Abstract
Sclerostin, a secreted glycoprotein, regulates osteoblast function. Using yeast two-hybrid and direct protein interaction analyses, we demonstrate that sclerostin binds the Wnt-modulating and Wnt-modulated, extracellular matrix protein, cysteine-rich protein 61 (Cyr61, CCN1), which regulates mesenchymal stem cell proliferation and differentiation, osteoblast and osteoclast function, and angiogenesis. Sclerostin was shown to inhibit Cyr61-mediated fibroblast attachment, and Cyr61 together with sclerostin increases vascular endothelial cell migration and increases osteoblast cell division. The data show that sclerostin binds to and influences the activity of Cyr61.
Keywords: Sclerostin, Cyr61, CCN1, osteoblasts, osteoclasts, high bone mass, LRP5, Wnt
Introduction
Sclerostin is a cystine-knot, osteocyte-derived, secreted protein that inhibits bone formation [1–7]. Mutations within the sclerostin gene (SOST) are associated with sclerosteosis (MIM # 269500) or with van Buchem’s disease (MIM # 239100) [7]. Both diseases are associated with a general progressive skeletal overgrowth and sclerosis of the axial and appendicular skeleton [7–9].
The mechanism of action of sclerostin is of intense interest since disruption of sclerostin action by drugs or antibodies would increase bone mass and be useful for the treatment of osteoporosis [10]. Sclerostin decreases BMP signaling and BMP-mediated mineralization in osteoblasts by competing with type I and II BMP receptors for binding to BMPs [3]. Recombinant sclerostin and the BMP antagonist, noggin, bind each other and the sclerostin-noggin complex is competitive with BMP for binding to BMP receptors [11]. Sclerostin reduces proliferation and alkaline phosphatase activity, and increases caspase activity and apoptosis in human mesenchymal stem cells [5]. Alkaline phosphatase activity altered by sclerostin in such cells is partially restored by BMP 6 treatment [5]. Sclerostin, however, may not function as a classical BMP antagonist in some bone-derived cells [2], suggesting that other pathways mediate sclerostin activity. Since the bone phenotype of sclerosteosis and van Buchem disease resembles that observed in the high bone mineral density syndrome (BMND1, MIM # 601884) [12–14] that is due to mutations in the LRP5 receptor, the role of the LRP5 receptor and Wnt signaling in sclerostin function has been investigated. Sclerostin antagonizes Wnt signaling in Xenopus embryos and mammalian cells by binding to the extracellular domain of LRP5 and LRP6, and by disrupting Wnt-induced frizzled-LRP complex formation [15]. LRP5 mutations linked to the high bone mass syndrome are associated with reduced binding of sclerostin to LRP5 and a concomitant reduction of sclerostin-induced inhibition of Wnt signaling [16–19]. Others have proposed that sclerostin blocks Wnt-induced cell differentiation indirectly through its modulation of BMP function [4]. Thus, current data suggest that sclerostin functions by at least two, possibly related, mechanisms in bone.
To investigate biochemical pathways that might play a role in sclerostin function, we examined the binding of sclerostin to other proteins using the yeast two-hybrid approach. We now report that in addition to interacting with BMP 6 and LRP5 with high affinity, sclerostin interacts with Cyr61 (CCN1), a protein which regulates bone cell function and angiogenesis. We show that sclerostin antagonizes Cyr61-mediated fibroblast attachment and that sclerostin and Cyr61 together increase endothelial cell migration and osteoblast proliferation. The data point to a novel and biologically relevant interaction between sclerostin, a secreted osteocyte-derived protein, and Cyr61.
Methods and Materials
Yeast Two-hybrid Experiments
Yeast two-hybrid experiments were performed using a Normalized Universal Human Mate & Plate Library and the Matchmaker Gold System (Clontech, Mountainview, CA). Human SOST cDNA was amplified by PCR methods with appropriate primers.
5′ primer: 5′GAGAGAATTCCAGGGGTGGCAGGCGTTCAAGAATGATGCC3′ and 3′ primer: 5′GAGAGGATCCCTAGTAGGCGTTCTCCAGCTCGGCCTGGTTGG3′; underlined sequences are EcoRI and BamHI restriction endonuclease sites.
