCR detection system (Bio-Rad Laboratories, Hercules, CA). Quantitative PCR reaction mixtures included 15 μl of IQ SYBR green Supermix (Bio-Rad, Laboratories, Hercules, CA), 3.75 μl of a 10-mg ml−1 mixture of bovine serum albumin (BSA), 0.5 μl of a 5-pmol μl−1 mixture of each primer, 1 μl of 1:10 diluted soil DNA solution, and H2O to a final volume of 30 μl. The primers used for 16S rRNA bacterial genes were EUB338 (32) and Eub518 (42) at an annealing temperature of 55°C and for fungal rRNA genes were nu-SSU-1196F and nu-SSU-1536R (6) at an annealing temperature of 53°C. The reaction was carried out under the following conditions: 95°C for 3.25 min, followed by 40 cycles of 95°C for 15 s, annealing temperature for 30 s, and 72°C for 30 s, followed in turn by 95°C for 1 min and then 80 cycles of 55.0°C for 10 s. Standard curves were prepared using known amounts of DNA extracted from pure cultures carried out concomitantly with experimental samples and exhibited a linear relationship between the log of the rRNA copy number and the calculated threshold (CT) value (R2 > 0.99). Amplification efficiencies varied but were >1.8, which is consistent with previously reported values (17). Differences in the quantitative PCR data were tested by using a split-plot analysis of variance with a combined random and fixed effects model, including the Kenward-Rogers adjustment for degrees of freedom (PROC MIXED; SAS Systems, Cary, NC).
Bacterial, acidobacterial, and fungal rRNA gene PCR amplification and cloning.
Bacterial 16S rRNA genes were amplified by using the universal 16S primers 8F and 1492R (32). The reaction mixture included 2 μl of 10× PCR buffer, 1.6 μl of 25 mM MgCl2, 2 μl of a 10-mg ml−1 BSA mixture, 1.6 μl of 25 mM deoxynucleoside triphosphates (dNTPs), 1 μl of a 20-pmol μl−1 concentration of each primer, 1 μl of 1:10-diluted soil DNA solution, 0.3 μl of Taq polymerase, and H2O to a final volume of 20 μl. The reaction was carried out under the following conditions: 94°C for 5 min; followed by 30 cycles of 94°C for 1 min, 55°C for 1 min, and 72°C for 2 min; and finally 72°C for 7 min in GeneAmp PCR system 9700 thermocyclers (PE Applied Biosystems, Norwalk, CT). Acidobacterial rRNA genes were amplified by using ACD31F (5) and 1492R using identical cycling conditions to the bacterial PCR amplifications. Fungal rRNA gene regions were amplified using ITS9F (13) and LSU1221R (49) primers that amplify a region including the ITS1, 5.8S rRNA, ITS2, and the 5′ end of the 28S rRNA gene. The reaction mixture contained 2 μl of 10× PCR buffer, 2.2 μl of 2.75 mM MgCl2, 2.5 μl of a 10-mg/ml concentration of BSA, 1 μl of 10-pmol μl−1 concentration of each primer, 1.6 μl of 25 mM dNTPs, 0.6 μl of Taq polymerase, and 1 μl of 1:10-diluted soil DNA solution and H2O to a final volume of 20 μl. The reaction was carried out under the following conditions: 95°C for 2 min; followed by 25 cycles of 94°C for 45 s, 57.5°C for 50 s, and 72°C for 1 min 45 s (+1 s each cycle); and finally 72°C for 5 min. Positive and negative controls were used in all PCRs. Electrophoresis was performed in 1% agarose in Tris-acetate EDTA buffer to confirm successful amplification, and PCR products were extracted from the agarose gel by using a gel purification kit (Qiagen, Valencia, CA). PCR products from the wet and dry sides of the three blocks for each treatment were then pooled, and the resulting eight PCR mixtures were cloned. A TOPO-TA PCR 2.1 cloning kit (Invitrogen, Carlsbad, CA) was used according to the manufacturer's recommendations, except that the salt solutions were omitted from the reaction and ligation was extended to 30 min. White Escherichia coli colonies were picked, and 16S cloned rRNA PCR products were lifted by using M13 primers (M13F, 5′-GTA AAA CGA CGG CCA G-3′; M13R, 5′-CAG GAA ACA GCT ATG AC-3′) in a second PCR under standard conditions. M13 PCR amplification was screened by using 1.5% agarose in Tris-acetate EDTA buffer electrophoresis and 5 μl of product. Amplifications were then purified with Millipore multiscreen HTS PCR filter plates (Millipore Corp., Bellerica, MA).
Sequencing, taxonomic assignment, and phylogenetic analysis.
