Abstract
This study reports the use of microfluidics, which intrinsically has a large surface-to-volume ratio, toward rapid antimicrobial susceptibility testing at the point of care. By observing the growth of uropathogenic E. coli in gas permeable polymeric microchannels with different dimensions, we demonstrate that the large surface-to-volume ratio of microfluidic systems facilitates rapid growth of bacteria. For microchannels with 250 micrometer or less in depth, the effective oxygenation can sustain the growth of E. coli to over 109 cfu/ml without external agitation or oxygenation, which eliminates the requirement of bulky instrumentation and facilitates rapid bacterial growth for antimicrobial susceptibility testing at the point of care. The applicability of microfluidic rapid antimicrobial susceptibility testing is demonstrated in culture media and in urine with clinical bacterial isolates that have different antimicrobial resistance profiles. The antimicrobial resistance pattern can be determined as rapidly as 2 hours compared to days in standard clinical procedures facilitating diagnostics at the point of care.
INTRODUCTION
Antimicrobial susceptibility test (AST) is often performed to determine the antibiotic sensitivity of bacterial pathogens in clinical samples such as urine, blood, sputum, or wound swabs1. The current clinical practice requires sample transportation to a centralized microbiology laboratory and overnight culture of the infectious agents. AST results are not available for days after sample collection. These aspects have limited the accessibility at the point of care. Rapid determination of antimicrobial susceptibility is especially crucial towards judicious management of infectious diseases in emergency situations and high-risk areas such as hospitals, intensive care units, and clinics established in response to disasters2–4. Without objective information of the drug resistance profile of the suspected pathogen, physicians have to select antibiotic therapy empirically based on the nature of the infection and antibiotic treatments are typically chosen based on the worst-case-scenario assumption. Injudicious use of broad spectrum antibiotics by clinicians, as a result of lack of objective diagnosis at the point of care and significant delay of standard procedures, has contributed to the emergence of resistant pathogens worldwide5, 6. A point-of-care device for rapid AST in resource limited settings is, therefore, highly desirable. It will lead to evidence-based, rather than empiric, management of infectious diseases and will allow more judicious use of antibiotics, which in turn will reduce the emergence of multidrug-resistant pathogens7.
While various genotypic markers have been identified for antibiotic resistance, measurement of the phenotypic response of bacteria to antibiotics is often superior to genotypic detection of antibiotic resistance genes due to the diverse resistance mechanisms and the continuous evolution of the pathogens1, 8. In particular, growth-based phenotyping AST methods are the current gold standard in clinical microbiology laboratory. Conventional techniques for determining antibiotic resistance include broth dilution and disc diffusion9, 10. For disc diffusion, the bacterial isolates are inoculated on the surface of an agar plate and a disc-shaped filter paper soaked with a standard amount of antibiotic is loaded onto the surface of the dish. With the diffusion of the antibiotic and the formation of an antibiotic concentration gradient into the adjacent medium, after 18~24 hours period of incubation, a zone of inhibition of bacterial growth appears depending on the effectiveness of the antibiotic. The size of the inhibition zone provides an indication of the potency of the antibiotic and is inversely proportional to the minimum inhibitory concentration1. The disk diffusion and agar diffusion methods, while ‘low-tech’ and labor-intensive, are well established and still commonly used, particularly in resource-limited settings. To automate the labor intensive procedures and provide quantitative assessment of the antimicrobial sensitivity, various techniques that directly measure the concentration of the pathogens (e.g., optical density and micromechanical oscillators) or their activities (e.g., microcalorimetry, bioluminescence, and radioactive CO2 release) have been developed to facilitate the identification and evaluation of the antimicrobial resistance of bacteria in microtiter plate or other formats11–16. All the current automated antimicrobial susceptibility techniques rely on first isolating the pathogens from the body fluid or tissue samples, which takes 18–24 hours, followed by phenotypic testing of the isolated bacteria for AST, which takes another 18–24 hours of incubation. Furthermore, these systems are expensive and have bulky footprints. Using a fluorescent viability indicator and an epifluorescence microscope, an emulsion-based microfluidic technique has been recently reported by observing the activity of individual bacteria confined in droplets17. AST results can be obtained in 7.5 hours. Nevertheless, all these systems are difficult to be adapted to a point-of-care setting. In particular, the major hurdles for these techniques toward rapid, point-of-care testing are the time consuming bacterial growth step and the requirement of bulky supporting instrumentation.
The advent of microfluidics has the potential to revolutionize the clinical management of infectious diseases and the implementation of AST at the point of care18, 19. An important requirement for rapid bacterial growth is sufficient oxygen in the microenvironment20. In conventional bacteria culture, vigorous shaking with an orbital shaker is typically applied to facilitate oxygenation in the media to sustain the bacterial growth. Oxygenator systems are often included in perfusion circuits and bioreactors to supply adequate oxygen for tissue and cell culture21–23. On the other hand, microfluidic devices have intrinsically a large surface-to-volume (S/V) ratio as a result of the small length scale. This provides a simple, yet effective, approach for oxygenation inside a microfluidic cell culture system24. At a given concentration of bacteria, the amount of oxygen requires for sustaining the bacterial growth is proportional to the volume of the culture media (i.e., number of bacteria) while the oxygen flux is proportional to the surface area. This implies relatively abundant oxygen is available for bacterial culture at the microscale. Figure 1 illustrates the effect of the dimension of the microchannel for bacterial culture. The large S/V ratio of microfluidics for facilitating effective oxygenation has been utilized in various chip-based cell studies25–29. Nevertheless, the relationship between the S/V ratio and bacterial growth has not been investigated systematically and the microfluidic approach has not been demonstrated for rapid AST.