The PCR construct was cloned in frame with the GAL4 DNA-BD of the pGBKT7 DNA-BD vector using EcoRI and BamHI restriction sites within the multiple cloning site. The sclerostin pGBKT7 plasmid was used to transform Y2H Gold yeast cells. Appropriate positive and negative control mating, auto-activation and toxicity experiments were performed. 1 mL of the Mate and Plate Library was combined with 5 mL of the bait strain and 45 mL of 2X YPDA medium (50 μg/mL kanamycin). Cells were incubated for 24 h at 30°C with shaking. The cells were pelleted, washed with 50 mL 0.5X YPDA (50 μg/mL kanamycin) and re-suspended in 10 mL 0.9% NaCl. From the mated culture, 100 μL of 1:10, 1:100, 1:1000; and 1: 10,000 dilutions were spread on 100-mm SD/-Trp, SD/-Leu and SD/-Trp/-Leu plates. The remainder of the culture was plated onto 53, 150-mm SD/-Trp/-Leu/X-αGal/Aureobasidin A plates. The plates were incubated at 30°C for 5 d. Positive colonies were re-analyzed by streaking onto quadruple dropout (SD/-Leu/-Trp/-His/-Ade) plates containing X-αGal/Aureobasidin A. Aliquots of blue colonies that had been serially selected were used for PCR with flanking primers specific for the pGAD-T7-RecAB plasmid, to generate insert DNA for sequencing. Plasmids were rescued from yeast and were used to transform E. coli grown on ampicillin containing plates to isolate the "prey" plasmid. Protein interactions were confirmed in transformed yeast cells. Yeast cells were grown in SD/-Leu/-His/-Trp medium (50 μg/mL kanamycin). Cells expressing both proteins were lysed in 100 mM Tris, pH 7.4, 100mM NaCl containing protease inhibitors (lysis buffer). Proteins were precipitated with HA or c-myc antibodies and protein A beads (40 μL). The washed precipitated complexes were analyzed by SDS-PAGE, and electrophoresed proteins were transferred to PVDF membranes. The presence of the partner protein was determined with a peroxidase-labeled c-myc antibody and chemiluminescence methods. To detect sclerostin in complexes, a monoclonal antibody directed against the N-terminus of sclerostin was used as probe [20]. Anti-mouse peroxidase-labeled IgG was used to detect bound sclerostin antibody with chemiluminescence methods.
Bacterial Expression of Sclerostin
Human sclerostin, aa 24–213, was expressed using the pMAL-p4E vector in E. coli Origami 2(DE3) cells as described [20]. 5′ and 3′ oligonucleotides were used to generate a PCR product using human SOST cDNA. The PCR product was cloned into pMAL-p4E (New England Biolabs, Beverly, MA). Expression of sclerostin in E. coli was induced at 37°C with 1 mM IPTG. 24–213 human sclerostin-maltose binding protein was purified on an amylose resin.
Sclerostin production in Trichoplusia ni cells
The method for expression has been described earlier [20]. 5′ and 3′ oligonucleotides were used to generate a PCR product using human SOST cDNA. The PCR product was purified and ligated into the pIB/V5-His vector (Invitrogen, Carlsbad, CA). Stably transfected BTI-TN-5B1-4 High Five (Trichoplusia ni) cells were used to produce sclerostin that was subsequently purified.
Production of hBMP6-maltose binding protein production in E. coli Origami 2 (DE3) cells
To express the processed, secreted form of human bone morphogenetic protein 6 (hBMP6), aa S375-H513, a 5′ oligonucleotide with a 5′ BamHI site, followed by PreScission protease cleavage site, and a 3′ oligonucleotide with a stop codon and with a SalI site, were used to generate a PCR product using human BMP6 cDNA as template.
The 5′ PCR primer sequence was: 5′GAGAGGATCCCTGGAAGTTCTGTTCCAGGGGCCCTCAGCCTCCAGCCGGCGCCGACAA CAGAGTCG3′. The 3′ PCR primer sequence was: 5′GAGAGTCGACCTAGTGGCATCCACAAGCTCTTACAACCATATTCCTG3′.
Restriction endonuclease sites are underlined and the 5′ PreScission protease site is in bold. The PCR product was cut with appropriate restriction endonucleases and cloned into BamHI and SalI cleaved pMAL-c4E vector (New England Biolabs). The BMP6-pMAL-c4E construct was used to transform competent E. coli Origami 2 (DE3) cells (Novagen/EMD, Gibbstown, NJ). For expression of protein, 600 mL 2xYT media (50 μg/mL ampicillin, 12.5 μg/mL tetracycline, and 50 μg/mL streptomycin sulfate) were inoculated with starter culture and grown at 37°C, 250 rpm, to an OD600nm of ~0.8. BMP6 production was induced at 20°C with 0.3 mM IPTG for 6 hr. Cells were lysed in 20 mM Tris, 200 mM NaCl, and 1 mM EDTA Na, pH 7.4, containing 4 mM phenylmethylsulfonyl fluoride (PMSF). The supernatant was applied to amylose resin, washed with 20 mM Tris, 200 mM NaCl, 1 mM EDTA Na, pH 7.4 (WB) and eluted with WB containing 10 mM maltose. Fractions were analyzed by SDS-PAGE and by immunoblots using anti-MBP HRP and chemiluminescent substrate (Roche).