BigDye Terminator v3.1 (Applied Biosystems, Foster City, CA) cycle sequencing reactions were performed using TA primers (48) for bacterial and acidobacterial libraries and LROF (55) for fungal libraries using the following master mix: 1 μl of 5× sequencing buffer, 1.7 μl of H2O, 1 μl of BigDye, 0.3 μl of 20-pmol μl−1 mixture of TA primer, and 1 μl of DNA template. Products were precipitated in ethanol and resuspended in 10 μl of Hi-Di Formamide (Applied Biosystems) and run on an ABI Prism 3730 DNA analyzer (Applied Biosystems). Sequences were edited in Sequencher program version 4.7 (Gene Codes Corp., Ann Arbor, MI), where the remnants of the vector and the low-quality ends of the sequences were removed. All sequences used in the analysis were partial sequences of a length of 400 to 500 bp. Bacterial and acidobacterial clones were taxonomically assigned by using the Ribosomal Database Project II website (http://rdp.cme.msu.edu/) (40). Fungal clone analyses were performed by using NCBI BLAST database (http://www.ncbi.nlm.nih.gov/BLAST/Blast.cgi) (59) using the PLAN system (24) to sort and manually classify sequences according to top identified matches. Sequences were aligned by CLUSTAL W version 1.4 (53) in BioEdit package (23). Phylogenetic trees were generated by using neighbor-joining method in the BioEdit package using the following sequences as outgroups: Methanospirillum hungatei (M60880) for bacterial libraries, Rhodobacter capsulatus (D16427) for acidobacterial libraries, and Thamnura ruffoi (AJ251734) for fungal libraries. Bootstrap analysis was performed with 100 replications. Edited sequences were submitted to GenBank (accession numbers GU374349 to GU376452). Rooted phylogenetic tree created with BioEdit were analyzed using the Unifrac interface which provides a suite of tools to compare microbial communities. Unifrac significance tests the differences among treatments in terms of the branch which is unique to each treatment. A P-test uses the phylogenetic tree to test whether two environments are significantly different using parsimony (41). Unifrac metrics were used in jackknife environment clustering which performs statistical resampling to test the confidence of clusters, and principal coordinate analyses (PCoA) were used to find clusters and the most important axes of variation among samples. Unifrac tests were performed using 1,000 permutations and calculated with the Unifrac web application (http://bmf2.colorado.edu/unifrac) (39). Unifrac and P-test significance values were corrected by using the Bonferroni correction for multiple comparisons. Diversity indexes and rarefaction curves were calculated by using the DOTUR program (51), and operational taxonomic units (OTUs) were defined at a distance level of 3% for bacterial and acidobacterial sequences and 1% for fungal sequences.
RESULTS
Treatment effects on bacterial and fungal abundance and community phylogenetic composition metrics.
Fungal abundance increase with elevated temperature, as measured by total copies of rRNA genes using quantitative PCR (P ≤ 0.09). Bacterial abundance decreased under ambient CO2, and elevated temperature decreased abundance. However, under elevated CO2 and elevated temperature, an increased bacterial abundance was observed (CO2 × temperature; P < 0.06) (see Fig. SA1 in the supplemental material and Table S1 in the supplemental material). The relative fungus/bacterium ratio was not significantly different among treatments, as measured by gene copy numbers, but they were within the range previously reported for similar soils (11, 17).
For the bacterial libraries, a total of 665 rRNA gene sequences were used in the analysis (349 clones for wet plots and 316 clones for dry plots) representing one clone library per treatment (about 70 to 90 clones per library). For the fungal libraries, a total of 291 clones were used in the analysis for the wet treatments and 310 clones for the dry treatments. In the case of acidobacterial 16S rRNA sequences, 195 clones for the wet plots and 160 clones for the dry plots were used in the analysis (see Table S2 in the supplemental material). Rarefaction curves for fungal libraries from wet plots reached saturation, and libraries from dry plots started to approach saturation (data not shown). For acidobacterial libraries a similar pattern was observed but was less marked than with the fungal libraries. Not surprisingly, bacterial rarefaction curves did not reach saturation, and a more exhaustive screening of clones would be needed. Although the recovered phylotypes reflect the major trends in the libraries, a more extensive clone screening would have been ideal. Although full sampling coverage of the diversity within the bacterial library samples was not achieved with the number of clones sequenced, clone distribution analysis performed in four individual 16S bacterial rRNA libraries from aCO2aTemp samples (technical replicates) revealed that the phylogenetic distribution and diversity metrics were consistent and reproducible between replicate libraries (see Table S2 in the supplemental material). In addition, because we were not able to saturate sampling, we restrict our discussion here to trends in high-level phylogenetic groups, rather than individual OTUs, which are likely to be undersampled.
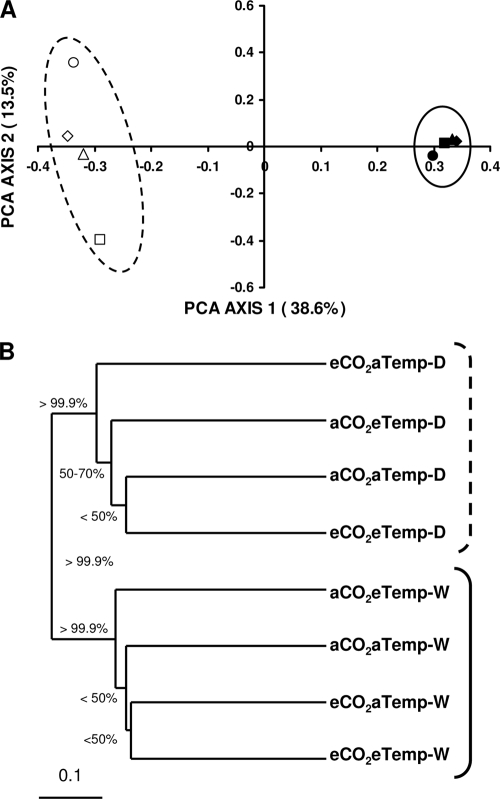
We detected consistent and significant changes among our treatments in the 16S bacterial and acidobacterial rRNA gene, and 28S fungal rRNA gene libraries composition using Unifrac tests, P-tests, Jackknife environment clusters, and PoCA. Using Unifrac metrics, bacterial communities were significantly different under wet relative to dry treatments (Unifrac and P-test significance, P ≤ 0.028). Similar to the bacterial response, the fungal communities under dry and wet treatments also differed significantly (P ≤ 0.028).