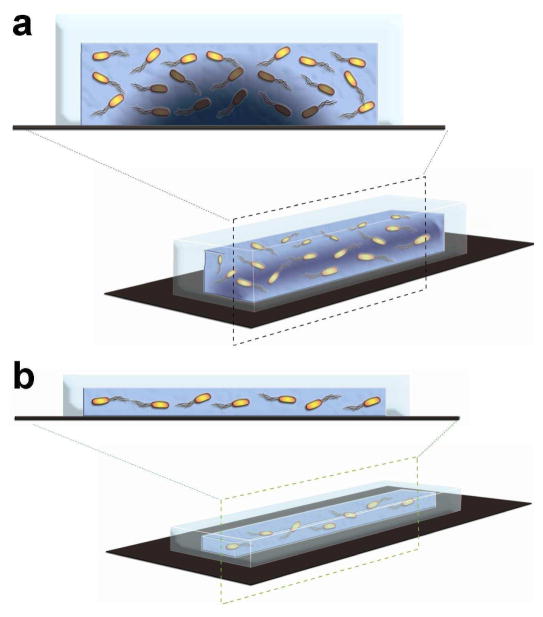
Figure 1.
Schematics illustrating the effect of the depth of a microchannel on the growth of E. coli. (a) For a microchannel with a large depth, the oxygen level is relatively low for supporting the growth of all E. coli as a result of the large S/V ratio. (b) For a microchannel with a small depth, the oxygen level is relatively abundant, which supports rapid growth of the pathogens.
In this study, we explore the use of gas permeable PDMS microchannels that has a large S/V ratio toward the implementation of rapid AST at the point of care. The growth of uropathogenic Escherichia coli (E. coli) in microfluidic channels was compared to other culture conditions, including an Erlenmeyer flask in an orbital shaker, a static Erlenmeyer flask, and a static Petri dish. The bacterial growth was investigated as a function of the S/V ratio of the apparatus by using laser-machined microchannels with different depths. Understanding the effect of the S/V ratio on bacterial growth helps to optimize the microfluidic design for rapid AST. Experimental results are presented to determine the dose dependence of ampicillin on an uropathogenic E. coli using microfluidic channels. Furthermore, the antimicrobial resistance profiles of four E. coli clinical isolates were also determined using the microfluidic channels with an optimized S/Vume ratio. These tests can be performed directly in urine mixed with bacteria culture media. The current study will potentially form the technological foundation of a microfluidic approach for performing rapid AST at the point of care without relying on a centralized clinical microbiology laboratory.
EXPREIMENTAL SECTION
Microchannel fabrication
To evaluate the possibility of rapid bacterial culture using microfluidic channels, we designed microfluidic channels with different depths, which adjust the overall S/V ratio. The channels were fabricated using a combination of laser-micromachining and a two-step polymer molding process. Polydimethylsiloxane (PDMS), which is gas permeable and transparent, was chosen as the channel structural material. PDMS is one of the silicone polymers that has a high oxygen diffusivity30. A two-stage molding process was developed to fabricate microchannels with different S/V ratios (Figure 2). Briefly, laser machining was performed to engrave polycarbonate substrates, which served as the master mold of the microchannels. A laser machining system (Versa, Universal Laser System Inc) was applied and the cutting depth (i.e., the final channel depth) was controlled by adjusting the laser power and duration. The laser-machined polycarbonate master was characterized using optical microscopy. Channel dimensions on the order of 100 μm can be easily created with this laser machining process. A reverse mold was then created by molding urethane on the polycarbonate master mold overnight at room temperature. After the urethane mold was solidified, it was peeled off and cut to appropriate size for PDMS molding. The PDMS channel layer was cured at 70°C overnight. The PDMS channel layer and a glass substrate were sterilized and bonded using an atmospheric (air) plasma system. To test the effect of surface coating on bacterial adhesion, the channel was incubated with 1% (w/v) bovine serum albumin (BSA) for 10 minutes and rinsed with phosphate buffered saline (PBS). Different channel depths can be created on the same mold using this process. The master mold had 12 channels. In this study, microchannels with depth of 114 to 2707 μm were fabricated. For the 200 μm channel, the length and width were 25.4 mm and 2 mm respectively. This resulted in a total volume of approximately 10 μl inside the channel.
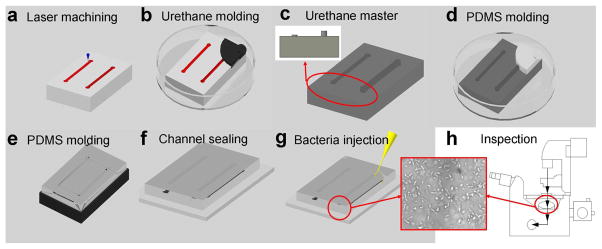
Figure 2.
Process flow for fabricating microfluidic channels with different S/V ratios for rapid AST. (a) A CO2 laser machining system was used to engrave microchannels on a polycarbonate substrate. Different channel depths can be generated simultaneously in the process. (b) Urethane molding on the polycarbonate master. (c) A reverse, urethane mold with microchannels of different depths. Only two channels are shown for simplicity. The insert shows the cross-section view. (d–e) PDMS channels were molded on the urethane mold. (f) The PDMS channel was sealed with a glass substrate using an atmospheric (air) plasma system. The plasma treatment step also sterilized the channel. (g) Injection of pathogens into the channel with a syringe. Microchannels were washed with PBS and incubated with or without BSA before the experiment. (H) Bacterial growths can be monitored by phase contrast microscopy or absorbance spectroscopy.
