Abstract
E-cadherin (E-cad) is an adhesion molecule associated with tumor invasion and metastasis. Its down-regulation is associated with poor prognosis for many epithelial tumor types. We have profiled E-cad in the NCI-60 cancer cell lines at the DNA, RNA, and protein levels using six different microarray platforms plus bisulfite sequencing. Here we consider the effects on E-cad expression of eight potential regulatory factors: E-cad promoter DNA methylation; the transcript levels of six transcriptional repressors (SNAI1, SNAI2, TCF3, TCF8, TWIST1, and ZFHX1B); and E-cad DNA copy number. Combined bioinformatic and pharmacological analyses indicate the following ranking of influence on E-cad expression: (i) E-cad promoter methylation appears predominant, is strongly correlated with E-cad expression, and shows a 20–30% threshold above which E-cad expression is silenced; (ii) TCF8 expression levels correlate with (−0.62) and predict (p<0.00001) E-cad expression; (iii) SNAI2 and ZFHX1B expression levels correlate positively with each other (+0.83) and also correlate with (−0.32 and −0.30, respectively) and predict (p=0.03 and 0.01, respectively) E-cad expression; (iv) TWIST1 correlates with (−0.34) but does not predict E-cad expression (v) SNAI1 expression, TCF3 expression, and E-cad DNA copy number do not correlate with or predict E-cad expression. Predictions of E-cad regulation based on the above factors were tested and verified by (i) demethylation studies using 5-aza-2’-deoxycytidine treatment (5-AC); (ii) siRNA knock-down of TCF8, SNAI2, or ZFHX1B expression; (iii) combined treatment with 5-AC and TCF8 siRNA. Finally, levels of cellular E-cad expression are associated with levels of cell-cell adhesion and response to drug treatment.
Keywords: E-cadherin, methylation, adhesion, transcriptional repressors, integromics
Introduction
E-cadherin (E-cad) is a transmembrane glycoprotein that functions to maintain stable cell-cell contacts in epithelial cell types (1). It forms Ca+2-dependent homodimers that bind to their counterparts in adjacent cells, resulting in the formation of intercellular adherens junctions (2). Down-regulation of E-cad has been described in multiple carcinoma types during tumor progression (3–6). Its down-regulation, a sign of poor prognosis for multiple types of epithelial carcinomas (7–9), is associated with increases in both invasion (3, 10, 11) and metastasis (8, 12). In melanocytes, E-cad down-regulation and a concurrent up-regulation of N-cadherin lead to altered cell-cell relationships; whereas normal melanocytes interact primarily with keratinocytes, melanoma cells interact more strongly with melanocytes and fibroblasts (13, 14).
Multiple single factors have been reported to regulate E-cad expression in one or another cancer type (3–6, 15–23). However, those factors have not been studied together in combination as a system and across the spectrum of cancers. Accordingly, to provide a integrative portrait of E-cad regulation within and across cancer cell types, we have used six different microarray platforms and bisulfite sequencing to assess eight potential E-cad regulatory factors in the NCI-60 human cancer cell line panel at the DNA, RNA, protein, and epigenetic levels. One very practical motivation for understanding the complexities of E-cad regulation is the potential for reversing down-regulation of E-cad and restoring its function. That might, in principle, be achieved by use of agents that reverse promoter region methylation or by knocking down relevant transcriptional repressors.
The NCI-60 panel consists of 60 diverse human cancer cell lines used by the National Cancer Institute’s Developmental Therapeutics Program (NCI-DTP) to screen compounds for anticancer activity (24). The panel includes leukemias, melanomas, and cancer cells of breast, central nervous system (glioma), colon, non-small cell lung, ovarian, prostate, and renal origin. It constitutes the most comprehensively profiled set of cells in existence, having been analyzed at the DNA, RNA, protein, chromosomal, metabolomic, and pharmacological levels (25). Profiling of the NCI-60 has been considered a forerunner of The Cancer Genome Atlas Project (http://cancergenome.nih.gov/), which is restricted to the nucleic acid level but in the more difficult context of clinical tumors.
To test whether the correlative relationships uncovered are causal at the molecular level and whether they provide the basis for strategies to up-regulate E-cad on a cell type-specific basis, we followed up with siRNA knockdown and 5-azacytidine demethylation experiments. This overall “integromic” (26) approach, supported by functional data, yields a picture of the multi-factorial, regulation of E-cad expression. It provides the ability to predict rationally and prospectively, independent of cancer tissue of origin type, whether E-cad will be successfully up-regulated by a given treatment.
Materials and Methods
Cell Lines and Cell Culture
The NCI-60 cells were obtained from the NCI-Frederick Cancer DCTD Tumor/Cell Line Repository1 and cultured as described previously (18, 27). All culture flasks were examined by microscope for anomalies, and the cells were harvested at ~80% confluence. It is important to note, as was stated in the Introduction, that all cell culture, harvests, and purifications were done by a single researcher (WCR) to maximize interoperability of the data.
RNA and DNA Isolation
RNA was isolated as we have described previously (18, 27). Briefly, total RNA was purified using the RNeasy Midi Kit (Qiagen Inc., Valencia, CA) according to manufacturer's instructions. Genomic DNA was purified using either the QIAamp DNA Blood Maxi Kit or the Blood & Cell Culture DNA Maxi Kit (Qiagen), according to manufacturer's instructions (18).
Sodium Bisulfite DNA Modification, PCR Amplification, and Sequencing
Our bisulfite-sequencing protocol for the minimal promoter region of E-cad has been described previously (18). In brief, genomic DNA (5 µg) from each cell line was treated with sodium bisulfite, amplified using nested PCR primers, and sequenced.
Quantitation of Transcript Expression Using Four Microarray Platforms
We have previously described our processing and normalization of NCI-60 transcript expression data from pin-spotted cDNA arrays (Incyte, Inc., Palo Alto, CA) (27, 28), Affymetrix Hu-6800 arrays (Affymetrix, Sunnyvale, CA) (29), Affymetrix HG-U95 arrays (30), and Affymetrix HG-U133 (30). The data from those and the other molecular profiling studies are available in a queryable relational database (CellMiner)2. Further information for these microarrays is available at http://www.broadinstitute.org/mpr/NCI60/NCI60.html for Hu-6800, and at the Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo/), identifiers GDS1761, GSE5949, and GSE5720, for the cDNA array, HG-U95, and HG-U133, respectively.
Quantitation of E-cad Protein Expression Using Reverse-phase Lysate Arrays (RPLA)
Our methods for quantitation of proteins using RPLAs have been described previously (31). Further information for this array is available at the Gene Expression Omnibus, identifier GSE5501.
Array CGH
The arrays comprised 450 cancer-related BAC, PAC, and P1 clones printed in quadruplicate (32). The clones and their genomic locations have been defined previously3. Further information for this array is available at Array Express at http://www.ebi.ac.uk/microarray-as/ae/browse.html?keywords=e-geod-5720.
5-aza-2’-deoxycytidine (5-AC) Inhibition of DNA Methylation
We modified a previously described protocol (33) to study inhibition of DNA methylation by 5-AC. Briefly, 4000–5000 cells were seeded in 96-well cell culture clusters on day 0, treated with 0.1 to 2 mg/ml 5-AC (SIGMA, St. Louis, MO) on days 1, 3 and 5, and washed to remove the drug on days 2 and 4. The cells were lysed on day 6 using Lysis Mixture (Panomics, Inc., Fremont, CA).
RNA Interference
We used synthetic small interfering RNAs (siRNA’s), siTCF8.1, siTCF8.2, siSNAI2.1, siSNAI2.2, siZFHX1B.1 and siZFHX1B.2 (Qiagen Inc., Valencia, CA) to inhibit expression of the transcription factors TCF8, SNAI2 and ZFHX1B, as described previously (34).
Branched-DNA Assay for quantitation of RNA Expression
E-cad, TCF8, ZFHX1B, SNAI2, and PPIB mRNA levels were assayed in 5-AC and siRNA studies using a Branched-DNA Assay (Panomics, Inc.) as described previously (Panomics Inc.) (34).
Statistical Analyses
95% confidence intervals were determined for the correlations between E-cad methylation, expression, and DNA copy number by bootstrap analysis with 10,000 re-samplings. Unless otherwise stated, all calculations were done using R4. Confidence intervals and p-values for prediction of relationships betw5een expression of E-cad and the transcriptional repressors were calculated by multivariate linear regression for the cell lines with ≥30% mean methylation. P values for all correlations were calculated to test the null hypothesis of no association.
Phase Contrast Microscopy
Cell images (at 25x magnification) were obtained using a Zeiss Axiovert 25 inverted phase contrast microscope with a halogen bulb and a Canon DS126071, Rebel XL digital camera. In some cases, the contrast was increased for better visibility.
Drug activity data
The “mechanism-of-action” drug set used in this study has been described previously (32). It comprises 118 compounds whose mechanism is presumptively known. The activities are growth inhibition 50% concentrations, expressed as –log(GI50) and determined as described previously (27, 28).
Results
Multiple Factors can Regulate E-cad Expression
Table 1 summarizes the molecular profiles used in this study. We classify MDA-MB-435 and its ERBB2-transfectant MDA-N as melanomas based on multiple molecular and pharmacological profiles from our laboratory and others (18, 27–29, 35–37). Similarly, we classify OVCAR8/ADR-RES (previously called MCF7/ADR-RES and NCI/ADR-RES) as ovarian based on compelling evidence from our spectral karyotyping (38), CGH (32), single nucleotide polymorphism analysis (39), and microsatellite fingerprinting (40) that it is a (drug-resistant) derivative of OVCAR-8.
Table 1.