The precipitation treatments (dry and wet) resulted in distinct clusters of bacterial, acidobacterial, and fungal clone libraries using Unifrac cluster analysis (Fig. 1). These results support our conclusions based on P-test and Unifrac analyses. Bacterial and acidobacterial sequences exhibited the highest level of Jackknife support for the distinct precipitation (dry and wet) clustering (>99.95). Unifrac PoCA analysis also supported these results, where PoCA axis 1 showed 39, 30, and 33% of the variation and PoCA axis 2 showed 14, 17, and 21% of the variation in bacterial, acidobacterial, and fungal communities, respectively (Fig. 1). These results also are supported by the observed changes in representative abundance in the major clades within the fungal data set discussed below. Overall, diversity metrics did not differ within or among treatments according to Chao1, Shannon, and Simpson metrics calculated by using the DOTUR program for bacterial or acidobacterial libraries, but did show changes within fungal libraries and indicated the wet soils were much less diverse than dry soils (see Table S1 in the supplemental material). A similar clustering of communities corresponding to precipitation treatment was observed for the fungal and acidobacterial community (Fig. 1, only patterns for bacterial community are presented for clarity since fungal and acidobacterial exhibited similar patterns).
FIG. 1.
PCoA analysis based on Unifrac of clone libraries (A) and UniFrac cluster analysis (B) for bacterial soil communities. Acidobacterial and fungal soil communities exhibited similar patterns (data not shown). Unifrac Jackknife supports for each node of the Unifrac cluster analysis are indicated (1,000 permutations). White and black symbols represent dry and wet plots, respectively. Dashed and solid lines were drawn to highlight dry and wet treatments, respectively. Diamonds represent aCO2aTemp treatments, squares represent eCO2aTemp treatments, circles represent aCO2eTemp treatments, and triangles represent eCO2eTemp treatments.
Climate change factor effects on specific bacterial or fungal phylogenetic groups.
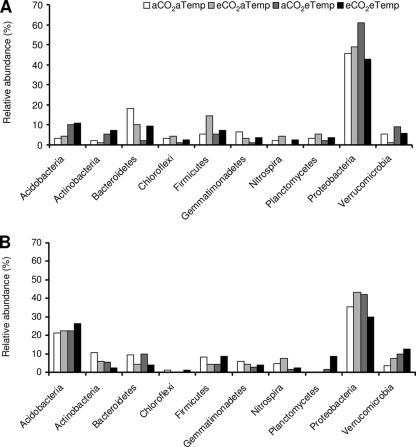
The bacterial 16S rRNA clone libraries revealed a great diversity of organisms present and a phylum-level distribution similar to previously reported soil 16S rRNA clone libraries (27). We taxonomically grouped the clone libraries of the PCR-amplified 16S bacterial rRNA genes according to RDP (Fig. 2). The majority of the sequences were related to the Proteobacteria phylum, followed by sequences related to Firmicutes, Bacterioidetes, Actinobacteria, Acidobacteria, and Planctomycetes. Minor components of the library were sequences related to the Verrucomicrobia, Chloroflexi, and Nitrospirae phyla. The relative abundance of certain groups within the bacterial community responded strongly to the precipitation treatment. Although caution is needed since the libraries are unreplicated, it seems the acidobacteria were more abundant under dry conditions—making up 26% of the community in dry plots relative to only 10% of the community in wet plots, whereas proteobacteria were more abundant under wet conditions —making up 43% of the community in dry plots relative to 61% of the community in wet plots.
FIG. 2.
Relative abundance of bacterial phyla within samples originating from wet plots (A) and dry plots (B). Phylogenetic affiliations of clones are based on 16S rRNA gene sequences assigned through the Classifier program within the Ribosomal Database Project.
The Proteobacteria phylum dominated the bacterial community across all treatments. Within the Proteobacteria, the alphaproteobacteria were dominated by members of the Rhizobiales, the betaproteobacteria were dominated by Burkholderiales, the deltaproteobacteria were dominated by Myxococcales, Syntrophobacterales, and Desulfobacterales, and the gammaproteobacteria were dominated by Chromatiales and Enterobacteriales. Within the Proteobacteria phyla, the subdivision deltaproteobacteria was present in high abundance in most of the treatments (see Fig. SA2 in the supplemental material), and it seems that the deltaproteobacteria increased under eCO2 treatment. In the overall bacterial libraries, the smaller proportion of clones recovered from phylogenetic groups other than Proteobacteria made their responses more difficult to interpret and more likely to be biased by the low sample numbers of cloned representatives.
Since overall acidobacteria were responsive to precipitation regimen and they are an understudied soil microbial group, we constructed additional clone libraries using the acidobacterial primers published by Barns et al. (4) and analyzed for changes in the relative abundance of the defined phylogenetic subgroups also presented in that study. The relative composition of different acidobacterial lineages defined by Barns et al. (4) varied with precipitation, but not other treatments (see Fig. SA3 in the supplemental material). In all treatments, however, the dominant acidobacterial clones were related to group Gp6, followed by Gp4, Gp5, and Gp1, a finding similar to previously reported soil distributions (5), including samples reported previously from subsurface soils near Oak Ridge (4). Gp6 tended to increase under elevated CO2 in wet plots and decrease under elevated CO2 in dry plots. Other group-specific acidobacteria responses were not detected or apparent.