Bacterial strains
Uropathogenic E. coli, which accounts for more than 80% of uncomplicated urinary tract infection (UTI), was selected as the model pathogen for this study. Four E. coli strains (EC137, EC132, EC462, and EC136) isolated from clinical urine samples of patients with UTI were used. These strains were isolated as part of a research protocol approved by the Stanford University Institutional Review Board. For each bacterial strain, four conditions were tested: no antibiotic control, ampicillin (Amp), ciprofloxacin (Cipro) and trimethoprim/sulfamethoxazole (T/S). All three antibiotics chosen are commonly used oral antibiotics for UTI treatment. The four E. coli strains have different antimicrobial resistance profiles, which were previously determined by the clinical microbiology laboratory. EC137 is sensitive to all 3 antibiotics (AmpS, CiproS, T/SS); EC136 is resistant to ampicillin (AmpR); EC132 is resistant to ampicillin and ciprofloxacin (AmpR, CiproR); and EC462 is resistant to ampicillin and trimethoprim/sulfamethoxazole (AmpR, T/SR). These clinical isolates were cultured in Mueller-Hinton medium, which is the standard laboratory media for MIC and AST tests31. Before the experiment, bacteria on agar plate were inoculated in Mueller-Hinton medium. The clinical isolates were grown to early-exponential phase and diluted to appropriate concentrations for the experiment. E. coli strain DH5α was also used in control experiments and was cultured in Lysogeny broth.
Bacterial growth in microchannels
The growth of uropathogenic E. coli inside a microchannel was compared to other culture conditions. The bacteria samples were injected into 200 μm depth channels by syringes. At the same time, 13 ml and 5 ml of bacteria samples were pipetted into an Erlenmeyer flask and a 60 mm Petri dish, respectively. The amounts of the solution were chosen that the height of the media is 3 mm in both apparatuses. The flasks, Petri dishes, and microchannels were incubated statically in a miniaturized incubator at 37°C. One of the flasks was loaded into an orbital shaker (BarnStead Inc) at 250 rpm and 37°C as positive control. For optical inspection, the microfluidic channels were directly mounted on a digital microscope equipped with phase contrast optics (Leica, DMI 4000B). The morphology and density of the bacteria were recorded by a CCD camera (Planetary Imaging, DMK 31AF03) and digitized into a video capture system (Image Source, IC Capture 2.0). The growth of the bacteria was monitored at regular time intervals using a micro-sample spectrophotometer (Nanodrop 2000) by withdrawing all the solution from the microchannel. The concentration of the bacteria was determined by observing the absorbance at 600 nm. Similar experiments were also performed using E. coli DH5α. To determine the effect of the S/V ratio on the bacterial growth rate, the clinical isolate EC137 was injected into channels from 114 μm to 2707 μm in depth. Different volumes of bacterial samples were also loaded to Erlenmeyer flasks, which resulted in different S/V ratios. The range of S/V ratio spanned from 0.05 to 94.4 cm−1 in our experiment. Each condition was repeated at least four times.
Oxygen supply and consumption
As PDMS is gas permeable, oxygen can diffuse into the medium inside a microchannel. The order of magnitude of the maximum oxygen flux Fmax by diffusion through the PDMS layer can be approximated by the diffusion equation32:
| (1) |
In equation 1, DPDMS, ΔC, and Δz denote the diffusivity of oxygen in PDMS, the difference of oxygen concentration across the PDMS layer, and the thickness of the PDMS layer, respectively. The diffusivity of oxygen in PDMS30 is 4.1×10−9 m2/s. The oxygen concentration in the atmosphere is 0.2 mole/m3 and is used to estimate the maximum oxygen difference32. The PDMS thickness Δz is 2 mm for the current microchannel design. If the channel width and length are large compared to the depth, the oxygen supply is dominated by the top surface and is approximately the same for channels with different depths. For a 200 μm depth channel, the total surface area of the gas permeable PDMS channel is 6.18×10−5 m2 and the maximum oxygen flux is estimated to be 25.3 pmol/s. On the other hand, the oxygen consumption rate depends on the particular bacteria being tested. The oxygen consumption rate of E. coli has been reported to be 1.78×10−18 mol/cell/s33. For the same concentration of E. coli, a deeper channel requires more oxygen than a shallower channel, as the total number of bacteria is proportional to the volume of the channel. At a bacteria concentration of 1.4×109 cfu/ml in a 200 μm channel, the oxygen consumption rate is the same as the maximum oxygen flux of 25.3 pmol/s.
Microfluidics based AST
Experiments were performed to determine the MIC of ampicillin using 200 μm microfluidic channels. The clinical isolate EC137 in Mueller-Hinton medium was used in the experiment. Furthermore, antimicrobial resistance profiling experiments were performed to evaluate the applicability for microfluidic AST. Stock solutions of antibiotics were freshly prepared before each experiment. The concentrations of Amp and Cipro were 100 μg/ml and 4 μg/ml, respectively. Trimethoprim and sulfamethoxazole were mixed at a 1:19 ratio (16 μg/ml and 304 μg/ml). These concentrations were selected based on standard AST protocols. Antibiotic samples were pre-warmed at 37°C for 20 mins. The antibiotics were then mixed with the clinical isolates and culture media before being injected into microchannels. The experiment was also performed by mixing urine and Mueller- Hinton medium at 1:1 ratio. In addition, E. coli DH5α and DH5α transformed with an ampicillin resistance gene were tested using the same procedure. In all AST experiments, microchannels with a depth of 200 μm were incubated at 37°C for 2 hours. The concentrations of bacteria were determined using the Nanodrop spectrophotometer. A pathogen was considered to be sensitive to an antibiotic if the bacterial concentration is less than 30% of the control value.