E-cadherin methylation, transcript, protein, and DNA copy number, and six E-cad transcriptional repressor transcript profiles.
| E-cadherin profiles | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Mean | Transcript levels | Protein | E-cad transcriptional repressors transcript levels a | ||||||||||
| Cell line b | methylation c | Hu6800d | 9706 cDNA e | U95 f | U133 g | levels h | aCGH i | SNAl1 j,k | SNal2 l,k | TCF3 m,k | TCF8 n,k | TWIST1 o | ZFHX1B p |
| BR:BT-549 | 54 | 4.91 | 0.51 | 4.32 | 4.82 | −1.46 | −0.15 | 5.75 | 7.09 | 6.28 | 7.59 | 6.82 | 3.68 |
| BR:HS578T | 47 | 4.58 | 0.47 | 4.32 | 4.91 | −0.69 | NAp | 5.59 | 10.11 | 5.95 | 8.32 | 6.27 | 4.42 |
| BR:MCF7 | 21 | 9.39 | 2.82 | 9.01 | 8.39 | 1.63 | 0.11 | 5.74 | 4.38 | 6.35 | 3.63 | 3.84 | 3.02 |
| BR:MDA-MB-231 | 13 | 4.32 | 0.21 | 4.32 | 4.87 | −2.62 | −0.37 | 5.41 | 8.16 | 5.91 | 7.42 | 3.49 | 3.32 |
| BR:T47D | 6 | 8.96 | 2.96 | 9.44 | 8.46 | 1.66 | −0.04 | 5.48 | 4.22 | 6.22 | 3.63 | 3.86 | 3.09 |
| CNS:SF-268 | 48 | 4.70 | 0.40 | 4.32 | 4.56 | −2.06 | −0.26 | 5.36 | 6.98 | 5.86 | 7.04 | 6.15 | 5.11 |
| CNS:SF-295 | 43 | 4.32 | 0.66 | 4.32 | 4.99 | −1.37 | 0.33 | 6.33 | 7.32 | 5.89 | 7.18 | 6.30 | 3.37 |
| CNS:SF-539 | 73 | 4.32 | 0.75 | 4.32 | 4.85 | −1.73 | −0.13 | 6.61 | 9.91 | 5.97 | 7.42 | 5.78 | 4.06 |
| CNS:SNB19 | 86 | 5.17 | 0.46 | 4.30 | 4.92 | −1.16 | 0.40 | 5.64 | 6.00 | 6.18 | 6.22 | 6.26 | 3.49 |
| CNS:SNB-75 | 16 | 4.39 | 0.62 | 4.32 | 4.81 | −1.92 | −0.03 | 6.57 | 7.94 | 6.37 | 7.01 | 5.65 | 4.93 |
| CNS:U251 | 93 | 5.29 | 0.39 | 4.30 | 4.83 | −1.74 | −0.10 | 5.51 | 6.39 | 5.98 | 5.61 | 5.43 | 3.84 |
| CO:COLO205 | 8 | 9.52 | 2.34 | 9.50 | 7.77 | 1.08 | −0.14 | 5.97 | 4.26 | 6.11 | 3.81 | 3.74 | 3.09 |
| CO:HCC-2998 | 11 | 7.81 | 1.46 | 8.58 | 6.13 | 1.56 | 0.33 | 5.98 | 4.37 | 6.50 | 3.65 | 4.02 | 2.98 |
| CO:HCT-116 | 9 | 6.29 | 0.60 | 5.90 | 4.97 | −0.16 | 0.03 | 6.19 | 4.44 | 6.42 | 4.52 | 4.16 | 3.12 |
| CO:HCT-15 | 11 | 8.08 | 2.22 | 8.99 | 6.47 | 1.10 | 0.01 | 5.65 | 4.21 | 6.30 | 3.57 | 3.66 | 3.17 |
| CO:HT29 | 8 | 5.75 | 1.53 | 8.50 | 6.05 | 1.51 | −0.01 | 5.72 | 4.36 | 6.26 | 3.70 | 3.64 | 3.19 |
| CO:KM12 | 6 | 7.55 | 1.27 | 8.04 | 6.33 | 0.32 | 0.03 | 5.83 | 4.56 | 6.36 | 4.17 | 3.72 | 3.09 |
| CO:SW-620 | 6.75 | 0.80 | 7.02 | 5.20 | 0.07 | 0.12 | 6.17 | 4.97 | 6.18 | 5.18 | 3.61 | 3.12 | |
| LC:A549-ATCC | 14 | 6.89 | 0.88 | 6.90 | 5.12 | −0.52 | 0.15 | 5.98 | 4.38 | 6.58 | 5.77 | 3.99 | 3.13 |
| LC:EKVX | 7 | 7.72 | 1.13 | 6.90 | 5.58 | −0.03 | 0.33 | 5.93 | 4.50 | 6.34 | 6.05 | 3.72 | 3.08 |
| LC:HOP-62 | 21 | 4.32 | 0.41 | 4.32 | 4.88 | −1.40 | −0.15 | 6.14 | 5.35 | 6.14 | 6.14 | 5.79 | 3.50 |
| LC:HOP-92 | 7 | 4.32 | 0.30 | 4.32 | 4.71 | −1.76 | 0.21 | 5.89 | 7.75 | 6.57 | 6.95 | 3.99 | 3.12 |
| LC:NCI-H226 | 12 | 6.11 | 0.46 | 4.30 | 4.86 | −1.44 | 0.00 | 5.69 | 7.32 | 6.14 | 6.37 | 5.55 | 3.74 |
| LC:NCI-H23 | 12 | 44.6 | 0.27 | 4.30 | 4.69 | −2.31 | 0.11 | 5.61 | 5.20 | 5.93 | 6.67 | 5.85 | 3.59 |
| LC:NCI-H322M | 9 | 8.08 | 2.36 | 9.25 | 6.63 | 1.80 | NA p | 5.55 | 4.84 | 6.16 | 3.51 | 4.36 | 3.07 |
| LC:NCI-H460 | 41 | 4.32 | 0.58 | 4.30 | 4.90 | −0.55 | 0.46 | 6.09 | 4.95 | 6.50 | 5.66 | 3.64 | 3.42 |
| LC:NCI-H522 | 15 | 4.46 | 0.34 | 4.32 | 4.94 | −1.17 | NA p | 6.07 | 4.32 | 6.24 | 6.74 | 5.09 | 3.17 |
| LE:CCRF-CEM | 88 | 4.75 | 0.39 | 4.32 | 4.99 | −2.58 | 0.22 | 5.62 | 4.39 | 6.39 | 8.17 | 4.08 | 3.19 |
| LE:HL-60 | 85 | 4.46 | 0.35 | 4.32 | 5.24 | −2.59 | 0.44 | 6.07 | 4.29 | 6.50 | 6.12 | 4.37 | 4.43 |
| LE:K-562 | 98 | 5.83 | 0.36 | 4.32 | 4.97 | −3.29 | 0.06 | 6.15 | 4.32 | 6.06 | 4.73 | 3.80 | 5.19 |
| LE:MOLT-4 | 97 | 4.32 | 0.47 | 4.32 | 5.03 | −2.13 | 0.00 | 5.61 | 4.48 | 6.36 | 7.42 | 3.97 | 3.16 |
| LE:RPMI-8226 | 39 | 5.39 | 0.84 | 4.32 | 5.06 | −1.48 | 0.40 | 5.94 | 5.62 | 6.38 | 5.34 | 3.69 | 3.15 |
| LE:SR | 95 | 4.32 | 0.36 | 4.32 | 4.84 | −1.51 | −0.02 | 5.41 | 4.73 | 6.03 | 6.43 | 5.54 | 4.35 |
| ME:LOXIMVI | 85 | 4.32 | 0.52 | 4.32 | 4.79 | −1.64 | −0.17 | 5.66 | 9.98 | 6.11 | 7.54 | 5.06 | 4.00 |
| ME:M14 | 20 | 4.32 | 0.69 | 4.32 | 5.03 | −2.52 | −0.05 | 5.74 | 8.90 | 6.22 | 3.72 | 5.97 | 4.93 |
| ME:MALME-3M | 12 | 7.28 | 1.00 | 9.04 | 6.23 | −0.28 | −0.44 | 5.81 | 8.14 | 6.32 | 3.88 | 4.48 | 4.78 |
| ME:MDA-MB-435 r | 84 | 4.32 | 0.33 | 4.32 | 5.14 | −1.44 | −0.42 | 5.95 | 9.57 | 6.16 | 5.08 | 5.70 | 4.60 |
| ME:MDA-N r | 84 | 4.32 | 0.35 | 4.32 | 4.80 | −1.49 | −0.46 | 5.73 | 9.65 | 5.92 | 5.71 | 6.03 | 5.69 |
| ME:SK-MEL-2 | 4.32 | 0.40 | 4.32 | 5.11 | −1.74 | −0.03 | 6.06 | 6.46 | 6.57 | 4.44 | 6.32 | 4.09 | |
| ME:SK-MEL-28 | 5.25 | 0.33 | 4.32 | 4.76 | −2.37 | −0.47 | 5.51 | 9.86 | 5.96 | 4.07 | 5.71 | 5.85 | |
| ME:SK-MEL-5 | 80 | 6.89 | 0.58 | 5.13 | 5.23 | −2.13 | −0.23 | 5.81 | 8.90 | 6.43 | 4.36 | 5.64 | 4.31 |
| ME:UACC-257 | 7 | 7.34 | 1.17 | 7.48 | 5.58 | −0.64 | −0.04 | 5.76 | 10.00 | 6.29 | 3.52 | 4.70 | 5.07 |
| ME:UACC-62 | 59 | 4.32 | 0.45 | 4.32 | 4.98 | −3.05 | −0.29 | 6.47 | 8.55 | 6.44 | 5.36 | 5.74 | 4.10 |
| OV:IGROV1 | 4.95 | 0.61 | 4.32 | 4.93 | −1.97 | 0.03 | 5.86 | 4.49 | 6.61 | 5.87 | 3.60 | 3.06 | |
| OV:OVCAR-3 | 8 | 7.20 | 0.99 | 6.83 | 5.47 | −1.70 | −0.60 | 6.23 | 4.42 | 6.28 | 3.85 | 4.98 | 3.06 |
| OV:OVCAR-4 | 5 | 7.22 | 1.20 | 7.43 | 5.56 | 1.80 | −0.48 | 6.19 | 4.27 | 6.48 | 3.75 | 5.09 | 3.19 |
| OV:OVCAR-5 | 13 | 5.70 | 0.41 | 4.32 | 4.81 | −1.91 | −0.20 | 5.69 | 5.42 | 6.37 | 6.40 | 3.67 | 3.29 |
| OV:OVCAR-8 | 85 | 5.21 | 0.32 | 4.32 | 4.82 | −1.78 | 0.22 | 5.81 | 4.97 | 6.18 | 6.53 | 3.69 | 3.57 |
| OV:OVCAR8/ADR-RES s | 89 | 6.19 | 0.32 | 4.32 | 4.96 | −2.03 | 0.24 | 6.14 | 5.95 | 6.24 | 5.29 | 5.77 | 3.46 |
| OV:SKOV3−3 | 8 | 5.36 | 0.72 | 4.32 | 5.32 | −0.92 | 0.15 | 5.86 | 5.95 | 6.46 | 5.50 | 5.09 | 3.13 |
| PR:DU-145 | 14 | 6.74 | 0.47 | 5.58 | 5.19 | −0.16 | 0.04 | 5.98 | 4.60 | 6.34 | 5.41 | 3.73 | 3.22 |
| PR:PC-3 | 15 | 6.75 | 0.69 | 4.83 | 5.21 | −2.18 | −0.07 | 5.89 | 7.23 | 6.52 | 5.48 | 5.67 | 3.33 |
