Abstract
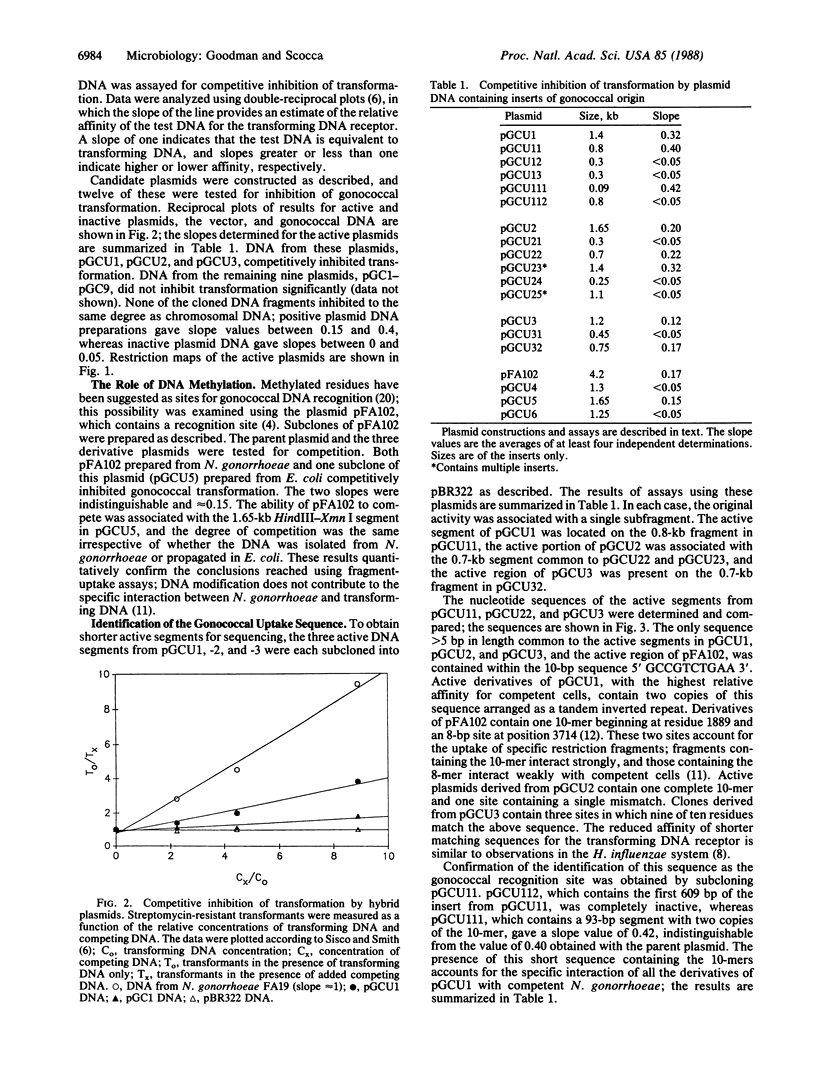
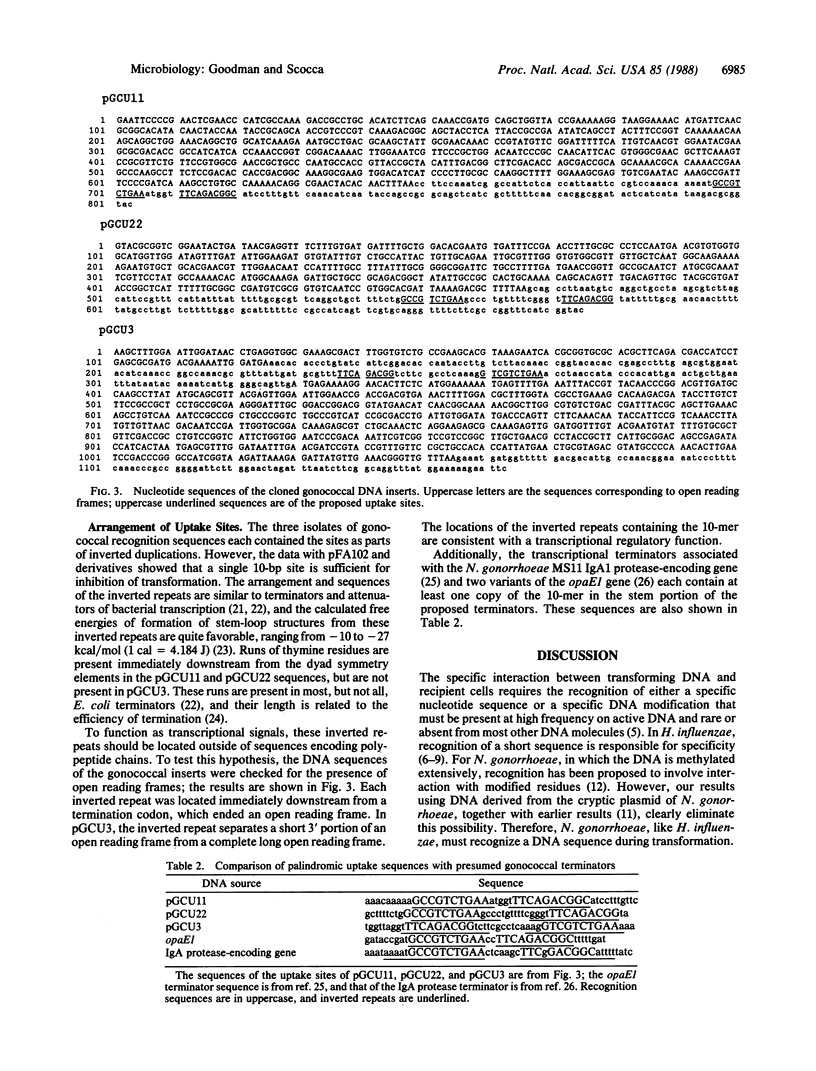
DNA segments from Neisseria gonorrhoeae, cloned and propagated in Escherichia coli, were tested for the ability to competitively inhibit gonococcal transformation. The nucleotide sequences of active segments were determined and compared; these sequences contained the sequence 5' GCCGTCTGAA 3' in common. Subcloning studies confirmed the identity of this sequence as the gonococcal DNA recognition site. The three instances of the recognition sequence isolated from N. gonorrhoeae chromosomal DNA contain the sequence in the immediate neighborhood of its inverted repeat. Because a single copy of the sequence functions as a recognition site, the inverted duplication is not required for specific binding. The dyad symmetric arrangements of the chromosomal recognition sequences may form stable stem-loop structures that can function as terminators or attenuators of transcription. These inverted repeats are located at the boundaries of long open reading frames. The recognition sequence also constitutes part of two other probable terminators of gonococcal genes. We conclude that the signal for recognition of transforming DNA by gonococci is a frequent component of transcriptional terminator sequences. This regulatory function might account for the origin and maintenance of recognition sequences in the chromosomes of Gram-negative transformable bacteria.
Full text
PDF


Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Brendel V., Hamm G. H., Trifonov E. N. Terminators of transcription with RNA polymerase from Escherichia coli: what they look like and how to find them. J Biomol Struct Dyn. 1986 Feb;3(4):705–723. doi: 10.1080/07391102.1986.10508457. [DOI] [PubMed] [Google Scholar]
- Burnstein K. L., Dyer D. W., Sparling P. F. Preferential uptake of restriction fragments from a gonococcal cryptic plasmid by competent Neisseria gonorrhoeae. J Gen Microbiol. 1988 Mar;134(3):547–557. doi: 10.1099/00221287-134-3-547. [DOI] [PubMed] [Google Scholar]
- Danner D. B., Deich R. A., Sisco K. L., Smith H. O. An eleven-base-pair sequence determines the specificity of DNA uptake in Haemophilus transformation. Gene. 1980 Nov;11(3-4):311–318. doi: 10.1016/0378-1119(80)90071-2. [DOI] [PubMed] [Google Scholar]
- Dougherty T. J., Asmus A., Tomasz A. Specificity of DNA uptake in genetic transformation of gonococci. Biochem Biophys Res Commun. 1979 Jan 15;86(1):97–104. doi: 10.1016/0006-291x(79)90386-3. [DOI] [PubMed] [Google Scholar]