RESULTS AND DISCUSSION
The effect of S/V ratio on bacterial growth
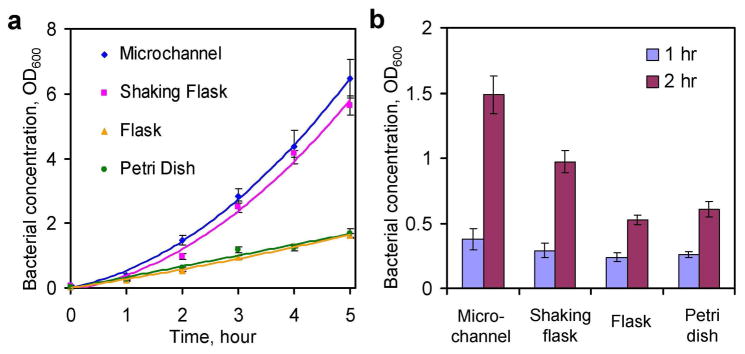
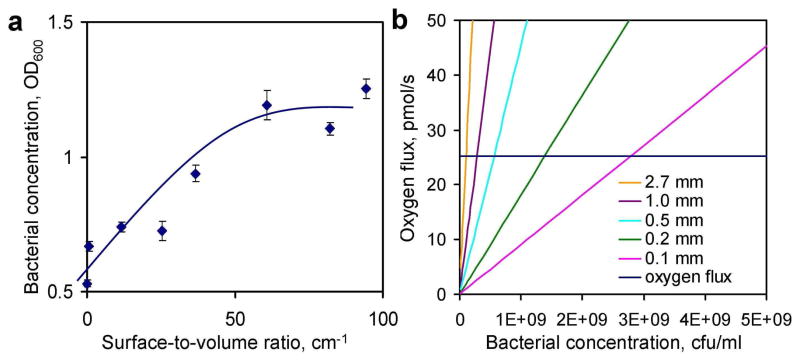
To evaluate the effectiveness of microfluidic bacteria culture, the growth of bacteria inside a 200 μm microchannel was compared to other apparatuses and culture conditions (Figure 3a). In the first hour, significant growths were observed in all culture conditions. The growth rates were similar initially between all different culturing conditions. As the bacteria continued to grow, the static Petri dish and flask displayed significantly slower growth rates compared to the microchannel. The growth rates could be clearly distinguished after two hours. Figure 3b compares the growth of EC137 after 1 and 2 hours of bacterial culture. Using the microchannel, the absorbance value reached ~1.5 in 2 hours while the static dish and flask required 5 hours to result in a similar value. The growth rate (i.e., the slope of the growth curve) of the E. coli in the microchannel was similar to the value in the shaking flask. A slight delay of the bacterial growth (the growth curve shifted to the right for ~ 5 mins) was consistently observed for the shaking flask when compared to the microchannel (Figure 3a). This can be understood by the difference in thermal time constants of the microchannel and the flask34. More importantly, the fast growth rate in the microchannel that is comparable to the shaker flask opens the possibility of eliminating the bulky culture equipment, which dramatically simplifies the system requirement for point-of-care AST. To further evaluate the effect of the S/V ratio on bacterial growth, the clinical isolate EC137 was cultured in channels with different depths. Figure 4a shows the concentration of the bacteria after 2 hours of inoculation and the growth rate generally increased with the S/V ratio. For 2 hours of bacterial culture, the absorbance value appeared to saturate when the S/V ratio was equal to or greater than 60 cm−1. This S/V ratio was corresponding to a microchannel with a depth of 250 μm. Further increase in the S/V ratio (i.e., channels with smaller depths) showed little effect on the growth of the bacteria. The same result was also observed using E. coli DH5α (data not shown).
Figure 3.
Comparison of bacterial growth in a 200 μm microchannel (microchannel), an Erlenmeyer flask inside a shaking incubator (shaking flask), a static Erlenmeyer flask (flask), and a static Petri dish (Petri dish). (a) The bacterial growth curves for E. coli EC137 in different conditions. Bacteria in the shaking flask grew at a similar rate compared to bacteria inside the microchannel. The bacteria in the static flask and Petri dish displayed significantly lower growth rates compared to the microchannel. (b) Initial growth rates were similar after one hour for all experimental conditions. After two hours of bacterial culture, the growth rates can be clearly distinguished. Each data point represents at least four experiments. Data represent absorbance ±standard deviation.
Figure 4.
(a) The effect of the S/V ratio on bacterial growth after culturing for 2 hours. The bacterial growth increased with the S/V ratio and saturated at approximately 60 cm−1 corresponding to a channel of 250 μm in depth. Error bars represent the standard deviation. (b) Comparison of the oxygen consumption rates for different channel depths. The blue line indicates the oxygen supply for the channel used in the experiment.
An order-of-magnitude calculation has been performed to estimate the oxygen supply and consumption during bacterial culture inside a microfluidic channel. Figure 4b compares the oxygen consumption rates for different channels, which is proportional to the volume of the channel and the concentration of the bacteria. If the surface area is fixed at 6.18×10−5 m2, the maximum oxygen flux is approximately 25.3 pmol/s. For a 200 μm channel, the maximum oxygen flux coincides with the oxygen consumption rate at a concentration of 1.4×109 cfu/ml. For a channel with a smaller depth (e.g., 100 μm channel), the curve intersects with the oxygen flux at a higher absorbance value (~2.8×109 cfu/ml). Oxygen will become limited for bacterial growth near the intersection points. For a channel with a larger depth, the interaction point occurs at a much lower bacterial concentration. The channel with 1 mm depth has 5 times more bacteria than the 200 μm channel; therefore, it consumes 5 times more oxygen.