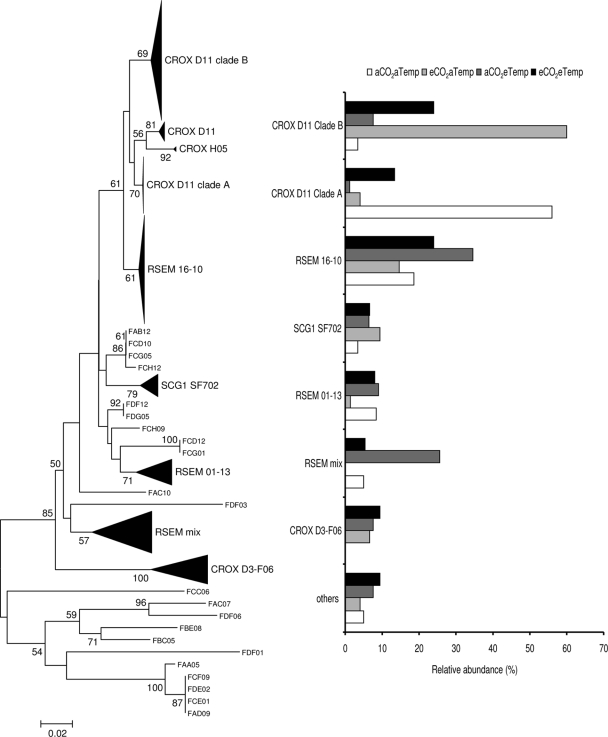
Specific fungal lineages also differed among the climate treatments (Fig. 3). In the wet plot libraries, the sequences were mostly related to uncultured fungal sequences in the fungal branch known as soil clone group I (SCGI) (47). These sequences represented almost 95% of the total recovered sequences in the wet treatments and exhibited some grouping based on the other climate drivers (CO2 or temperature). Although most of the SCGI subgroups were uniformly distributed among treatments, sequences related to uncultured SCGI clone CROX-D11 (EU179550) were surprisingly dominant in the aTemp treatments. In this clade, two clear subclades were present: clade A was dominant under aCO2 treatments, and clade B was dominant under eCO2 treatments. Within the dry treatments, sequences related to SCGI represented 73% of the clone libraries but showed no detectable differences in the relative distribution of clones among the temperature and CO2 treatments.
FIG. 3.
Fungal rRNA neighbor-joining tree of SCGI clones and distribution of different subclades recovered from each treatment for wet plots. Scale bars represent 2% changes. Numbers at the nodes represent bootstrap resampling support based on 100 replicates; only values >50 are presented.
DISCUSSION
Atmospheric and climate change drivers and their interactions may select for distinct soil microbial communities, and these community changes may shape the way ecosystems function in the future. We took advantage of an ongoing, multifactor climate change experiment to investigate how multiple climatic change factors and their interactions impact both the abundance and the diversity of soil bacterial and fungal communities. Using molecular techniques, we targeted changes in the abundance and community composition of the bacterial, acidobacterial, and fungal components of the soil microbial community. Our treatments did not impact fungal to bacteria relative ratio, but bacterial abundance was affected by an interactive effect of CO2 and temperature, and fungal abundance was affected by temperature but not precipitation or CO2 regime. In addition, there were changes in the relative composition of bacterial and fungal groups, and these shifts were largely shaped by the precipitation regime.
Do climatic change factors interact to affect bacterial and fungal abundance and community diversity metrics?
Fungal abundance was responsive to elevated temperature, and bacterial abundance was responsive to interactive effect of CO2 and temperature. However, there were no detectable direct or interactive impacts on the fungus/bacterium ratio among our climate change treatments. Our results are in agreement with those of previous reports. For example, researchers have found that changes in CO2 do not predictably alter microbial biomass (35, 62). Similarly, changes in soil moisture and ecosystem warming do not always lead to predictable or significant changes in bacterial and fungal abundance (8, 19, 29, 58). Previous reports in this site have shown a response of some plants to a warming effect, in particular Solidago and Andropogon, with some other plant productivity being reduced; however, the response is complicated by time of sampling (15). Therefore, our quantitative PCR data may be revealing a fungal communities response to temperature that can be the result of direct effects of temperature in the fungal community and/or an indirect effect by responding to plant community changes. The bacterial response to interactive effects of CO2 and temperature may represent a more complex scenario. It is possible that a lack of labile substrates under elevated temperature alone favors fungal over bacterial community members, while increased substrate availability from plants under elevated CO2 allows increases in bacterial abundance with increased temperature.
Major changes in the bacterial and fungal libraries were observed in the distribution of phylogenetic affiliation of clones, and these changes were mostly influenced by the precipitation treatment rather than CO2 or temperature treatments. The precipitation treatment also had the largest influence relative to temperature and atmospheric [CO2] on plant community biomass and plant community composition in these experimental ecosystems (30). Precipitation-driven shifts in plant community composition may thus alter the amount and quality of litter inputs into the soil ecosystem, and these changes may indirectly alter the soil microbial community. Our precipitation treatment had the largest impact on microbial community composition, and this result was confirmed by using several phylogeny-based comparison tools in Unifrac (38). Soil moisture can impact the physiological status of soil microorganisms, physicochemical properties of soils, and plant productivity. Increased plant productivity may result in greater inputs of plant-derived carbon in soils, which may alter soil microbial communities. The quality and quantity of the plant-derived carbon substrates (litter, dead roots, and roots exudates) may be affected by moisture resulting in different microbial communities. Alternatively, microbial communities could be responding directly to soil moisture, which was quite different between the wet and dry treatments (10). The response to elevated CO2 or temperature was relatively small and insignificant for community composition metrics; however, minor responses were observed for particular groups such as deltaproteobacteria or fungal sequences related to the uncultured SCGI clone CROX-D11 discussed below, indicating that perhaps temperature and CO2 do not play the major role in affecting microbial composition, which is in agreement with previous studies (8, 19, 29, 36, 58, 60). Unfortunately, due to a lack of available resources, we were not able to take full advantage of the replication of this multifactor experiment and were forced to pool replicates across the treatments in order to sample the microbial community broadly (Bacteria, Fungi, and Acidobacteria) so our ability to use more traditional statistical methods across treatments is somewhat limited beyond the phylogenetic-distance-based methods described above.
Do climatic change factors differentially affect bacterial or fungal phylogenetic groups?
The relative abundance of bacterial phyla was clearly impacted by the precipitation treatment, which led to shifts in the relative abundance of Proteobacteria and Acidobacteria. The other two factors, CO2 and temperature, did not have a major impact on the distribution of these groups. The relative abundance of Proteobacteria was greater in the wet relative to the dry treatments, whereas Acidobacteria abundance was greater in dry treatments.