- Fitzmaurice W. P., Benjamin R. C., Huang P. C., Scocca J. J. Characterization of recognition sites on bacteriophage HP1c1 DNA which interact with the DNA uptake system of Haemophilus influenzae Rd. Gene. 1984 Nov;31(1-3):187–196. doi: 10.1016/0378-1119(84)90209-9. [DOI] [PubMed] [Google Scholar]
- Gotschlich E. C., Blake M. S., Koomey J. M., Seiff M., Derman A. Cloning of the structural genes of three H8 antigens and of protein III of Neisseria gonorrhoeae. J Exp Med. 1986 Sep 1;164(3):868–881. doi: 10.1084/jem.164.3.868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graves J. F., Biswas G. D., Sparling P. F. Sequence-specific DNA uptake in transformation of Neisseria gonorrhoeae. J Bacteriol. 1982 Dec;152(3):1071–1077. doi: 10.1128/jb.152.3.1071-1077.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagblom P., Korch C., Jonsson A. B., Normark S. Intragenic variation by site-specific recombination in the cryptic plasmid of Neisseria gonorrhoeae. J Bacteriol. 1986 Jul;167(1):231–237. doi: 10.1128/jb.167.1.231-237.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korch C., Hagblom P., Normark S. Sequence-specific DNA modification in Neisseria gonorrhoeae. J Bacteriol. 1983 Sep;155(3):1324–1332. doi: 10.1128/jb.155.3.1324-1332.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korch C., Hagblom P., Ohman H., Göransson M., Normark S. Cryptic plasmid of Neisseria gonorrhoeae: complete nucleotide sequence and genetic organization. J Bacteriol. 1985 Aug;163(2):430–438. doi: 10.1128/jb.163.2.430-438.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynn S. P., Kasper L. M., Gardner J. F. Contributions of RNA secondary structure and length of the thymidine tract to transcription termination at the thr operon attenuator. J Biol Chem. 1988 Jan 5;263(1):472–479. [PubMed] [Google Scholar]
- Mandel M., Higa A. Calcium-dependent bacteriophage DNA infection. J Mol Biol. 1970 Oct 14;53(1):159–162. doi: 10.1016/0022-2836(70)90051-3. [DOI] [PubMed] [Google Scholar]
- Mathis L. S., Scocca J. J. Haemophilus influenzae and Neisseria gonorrhoeae recognize different specificity determinants in the DNA uptake step of genetic transformation. J Gen Microbiol. 1982 May;128(5):1159–1161. doi: 10.1099/00221287-128-5-1159. [DOI] [PubMed] [Google Scholar]