In general, our results indicate that the S/V ratio plays an important role in the bacterial growth and that a microchannel with a large S/V ratio can facilitate rapid bacterial growth. The experimental observations can be understood by considering the oxygen available to the bacteria in the microchannel. For an apparatus with a small S/V ratio (e.g., the static flask and Petri dish), the oxygen flux is only enough to support the growth of the bacteria at low concentration. This was likely to be the case during the first hour of bacteria culture when the bacterial concentration was relatively low. Indeed, the observed growth rates were similar in the first hour when the bacterial concentrations were relative low in different culture conditions. At a higher bacteria concentration, the oxygen was insufficient for sustaining the growth of the bacteria and the bacteria displayed slower growth rates in the static flask and Petri dish after 2 hours of inoculation (Figure 3). For an apparatus with a large S/V ratio, an excessive amount of oxygen is available for the bacteria and bacterial growth. This provides an explanation for the maximum absorbance value observed after two hours of inoculation (Figure 4a). In this condition, further reduction of the channel depth could not improve the bacterial growth. As demonstrated in our data, a microchannel with 250 μm or less in depth sustained the growth of E. coli to over 109 cfu/ml. This value is in good agreement with our order of magnitude estimation considering the oxygen consumption rate inside the channel and the bacterial growth rate generally correlates with the oxygen supply and the S/V ratio of the apparatus.
In addition to effective oxygenation, the large S/V ratio could also result in other effects. For instance, the bacteria can adhere to the surface of the microchannel, which may affect its growth rate. For PDMS microchannel, the surface is known to promote bacteria adhesion. In order to reduce bacterial adhesion on the channel surface, BSA was incubated in the channel Figure 5 shows bacterial adhesion on the surfaces of 200 μm depth channels with and without BSA coating after 2 hours of inoculation. A significant amount of E. coli was attached to the surface while the BSA coating significantly reduced the adhesion (Figure 5). Nevertheless, the values can not be distinguished based on the absorbance measurement. This is likely due to the fact that the number of bacteria adhered on the surface is relatively small compared to the bacteria in suspension. This shows that bacterial adhesion has an insignificant effect on the measurement in the current experimental condition. The volume of the sample is another consideration for optimizing the S/V ratio especially at a low pathogen concentration. At low concentration (e.g., less than 10 bacteria in the channel), random variation of the initial number of bacterial could introduce uncertainty in the measurement. If necessary, the channel length and width should be increased to obtain a large volume of sample while maintaining the S/V ratio by adjusting the depth of the channel. Additionally, extra incubation time can be applied in microfluidic AST to eliminate the uncertainty due to the variation in the initial concentration. Based on these results and considerations, a channel depth of 200 μm was designed in all the successive AST experiments and the culture time was chosen to be 2 hours.
Figure 5.
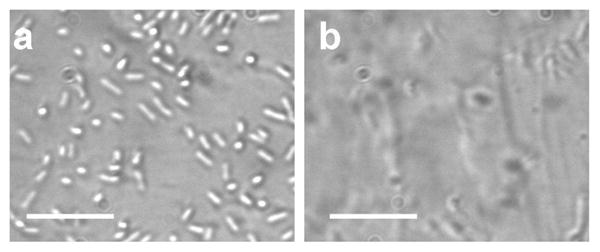
The effect of BSA coating on bacteria adhesion. (a) Bacteria adhesion on the channel surface without BSA coating. (b) The number of E. coli adhered to the surface can be reduced with 10 mins incubation of BSA prior to the experiment. Images were taken after 4 hours of bacterial culture and removal of the suspension. Scale bars represent 20 μm.
Dose dependence of antibiotics
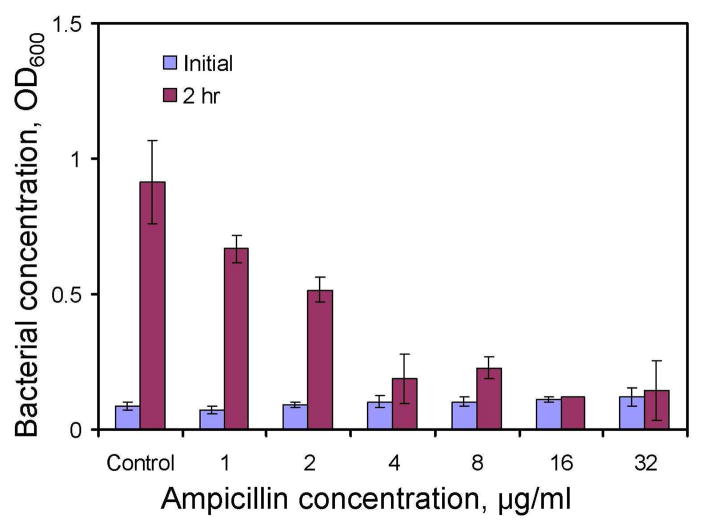
The parallel processing nature of microfluidics provides a useful tool for performing multiplexed tests simultaneously. For instance, we have determined the dose dependence of ampicillin for the uropathogenic E. coli EC137 using microfluidic channels. The pathogens were inoculated with different concentrations of ampicillin inside the microchannels. Figure 6 shows the result of the dose dependence test using 200 μm microfluidic channels. Without ampicillin, the bacteria grow to an optical density over 1 within 2 hour. At a lower concentration (e.g., 1 or 2 μg/ml), a slight decrease in the bacterial grow was observed compared to the control. Nevertheless, the concentration of the pathogen was significant compared to the control (over 50%). At an ampicillin concentration equal to or greater than 4 μg/ml, the growth of the bacteria was less than 30% of the control value. Total inhibition of the bacterial was observed at 16 μg/ml or higher. This suggests the lowest concentration of ampicillin to inhibit the growth of the E. coli ES 137 is 16 μl.