Surprisingly, within the Proteobacteria phylum, the predominant class was the deltaproteobacteria, a surprising result since most of soil proteobacteria are dominated by the α, β, and γ classes. Deltaproteobacteria recovered from these soils consisted mainly of members of the Myxococcales and Syntrophobacterales orders. Myxococcales are typically found in soils and exhibit an aerobic organotrophic metabolism with some representatives able to utilize cellulose (9); however, their dominance is unexpected. The presence of Syntrophobacterales in these aerobic soils is also surprising, since their metabolism is usually anaerobic (31). The dominance of the deltaproteobacterial class could be related to a specific microorganism-plant interaction in this old field design, but the reasons still remains unclear. Acidobacteria, a very ubiquitous phylum in soils, were most predominant within the dry treatment soils. Despite their ubiquity in soils, very little is known about their metabolism, and very few members of this group are cultured or studied (5). Recently, 26 subdivisions for the Acidobacteria phyla were proposed (4), and three genomes from cultured representatives have been sequenced (57). According to the genome data, these organisms possess the ability to utilize a broad range of substrates and exhibit genetic traits typical of microorganisms able to survive desiccation stress and withstand fluctuations in soil moisture(57). Moreover, acidobacteria tend to prefer oligotrophic niches (16), suggesting that they may be favored in dry soils, where a lower plant metabolism and productivity may result in lower availability of plant-derived carbon sources, possibly decreased dissolved organic carbon levels, and generally more oligotrophic conditions. Fierer et al. (16) reported that members of the Acidobacteria tend to be favored in oligotrophic soils with lower carbon availability, and betaproteobacteria were favored in copiotrophic conditions, but these authors did not study the behavior of deltaproteobacteria. Moreover, it has been proposed that the ratio of acidobacteria to proteobacteria could be used as indicator of the trophic level of soils (52), and the ratios we observed would thus suggest that the dry treatments are indeed more oligotrophic than the wet treatments. Garten et al. (20) showed that in these sites reduced soil moisture slowed carbon turnover in the particulate organic matter pool, which can reduce substrate availability and result in more oligotrophic conditions. In another multifactorial global change experiment at Jasper Ridge in central California, ammonia-oxidizing bacteria, betaproteobacteria responsible for the oxidization of ammonia in nitrification, were shown to decrease in response to elevated CO2 and the decrease was more pronounced with elevated precipitation (25). In our study, deltaproteobacteria also seemed to respond positively to elevated CO2 in the dry and wet treatments, similarly to previous studies in chaparral ecosystems (36).
Although the total number of acidobacterial clones in the overall 16S bacterial libraries differed among wet and dry treatments, the distribution of the clones within separate specific acidobacterial libraries was not affected by climate treatments, which may indicate shifts in overall abundance but not particular selection for any subdivision of Acidobacteria among the different climate treatments. Clearly, acidobacteria are affected by soil moisture and, given their high abundance in many environments worldwide, should be further studied with methods specific to this group to better understand their participation and roles in soil processes.
Within the fungal libraries, most of the sequences were associated to the uncultured fungal SCGI that is known only from rRNA gene sequence data but represents a major subphylum level lineage within the Ascomycotina (47). SCGI sequences were more abundant in wet treatments (95%) than in dry treatments (73%) in the present study. These relative abundances are much higher than the range of previous reported values (7 to 27%) (47, 54) despite the use of PCR and other laboratory methods identical to our previous studies. We are unsure how exactly to interpret this extremely high abundance given the unknown nature of the physiology and ecology of these organisms; however, such high levels of SCGI suggest a specific relationship of SCGI taxa with these soil types or perhaps a specific interaction with one or more of the plants present in our experiment. This group has always previously been found to be associated with active plant rhizosphere (Porter et al. [47]), and since rhizosphere dynamics such as carbon exudation, root proliferation, and turnover, and other belowground processes may be impacted by climatic change, this fungal group could be an important indicator species and should be targeted in future research.
Conclusions.
Our results demonstrate that the direct and interactive impacts of climatic change will likely reshape bacterial and fungal soil communities and that changes in precipitation in particular will be important in dictating the response of microbial community composition in the future. Further, we found that while fungal abundance overall responds to temperature and bacterial abundance to interactive effects of CO2 and temperature and not to direct or indirect effects of precipitation regime, changes in bacterial and fungal community composition were readily apparent with precipitation regime. Our data highlight the need to know more about the physiology and ecology of the various uncultured, dominant, and responsive soil community members, such as SCGI and acidobacteria, and how these organisms will potentially be affected by, and feedback to, the response of ecosystems to climatic change.
Supplementary Material
Acknowledgments
We thank C. Engel, E. Felker-Quinn, S. Kortbein, K. Sides, C. Campany, J. Childs, M. Kerley, and L. Gunter for assisting with field and laboratory work. Members of the Ecosystem Ecology Lab group at Oak Ridge National Laboratory (ORNL) and the University of Tennessee, the Molecular Microbial Ecology Group at ORNL, and the anonymous reviewers gave insightful comments and suggestions on the manuscript. J. Weltzin was integral in establishing the experiment.
Research was sponsored by the U.S. Department of Energy, Office of Science, Biological, and Environmental Research Program (grant DE-FG02-02ER63366) and by the Laboratory Directed Research and Development Program of Oak Ridge National Laboratory. Oak Ridge National Laboratory is managed by UT Battelle, LLC, for the U.S. Department of Energy under contract DE-AC05-00OR22725.