| RE:786-0 | 87 | 4.58 | 0.44 | 4.32 | 4.69 | −1.87 | 0.10 | 6.25 | 6.57 | 6.18 | 5.97 | 3.58 | 3.40 |
| RE:A498 | 93 | 4.46 | 0.53 | 4.32 | 4.61 | −1.61 | 0.34 | 6.49 | 4.73 | 6.43 | 5.50 | 3.64 | 3.52 |
| RE:ACHN | 10 | 5.81 | 0.58 | 5.51 | 5.01 | −2.16 | 0.35 | 5.61 | 4.73 | 6.20 | 5.76 | 3.56 | 3.70 |
| RE:CAKI-1 | 33 | 6.74 | 0.78 | 4.64 | 4.97 | −2.30 | 0.66 | 6.27 | 4.48 | 6.34 | 5.81 | 5.10 | 3.36 |
| RE:RXF-393 | 4.32 | 0.33 | 4.32 | 4.73 | −1.62 | −0.14 | 6.06 | 7.00 | 6.12 | 7.04 | 3.50 | 4.61 | |
| RE:SN12C | 74 | 5.55 | 0.29 | 4.32 | 4.96 | −1.25 | 0.72 | 5.76 | 5.42 | 6.43 | 7.03 | 5.58 | 3.39 |
| RE:TK-10 | 99 | 4.32 | 0.27 | 4.32 | 5.02 | −2.25 | −0.03 | 6.25 | 5.41 | 6.52 | 4.68 | 3.83 | 3.30 |
| RE:UO-31 | 12 | 5.78 | 0.43 | 4.32 | 4.07 | −1.73 | 0.29 | 5.86 | 5.16 | 6.19 | 5.95 | 4.68 | 3.46 |
U133 transcript measurement, log2-transformed. Data processed using the RMA algorithm. Gray blocks indicate levels of transcriptional associated with low levels of E-cad expression (see Figure 1).
Tissues of origin are breast (BR), central nervous system (CNS), colon (CO), non-small cell lung cancer (LC). leukemia (LE), melanoma (ME), ovarian (OV), prostate (PR). and renal (RE).
Mean methylation profile of E-cad promoter. The gray and cross hatched blocks are above or within the 20–30% E-cad expression threshold. respectively.
Gene accession number Z35402. Hu6800 oligonucleotide arrays.
Clone ID 416386. cDNA microarrays. Values are ratios from co-hybridization of an individual cell type and a 12-cell line pool.
Fragment name 977_s_at. Affymetrix HG-U95 oligonucleotide microarray, log2-transformed.
Affymetrix fragment identifier 201130_s_at.
Protein levels expressed as log2 of relative protein concentration from reverse phase protein lysate microarrays.
Ploidy-relative DNA content. Values are log2-transformed. Array spot number 1485. BAG number RMC16P004.
Common name Snail, Affymetrix identifier 219480_at. Exemplar Sequence Accession Number NM_005985. Data
Transcriptional repressors chosen as being representative from multiple probe sets.
Common name SLUG. Affymetrix identifier 213139_at, Exemplar Sequence Accession Number AI572079.
Common name E12/E47 gene product, Affymetrix identifier 216647_at, Exemplar Sequence Accession Number AL117663. HG-U133 transcript measurements, log2-transformed.
Common name ZEB1, Affymetrix identifier 212764 at, Exemplar Sequence Accession Number AI806174.
Common name ACS3, Affymetrix identifier 213943_at, Exemplar Sequence Accession Number NM_000474.
Common name ZEB2, Affymetrix identifier 203603_s_at, Exemplar Sequence Accession Number NM_014795
Data not available.
Cell lines considered to be melanomas. See Results section.
Cell line considered to be a doxorubirin-selected, resistant derivative of OVCAR-8. Previously named MCF7/ADR-RES and NCI/ADR-RES. See results section.
The E-cad promoter methylation levels in Table 1 are expressed as mean methylationpercentage for all 29 CpG cytosines in the minimal promoter region (18). The E-cad mRNA levels were determined using our data from 9,706-clone cDNA arrays (27, 28) and three different Affymetrix chip types, HU-6800 (29), HG-U95 (30), and HG-U133 (30). E-cad protein levels were determined using RPLAs (31). E-cad expression levels were generally higher for epithelial cell types (breast, colon, non-small cell lung, ovarian, prostate, and renal) than for non-epithelial ones (gliomas, leukemias, and melanomas).
Table 1 also contains our array CGH DNA copy number data (ploidy-relative) for E-cad (32) and mRNA expression levels from HG-U133 Affymetrix arrays for the E-cad transcriptional regulatory factors SNAI1, SNAI2, TCF3, TCF8, TWIST1, and ZFHX1B (30). Gray blocks indicate DNA methylation levels (18) and transcriptional repressor transcript levels associated at statistically significant levels (two-tailed p<0.05 by bootstrap with 10,000 iterations) with E-cad down-regulation.
E-cad Methylation is Inversely Correlated with E-cad mRNA and Protein Expression but not with E-cad DNA Copy Number
The mean methylation pattern (Table 1) for E-cad correlated inversely with E-cad transcript levels measured using all four microarray types (r=−0.49, −0.45, −0.52, and −0.38 for the Table 1 Hu6800, cDNA, HG-U95, and HG-U133 microarrays, respectively). It also correlated inversely with the protein (r=−0.49) pattern (data from Table 1). All of those correlations were statistically significant (bootstrap two-tailed p<0.05; data not shown). The ploidy-relative DNA copy number (Table 1), however, showed no significant association with mean methylation, transcript expression, or protein expression. Transcript level data from the four array types were correlated with each other at statistically significant levels, with mean r = +0.86. That is, the four transcript expression platforms corroborated each other, indicating that the measurements were generally robust. The protein and transcript levels also correlated with each other (r = +0.72, +0.81, +0.86, and +0.75 for the Hu6800, cDNA, HG-U95, and HG-U133 microarrays, respectively) at statistically significant levels.
mRNA Levels of Some Transcriptional Regulators are Inversely Correlated with E-cad mRNA Levels
Pearson’s correlation coefficients relating E-cad and E-cad transcriptional repressor expression (Table 1) in the NCI-60 are shown in Table 2. Bold type indicates correlations with p<0.05 (without multiple comparisons correction). Significant negative correlations, consistent across the five E-cad expression platforms, were seen for the NCI-60 for SNAI2, TCF8, TWIST1, and ZFHX1B. SNAI2 and E-cad expression were negatively correlated for breast cancer. TCF8 and E-cad were negatively correlated for breast, lung, and ovarian cancers. TWIST1 and E-cad expression were negatively correlated for the melanomas. All of those sub-panel relationships, however, were based on small numbers of cell types, and they must be considered in the light of multiple comparisons issues.
Table 2.