Figure 6.
Minimum inhibitory concentration (MIC) determination using microchannels. The MIC of ampicillin to EC137 was determined using high S/V ratio microchannels. The growth of the bacteria was observed to be insignificant at concentration higher than 4 μg/ml. The experiments were performed in Mueller-Hinton broth. Data represent mean ±standard deviation.
Antimicrobial resistance profiling of clinical isolates
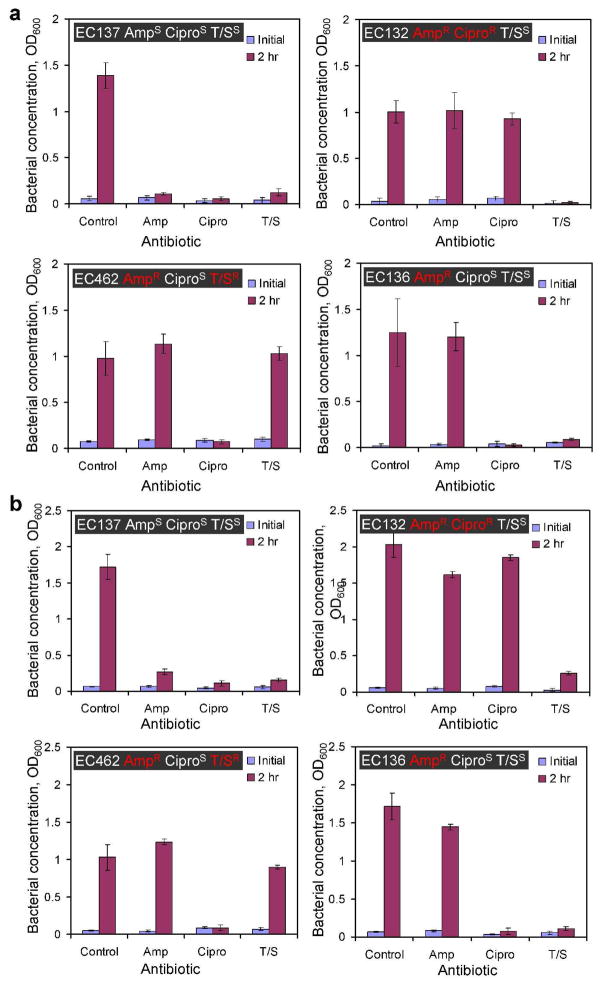
Antimicrobial resistance profiling is required to determine the antibiotic susceptibility pattern of a pathogen. This provides clinically relevant information for selecting the type of antibotic that a patient will receive. To demonstrate the applicability of microfluidic devices for AST, we cultured four E. coli clinical isolates in four different conditions (no antibiotics, Amp, Cipri, and T/S) and observed their growths after two hours. Figure 7a shows the result of microfluidic AST in Mueller-Hinton media. The absorbance value of the control set for EC137 strain is 1.4 and the value is significantly higher than other conditions with antibiotics. This indicates EC137 strain is sensitive to these antibiotics. EC132, which is sensitive to T/S and resistant to Amp and Cipro, displayed significant growth with Amp and Cipro, but insignificant growth with T/S compared to the control. The antibiotic resistance profiles of EC462 and EC136 strains were also determined. These results agree with the antibiotic resistance profiles previously determined by the clinical microbiology laboratory. Therefore, the antimicrobial resistance profiles of all these four clinical isolates were determined successfully using microfluidic AST in 2 hours. To explore the feasibility of directly performing microfluidic AST in biological fluids, similar experiments were also performed by mixing the culture media with the pathogens in urine (Figure 7b). Interestingly, we observed faster growth rates of the uropathogens in urine mixed with culture media (except EC462). The absorbance of EC132 in urine was almost twice of the value in culture media. We also observed a larger variation in the bacterial growth with the antibiotics in urine. Nevertheless, the antibiotic resistance profiles for all four clinical isolates were correctly identified using microfluidic AST. This supports that the microfluidic AST approach can be directly applied for AST of uropathogens in urine. In addition, we have also demonstrated rapid AST using standard laboratory strain E. coli DH5α genetically engineered with and without ampicillin resistance (data not shown). Together, our data demonstrate the general applicability of using microfluidics for rapid AST and the approach is compatible with urine and potentially other biological fluids.
Figure 7.
Antimicrobial susceptibility testing (AST) using microchannels. (a) Four clinical isolates (EC137, EC132, EC462, and EC136) were tested for their antimicrobial resistance patterns in Mueller-Hinton media. The antimicrobial resistance patterns for Ampicillin (Amp), Ciprofloxacin (Cipro) and Trimethoprim/Sulfamethoxazole (T/S) of the strains are highlighted in the figure. The superscripts “S” and “R” indicate sensitive and resistance of the strain to the antibiotics. (b) Antimicrobial resistance profiling of the same clinical isolates in urine using 200 μm microchannels. The experiments were performed in Mueller-Hinton media mixed with urine at a 1:1 ratio. Data represent mean ±standard deviation.