Expression of the 1st beta propeller of LRP5 in insect cells
To express the secreted form of the 1st beta propeller of human LRP5, amino acids Ser32-Ala338, in Trichoplusia ni insect cells (Invitrogen) using a pIB/V5-His vector, a 5′ oligonucleotide with a 5′HindIII site, Kozak consensus sequence and a melettin secretory signal sequence and a 3′ oligonucleotide with a SacII site, without a stop codon, with a StrepTagII sequence and PreScission protease sequence were used to generate PCR products using human LRP5 cDNA as template.
The 5′ PCR primer sequence is as follows (the restriction site is underlined and the melettin secretory sequence is in bold type): 5′TGACTGAAGCTTACCATGAAATTCTTAGTCAACGTTGCCCTTGTTTTTATGGTCGTATAC ATTTCTTACATCTATGCCTCGCCGCTCCTGCTATTTGCCAACCGC3′.
The 3′ PCR primer is as follows (restriction site is underlined, the StrepTagII sequence is italicized and PreScission protease sequence is in bold type): 5′ATATATCCGCGGCTTCTCAAATTGAGGATGAGACCAGGGCCCCTGGAACAGAACTTCCA GTGCCTTACACGTCCTGCCGTTGTC3′
The PCR product was ligated into HindIII, SacII cut pIB/V5-His vector. The plasmid was used to transfect BT1-TN-5B1-4 High Five (Trichoplusia ni) insect cells. Secreted proteins were isolated from the conditioned media by the addition of Ni Sepharose (GE-Amersham). The resin was pelleted and washed with 1.5 L 1M NaCl, 20 mM Na2HPO4 pH 7.0 wash buffer (WB). LRP5 protein was eluted with 40 mL WB with 1 M imidazole. Eluted LRP5 protein was further purified by His-trap chromatography (GE-Amersham) and elution with a gradient of 0–1 M imidazole in WB. Immunoblot analyses were carried out using anti-V5 HRP antibody (Invitrogen) to identify fractions containing the human LRP5 β–propeller 1 protein.
Determination of BMP 6 and LRP5 affinity for sclerostin using surface plasmon resonance spectroscopy
Recombinant insect cell-produced human sclerostin (~1200 RU) was coupled to a CM5 Biacore chip (Amine Coupling Kit, Biacore Life Sciences) [20]. Human sclerostin was diluted in 10 mM sodium acetate buffer, pH 5.0 to 30 μg/mL prior to coupling to a CM5 Biacore chip. BMP6, MBP, or LRP5 proteins were diluted in 10 mM HEPES, 0.15 M NaCl, 3 mM EDTA, 0.005% surfactant P20, pH 7.4, to give concentrations between 500 nM and 0.05 nM and used for sensorgram experiments. A sensorgram control signal from a separate Biacore channel was subtracted from the signal obtained from the respective proteins. Chips were regenerated after each sensorgram run with 15–30 μL additions of 1 M NaCl in running buffer followed by 6–12 sec additions of 10 mM glycine pH 2.5 at a flow rate of 30 μL /min. Data were analyzed using Biacore 3000 Control Software version 3.2.
Cell attachment assay
Media containing Cyr61/CCN1-Fc chimera (flcyr61, 4 μg/mL) cyr61-Fc alone, or containing Cyr61/CCN1-Fc chimera (flcyr61, 4 μg/mL) plus insect-cell recombinant sclerostin (4 μg/mL) or sclerostin (4 μg/mL) alone were prepared, and added to 96-well plates (50 μL/well). Plates were incubated at 37°C for 30–60 min before the addition of cells. Swiss 3T3 mouse cells (ATCC #CCL-92) were grown to confluence in T75 flasks in Dulbecco’s high glucose, modified Eagle medium (D-MEM, #11965, Invitrogen), 10% fetal bovine serum (FBS), 100 units/mL penicillin, and 100 μg/mL streptomycin. Cells (4, 10 or 20 × 104 in 50–70 μL medium) were added to each well and incubated in the presence of test proteins. After attachment (40–85 minutes), non-adherent cells were removed, and adherent cells were washed once with 200 μL medium. 100 μL fresh medium containing 1–3.5% FBS was added to the cells. 10–12 μL WST-1 proliferation reagent (Roche) was added to each well and the plates were incubated at 37°C in a 5% CO2 atmosphere for 6–15 hr. Absorbance at 440 nm was measured to determine relative numbers of adherent cells.