Correlations between E-cad expression and melhylation parameters, and transcript factor expression levels from HG-U133 DNA microarray analyses.a
| SNAI1 | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| NCI-60 | BR | CNS | CO | LC | LE | ME | OV | RE | |
| methylation | 0.07 | 0.62 | −0.49 | 0.50 | 0.51 | −0.27 | 0.12 | −0.05 | 0.62 |
| Hu6800 | −0.12 | 0.21 | −0.81 | −0.13 | −0.43 | 0.69 | −0.25 | 0.85 | −0.40 |
| 9706 | −0.13 | 0.15 | 0.96 | −0.68 | −0.45 | 0.14 | −0.15 | 0.63 | 0.17 |
| U95 | −0.13 | 0.07 | 0.59 | −0.69 | −0.40 | na b | −0.13 | 0.76 | −0.54 |
| U133 | −0.17 | 0.09 | 0.47 | −0.48 | −0.43 | 0.62 | 0.05 | 0.73 | −0.41 |
| Protein | −0.11 | 0.27 | 0.02 | −0.64 | −0.21 | −0.58 | −0.34 | 0.42 | −0.18 |
| SNAI2 | |||||||||
| NCI-60 | BR | CNS | CO | LC | LE | ME | OV | RE | |
| methylation | 0.17 | 0.59 | −0.26 | 0.76 | −0.23 | −0.90 | 0.31 | 0.37 | 0.11 |
| Hu6800 | −0.39 | −0.91 | −0.77 | −0.39 | −0.28 | 0.26 | 0.07 | −0.39 | −0.63 |
| 9706 | −0.32 | −0.90 | 0.84 | −0.71 | −0.35 | 0.93 | 0.06 | −0.63 | −0.60 |
| U95 | −0.34 | −0.90 | 0.68 | −0.58 | −0.42 | na b | −0.06 | −0.69 | −0.43 |
| U133 | −0.32 | −0.90 | 0.01 | −0.60 | −0.37 | −0.08 | −0.28 | −0.38 | −0.37 |
| Protein | −0.35 | −0.76 | −0.33 | −0.63 | −0.44 | 0.75 | 0.05 | −0.39 | 0.34 |
| TCF3 | |||||||||
| NCI-60 | BR | CNS | CO | LC | LE | ME | OV | RE | |
| methylation | −0.17 | 0.00 | −0.30 | −0.29 | 0.21 | −0.40 | −0.26 | −0.66 | 0.69 |
| Hu6800 | 0.19 | 0.72 | 0.03 | −0.29 | 0.04 | −0.28 | 0.24 | −0.18 | −0.07 |
| 9706 | 0.10 | 0.69 | 0.12 | −0.37 | −0.02 | 0.30 | 0.27 | 0.43 | −0.14 |
| U95 | 0.15 | 0.64 | −0.16 | −0.30 | 0.08 | na b | 0.21 | 0.09 | −0.24 |
| U133 | 0.12 | 0.65 | 0.22 | −0.42 | −0.09 | 0.84 | 0.41 | 0.24 | 0.39 |
| Protein | 0.14 | 0.67 | 0.08 | 0.05 | 0.14 | 0.03 | −0.05 | 0.36 | −0.05 |
| TCF8 | |||||||||
| NCI-60 | BR | CNS | CO | LC | LE | ME | OV | RE | |
| methylation | 0.36 | 0.69 | −0.64 | 0.70 | −0.04 | 0.32 | 0.77 | 0.44 | −0.35 |
| Hu6800 | −0.66 | −0.98 | −0.94 | −0.37 | −0.67 | −0.70 | −0.55 | −0.90 | 0.11 |
| 9706 | −0.62 | −0.98 | 0.74 | −0.77 | −0.94 | −0.27 | −0.48 | −0.88 | −0.19 |
| U95 | −0.70 | −0.99 | 0.94 | −0.77 | −0.84 | na b | −0.46 | −0.91 | −0.13 |
| U133 | −0.62 | −0.99 | −0.11 | −0.65 | −0.91 | −0.07 | −0.52 | −0.92 | −0.19 |
| Protein | −0.53 | −0.87 | −0.24 | −0.85 | −0.92 | 0.13 | −0.15 | −0.62 | 0.71 |
| TWIST1 | |||||||||
| NCI-60 | BR | CNS | CO | LC | LE | ME | OV | RE | |
| methylation | 0.20 | 0.97 | −0.12 | −0.26 | −0.14 | 0.38 | 0.29 | 0.06 | −0.10 |
| Hu6800 | −0.35 | −0.52 | −0.17 | −0.10 | −0.37 | −0.61 | −0.72 | 0.61 | 0.68 |
| 9706 | −0.32 | −0.52 | 0.02 | −0.41 | −0.38 | −0.47 | −0.78 | 0.34 | 0.10 |
| U95 | −0.40 | −0.59 | 0.17 | −0.48 | −0.43 | na b | −0.84 | 0.37 | −0.19 |
| U133 | −0.32 | −0.59 | 0.12 | −0.33 | −0.33 | −0.46 | −0.70 | 0.60 | 0.35 |
| Protein | −0.30 | −0.30 | 0.51 | −0.20 | −0.52 | 0.41 | −0.69 | 0.33 | 0.26 |
| ZFHX1B | |||||||||
| NCI-60 | BR | CNS | CO | LC | LE | ME | OV | RE | |
| methylation | 0.28 | 0.77 | −0.57 | −0.02 | 0.33 | 0.45 | −0.26 | 0.87 | −0.44 |
| Hu6800 | −0.35 | −0.71 | −0.25 | −0.42 | −0.43 | 0.32 | 0.13 | −0.23 | −0.33 |
| 9706 | −0.34 | −0.68 | −0.15 | 0.07 | −0.51 | −0.58 | 0.01 | −0.69 | −0.18 |
| U95 | −0.31 | −0.72 | 0.49 | −0.08 | −0.62 | na b | 0.09 | −0.41 | 0.04 |
| U133 | −0.30 | −0.71 | −0.86 | −0.14 | −0.53 | −0.10 | −0.09 | −0.60 | −0.28 |
| Protein | −0.42 | −0.44 | −0.90 | −0.13 | −0.57 | −0.53 | 0.14 | −0.23 | 0.25 |
Significance of correlations at p<0.05, without comparisons correction, for blocks with bold type. All data profiles are from Table 1.
Correlation not available due to an absence of pattern for E-cad expression in HG-U95 LE’s
Figure 1 shows the statistically significant relationships between E-cad and transcriptional repressor transcript expression levels (Table 1). We previously noted a threshold of 20–30% mean E-cad methylation above which E-cad expression was not detected (18). Four of the repressors, SNAI2, TCF8, TWIST1, and ZFHX1B (Figures 1 A, B, C, and D), showed “L-shaped” relationships with E-cad expression. Brackets indicate the approximate levels of repressor (>5.16, 5.18, 5.25, and 3.30, respectively) above which E-cad expression was predominately at background levels. SNAI1 and TCF3 (data not shown) did not reliably predict E-cad repression. The SNAI2, TCF8, and ZFHX1B E-cad down-regulation regions (as demarcated by brackets) had measurable E-cad expression in two, one, and two cell lines, respectively. Those three E-cad expressing cell lines (MALME-3M and UACC-257 in Figure 1A and D; EKVX in B) indicate a lack of complete repression of E-cad by the transcriptional repressors when expressed within the (bracketed) ranges.
Figure 1.
Distribution of E-cad expression as a function of four transcriptional represser expression levels for the NCI-60. E-cad expression vs. A. SNAI2 expression; B. TCF8 expression; C. TWIST1 expression; D. ZFHX1B expression. In all panels, the X-axis is the log2 transcriptional repressor transcript level obtained from Affymetrix HG-U133 microarrays; the Y-axis is the log2 E-cad expression level from the same arrays (data from Table 1). Diamonds, circles, and squares represent cell lines with <20%, 20–30%, and >30% methylation of the E-cad promoter region, respectively. Pearson’s correlation coefficients (R, from Table 2), p values (P), and confidence intervals (CI) appear in the upper right for each dataset.
TCF8, ZFHX1B and SNAI2 Predict E-cad Expression
For reasons considered in the Discussion section, it appears likely that the transcription factor effects are secondary to methylation status in regulating E-cad expression. Hence, for the following calculation, we adopted a model in which the transcriptional repressors cause differences in transcription rates only when mean methylation is low enough to permit E-cad expression, i.e., <30% (18). When we then tested for a linear relationship between TCF8 and E-cad transcript levels for the cell lines with permissive methylation levels, we found p < 0.00001 and slope (effect size) 0.50 when both variables were represented on a log2 scale. After fitting that model, we tested the residuals for additional effects of SNAI2 (p =0.03; effect size =0.16) and ZFHX1B (p =0.01; effect size =0.4) when methylation is permissive. An F-test was used to check whether addition of SNAI2 or ZFHX1B leads to a significant decrease in the unexplained variance of E-cad expression. Because the levels of SNAI2 and ZFHX1B were significantly correlated (+0.83) over the cell lines with <30% methylation, effects of the two on E-cad expression were not linearly separable. This significant (p<0.0001) correlation suggests coordinate regulation. The statistical calculations were based on standard parametric t-tests for the null hypothesis that the true regression slope was 0. The p-values were not significant for the other transcriptional regulator combinations. The relationships between E-cad expression and TCF8, SNAI2, and ZFHX1B (with SNAI2 and ZFHX1B not being separable) were also maintained qualitatively when all cell lines, regardless of mean methylation level, were included (not shown).
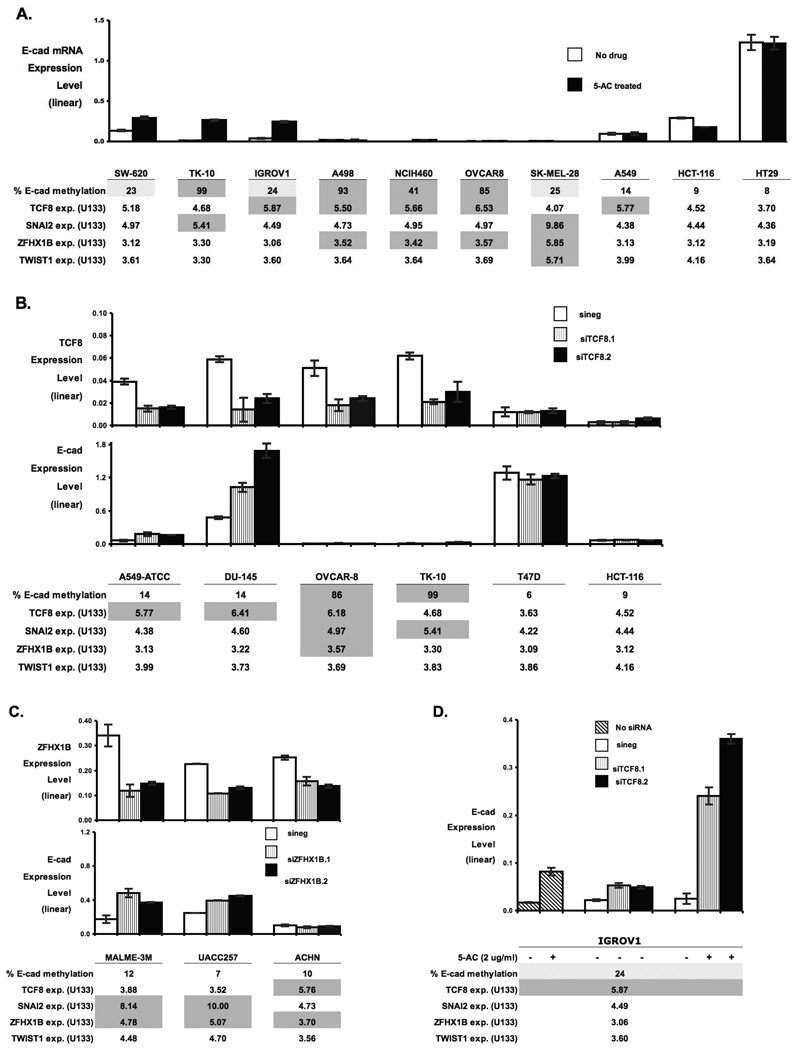
5-AC can up-regulate E-cad Expression
As promoter methylation of E-cad is strongly associated with it’s down-regulation, we choose the DNA demethylation agent 5-AC to determine if we could up-regulate E-cad expression. Figure 2A shows the expression levels of E-cad transcript in ten cell lines as measured by branched-DNA assay, with or without 5-AC treatment. The lines are ordered based on a combination of their actual and predicted levels of E-cad up-regulation following 5-AC treatment. E-cad was up-regulated by 5-AC in the three cell lines (SW-620, TK-10, and IGROV1) that exhibited a combination of (i) repressive levels of methylation (≥20%) and (ii) zero or one transcriptional repressor at levels associated with low levels of E-cad (Figure 1). E-cad was not up-regulated by 5-AC in any of the four cell lines (A498, NCI-H460, OVCAR-8, and SK-MEL-28) with repressive levels of methylation plus two or more transcriptional repressors at levels associated with low E-cad expression (Figure 1). It was also not up-regulated in any of the three cell lines (A549-ATCC, HCT-116, and HT29) with non-repressive levels of E-cad methylation. The latter three cells lines served as controls for secondary effects due to the non-specificity of 5-AC’s demethylating activity.