In this study, we investigated the effect of the S/V ratio to bacterial growth using laser-machined microchannels with different dimensions. Our data suggest that microchannels with 250 μm or less in depth can provide sufficient oxygen for the growth of uropathogenic E. coli to over 109 cfu/ml. The experimental observation is in good agreement with the order-of-magnitude calculation based on oxygen balance. The value is also consistent with a previous bioreactor study, which reports that a reactor depth of 300 μm can support the growth of 109 cfu/ml27. It should be noted that this value depends on the particular strain of pathogens. In general, our results will serve as guidelines for optimizing the channel design for rapid AST. For instance, the PDMS thickness and channel depth can be further reduced to facilitate oxygen transportation if necessary. The bottom substrate can also be replaced by gas permeable materials to further increase the surface area. Another major finding in this study is that microfluidic AST can be finished in a two hour time frame, which is 1–2 orders of magnitude faster than the standard AST procedures currently in clinical practice. The reported plug-based stochastic confinement technique, which is not optimized by oxygen transportation, requires 7 hours for MIC determination and AST17. Furthermore, we have shown that the microfluidic AST technique is directly applicable to urine. This may potentially simplify the labor-intensity and time-consuming sample preparation steps in the AST procedure, and facilitates point-of-care AST in non-traditional settings. In addition, the microfluidic approach offers several other advantages for rapid AST at the point of care. Firstly, the microfluidic platform can be easily integrated with other detection systems for monitoring the growth of the bacteria. For instance, microelectrode arrays can be easily deposited on the glass substrate for performing microscale impedimetric analysis35, 36 or electrochemical sensing2, 19, 37, 38. Onchip monitoring of the bacterial activity may improve the accuracy of the assay by eliminating the uncertainty due to the lag phase of the bacteria. Secondly, various microfluidic techniques, such as surface modification39, 40, gradient generation41, 42 and electrokinetic manipulation43, 44, can be combined to enhance the functionality of the system for automated AST and other drug screening applications.
CONCLUSION
We have demonstrated a microfluidic device for bacterial culture and rapid AST in resource limited settings. The large S/V ratio of microfluidic systems facilitates effective oxygenation for bacterial culture, and provides a simple and effective platform for rapid AST at the point of care. The microfluidic approach allows antimicrobial resistance profiling to be finished as rapidly as two hours. In the future, high S/V ratio microchannels can be combined with other detection and microfluidic strategies to enhance the functionality of the system. We envision that the microfluidic, point-of-care AST system will improve the clinical management of infectious diseases by allowing more judicious use of antibiotics, which reduces the emergence of multidrug-resistant pathogens.
Acknowledgments
The authors thank Dr. Zheng Sun for valuable discussion. This work is supported by NIH NIAID (1U01AI082457), NSF ECCS (0900899), and VA RR&D Merit Review (B4872) awarded to J.C.L..
References
- 1.Balows A Canalco inc. Current techniques for antibiotic susceptibility testing. Thomas; Springfield, Ill: 1974. [Google Scholar]
- 2.Liao JC, Mastali M, Gau V, Suchard MA, Moller AK, Bruckner DA, Babbitt JT, Li Y, Gornbein J, Landaw EM, McCabe ERB, Churchill BM, Haake DA. Journal of Clinical Microbiology. 2006;44:561–570. doi: 10.1128/JCM.44.2.561-570.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fridkin SK, Edwards JR, Tenover FC, Gaynes RP, McGowan JE, Project I, Hosp NS. Clinical Infectious Diseases. 2001;33:324–329. doi: 10.1086/321893. [DOI] [PubMed] [Google Scholar]
- 4.Kollef MH, Fraser VJ. Annals of Internal Medicine. 2001;134:298–314. doi: 10.7326/0003-4819-134-4-200102200-00014. [DOI] [PubMed] [Google Scholar]
- 5.Infectious Disease Society of America Task Force on Antimicrobial Availability. 2004 [Google Scholar]
- 6.Kardas P, Devine S, Golembesky A, Roberts C. International Journal of Antimicrobial Agents. 2005;26:106–113. doi: 10.1016/j.ijantimicag.2005.04.017. [DOI] [PubMed] [Google Scholar]
- 7.Hawkey PM. Journal of Antimicrobial Chemotherapy. 2008;62:I1–I9. doi: 10.1093/jac/dkn241. [DOI] [PubMed] [Google Scholar]
- 8.Sommer MO, Dantas G, Church GM. Science. 2009;325:1128–1131. doi: 10.1126/science.1176950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bauer AW, Kirby WMM, Sherris JC, Turck M. American Journal of Clinical Pathology. 1966;45:493. [PubMed] [Google Scholar]