Cell growth assay
MC3T3-E1 mouse osteoblast cells (ATCC #CRL-2593) were grown in α-MEM (#10-022-CV, Mediatech Inc., Herndon, VA), 10% FBS, 100 units/mL penicillin, and 100 μg/mL streptomycin. Cells were plated in 96-well plates (5000 or 800 cells/well) and allowed to grow for 27 hr at 37°C, in a 5% CO2 atmosphere. Serum-containing medium was replaced with serum-free medium. After 24 hr the medium was replaced with 150 μL serum-free medium containing human recombinant Cyr61/CCN1-Fc chimera (flcyr61, 0.5 μg/mL) (R&D Systems), or 24–213 insect cell human sclerostin (0.5 μg/mL), or both proteins. Cells were grown an additional 32 hr at 37 °C. 15 μL of WST-1 cell proliferation reagent was then added. Cells were incubated 4 hr in a 37°C in a 5% CO2 incubator. After 4 h absorbance at 440 nm was measured.
Vascular cell migration [21]
Human umbilical vein endothelial cells (HUVECs) were isolated and grown in collagen I-coated, 6-well plates. The cells were serum-starved overnight in Clonetics® EGM® Endothelial Cell Growth Medium (EBM Basal Medium, Fisher Scientific, Pittsburgh, PA) without serum. A scratch was made across each well with a pipette tip. Cells were grown in 2 mL medium containing sclerostin buffer (20 mM Na2HPO4, 1 M NaCl, 110 mM imidazole pH 7.0, negative control), vascular endothelial growth factor (VEGF, 10 ng/mL, positive control), cyr61 in varying amounts (0.1, 1.7 or 10 μg/mL), cyr61 (0.1, 1.7 or 10 μg/mL) plus sclerostin (0.1, 1.7 or 10 μg/mL), or sclerostin alone (0.1, 1.7 or 10 μg/mL). Thymidine (10 μM; Sigma) was included during the incubation to inhibit cell proliferation. Cells were imaged using bright field optics and photographed prior to and after overnight incubation at 37°C. Cells migrating into the cell-free scratched area were counted.
Results and Discussion
Sclerostin interacts with BMP 6 and the first β-propeller of LRP5
Using surface plasmon resonance spectroscopy and recombinant proteins, we confirmed that sclerostin interacts with BMP6 and with the first β-propeller of LRP5. Insect cell sclerostin bound to a CM5 Biacore chip interacted with MBP-BMP6 with a Kd of 0.5 × 10−9M. MBP alone did not significantly bind to sclerostin. Similar findings were observed with the first β-propeller of LRP5.
Sclerostin interacts with Cyr61
Yeast two-hybrid experiments demonstrated that sclerostin used as “bait” bound the extracellular matrix protein, Cyr61 (CCN1). Yeast colonies in which the sclerostin “bait” associated with expressed “prey” protein were identified by their blue color and their ability to grow on quadruple drop-out medium containing antibiotic (SD/-Leu/-Trp/-His/-Ade with X-αGal/Aureobasidin A). Two such colonies contained an in-frame DNA insert for the Cyr61 protein. Ninety, C-terminal amino acid residues (aa 292–381, beginning with aa 292SPEPV- through to the C- terminal, 381D) encompassing almost the entire cystine-knot domain were encoded by the DNA insert. Interactions were confirmed by co-expression of proteins in yeast cells followed by immunoprecipitation of one of the binding partners with a specific antibody, and identification of the second member of the binding pair with another specific antibody (Figure 1). In lane A, proteins expressed in yeast cells were immuno-precipitated with c-myc antibody; in lane B, proteins were precipitated with HA antibody; whereas in lane C, no antibodies were used for precipitation. The immunoblot with lanes A-C was probed with a c-myc antibody that is expressed as an N-terminal fusion with sclerostin from the pGBKT7 plasmid. As expected, in lane A, the immuno-precipitating c-myc antibody precipitated the sclerostin fusion which was subsequently detected with c-myc antibody. In lane B, the immuno-precipitating HA antibody precipitated the HA-Cyr61 fusion protein to which sclerostin fusion protein was bound. The c-myc-sclerostin fusion protein was subsequently detected by the c-myc antibody. In lane D, the immuno-precipitating c-myc antibody precipitated the c-myc-sclerostin fusion that was subsequently detected by a specific monoclonal antibody directed against the N-terminal portion of sclerostin. In lane D, the immuno-precipitating HA antibody precipitated the HA-Cyr61 fusion protein to which sclerostin fusion protein was bound. The sclerostin fusion protein was subsequently detected by the specific sclerostin monoclonal antibody.
Figure 1.