Footnotes
Published ahead of print on 18 December 2009.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Austin, E. E., H. F. Castro, K. E. Sides, C. W. Schadt, and A. T. Classen. 2009. Assessment of 10 years of CO2 fumigation on soil microbial communities and function in a sweetgum plantation. Soil Biol. Biochem. 41:514-520. [Google Scholar]
- 2.Bakkenes, M., J. R. M. Alkemade, F. Ihle, R. Leemans, and J. B. Latour. 2002. Assessing effects of forecasted climate change on the diversity and distribution of European higher plants for 2050. Global Change Biol. 8:390-407. [Google Scholar]
- 3.Bardgett, R. D., C. Freeman, and N. J. Ostle. 2008. Microbial contributions to climate change through carbon cycle feedbacks. ISME J. 2:805-814. [DOI] [PubMed] [Google Scholar]
- 4.Barns, S. M., E. C. Cain, L. Sommerville, and C. R. Kuske. 2007. Acidobacteria phylum sequences in uranium-contaminated subsurface sediments greatly expand the known diversity within the phylum. Appl. Environ. Microbiol. 73:3113-3116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barns, S. M., S. L. Takala, and C. R. Kuske. 1999. Wide distribution and diversity of members of the bacterial kingdom Acidobacterium in the environment. Appl. Environ. Microbiol. 65:1731-1737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Borneman, J., and R. J. Hartin. 2000. PCR primers that amplify fungal rRNA genes from environmental samples. Appl. Environ. Microbiol. 66:4356-4360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bradford, M. A., C. A. Davies, S. D. Frey, T. R. Maddox, J. M. Melillo, J. E. Mohan, J. F. Reynolds, K. K. Treseder, and M. D. Wallenstein. 2008. Thermal adaptation of soil microbial respiration to elevated temperature. Ecol. Lett. 11:1316-1327. [DOI] [PubMed] [Google Scholar]
- 8.Chen, M. M., Y. G. Zhu, Y. H. Su, B. D. Chen, B. J. Fu, and P. Marschner. 2007. Effects of soil moisture and plant interactions on the soil microbial community structure. Eur. J. Soil Biol. 43:31-38. [Google Scholar]
- 9.Dawid, W. 2000. Biology and global distribution of myxobacteria in soils. FEMS Microbiol. Rev. 24:403-427. [DOI] [PubMed] [Google Scholar]
- 10.Dermody, O., J. F. Weltzin, E. C. Engel, P. Allen, and R. J. Norby. 2007. How do elevated [CO2], warming, and reduced precipitation interact to affect soil moisture and LAI in an old field ecosystem? Plant Soil. 301:255-266. [Google Scholar]
- 11.Drigo, B., G. A. Kowalchuk, E. Yergeau, T. M. Bezemer, H. T. S. Boschker, and J. A. Van Veen. 2007. Impact of elevated carbon dioxide on the rhizosphere communities of Carex arenaria and Festuca rubra. Global Change Biol. 13:2396-2410. [Google Scholar]
- 12.Edwards, N. T., and R. J. Norby. 1999. Below-ground respiratory responses of sugar maple and red maple saplings to atmospheric CO2 enrichment and elevated air temperature. Plant Soil 206:85-97. [Google Scholar]
- 13.Egger, K. N. 1995. Molecular analysis of ectomycorrhizal fungal communities. Can. J. Bot. 1995:S1415-S1422. [Google Scholar]
- 14.Emmett, B. A., C. Beier, M. Estiarte, A. Tietema, H. L. Kristensen, D. Williams, J. Penuelas, I. Schmidt, and A. Sowerby. 2004. The response of soil processes to climate change: results from manipulation studies of shrublands across an environmental gradient. Ecosystems 7:625-637. [Google Scholar]
- 15.Engel, E. C., J. F. Weltzin, R. J. Norby, and A. T. Classen. 2009. Responses of an old-field plant community to interacting factors of elevated [CO2], warming, and soil moisture. J. Plant Ecol. 2:1-11. [Google Scholar]
- 16.Fierer, N., M. A. Bradford, and R. B. Jackson. 2007. Toward an ecological classification of soil bacteria. Ecology 88:1354-1364. [DOI] [PubMed] [Google Scholar]
- 17.Fierer, N., J. A. Jackson, R. Vilgalys, and R. B. Jackson. 2005. Assessment of soil microbial community structure by use of taxon-specific quantitative PCR assays. Appl. Environ. Microbiol. 71:4117-4120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Franklin, O., R. E. McMurtrie, C. M. Iversen, K. Y. Crous, A. C. Finzi, D. T. Tissue, D. S. Ellsworth, R. Oren, and R. J. Norby. 2009. Forest fine-root production and nitrogen use under elevated CO2: contrasting responses in evergreen and deciduous trees explained by a common principle. Global Change Biol. 15:132-144. [Google Scholar]
- 19.Frey, S. D., R. Drijber, H. Smith, and J. Melillo. 2008. Microbial biomass, functional capacity, and community structure after 12 years of soil warming. Soil Biol. Biochem. 40:2904-2907. [Google Scholar]
- 20.Garten, C. T., A. T. Classen, and R. J. Norby. 2009. Soil moisture surpasses elevated CO2 and temperature as a control on soil carbon dynamics in a multi-factor climate change experiment. Plant Soil 319:85-94. [Google Scholar]
- 21.Garten, C. T., A. T. Classen, R. J. Norby, D. J. Brice, J. F. Weltzin, and L. Souza. 2008. Role of N-2-fixation in constructed old-field communities under different regimes of [CO2], temperature, and water availability. Ecosystems 11:125-137. [Google Scholar]
- 22.Haase, S., L. Philippot, G. Neumann, S. Marhan, and E. Kandeler. 2008. Local response of bacterial densities and enzyme activities to elevated atmospheric CO2 and different N supply in the rhizosphere of Phaseolus vulgaris L. Soil Biol. Biochem. 40:1225-1234. [Google Scholar]