- 10.Stalons DR, Thornsberry C. Antimicrobial Agents and Chemotherapy. 1975;7:15–21. doi: 10.1128/aac.7.1.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ljungholm K, Wadso I, Mardh PA. Journal of General Microbiology. 1976;96:283–288. doi: 10.1099/00221287-96-2-283. [DOI] [PubMed] [Google Scholar]
- 12.Deblanc HJ, Wagner HN, Charache P. Antimicrobial Agents and Chemotherapy. 1972;22:360. doi: 10.1128/aac.2.5.360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Isenberg HD, Maclowry JD. Annual Review of Microbiology. 1976;30:483–505. doi: 10.1146/annurev.mi.30.100176.002411. [DOI] [PubMed] [Google Scholar]
- 14.Gfeller KY, Nugaeva N, Hegner M. Biosensors & Bioelectronics. 2005;21:528–533. doi: 10.1016/j.bios.2004.11.018. [DOI] [PubMed] [Google Scholar]
- 15.Ertl P, Robello E, Battaglini F, Mikkelsen SR. Analytical Chemistry. 2000;72:4957–4964. doi: 10.1021/ac0003596. [DOI] [PubMed] [Google Scholar]
- 16.Mann TS, Mikkelsen SR. Analytical Chemistry. 2008;80:843–848. doi: 10.1021/ac701829c. [DOI] [PubMed] [Google Scholar]
- 17.Boedicker JQ, Li L, Kline TR, Ismagilov RF. Lab on a Chip. 2008;8:1265–1272. doi: 10.1039/b804911d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kim J, Junkin M, Kim DH, Kwon S, Shin YS, Wong PK, Gale BK. Microfluidics and Nanofluidics. 2009;7:149–167. [Google Scholar]
- 19.Liao JC, Mastali M, Li Y, Gau V, Suchard MA, Babbitt J, Gornbein J, Landaw EM, McCabe ERB, Churchill BM, Haake DA. Journal of Molecular Diagnostics. 2007;9:158–168. doi: 10.2353/jmoldx.2007.060052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schulze KL, Lipe RS. Archiv Fur Mikrobiologie. 1964;48:1. doi: 10.1007/BF00406595. [DOI] [PubMed] [Google Scholar]
- 21.Borenstein JT, Terai H, King KR, Weinberg EJ, Kaazempur-Mofrad MR, Vacanti JP. Biomedical Microdevices. 2002;4:167–175. [Google Scholar]
- 22.Knazek RA, Kohler PO, Gullino PM, Dedrick RL. Science. 1972;178:65. doi: 10.1126/science.178.4056.65. [DOI] [PubMed] [Google Scholar]
- 23.Maharbiz MM, Holtz WJ, Sharifzadeh S, Keasling JD, Howe RT. Journal of Microelectromechanical Systems. 2003;12:590–599. [Google Scholar]
- 24.Kim DH, Wong PK, Park J, Levchenko A, Sun Y. Annual Review of Biomedical Engineering. 2009;11:203–233. doi: 10.1146/annurev-bioeng-061008-124915. [DOI] [PubMed] [Google Scholar]
- 25.Gomez-Sjoberg R, Leyrat AA, Pirone DM, Chen CS, Quake SR. Analytical Chemistry. 2007;79:8557–8563. doi: 10.1021/ac071311w. [DOI] [PubMed] [Google Scholar]
- 26.Balagadde FK, You LC, Hansen CL, Arnold FH, Quake SR. Science. 2005;309:137–140. doi: 10.1126/science.1109173. [DOI] [PubMed] [Google Scholar]
- 27.Zanzotto A, Szita N, Boccazzi P, Lessard P, Sinskey AJ, Jensen KF. Biotechnology and Bioengineering. 2004;87:243–254. doi: 10.1002/bit.20140. [DOI] [PubMed] [Google Scholar]
- 28.Wong PK, Yu FQ, Shahangian A, Cheng GH, Sun R, Ho CM. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:5105–5110. doi: 10.1073/pnas.0800823105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Walker GM, Zeringue HC, Beebe DJ. Lab on a Chip. 2004;4:91–97. doi: 10.1039/b311214d. [DOI] [PubMed] [Google Scholar]
- 30.Charati SG, Stern SA. Macromolecules. 1998;31:5529–5535. [Google Scholar]
- 31.Atlas RM, Parks LC. Handbook of microbiological media. 2. CRC Press; Boca Raton: 1997. [Google Scholar]
- 32.Leclerc E, Sakai Y, Fujii T. Biomedical Microdevices. 2003;5:109–114. [Google Scholar]
- 33.Geckil H, Stark BC, Webster DA. Journal of Biotechnology. 2001;85:57–66. doi: 10.1016/s0168-1656(00)00384-9. [DOI] [PubMed] [Google Scholar]
- 34.Figliola RS, Beasley DE. Theory and design for mechanical measurements. 4. John Wiley; Hoboken, N.J: 2006. [Google Scholar]
- 35.Yang LJ. Talanta. 2008;74:1621–1629. doi: 10.1016/j.talanta.2007.10.018. [DOI] [PubMed] [Google Scholar]
- 36.Cheng X, Liu YS, Irimia D, Demirci U, Yang LJ, Zamir L, Rodriguez WR, Toner M, Bashir R. Lab on a Chip. 2007;7:746–755. doi: 10.1039/b705082h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gau V, Ma SC, Wang H, Tsukuda J, Kibler J, Haake DA. Methods. 2005;37:73–83. doi: 10.1016/j.ymeth.2005.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mach KE, Du CB, Phull H, Haake DA, Shih MC, Baron EJ, Liao JC. J Urol. 2009;182:2735–2741. doi: 10.1016/j.juro.2009.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Keyes J, Junkin M, Cappello J, Wu X, Wong PK. Applied Physics Letters. 2008;93:023120. [Google Scholar]
- 40.Junkin M, Watson J, Geest JPV, Wong PK. Advanced Materials. 2009;21:1247–1251. [Google Scholar]
- 41.Jeon NL, Dertinger SKW, Chiu DT, Choi IS, Stroock AD, Whitesides GM. Langmuir. 2000;16:8311–8316. [Google Scholar]
- 42.Irimia D, Geba DA, Toner M. Analytical Chemistry. 2006;78:3472–3477. doi: 10.1021/ac0518710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wong PK, Chen CY, Wang TH, Ho CM. Analytical Chemistry. 2004;76:6908–6914. doi: 10.1021/ac049479u. [DOI] [PubMed] [Google Scholar]
- 44.Sin MLY, Shimabukuro Y, Wong PK. Nanotechnology. 2009;20:165701. doi: 10.1088/0957-4484/20/16/165701. [DOI] [PubMed] [Google Scholar]