Figure 2.
Branched-DNA assay measurement of the effect of 5-AC or siRNA treatment on E-cad transcript expression. In all cases, the bar graphs depict branched DNA assay measurements of transcript levels. The Y-axis of each bar is a cyclophilin-normalized expression. The error bars indicate one standard deviation for duplicate measurements. Factors that are proposed as suppressive of E-cad expression are shown in tabular form (log2 data from Table 1), with grayed or cross-hatched blocks indicating proposed repressive levels (as described in Table 1). Because of variations in the number of cells harvested, we do not consider the branched-DNA assay expression levels to be accurate for cross-cell line comparisons. A. The effect of 5-AC on E-cad expression levels. The open bars indicate E-cad expression in the absence of drug. The black bars indicate E-cad expression after 5-AC treatment for 24 hours at 2, 0.5, 0.5, 2, 2, 0.5, 2, 2, 0.1, and 1 mg/ml (for SW-620, TK-10, IGROV1, A498, NCI-H460, OVCAR-8, SK-MEL-28, A549-ATCC, HCT-116, and HT29, respectively). B. The effect of TCF8 down-regulation by siRNA on E-cad expression levels. Six cell lines were treated with siNeg (open bars), siTCF8.1 (vertically-lined bars), or siTCF8.2 (black bars), and their TCF8 and E-cad expression levels determined. C. The effect of ZFHX1B down-regulation by siRNA on E-cad expression levels. Three cell lines were treated with siNeg (open bars), siZFHX1B.1 (vertically-lined bars), or siZFHX1B.2 (black bars), and their ZFHX1B and E-cad expression levels determined. D. The effect of combined 5-AC treatment and TCF8 down-regulation by siRNA on E-cad expression levels. IGROV1 was treated with no siRNA (left-diagonal strips), siNeg (open bars), siTCF8.1 (vertically lined bars), or siTCF8.2 (black bars). The cells were treated with 5-AC treatment (+) or no drug (−) for 24 hours at 2 µg/ml.
Effects of TCF8-siRNA on E-cad Expression
The transcript expression of TCF8 had (i) the highest correlation with the E-cad expression profiles (Table 2), (ii) a striking L-shaped data distribution in the plot of E-cad versus TCF8 expression (Figure 1B), and (iii) the strongest predictive value among the six transcriptional repressors for E-cad expression (by parametric t-test, p<0.00001). We therefore used siRNAs (siTCF8.1 and siTCF8.2) to test the effect of TCF8 down-regulation on E-cad expression (Figure 2B). The first two cell lines in the figure (A549-ATCC and DU-145) were among the best candidates for E-cad up-regulation, and both displayed E-cad up-regulation when TCF8 expression was knocked down. As predicted, OVCAR-8 and TK-10, which both have E-cad DNA methylation levels above the 30% threshold and levels of SNAI2 and ZFHX1B associated with low-level E-cad expression (Figure 1), failed to up-regulate E-cad when TCF8 was knocked down. T47D and HCT-116 both had levels of TCF8 not associated with low E-cad expression, and thus provided controls for non-specific effects. As predicted, neither cell line showed any change in either TCF8 or E-cad expression levels.
Effects of ZFHX1B-siRNA and SNAI2-siRNA on E-cad Expression
The transcript expression levels of ZFHX1B and SNAI2 were (i) significantly correlated with E-cad expression (Table 2), (ii) L-shaped in distribution when plotted against E-cad expression (Figure 1D and A, respectively), (iii) significantly correlated with one another in expression (r=0.83), and (iv) predictive of E-cad expression (by parametric two-tailed t-test, p=0.03 and 0.01, respectively). However, when we tried to identify candidate cell lines in our set with which to test whether ZFHX1B or SNAI2 down-regulation was independently sufficient for up-regulation of E-cad expression, there were none. That is, none of the cells showed ZFHX1B or SNAI2 expression in the range associated with low E-cad expression (Figure 1, Table 1), E-cad DNA methylation levels less than 20%, and levels of ZFHX1B or SNAI2 associated with low E-cad expression. We therefore tested the effects on E-cad expression of ZFHX1B and SNAI2 knock-down by siRNA using the next best possibilities: MALME-3M, UACC-257, and ACHN for ZFHX1B (Figure 2C), and MALME-3M and UACC-257 for SNAI2 (data not shown). Each of those cell lines (for both ZFHX1B and SNAI2) has one additional transcription factor in the range associated with low E-cad expression. MALME-3M and UACC-257, but not ACHN, displayed E-cad up-regulation in the presence of ZFHX1B down-regulation (Figure 2C). MALME-3M and UACC-257 failed to up-regulate E-cad when SNAI2 expression was knocked down.
Combining 5-AC with siTCF8 on TCF8 and E-cad Expression
We tested the effects on E-cad expression of combination treatment directed on the two factors at the top of our proposed ranking of influence on E-cad, DNA methylation and TCF8 (Figure 2D). IGROV1, which has both E-cad methylation and TCF8 at levels associated with low E-cad expression, displayed E-cad up-regulation following either 5-AC or siTCF8 treatment. With combined treatment, the level of E-cad up-regulation was more than additive.
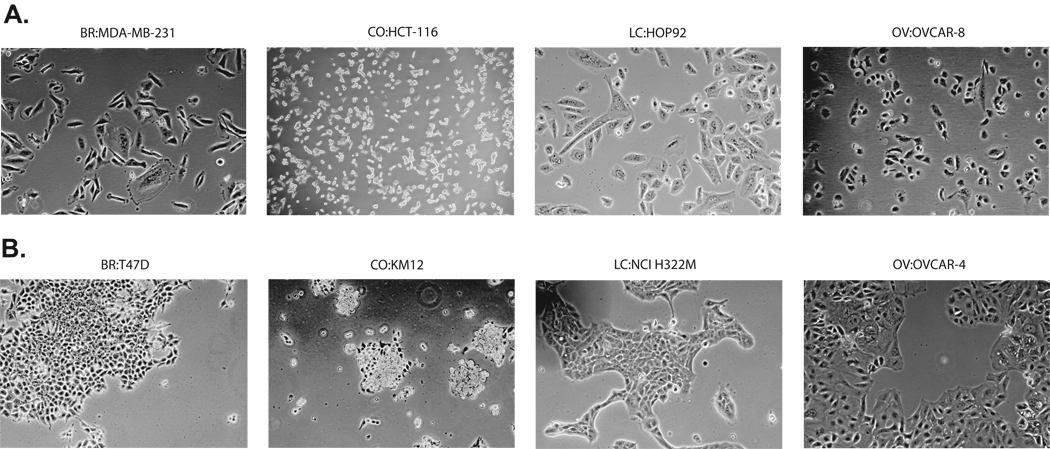
Epithelial Cell Lines with Elevated Levels of E-cad show Increased Cell-cell Adhesion
Using the HG-U133 E-cad expression data in Table 1, Figure 3A shows that cell lines with undetectable (<5.40) E-cad expression displayed lower levels of cell-cell adhesion than did those with detectable levels (Figure 3B). In addition to the obvious increase in cell-cell contact in Figure 3B, T47D, KM12, NCI-H322M, and OVCAR-4 also showed vertical cell stacking. Stacking occurred in none of the panel A cells. An exception to the general trend was the unusual cell line COLO205, which expresses detectable levels of E-cad (transcript and protein) but has an almost leukemic appearance and is only weakly adherent to plastic (image not shown).
Figure 3.
Variations in cell-cell adhesion as observed by phase contrast microscopy. A. Cells with undetectable levels of E-cad expression (<5.40 units on HG-U133 arrays; Table 1). B. Cells with detectable levels of E-cad (≥ 5.4 units on HG-U133 arrays; Table 1).
E-cad Expression in the 54 attached NCI-60 Cell Lines is Correlated with Drug Activities
We next examined the relationship between growth inhibitory 50% (GI50) measurements of drug activity for 118 drugs (27, 28) and E-cad measurements of mean methylation, transcript level, protein level and DNA content (Table 3). The mean of the correlation coefficients of the five E-cad expression measurements for the 54 attached (non-leukemic) cell lines reached significant levels (p<0.02 without multiple comparisons correction) for six alkylating agents, three topoisomerase I inhibitors, and one DNA antimetabolite. Of those ten drugs, all but the DNA antimetabolite showed negative correlations, indicating that as E-cad expression increases, the drug activity decreases. Adding back the leukemias to the correlation calculations (Table 3) yielded consistent, or slightly stronger, correlations for all but the DNA antimetabolite.