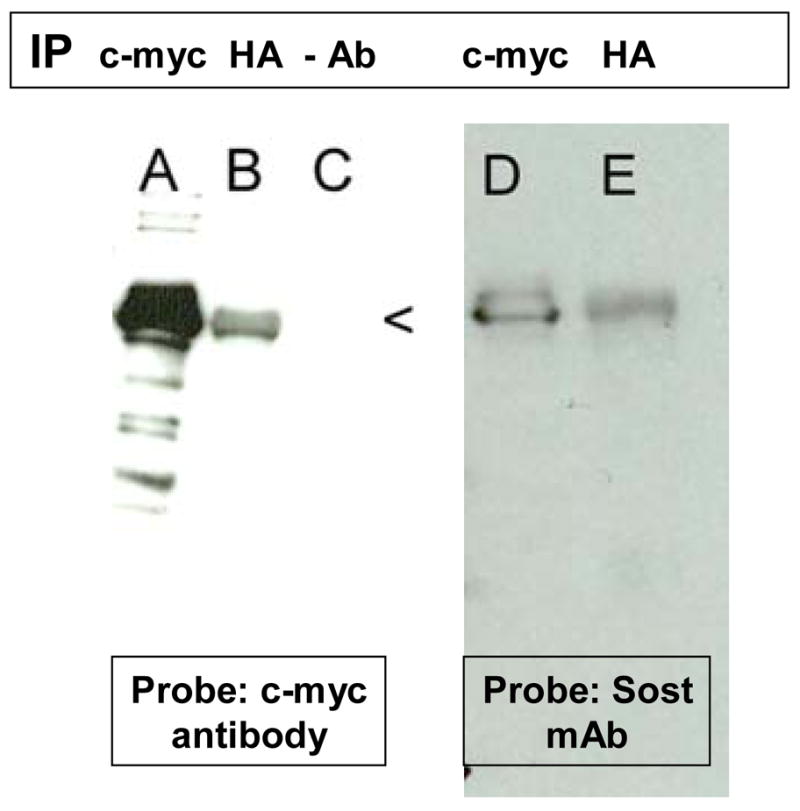
Yeast cells containing c-myc sclerostin and HA Cyr61 expression plasmids were grown for 3 days at 30°C. Cells were lysed and immunoprecipitation was performed using c-myc (lanes A and D) or HA (lanes B and E) antibodies or no antibodies (lane C). The antibody complexes were precipitated by protein A beads. The precipitates were subjected to SDS-PAGE and separated proteins were transferred to PVDF membranes. The membranes were probed with c-myc or sclerostin (SOST) monoclonal antibodies and chemiluminescence methods.
Sclerostin modulates the activity of CYR61
To determine whether the interaction between sclerostin and Cyr61 is biologically relevant, we determined whether sclerostin alters cellular functions such as cell adhesion, vascular migration and cellular growth that are regulated by CYR61. The results of such essays are shown in table 1. When Swiss 3T3 fibroblasts are allowed to attach to cell culture wells, Cyr61 increases the numbers of cells adhering to the surface of the culture wells. The addition of sclerostin alone does not influence cell attachment. However, in the presence of sclerostin and Cyr61, cell attachment was either abolished or considerably reduced. These data indicate that sclerostin alters the biological properties of Cyr61 by influencing cellular adhesion. Cyr61 is known to play a significant role in angiogenesis and vascular cell migration. To test the influence of sclerostin on Cyr61-mediated vascular cell migration, we examined the migration of HUVECs in the presence of Cyr61, sclerostin and a combination of the two proteins (table 1, middle panel). Migration of HUVECs was increased in the presence of both Cyr61 and sclerostin but not by either protein alone. Thus sclerostin is likely to potentiate the pro-angiogenic properties of Cyr61. As expected, VEGF increased HUVEC cell migration. Since sclerostin and Cyr61 are known to influence osteoblast function, we determined the effects of each protein on MC3T3 osteoblast proliferation. Shown in table 1, right panel, are the results of the addition of sclerostin, Cyr61 or both on osteoblast proliferation. Neither sclerostin nor Cyr61 directly influenced osteoblast cell proliferation. Of note, however, a combination of the two proteins increased osteoblast proliferation. This data suggests that sclerostin potentiates Cyr61 function in osteoblasts.
Table 1.
Effect of Cyr61, sclerostin or Cyr61 plus sclerostin on cellular attachement of Swiss 3T3 cells, migration of HUVEC or proliferation of MC3T3 osteoblasts.