- Mathis L. S., Scocca J. J. On the role of pili in transformation of Neisseria gonorrhoeae. J Gen Microbiol. 1984 Dec;130(12):3165–3173. doi: 10.1099/00221287-130-12-3165. [DOI] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- Meyer T. F., Billyard E., Haas R., Storzbach S., So M. Pilus genes of Neisseria gonorrheae: chromosomal organization and DNA sequence. Proc Natl Acad Sci U S A. 1984 Oct;81(19):6110–6114. doi: 10.1073/pnas.81.19.6110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukhopadhyay M., Mandal N. C. A simple procedure for large-scale preparation of pure plasmid DNA free from chromosomal DNA from bacteria. Anal Biochem. 1983 Sep;133(2):265–270. doi: 10.1016/0003-2697(83)90080-5. [DOI] [PubMed] [Google Scholar]
- Pohlner J., Halter R., Beyreuther K., Meyer T. F. Gene structure and extracellular secretion of Neisseria gonorrhoeae IgA protease. 1987 Jan 29-Feb 4Nature. 325(6103):458–462. doi: 10.1038/325458a0. [DOI] [PubMed] [Google Scholar]
- Richards G. M. Modifications of the diphenylamine reaction giving increased sensitivity and simplicity in the estimation of DNA. Anal Biochem. 1974 Feb;57(2):369–376. doi: 10.1016/0003-2697(74)90091-8. [DOI] [PubMed] [Google Scholar]
- Roberts M., Piot P., Falkow S. The ecology of gonococcal plasmids. J Gen Microbiol. 1979 Oct;114(2):491–494. doi: 10.1099/00221287-114-2-491. [DOI] [PubMed] [Google Scholar]
- Rosenberg M., Court D. Regulatory sequences involved in the promotion and termination of RNA transcription. Annu Rev Genet. 1979;13:319–353. doi: 10.1146/annurev.ge.13.120179.001535. [DOI] [PubMed] [Google Scholar]
- Scocca J. J., Poland R. L., Zoon K. C. Specificity in deoxyribonucleic acid uptake by transformable Haemophilus influenzae. J Bacteriol. 1974 May;118(2):369–373. doi: 10.1128/jb.118.2.369-373.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sisco K. L., Smith H. O. Sequence-specific DNA uptake in Haemophilus transformation. Proc Natl Acad Sci U S A. 1979 Feb;76(2):972–976. doi: 10.1073/pnas.76.2.972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sparling P. F. Genetic transformation of Neisseria gonorrhoeae to streptomycin resistance. J Bacteriol. 1966 Nov;92(5):1364–1371. doi: 10.1128/jb.92.5.1364-1371.1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stern A., Brown M., Nickel P., Meyer T. F. Opacity genes in Neisseria gonorrhoeae: control of phase and antigenic variation. Cell. 1986 Oct 10;47(1):61–71. doi: 10.1016/0092-8674(86)90366-1. [DOI] [PubMed] [Google Scholar]
- Tinoco I., Jr, Borer P. N., Dengler B., Levin M. D., Uhlenbeck O. C., Crothers D. M., Bralla J. Improved estimation of secondary structure in ribonucleic acids. Nat New Biol. 1973 Nov 14;246(150):40–41. doi: 10.1038/newbio246040a0. [DOI] [PubMed] [Google Scholar]