Table 3.
Correlations of drug activities versus E-cad transcript, protein and DNA copy numbers a
| Mechanism of action type | Alkylating agents | Topoisomerase 1 inhibitors | DNA antimetaboliteb | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Drug name | Carmustine | Chlorozotocin | PCNUc | Cisplatin | DABQd | Spiromustine | CPT,9-MeOe | CPT,10-OHe | CPT,11-HOMe (RS)e | Inosine-glycodialdehyde |
| NSC | 409962 | 178248 | 95466 | 119875 | 182986 | 172112 | 176323 | 107124 | 606173 | 118994 |
| 56 attached cell lines g | ||||||||||
| Mean methylation | 0.36 | 0.44 | 0.53 | 0.13 | 0.17 | 0.15 | 0.13 | 0.42 | 0.28 | −0.20 |
| Hu6800h | −0.39 | −0.43 | −0.45 | −0.33 | −0.33 | −0.41 | −0.36 | −0.35 | −0.35 | 0.31 |
| cDNA h | −0.22 | −0.24 | −0.25 | −0.29 | −0.26 | −0.23 | −0.39 | −0.39 | −0.32 | 0.36 |
| U95 h | −0.28 | −0.33 | −0.35 | −0.31 | −0.35 | −0.37 | −0.37 | −0.49 | −0.39 | 0.35 |
| U133 h | −0.25 | −0.27 | −0.26 | −0.34 | −0.28 | −0.23 | −0.47 | −0.36 | −0.35 | 0.41 |
| Protein levels | −0.34 | −0.32 | −0.33 | −0.29 | −0.28 | −0.32 | −0.35 | −0.41 | −0.32 | 0.28 |
| DNA copy number | −0.07 | −0.04 | 0.05 | 0.02 | 0.31 | 0.24 | 0.29 | 0.22 | 0.17 | −0.27 |
| NCI-60 | ||||||||||
| Mean methylation | 0.53 | 0.48 | 0.59 | 0.26 | 0.29 | 0.33 | 0.21 | 0.41 | 0.44 | 0.10 |
| Hu6800h | −0.42 | −0.45 | −0.48 | −0.38 | −0.39 | −0.46 | −0.39 | −0.35 | −0.41 | 0.13 |
| cDNA h | −0.27 | −0.26 | −0.28 | −0.31 | −0.30 | −0.27 | −0.40 | −0.39 | −0.35 | 0.19 |
| U95 h | −0.32 | −0.35 | −0.37 | −0.33 | −0.37 | −0.38 | −0.40 | −0.45 | −0.40 | 0.15 |
| U133 h | −0.24 | −0.27 | −0.27 | −0.32 | −0.28 | −0.22 | −0.47 | −0.33 | −0.34 | 0.27 |
| Protein levels | −0.40 | −0.35 | −0.35 | −0.31 | −0.32 | −0.38 | −0.38 | −0.38 | −0.34 | 0.04 |
| DNA copy number | 0.09 | −0.03 | 0.07 | 0.08 | 0.32 | 0.29 | 0.32 | 0.17 | 0.14 | −0.06 |
Drugs selected from a 118 drug set of known mechanism of action based on level of correlation to the 54 attached cell lines. Bold numbers indicate statistically significant correlations at p<0.02 without multiple comparisons correction.
Classifications as a DNA antimetabolite from by DTP.
Polycarbonate-based polyurethane
Diaziridinylbenzoquinoue
Camptothecin.
National Service Center numbers.
The non-leukemic subset of the NCI-60 that grow in tissue cultures attached cells.
Discussion
A number of factors, generally studied in isolation in previous studies, have been shown to influence E-cad expression (4, 15, 16, 18–23). In the present study, we assessed regulation of E-cad expression levels in the NCI-60 cell panel by eight of the potential effectors (Table 1 and Figure 2A–D) using (i) our E-cad promoter DNA methylation data; (ii) data from six different microarray platforms; and (iii) data from pharmacological assays employing RNAi and 5-AC treatment. These factors were assessed in combination, yielding a coherent picture of multi-factorial network regulation of E-cad expression. Included were the effects of DNA methylation of the E-cad promoter region, DNA copy number, and expression levels of the transcriptional repressors SNAI1, SNAI2, TCF3, TCF8, TWIST1, and ZFHX1B. In addition, levels of E-cad were shown to be associated with cell-cell adhesion and to correlate to the potencies of alkylating agents, topoisomerase 1 inhibitors, and a DNA-antimetabolite.
Since transcript and protein levels often do not correlate over diverse cell types (32), it was not obvious that they would in this case. However, we found that the E-cad transcript and protein expression data do correlate with each at statistically significant levels (mean r = 0.83; p-values all <0.05). There was no correlation for DNA copy number; hence no evidence that copy number regulates E-cad expression.
Next, we found that E-cad expression correlates negatively with E-cad DNA promoter region methylation in a consistent and significant manner (correlation coefficient of the mean is −.47, p <.05). In modeling the influences on E-cad expression, we treated methylation as an independent variable because of two lines of evidence. First, there are multiple prior examples of transcription factor binding being blocked by the presence of DNA methylation, (41–43). Second, our data from multiple microarray platforms, as well as the branched DNA assay, indicated that E-cad is not expressed in any of the NCI-60 if the methylation level is greater than approximately 30% (18). That is, DNA methylation (but not the other factors studied) appeared sufficient by itself for down-regulation of E-cad. We, therefore, define a “non-permissive level” of methylation as >30% for purposes of the regulatory and statistical modeling. Down-regulation of E-cad in cancer by promoter region methylation (but not the threshold at which it occurs) has been well documented, (3–6, 17, 18, 44, 45).
Three additional factors found to be associated with and statistically predict E-cad expression in the NCI-60, were SNAI2, TCF8, and ZFHX1B. All showed significant negative correlations across the panel (p<0.05, two-tailed, no multiple comparisons correction) with the five measurements of E-cad expression (Table 2). Additionally, Figures 1A, B, and D show that high expression levels of SNAI2, TCF8, or ZFHX1B are associated with low levels of E-cad expression. Although high levels of TWIST1 were also associated with low E-cad expression (p=0.01), they were not independently predictive once the confounding effects of methylation, TCF8, SNAI2, and ZFHX1B had been removed. The bracketed expression ranges of SNAI2, TCF8, and ZFHX1B (Figures 1A, 1B, and 1D, respectively) associated with low E-cad expression all show “exception” cell lines with detectable E-cad expression (MALME-3M and UACC-257 for SNAI2 and ZFHX1B; EKVX for TCF8), indicating that those ranges do not absolutely prevent E-cad expression. SNAI1 and TCF3 expression were uncorrelated with E-cad expression.
The correlations shown in Table 2 between the six E-cad measurements and the six transcriptional repressor expression levels for the NCI-60 and the eight tissues of origin (excluding prostate for which there are only two lines) provides additional information that both confirms prior and indicates novel relationships. The significant correlations with E-cad expression for SNAI2 in breast and TCF8 in non-small cell lung cancer support prior findings (46, 47). The correlations between E-cad expression and TCF8 in breast and ovarian lines and TWIST1 in melanomas are novel. The correlations with SNAI1, TCF3, and ZFHX1B were not consistently statistically significant for any of the tissue-of-origin types.
Based on this bioinformatic and statistical analysis, we propose for the regulatory factors analyzed that the top three levels of influence on E-cad expression in the NCI-60 are (i) E-cad promoter methylation, (ii) expression of TCF8, and (iii) expression of ZFHX1B or SNAI2 (whose profiles are not linearly separable statistically). E-cad is not expressed at E-cad methylation levels >30%, and is repressed at SNAI2, ZFHX1B and TCF8 HG-U133 intensity levels >5.16, 3.30, and 5.18, respectively (Figures 1 A, D, and B, respectively). In this model, DNA methylation is sufficient for down-regulation of E-cad, whereas the transcriptional repressors are conditionally sufficient (i.e., sufficient in some cell types but not in others).
To test this regulatory ranking and also assess our ability to predict treatment combinations that would result in E-cad up-regulation, we designed functional assays (Figure 2A – D and data for SNAI2 not shown) in which we manipulated the four statistically predictive factors. In each case, the optimal test case for E-cad up-regulation was a cell line with that repressor in its proposed repressive range, levels of the other three repressive factors that were not, and undetectable levels of E-cad expression (i.e., <5.40 by HG-U133 array in Table 1). Quantitation of RNA expression was provided by the branched DNA assay, which proved to be more sensitive than the HG-U133 microarrays.
When choosing cells in which we predict that E-cad would be up-regulated by DNA demethylation, none fit our optimal test case parameters. The flaws in the three best candidates were that TK-10 expresses SNAI2 in its repressive range, and SW-620 and IGROV1 have methylation levels slightly below the 30% cutoff. Still, up-regulation of E-cad followed treatment with 5-AC (for SW-620, TK-10, and IGROV1) by 2.2-, 22-, and 6.1-fold, respectively (Figure 2A), providing functional evidence that (i) the 23 and 24% E-cad DNA methylation levels (in SW-620 and IGROV1, respectively), although repressive, are insufficient to silence E-cad expression completely (see Figure 2A, no drug); (ii) and that the association between E-cad methylation and expression might be causal. E-cad was re-expressed in TK-10 in the presence of repressive levels of SNAI2. We were unable to cause E-cad up-regulation in the presence of multiple transcriptional repressors in their repressive ranges (for A498, NCI-H460, OVCAR-8, and SK-MEL-28). E-cad is not up-regulated in the secondary affect control cell lines, A549, HCT-116 or HT29, in the presence of low (baseline) levels of methylation.