| Treatment | Cell Attachment Assay Absorbance at 440 nm, mean (SEM) | Vascular Endothelial Cell Migration Assay # cells migrating into clear area, mean (SEM) | Cell Proliferation Assay Absorbance at 440 nm, mean (SEM) | |||
|---|---|---|---|---|---|---|
| # cells plated × 103 | # cells plated × 103 | |||||
| 4 | 10 | 20 | 0.8 | 4 | ||
| Medium | 0.41 (0.002) | 0.42 (0.003) | 0.64 (0.018) | 10.3 (2.73) | 0.186 (0.003) | 0.49 (0.014) |
| Cyr61 | 0.46 (0.006)**** | 0.45 (0.012)*** | 0.94 (0.128)* | 24.7 (3.48) | 0.195 (0.003) | 0.50 (0.008) |
| Sclerostin | 0.42 (0.004) | 0.40 (0.007) | 0.85 (0.04) | 21.7 (3.53) | 0.192 (0.003) | 0.53 (0.019) |
| Cyr61 + Sclerostin | 0.43 (0.004)** | 0.44 (0.004) | 0.90 (0.105) | 30.3 (6.74)* | 0.202 (0.002)**** | 0.57 (0.009)**** |
| VEGF | - | - | - | 36.7 (5.04)** | ||
p<0.05
p<0.02,
p<0.005,
p<0.001.
Cysteine-rich protein 61 is an extracellular matrix-associated signaling protein that belongs to the CCN family of proteins that include connective tissue growth factor (CTGF), nephroblastoma overexpressed (NOV) and the Wnt-induced secreted proteins (WISP) proteins [22–28]. CYR61 is a potent stimulator of angiogenesis and mesenchymal stem cell expansion and differentiation [26]. VEGF stimulates CYR61 mRNA and protein expression in osteoblasts and increases endothelial cell migration [22]. CCN1/Cyr61 is up-regulated at the early stage of Wnt3A stimulation and RNA interference-mediated knockdown of CCN1/Cyr61 expression diminished Wnt3A-induced osteogenic differentiation [27]. Additionally, Cyr61 also modulates Wnt activity in Xenopus oocytes by binding to the Wnt co-receptor, LRP6 [29, 30]. Cyr61 also inhibits osteoclastogenesis [24]. Additionally, factors which regulate bone turnover and formation such as TGFβ, BMP2, and cortisol increase CYR61 mRNA and protein levels [31]. Our data suggest that sclerostin potentiates Cyr61-mediated cell growth and vascular migration and alters Cyr61-mediated cellular adhesion.
Cyr61 and other members of the CCN family of proteins are multi-domain proteins comprised of (from the N-terminus to the C-terminus) an insulin-like growth factor binding protein-like domain, a Von Willebrand’s factor type C repeat module, a thrombospondin type 1 module and a cystine knot-containing module [28]. The amino acid residues/domains of sclerostin that are responsible for binding to Cyr61 are not known. Since the cDNA inserts contained within the “prey” plasmids express the C-terminal cystine-knot region of Cyr 61, it is likely that this domain of Cyr61 is responsible, at least in part, for the binding of Cyr61 to sclerostin. Sclerostin might regulate bone formation by binding to the cystine knot-containing module of CYR61.
In summary, we have demonstrated that sclerostin binds to Cyr61, a Wnt-regulated and Wnt-regulating protein, and regulates its activity with respect to cellular adhesion, vascular migration and cell growth. We also confirm that sclerostin is bound by rBMP6 and LRP5. Thus, it is likely that sclerostin has multiple pathways of action that converge on Wnt-mediated signaling.
Acknowledgments
Supported by NIH grants DK 76829 and DK 77669 (to RK) and HL70567-07 (DM).
References
- 1.ten Dijke P, Krause C, de Gorter DJ, Lowik CW, van Bezooijen RL. Osteocyte-derived sclerostin inhibits bone formation: its role in bone morphogenetic protein and Wnt signaling. J Bone Joint Surg Am. 2008;90(Suppl 1):31–35. doi: 10.2106/JBJS.G.01183. [DOI] [PubMed] [Google Scholar]
- 2.van Bezooijen RL, Roelen BA, Visser A, van der Wee-Pals L, de Wilt E, Karperien M, Hamersma H, Papapoulos SE, ten Dijke P, Lowik CW. Sclerostin is an osteocyte-expressed negative regulator of bone formation, but not a classical BMP antagonist. J Exp Med. 2004;199:805–814. doi: 10.1084/jem.20031454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Winkler DG, Sutherland MK, Geoghegan JC, Yu C, Hayes T, Skonier JE, Shpektor D, Jonas M, Kovacevich BR, Staehling-Hampton K, Appleby M, Brunkow ME, Latham JA. Osteocyte control of bone formation via sclerostin, a novel BMP antagonist. Embo J. 2003;22:6267–6276. doi: 10.1093/emboj/cdg599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Winkler DG, Sutherland MS, Ojala E, Turcott E, Geoghegan JC, Shpektor D, Skonier JE, Yu C, Latham JA. Sclerostin inhibition of Wnt-3a-induced C3H10T1/2 cell differentiation is indirect and mediated by bone morphogenetic proteins. J Biol Chem. 2005;280:2498–2502. doi: 10.1074/jbc.M400524200. [DOI] [PubMed] [Google Scholar]