- 23.Hall, T. A. 1999. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids. Symp. Ser. 41:95-98. [Google Scholar]
- 24.He, J., X. B. Dai, and X. C. Zhao. 2007. PLAN: a web platform for automating high-throughput BLAST searches and for managing and mining results. BMC Bioinform. 8:10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Horz, H. P., A. Barbrook, C. B. Field, and B. J. M. Bohannan. 2004. Ammonia-oxidizing bacteria respond to multifactorial global change. Proc. Natl. Acad. Sci. U. S. A. 101:15136-15141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hungate, B. A., D. W. Johnson, P. Dijkstra, G. Hymus, P. Stiling, J. P. Megonigal, A. L. Pagel, J. L. Moan, F. Day, J. H. Li, C. R. Hinkle, and B. G. Drake. 2006. Nitrogen cycling during seven years of atmospheric CO2 enrichment in a scrub oak woodland. Ecology 87:26-40. [DOI] [PubMed] [Google Scholar]
- 27.Janssen, P. H. 2006. Identifying the dominant soil bacterial taxa in libraries of 16S rRNA and 16S rRNA genes. Appl. Environ. Microbiol. 72:1719-1728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Janus, L. R., N. L. Angeloni, J. McCormack, S. T. Rier, N. C. Tuchman, and J. J. Kelly. 2005. Elevated atmospheric CO2 alters soil microbial communities associated with trembling aspen (Populus tremuloides) roots. Microb. Ecol. 50:102-109. [DOI] [PubMed] [Google Scholar]
- 29.Kandeler, E., A. R. Mosier, J. A. Morgan, D. G. Milchunas, J. Y. King, S. Rudolph, and D. Tscherko. 2006. Response of soil microbial biomass and enzyme activities to the transient elevation of carbon dioxide in a semi-arid grassland. Soil Biol. Biochem. 38:2448-2460. [Google Scholar]
- 30.Kardol, P., M. A. Cregger, C. E. Campany, and A. T. Classen. 2010. Changes in plant community composition affect multifactor climate change effects on soil ecosystem functioning. Ecology 91:767-781. [DOI] [PubMed] [Google Scholar]
- 31.Kuever, J., F. A. Rainey, and F. Widdel. 2005. The deltaproteobacterial orders Desulfovibrionales, Desulfobacterales, Desulfarcales, and Syntrophobacterales, p. 925-1040. In G. Garrity (ed.), Bergey's manual of systematic bacteriology. Springer, New York, NY.
- 32.Lane, D. J. 1991. 16S/23S rRNA sequencing, p. 115-175. In E. Stackebrandt and M. Goodfellow (ed.), Nucleic acid techniques in bacterial systematics. John Wiley & Sons, Chichester, United Kingdom.
- 33.Larson, J. L., D. R. Zak, and R. L. Sinsabaugh. 2002. Extracellular enzyme activity beneath temperate trees growing under elevated carbon dioxide and ozone. Soil Sci. Soc. Am. J. 66:1848-1856. [Google Scholar]
- 34.Lesaulnier, C., D. Papamichail, S. McCorkle, B. Ollivier, S. Skiena, S. Taghavi, D. Zak, and D. van der Lelie. 2008. Elevated atmospheric CO2 affects soil microbial diversity associated with trembling aspen. Environ. Microbiol. 10:926-941. [DOI] [PubMed] [Google Scholar]
- 35.Lipson, D. A., M. Blair, G. Barron-Gafford, K. Grieve, and R. Murthy. 2006. Relationships between microbial community structure and soil processes under elevated atmospheric carbon dioxide. Microb. Ecol. 51:302-314. [DOI] [PubMed] [Google Scholar]
- 36.Lipson, D. A., R. F. Wilson, and W. C. Oechel. 2005. Effects of elevated atmospheric CO2 on soil microbial biomass, activity, and diversity in a chaparral ecosystem. Appl. Environ. Microbiol. 71:8573-8580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lloret, F., J. Penuelas, P. Prieto, L. Llorens, and M. Estiarte. 2009. Plant community changes induced by experimental climate change: seedling and adult species composition. Perspect. Plant Ecol. Evol. Syst. 11:53-63. [Google Scholar]
- 38.Lozupone, C., M. Hamady, and R. Knight. 2006. UniFrac: an online tool for comparing microbial community diversity in a phylogenetic context. BMC Bioinform. 7:14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lozupone, C., and R. Knight. 2005. UniFrac: a new phylogenetic method for comparing microbial communities. Appl. Environ. Microbiol. 71:8228-8235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Maidak, B. L., J. R. Cole, T. G. Lilburn, C. T. Parker, P. R. Saxman, R. J. Farris, G. M. Garrity, G. J. Olsen, T. M. Schmidt, and J. M. Tiedje. 2001. The RDP-II (Ribosomal Database Project). Nucleic Acids Res. 29:173-174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Martin, A. P. 2002. Phylogenetic approaches for describing and comparing the diversity of microbial communities. Appl. Environ. Microbiol. 68:3673-3682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Muyzer, G., E. C. Dewaal, and A. G. Uitterlinden. 1993. Profiling of complex microbial populations by denaturing gradient gel-electrophoresis analysis of PCR-amplified genes coding for 16S ribosomal RNA. Appl. Environ. Microbiol. 59:695-700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Norby, R. J., M. F. Cotrufo, P. Ineson, E. G. O'Neill, and J. G. Canadell. 2001. Elevated CO2, litter chemistry, and decomposition: a synthesis. Oecologia 127:153-165. [DOI] [PubMed] [Google Scholar]
- 44.Norby, R. J., J. Ledford, C. D. Reilly, N. E. Miller, and E. G. O'Neill. 2004. Fine-root production dominates response of a deciduous forest to atmospheric CO2 enrichment. Proc. Natl. Acad. Sci. U. S. A. 101:9689-9693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Norby, R. J., and Y. Q. Luo. 2004. Evaluating ecosystem responses to rising atmospheric CO2 and global warming in a multi-factor world. New Phytol. 162:281-293. [Google Scholar]