To test E-cad up-regulation following TCF8 down-regulation (Figure 2B) we used two cells lines, A549-ATCC and DU-145, with optimal profiles. E-cad was successfully up-regulated 2.4 and 2.8-fold, respectively, by TCF8 knock-down in those lines. Those findings: (i) support our predictive criteria for identifying cells that will up-regulate E-cad expression following TCF8 down-regulation and (ii) indicate that the E-cad/TCF8 expression association is potentially causal. Our model predicted successfully that E-cad up-regulation would fail in OVCAR-8 because of high DNA methylation. E-cad expression is not re-expressed in TK-10, T47D, and HCT-116 in the absence of repressive TCF-8 levels.
There were no cell lines ideal for testing E-cad up-regulation following down-regulation of ZFHX1B. Still, by using our criteria to select “next best” cell lines, we were able to up-regulate E-cad following ZFHX1B down-regulation in MALME-3M and UACC-257 by 2.4 and 1.7 fold (mean values), respectively (Figure 2C). Those findings (i) support our predictive criteria for identifying cells that can re-express E-cad following ZFHX1B down-regulation and (ii) indicate that the E-cad/ZFHX1B expression association is potentially causal. E-cad is not re-expressed in ACHN, in the presence of repressive levels of TCF8.
There were also no cell lines ideal for testing E-cad up-regulation following SNAI2 down-regulation. We were unable to re-express E-cad in either of our “next best” cell lines, MALME-3M or UACC-257, following SNAI2 down-regulation in the presence of repressive levels of ZFHX1B (data not shown).
Most of the cell lines that showed no detectable E-cad by U133 array have multiple repressive factors in their proposed repressive ranges, implying that multiple interventions might be required for substantial E-cad up-regulation in many cancer cell lines. Therefore, we did an initial test of the effects of siRNA and 5-AC treatment in combination in IGROV1, a cell line that combines the presence of the top two repressive factors in our regulatory ranking, E-cad DNA methylation and TCF8 expression. The results (Figure 2D) were increases in E-cad expression of 4.8-fold for 5-AC alone, a mean of 2.3-fold following siTCF8 down-regulation, and a mean of 12.0-fold for the combination of 5-AC with siTCF8. Those results (i) support our predictive criteria for identifying cells that can be made to re-express E-cad by down-regulating both DNA methylation and TCF8 expression; (ii) indicate that both associations are potentially causal; (iii) suggest that, in the presence of multiple repressive factors, a targeted combination of treatments is likely the optimal approach for up-regulation of E-cad expression.
Both TCF8 and ZFHX1B have previously been described as being targeted by the miR-200 family (48, 49). Consistent with these results, we find significant negative correlations between TCF8 expression and hsa-miR-200a and 200b of -0.49, -0.55, respectively, as well as between ZFHX1B and hsa-miR-200a, 200b, and 200c of -0.56, -0.67, and -0.34, respectively. Levels for the miRNA are from our prior study (50) and are available at http://discover.nci.nih.gov/cellminer/queryLoad.do.
In the present study, we also defined two additional important factors that correlate with E-cad expression in the NCI-60: cell-cell adhesion and the potencies of a variety of drugs. Within the subset of 54 (non-leukemic) attached cell lines, the high expressers of E-cad tend to exhibit higher levels of cell-cell adhesion (Figure 3A and B), consistent with prior reports (3, 10, 11) (19, 20, 22). The leukemias, with no detectable E-cad, grow as spherical detached cells and exhibit limited cell-cell adhesion (data not shown). The drug activity patterns (Table 3) have significant negative correlations to E-cad expression in 9/10 drugs, for both the attached cell subset and the NCI-60.
In conclusion, we report an ‘integromic’ analysis of multiple factors with the potential, either individually or in combination, to regulate E-cad expression. We relate those factors to E-cad expression at the transcript and protein levels. Statistical analysis of this data allows the prediction of which cell lines would show up-regulation of E-cad expression in pharmacological assays using 5-AC or siRNA’s against TCF8, ZFHX1B, or SNAI2. Among those regulatory factors, methylation status is proposed to be non-permissive and sufficient by itself to down-regulate E-cad expression when above a 30% threshold (18). TCF8 expression, SNAI2 and ZFHX1B are proposed to be conditionally sufficient, and repressive ranges are proposed for each of those factors. TWIST1 is correlated with E-cad down-regulation while not being shown to be predictive. SNAI1, TCF3, and DNA copy number show no obvious effect on E-cad. The functional assays done either confirmed or extended the proposed regulatory ranking, leading us to conjecture causality for E-cad regulation for promoter methylation, TCF8, and ZFHX1B. The data thus provide a rational basis for prospectively predicting what pharmacological combinations of DNA demethylation and down-regulation of transcriptional repressors would yield E-cad up-regulation in particular cancer cell types. The findings thus have implications for strategies to suppress cancer invasion and metastasis associated with E-cad loss.
Acknowledgements
This research was supported by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research. We are grateful to the many members of DTP for their work on the screen and Molecular Targets Program. We particularly acknowledge the contributions of Bruce Chabner and Michael Boyd, who led development of the NCI-60, and Kenneth Paull, who pioneered the associated informatics.
Abbreviations
- E-cad
E-cadherin
- SNAI1
snail 1
- SNAI2
snail 2
- TCF3
transcription factor 3
- TCF8
transcription factor 8
- TWIST1
twist
- ZFHX1B
zinc finger homeobox 1b
- 5-AC
5-aza-2’-deoxycytidine treatment
- NCI-DTP
National Cancer Institutes-Developmental Therapeutics Program
- NCI-60
National Cancer Institutes of Health 60 cancer cell lines
- aCGH
array comparative genomic hybridization
- RPLA
reverse-phase lysate microarrays
- HG-U95
Human Genome U95
- HG-U133
Human Genome U133
- BR
breast
- CO
colon
- CNS
central nervous system
- LC
non-small cell lung cancer
- LE
leukemia
- ME
melanoma
- OV
ovarian
- PR
prostate
- RE
renal
Footnotes
See www.r-project.org.
References
- 1.Huber O, Bierkamp C, Kemler R. Cadherins and catenins in development. Curr Opin Cell Biol. 1996;8:685–691. doi: 10.1016/s0955-0674(96)80110-4. [DOI] [PubMed] [Google Scholar]
- 2.Overduin M, Harvey T, Bagby S, et al. Solution structure of the epithelial cadherin domain responsible for selective cell adhesion. Science. 1995;267:386–389. doi: 10.1126/science.7824937. [DOI] [PubMed] [Google Scholar]
- 3.Matsumura T, Makino R, Mitamura K. Frequent down-regulation of E-cadherin by genetic and epigenetic changes in the malignant progression of hepatocellular carcinomas. Clin Cancer Res. 2001;7:594–599. [PubMed] [Google Scholar]
- 4.Li L, Zhao H, Nakajima K, et al. Methylation of the E-cadherin gene promoter correlates with progression of prostate cancer. J Urol. 2001;166:705–709. [PubMed] [Google Scholar]
- 5.Nojima D, Nakajima K, Li L, et al. CpG methylation of promoter region inactivates E-cadherin gene in renal cell carcinoma. Mol Carcinog. 2001;32:19–27. doi: 10.1002/mc.1060. [DOI] [PubMed] [Google Scholar]
- 6.Yoshiura K, Kanai Y, Ochiai A, Shimoyama Y, Sugimura T, Hirohashi S. Silencing of the E-cadherin invasion-suppressor gene by CpG methylation in human carcinomas. Proc Natl Acad Sci U S A. 1995;92:7416–7419. doi: 10.1073/pnas.92.16.7416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Siitonen S, Kononen J, Helin H, Rantala I, Holli K, Isola J. Reduced E-cadherin expression is associated with invasiveness and unfavorable prognosis in breast cancer. Am J Clin Pathol. 1996;105:394–402. doi: 10.1093/ajcp/105.4.394. [DOI] [PubMed] [Google Scholar]
- 8.Paul R, Ewing C, Jarrard D, Isaacs W. The cadherin cell-cell adhesion pathway in prostate cancer progression. Br J Urol. 1997;79:37–43. doi: 10.1111/j.1464-410x.1997.tb00799.x. [DOI] [PubMed] [Google Scholar]
- 9.Dunsmuir W, Gillett C, Meyer L, et al. Molecular markers for predicting prostate cancer stage and survival. BJU Int. 2000;86:869–878. doi: 10.1046/j.1464-410x.2000.00916.x. [DOI] [PubMed] [Google Scholar]
- 10.Berx G, Cleton-Jansen A, Nollet F, et al. E-cadherin is a tumour/invasion suppressor gene mutated in human lobular breast cancers. EMBO J. 1995;14:6107–6115. doi: 10.1002/j.1460-2075.1995.tb00301.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Christofori G, Semb H. The role of the cell-adhesion molecule E-cadherin as a tumour-suppressor gene. Trends Biochem Sci. 1999;24:73–76. doi: 10.1016/s0968-0004(98)01343-7. [DOI] [PubMed] [Google Scholar]
- 12.Nass S, Herman J, Gabrielson E, et al. Aberrant methylation of the estrogen receptor and E-cadherin 5' CpG islands increases with malignant progression in human breast cancer. Cancer Res. 2000;60:4346–4348. [PubMed] [Google Scholar]