- 5.Sutherland MK, Geoghegan JC, Yu C, Turcott E, Skonier JE, Winkler DG, Latham JA. Sclerostin promotes the apoptosis of human osteoblastic cells: a novel regulation of bone formation. Bone. 2004;35:828–835. doi: 10.1016/j.bone.2004.05.023. [DOI] [PubMed] [Google Scholar]
- 6.Veverka V, Henry AJ, Slocombe PM, Ventom A, Mulloy B, Muskett FW, Muzylak M, Greenslade K, Moore A, Zhang L, Gong J, Qian X, Paszty C, Taylor RJ, Robinson MK, Carr MD. Characterization of the structural features and interactions of sclerostin: molecular insight into a key regulator of Wnt-mediated bone formation. J Biol Chem. 2009;284:10890–10900. doi: 10.1074/jbc.M807994200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Balemans W, Ebeling M, Patel N, Van Hul E, Olson P, Dioszegi M, Lacza C, Wuyts W, Van Den Ende J, Willems P, Paes-Alves AF, Hill S, Bueno M, Ramos FJ, Tacconi P, Dikkers FG, Stratakis C, Lindpaintner K, Vickery B, Foernzler D, Van Hul W. Increased bone density in sclerosteosis is due to the deficiency of a novel secreted protein (SOST) Hum Mol Genet. 2001;10:537–543. doi: 10.1093/hmg/10.5.537. [DOI] [PubMed] [Google Scholar]
- 8.Loots GG, Kneissel M, Keller H, Baptist M, Chang J, Collette NM, Ovcharenko D, Plajzer-Frick I, Rubin EM. Genomic deletion of a long-range bone enhancer misregulates sclerostin in Van Buchem disease. Genome Res. 2005;15:928–935. doi: 10.1101/gr.3437105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brunkow ME, Gardner JC, Van Ness J, Paeper BW, Kovacevich BR, Proll S, Skonier JE, Zhao L, Sabo PJ, Fu Y, Alisch RS, Gillett L, Colbert T, Tacconi P, Galas D, Hamersma H, Beighton P, Mulligan J. Bone dysplasia sclerosteosis results from loss of the SOST gene product, a novel cystine knot-containing protein. Am J Hum Genet. 2001;68:577–589. doi: 10.1086/318811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chan A, van Bezooijen RL, Lowik CW. A new paradigm in the treatment of osteoporosis: Wnt pathway proteins and their antagonists. Curr Opin Investig Drugs. 2007;8:293–298. [PubMed] [Google Scholar]
- 11.Winkler DG, Yu C, Geoghegan JC, Ojala EW, Skonier JE, Shpektor D, Sutherland MK, Latham JA. Noggin and sclerostin bone morphogenetic protein antagonists form a mutually inhibitory complex. J Biol Chem. 2004;279:36293–36298. doi: 10.1074/jbc.M400521200. [DOI] [PubMed] [Google Scholar]
- 12.Boyden LM, Mao J, Belsky J, Mitzner L, Farhi A, Mitnick MA, Wu D, Insogna K, Lifton RP. High bone density due to a mutation in LDL-receptor-related protein 5. N Engl J Med. 2002;346:1513–1521. doi: 10.1056/NEJMoa013444. [DOI] [PubMed] [Google Scholar]
- 13.Johnson ML, Gong G, Kimberling W, Recker SM, Kimmel DB, Recker RB. Linkage of a gene causing high bone mass to human chromosome 11 (11q12–13) Am J Hum Genet. 1997;60:1326–1332. doi: 10.1086/515470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Patel MS, Karsenty G. Regulation of bone formation and vision by LRP5. N Engl J Med. 2002;346:1572–1574. doi: 10.1056/NEJM200205163462011. [DOI] [PubMed] [Google Scholar]
- 15.Semenov M, Tamai K, He X. SOST is a ligand for LRP5/LRP6 and a Wnt signaling inhibitor. J Biol Chem. 2005;280:26770–26775. doi: 10.1074/jbc.M504308200. [DOI] [PubMed] [Google Scholar]
- 16.Semenov MV, He X. LRP5 mutations linked to high bone mass diseases cause reduced LRP5 binding and inhibition by SOST. J Biol Chem. 2006;281:38276–38284. doi: 10.1074/jbc.M609509200. [DOI] [PubMed] [Google Scholar]