- 46.Pettersson, M., and E. Baath. 2003. Temperature-dependent changes in the soil bacterial community in limed and unlimed soil. FEMS Microbiol. Ecol. 45:13-21. [DOI] [PubMed] [Google Scholar]
- 47.Porter, T. M., C. W. Schadt, L. Rizvi, A. P. Martin, S. K. Schmidt, L. Scott-Denton, R. Vilgalys, and J. M. Moncalvo. 2008. Widespread occurrence and phylogenetic placement of a soil clone group adds a prominent new branch to the fungal tree of life. Mol. Phylogenet. Evol. 46:635-644. [DOI] [PubMed] [Google Scholar]
- 48.Qiu, X. Y., L. Y. Wu, H. S. Huang, P. E. McDonel, A. V. Palumbo, J. M. Tiedje, and J. Z. Zhou. 2001. Evaluation of PCR-generated chimeras: mutations, and heteroduplexes with 16S rRNA gene-based cloning. Appl. Environ. Microbiol. 67:880-887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schadt, C. W., A. P. Martin, D. A. Lipson, and S. K. Schmidt. 2003. Seasonal dynamics of previously unknown fungal lineages in tundra soils. Science 301:1359-1361. [DOI] [PubMed] [Google Scholar]
- 50.Schimel, J. P., J. M. Gulledge, J. S. Clein-Curley, J. E. Lindstrom, and J. F. Braddock. 1999. Moisture effects on microbial activity and community structure in decomposing birch litter in the Alaskan taiga. Soil Biol. Biochem. 31:831-838. [Google Scholar]
- 51.Schloss, P. D., and J. Handelsman. 2005. Introducing DOTUR, a computer program for defining operational taxonomic units and estimating species richness. Appl. Environ. Microbiol. 71:1501-1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Smit, E., P. Leeflang, S. Gommans, J. van den Broek, S. van Mil, and K. Wernars. 2001. Diversity and seasonal fluctuations of the dominant members of the bacterial soil community in a wheat field as determined by cultivation and molecular methods. Appl. Environ. Microbiol. 67:2284-2291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position specific gap penalties, and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Urban, A., M. Puschenreiter, J. Strauss, and M. Gorfer. 2008. Diversity and structure of ectomycorrhizal and co-associated fungal communities in a serpentine soil. Mycorrhiza 18:339-354. [DOI] [PubMed] [Google Scholar]
- 55.Vilgalys, R., and M. Hester. 1990. Rapid genetic identification and mapping of enzymatically amplified ribosomal DNA from several Cryptococcus species. J. Bacteriol. 172:4238-4246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wan, S., R. J. Norby, J. Ledford, and J. F. Weltzin. 2007. Responses of soil respiration to elevated CO2, air warming, and changing soil water availability in a model old-field grassland. Global Change Biol. 13:2411-2424. [Google Scholar]
- 57.Ward, N. L., J. F. Challacombe, P. H. Janssen, B. Henrissat, P. M. Coutinho, M. Wu, G. Xie, D. H. Haft, M. Sait, J. Badger, R. D. Barabote, B. Bradley, T. S. Brettin, L. M. Brinkac, D. Bruce, T. Creasy, S. C. Daugherty, T. M. Davidsen, R. T. DeBoy, J. C. Detter, R. J. Dodson, A. S. Durkin, A. Ganapathy, M. Gwinn-Giglio, C. S. Han, H. Khouri, H. Kiss, S. P. Kothari, R. Madupu, K. E. Nelson, W. C. Nelson, I. Paulsen, K. Penn, Q. Ren, M. J. Rosovitz, J. D. Selengut, S. Shrivastava, S. A. Sullivan, R. Tapia, L. S. Thompson, K. L. Watkins, Q. Yang, C. Yu, N. Zafar, L. Zhou, and C. R. Kuske. 2009. Three genomes from the phylum Acidobacteria provide insight into the lifestyles of these microorganisms in soils. Appl. Environ. Microbiol. 75:2046-2056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Williams, M. A. 2007. Response of microbial communities to water stress in irrigated and drought-prone tallgrass prairie soils. Soil Biol. Biochem. 39:2750-2757. [Google Scholar]
- 59.Ye, J., S. McGinnis, and T. L. Madden. 2006. BLAST: improvements for better sequence analysis. Nucleic Acids Res. 34:W6-W9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zak, D. R., W. E. Holmes, N. W. MacDonald, and K. S. Pregitzer. 1999. Soil temperature, matrix potential, and the kinetics of microbial respiration and nitrogen mineralization. Soil Sci. Soc. Am. J. 63:575-584. [Google Scholar]
- 61.Zak, D. R., K. S. Pregitzer, P. S. Curtis, and W. E. Holmes. 2000. Atmospheric CO2 and the composition and function of soil microbial communities. Ecol. Appl. 10:47-59. [Google Scholar]
- 62.Zak, D. R., K. S. Pregitzer, J. S. King, and W. E. Holmes. 2000. Elevated atmospheric CO2, fine roots and the response of soil microorganisms: a review and hypothesis. New Phytol. 147:201-222. [Google Scholar]
- 63.Zak, D. R., D. B. Ringelberg, K. S. Pregitzer, D. L. Randlett, D. C. White, and P. S. Curtis. 1996. Soil microbial communities beneath Populus grandidentata crown under elevated atmospheric CO2. Ecol. Appl. 6:257-262. [Google Scholar]
- 64.Zhang, W., K. M. Parker, Y. Luo, S. Wan, L. L. Wallace, and S. Hu. 2005. Soil microbial responses to experimental warming and clipping in a tallgrass prairie. Global Change Biol. 11:266-277. [Google Scholar]
- 65.Zogg, G. P., D. R. Zak, D. B. Ringelberg, N. W. MacDonald, K. S. Pregitzer, and D. C. White. 1997. Compositional and functional shifts in microbial communities due to soil warming. Soil Sci. Soc. Am. J. 61:475-481. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.