- 13.Hsu M, Andl T, Li G, Meinkoth J, Herlyn M. Cadherin repertoire determines partner-specific gap junctional communication during melanoma progression. J Cell Sci. 2000;113:1535–1542. doi: 10.1242/jcs.113.9.1535. [DOI] [PubMed] [Google Scholar]
- 14.Li G, Fukunaga M, Herlyn M. Reversal of melanocytic malignancy by keratinocytes is an E-cadherin-mediated process overriding beta-catenin signaling. Exp Cell Res. 2004;297:142–151. doi: 10.1016/j.yexcr.2004.03.012. [DOI] [PubMed] [Google Scholar]
- 15.Richards F, McKee S, Rajpar M, et al. Germline E-cadherin gene (CDH1) mutations predispose to familial gastric cancer and colorectal cancer. Hum Mol Genet. 1999;8:607–610. doi: 10.1093/hmg/8.4.607. [DOI] [PubMed] [Google Scholar]
- 16.Hiraguri S, Godfrey T, Nakamura H, et al. Mechanisms of inactivation of E-cadherin in breast cancer cell lines. Cancer Res. 1998;58:1972–1977. [PubMed] [Google Scholar]
- 17.Tsutsumida A, Hamada J, Tada M, et al. Epigenetic silencing of E- and P-cadherin gene expression in human melanoma cell lines. Int J Oncol. 2004;25:1415–1421. [PubMed] [Google Scholar]
- 18.Reinhold W, Reimers M, Maunakea A, et al. Detailed DNA methylation profiles of the E-cadherin promoter in the NCI-60 cancer cells. Mol Cancer Ther. 2007;6:391–403. doi: 10.1158/1535-7163.MCT-06-0609. [DOI] [PubMed] [Google Scholar]
- 19.Batlle E, Sancho E, Franci C, et al. The transcription factor snail is a repressor of E-cadherin gene expression in epithelial tumour cells. Nat Cell Biol. 2000;2:84–89. doi: 10.1038/35000034. [DOI] [PubMed] [Google Scholar]
- 20.Bolos V, Peinado H, Perez-Moreno M, Fraga M, Esteller M, Cano A. The transcription factor Slug represses E-cadherin expression and induces epithelial to mesenchymal transitions: a comparison with Snail and E47 repressors. J Cell Sci. 2003;116:499–511. doi: 10.1242/jcs.00224. [DOI] [PubMed] [Google Scholar]
- 21.Comijn J, Berx G, Vermassen P, et al. The two-handed E box binding zinc finger protein SIP1 downregulates E-cadherin and induces invasion. Mol Cell. 2001;7:1267–1278. doi: 10.1016/s1097-2765(01)00260-x. [DOI] [PubMed] [Google Scholar]
- 22.Perez-Moreno M, Locascio A, Rodrigo I, et al. A new role for E12/E47 in the repression of E-cadherin expression and epithelial-mesenchymal transitions. J Biol Chem. 2001;276:27424–27431. doi: 10.1074/jbc.M100827200. [DOI] [PubMed] [Google Scholar]
- 23.Yang J, Mani SA, Donaher JL, et al. Twist, a master regulator of morphogenesis, plays an essential role in tumor metastasis. Cell. 2004;117:927–939. doi: 10.1016/j.cell.2004.06.006. [DOI] [PubMed] [Google Scholar]
- 24.Stinson S, Alley M, Kopp W, et al. Morphological and immunocytochemical characteristics of human tumor cell lines for use in a disease-oriented anticancer drug screen. Anticancer Res. 1992;12:1035–1053. [PubMed] [Google Scholar]
- 25.Weinstein J. Spotlight on molecular profiling: ‘integromic’ analysis of the NCI-60 cancer cell lines. Mol Cancer Ther. 2006;5:2601–2605. doi: 10.1158/1535-7163.MCT-06-0640. [DOI] [PubMed] [Google Scholar]
- 26.Weinstein J. 'Omic' and hypothesis-driven research in the molecular pharmacology of cancer. Curr Opinion in Pharmacology. 2002;2:361–365. doi: 10.1016/s1471-4892(02)00185-6. [DOI] [PubMed] [Google Scholar]
- 27.Scherf U, Ross D, Waltham M, et al. A gene expression database for the molecular pharmacology of cancer. Nat Genet. 2000;24:236–244. doi: 10.1038/73439. [DOI] [PubMed] [Google Scholar]
- 28.Ross D, Scherf U, Eisen M, et al. Systematic variation in gene expression patterns in human cancer cell lines. Nat Genet. 2000;24:227–235. doi: 10.1038/73432. [DOI] [PubMed] [Google Scholar]
- 29.Staunton J, Slonim D, Coller H, et al. Chemosensitivity prediction by transcriptional profiling. Proc Natl Acad Sci U S A. 2001;98:10787–10792. doi: 10.1073/pnas.191368598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shankavarum U, Reinhold W, Nishizuka S, et al. Transcript and protein expression profiles of the NCI-60 cancer cell panel: an integromic microarray study. Mol Cancer Ther. 2007;6:820–832. doi: 10.1158/1535-7163.MCT-06-0650. [DOI] [PubMed] [Google Scholar]
- 31.Nishizuka S, Charboneau L, Young L, et al. Proteomic profiling of the NCI60 cancer cell lines using new high-density ‘reverse-phase’ lysate microarrays. Proc Natl Acad Sci U S A. 2003;100:14229–14234. doi: 10.1073/pnas.2331323100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bussey KJ, Chin K, Lababidi S, et al. Integrating data on DNA copy number with gene expression levels and drug sensitivities in the NCI-60 cell line panel. Mol Cancer Ther. 2006;5:853–867. doi: 10.1158/1535-7163.MCT-05-0155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gallagher WM, Bergin OE, Rafferty M, et al. Multiple markers for melanoma progression regulated by DNA methylation: insights from transcriptomic studies. Carcinogenesis. 2005;26:1856–1867. doi: 10.1093/carcin/bgi152. [DOI] [PubMed] [Google Scholar]
- 34.Lorenzi P, Reinhold W, Rudelius M, et al. Asparagine synthetase as a causal, predictive biomarker for L-asparaginase activity in ovarian cancer cells. Mol Cancer Ther. 2006;5:2613–2623. doi: 10.1158/1535-7163.MCT-06-0447. [DOI] [PubMed] [Google Scholar]
- 35.Annereau J, Szakacs G, Tucker CJ, et al. Analysis of ATP-binding cassette transporter expression in drug-selected cell lines by a microarray dedicated to multidrug resistance. Mol Pharmacol. 2004;66:1397–1405. doi: 10.1124/mol.104.005009. [DOI] [PubMed] [Google Scholar]
- 36.Huang Y, Anderle P, Bussey K, et al. Membrane transporters and channels: role of the transportome in cancer chemosensitivity and chemoresistance. Cancer Res. 2004;64:4294–4301. doi: 10.1158/0008-5472.CAN-03-3884. [DOI] [PubMed] [Google Scholar]
- 37.Ellison G, Klinowska T, Westwood R, Docter E, French T, Fox J. Further evidence to support the melanocytic origin of MDA-MB-435. Mol Pathol. 2002;55:294–299. doi: 10.1136/mp.55.5.294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Roschke A, Tonon G, Gehlhaus K, et al. Karyotypic Complexity of the NCI-60 Drug-Screening Panel. Cancer Research. 2003;63:8634–8647. [PubMed] [Google Scholar]
- 39.Ikediobi O, Davies H, Bignell G, et al. Mutation analysis of twenty-four known cancer genes in the NCI-60 cell line set. Mol Cancer Ther. 2006;5:2606–2612. doi: 10.1158/1535-7163.MCT-06-0433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lorenzi P, Reinhold W, Varma S, et al. DNA fingerprinting of the NCI-60 cell line panel. Mol Cancer Ther. 2009;8:713–724. doi: 10.1158/1535-7163.MCT-08-0921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hark A, Schoenherr C, Katz D, Ingram R, Levorse J, Tilghman S. CTCF mediates methylation-sensitive enhancer-blocking activity at the H19/Igf2 locus. Nature. 2000;405:486–489. doi: 10.1038/35013106. [DOI] [PubMed] [Google Scholar]
- 42.Kim J, Kollhoff A, Bergmann A, Stubbs L. Methylation-sensitive binding of transcription factor YY1 to an insulator sequence within the paternally expressed imprinted gene, Peg3. Hum Mol Genet. 2003;12:233–245. doi: 10.1093/hmg/ddg028. [DOI] [PubMed] [Google Scholar]
- 43.Mancini D, Singh S, Archer T, Rodenhiser D. Site-specific DNA methylation in the neurofibromatosis (NF1) promoter interferes with binding of CREB and SP1 transcription factors. Oncogene. 1999;18:4108–4119. doi: 10.1038/sj.onc.1202764. [DOI] [PubMed] [Google Scholar]
- 44.Kawakami T, Okamoto K, Ogawa O, Okada Y. Multipoint methylation and expression analysis of tumor suppressor genes in human renal cancer cells. Urology. 2003;61:226–230. doi: 10.1016/s0090-4295(02)02110-6. [DOI] [PubMed] [Google Scholar]
- 45.Ribeiro-Filho L, Franks J, Sasaki M, et al. CpG hypermethylation of promoter region and inactivation of E-cadherin gene in human bladder cancer. Mol Carcinog. 2002;34:187–198. doi: 10.1002/mc.10064. [DOI] [PubMed] [Google Scholar]
- 46.Hajra K, Chen D, Fearon E. The SLUG zinc-finger protein represses E-cadherin in breast cancer. Cancer Res. 2002;62:1613–1618. [PubMed] [Google Scholar]
- 47.Ohira T, Gemmill R, Ferguson K, et al. WNT7a induces E-cadherin in lung cancer cells. Proc Natl Acad Sci U S A. 2003;100:10429–10434. doi: 10.1073/pnas.1734137100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gregory PA, Bert AG, Paterson EL, et al. The miR-200 family and miR-205 regulate epithelial to mesenchymal transition by targeting ZEB1 and SIP1. Nat Cell Biol. 2008;10:593–601. doi: 10.1038/ncb1722. [DOI] [PubMed] [Google Scholar]
- 49.Park SM, Gaur AB, Lengyel E, Peter ME. The miR-200 family determines the epithelial phenotype of cancer cells by targeting the E-cadherin repressors ZEB1 and ZEB2. Genes Dev. 2008;22:894–907. doi: 10.1101/gad.1640608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Blower PE, Verducci JS, Lin S, et al. MicroRNA expression profiles for the NCI-60 cancer cell panel. Mol Cancer Ther. 2007;6:1483–1491. doi: 10.1158/1535-7163.MCT-07-0009. [DOI] [PubMed] [Google Scholar]