- 17.Balemans W, Piters E, Cleiren E, Ai M, Van Wesenbeeck L, Warman ML, Van Hul W. The binding between sclerostin and LRP5 is altered by DKK1 and by high-bone mass LRP5 mutations. Calcified tissue international. 2008;82:445–453. doi: 10.1007/s00223-008-9130-9. [DOI] [PubMed] [Google Scholar]
- 18.Ellies DL, Viviano B, McCarthy J, Rey JP, Itasaki N, Saunders S, Krumlauf R. Bone density ligand, Sclerostin, directly interacts with LRP5 but not LRP5G171V to modulate Wnt activity. J Bone Miner Res. 2006;21:1738–1749. doi: 10.1359/jbmr.060810. [DOI] [PubMed] [Google Scholar]
- 19.Li X, Zhang Y, Kang H, Liu W, Liu P, Zhang J, Harris SE, Wu D. Sclerostin binds to LRP5/6 and antagonizes canonical Wnt signaling. J Biol Chem. 2005;280:19883–19887. doi: 10.1074/jbc.M413274200. [DOI] [PubMed] [Google Scholar]
- 20.Craig TA, Sommer SL, Beito TG, Kumar R. Production and characterization of monoclonal antibodies to human sclerostin. Hybridoma (Larchmt) 2009;28:377–381. doi: 10.1089/hyb.2009.0036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bhattacharya R, Kwon J, Li X, Wang E, Patra S, Bida JP, Bajzer Z, Claesson-Welsh L, Mukhopadhyay D. Distinct role of PLCbeta3 in VEGF-mediated directional migration and vascular sprouting. J Cell Sci. 2009;122:1025–1034. doi: 10.1242/jcs.041913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Athanasopoulos AN, Schneider D, Keiper T, Alt V, Pendurthi UR, Liegibel UM, Sommer U, Nawroth PP, Kasperk C, Chavakis T. Vascular endothelial growth factor (VEGF)-induced up-regulation of CCN1 in osteoblasts mediates proangiogenic activities in endothelial cells and promotes fracture healing. J Biol Chem. 2007;282:26746–26753. doi: 10.1074/jbc.M705200200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Brigstock DR. The CCN family: a new stimulus package. The Journal of endocrinology. 2003;178:169–175. doi: 10.1677/joe.0.1780169. [DOI] [PubMed] [Google Scholar]
- 24.Crockett JC, Schutze N, Tosh D, Jatzke S, Duthie A, Jakob F, Rogers MJ. The matricellular protein CYR61 inhibits osteoclastogenesis by a mechanism independent of alphavbeta3 and alphavbeta5. Endocrinology. 2007;148:5761–5768. doi: 10.1210/en.2007-0473. [DOI] [PubMed] [Google Scholar]
- 25.Schutze N, Noth U, Schneidereit J, Hendrich C, Jakob F. Differential expression of CCN-family members in primary human bone marrow-derived mesenchymal stem cells during osteogenic, chondrogenic and adipogenic differentiation. Cell Commun Signal. 2005;3:5. doi: 10.1186/1478-811X-3-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schutze N, Schenk R, Fiedler J, Mattes T, Jakob F, Brenner RE. CYR61/CCN1 and WISP3/CCN6 are chemoattractive ligands for human multipotent mesenchymal stroma cells. BMC cell biology. 2007;8:45. doi: 10.1186/1471-2121-8-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Si W, Kang Q, Luu HH, Park JK, Luo Q, Song WX, Jiang W, Luo X, Li X, Yin H, Montag AG, Haydon RC, He TC. CCN1/Cyr61 is regulated by the canonical Wnt signal and plays an important role in Wnt3A-induced osteoblast differentiation of mesenchymal stem cells. Molecular and cellular biology. 2006;26:2955–2964. doi: 10.1128/MCB.26.8.2955-2964.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Holbourn KP, Acharya KR, Perbal B. The CCN family of proteins: structure-function relationships. Trends in biochemical sciences. 2008;33:461–473. doi: 10.1016/j.tibs.2008.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Latinkic BV, Mercurio S, Bennett B, Hirst EM, Xu Q, Lau LF, Mohun TJ, Smith JC. Xenopus Cyr61 regulates gastrulation movements and modulates Wnt signalling. Development (Cambridge, England) 2003;130:2429–2441. doi: 10.1242/dev.00449. [DOI] [PubMed] [Google Scholar]
- 30.Mercurio S, Latinkic B, Itasaki N, Krumlauf R, Smith JC. Connective-tissue growth factor modulates WNT signalling and interacts with the WNT receptor complex. Development (Cambridge, England) 2004;131:2137–2147. doi: 10.1242/dev.01045. [DOI] [PubMed] [Google Scholar]
- 31.Parisi MS, Gazzerro E, Rydziel S, Canalis E. Expression and regulation of CCN genes in murine osteoblasts. Bone. 2006;38:671–677. doi: 10.1016/j.bone.2005.10.005. [DOI] [PubMed] [Google Scholar]


