Abstract
A proof-of-concept demonstration of the use of label-free quantitative glycoproteomics for biomarker discovery workflow is presented here, using a mouse model for skin cancer as an example. Blood plasma was collected from 10 control mice, and 10 mice having a mutation in the p19ARF gene, conferring them high propensity to develop skin cancer after carcinogen exposure. We enriched for N-glycosylated plasma proteins, ultimately generating deglycosylated forms of the modified tryptic peptides for liquid chromatography mass spectrometry (LC-MS) analyses. LC-MS runs for each sample were then performed with a view to identifying proteins that were differentially abundant between the two mouse populations. We then used a recently developed computational framework, Corra, to perform peak picking and alignment, and to compute the statistical significance of any observed changes in individual peptide abundances. Once determined, the most discriminating peptide features were then fragmented and identified by tandem mass spectrometry with the use of inclusion lists. We next assessed the identified proteins to see if there were sets of proteins indicative of specific biological processes that correlate with the presence of disease, and specifically cancer, according to their functional annotations. As expected for such sick animals, many of the proteins identified were related to host immune response. However, a significant number of proteins also directly associated with processes linked to cancer development, including proteins related to the cell cycle, localisation, trasport, and cell death. Additional analysis of the same samples in profiling mode, and in triplicate, confirmed that replicate MS analysis of the same plasma sample generated less variation than that observed between plasma samples from different individuals, demonstrating that the reproducibility of the LC-MS platform was sufficient for this application. These results thus show that an LC-MS-based workflow can be a useful tool for the generation of candidate proteins of interest as part of a disease biomarker discovery effort.
Keywords: Skin cancer, LC-MS, Label-free protein quantification, Biomarker discovery, Systems biology, Targeted peptide sequencing, Glycoproteomics, Plasma
INTRODUCTION
Cancer is a leading cause of mortality in the United States [1] and other developed countries. Years of research have revealed cancer to be a complex disease typically involving both genetic and environmental factors, which results in the molecularly heterogeneous disease that we know [2]. Collectively, the various genetic and environmental factors combine to cause activation and inhibition of multiple cellular pathways, resulting in a range of pathophysiologies, such as angiogenesis, immune system evasion, metastasis, altered cell growth, death and metabolism [3]. In turn, the heterogeneous nature of cancer has presented a significant challenge for finding new biomarkers for the disease, a topic which thus continues to generate significant research interest [4–6].
There are many types of biomarkers, each fulfilling a particular function. These would include diagnostic, prognostic, predictive and pharmacodynamic markers [6]. However, regardless of the purpose for which a biomarker is used, it relies on our ability to measure a change in abundance for one or more molecules of interest. Thus in any new biomarker discovery effort, analytical methods need to be able to robustly and reproducibly measure changes in biomolecules, and be able to identify and quantify with confidence the molecular species that are changing as a result of disease.
In this study, we demonstrate the feasibility of using an LC-MS-based approach to identifying protein changes in the blood, as a result of disease, using plasma obtained a mouse model of skin cancer and controls, and show how the use of protein interaction networks and functional analyses can further inform a biomarker discovery effort. This approach is not just valid for skin cancer, or even cancer in general, but should be equally applicable to most other diseases, if suitable biospecimens are available for analysis.
Blood, perfusing the whole body, moving molecules between organs is a rich body fluid for biomarker discovery. It is assumed that blood and plasma, where the blood cells have been removed, contains clues about the health status of most organs and tissues of the body. Because it is readily accessible, blood plasma is very attractive for biomarker discovery efforts. The main hurdle with plasma is its high degree of complexity, containing probably millions of distinct molecular species, spanning more than 10 orders of magnitude in concentration [7]. Since it is assumed that potential protein biomarkers are most likely present in the lower abundance range, and whole plasma analyses overwhelm even the most powerful LC-MS system, the adoption of one of many available strategies for reduction of sample complexity is additionally required.
Sample fractionation seeks to divide one complex sample into many samples of lesser complexity, and takes advantage of separation techniques that are orthogonal or complimentary to the C18 chromatography used in LC-MS. Protein separation [8], strong cation exchange [9, 10], 1D [11] and 2D gel electrophoresis [12, 13] are among the most common fractionation methods used to date for complex samples in proteomics. Although fractionation is an effective way to perform an extensive inventory of a complex sample such as plasma, the multiplication of samples it creates becomes a limitation for analyzing large sample populations, as one would do for biomarker discovery.
Another way to reduce complexity is to remove the proteins that are likely not relevant to the disease, such as highly abundant and ubiquitous proteins, which are generally dismissed as biomarker candidates. This is a particularly important consideration for plasma protein analyses since albumin alone accounts for over half of the total protein content [7], and just the top 22 most abundant make up about 99%. Immunoaffinity subtraction [14, 15] columns have thus been used to selectively deplete these most abundant proteins from plasma samples, opening up the dynamic range for potentially detecting more interesting proteins. One problem with this approach is that some of the potential biomarkers might bind to abundant proteins and be removed either by non-specific binding or by binding to proteins that are removed.
An alternative strategy to complexity reduction involves specific enrichment for a subfraction of analytes, rather than removing some of them as above. Such approaches typically focus on taking advantage of unique chemical properties of a subfraction of the proteome to enrich for it. Examples of this approach include selective enrichment of cysteinyl-containing peptides [16], phosphorylated peptides [17] and N-glycosylated peptides [18]. Glycosylated proteins are particularly attractive as potential candidate biomarkers that might be detectable in plasma since, by definition, they almost exclusively exist as secreted proteins or are embedded in cell surface membranes. They may thus represent a sub-proteome more likely to be detectable on the blood, either through direct secretion, or via deposit from cell membranes via a process of cell shedding, leaching, or cell death. It is also known that the oncogenic process itself causes altered cell surface protein expression patterns in tumor cells [19, 20], making glycoproteins also interesting from the disease perspective, since most cell surface proteins are glycosylated. Additionally, their hydrophilic sugar moieties tend to make them quite soluble, further increasing our chance of detecting them in blood. Finally, protein N-glycan structures provide a convenient chemical ‘handle’ that can readily be used for their selective enrichment.
Since this approach typically generates only one or a few peptides per protein (more than enough to identify most proteins), the significant reduction in complexity it affords results in a sample that can be directly analyzed by LC-MS without the need for additional fractionation, though this can still be an option in some workflows. This allows us to analyze only one sample per individual, making the approach ideal for population-based studies, as typically required for biomarker discovery. Another advantage of selective enrichment of N-glycopeptides comes at the data analysis stage, where the known consensus sequence for N-glycosylation, N-X-S/T (where X is any amino acid but proline, and S/T is serine or threonine), serves as a useful confirmation of peptide identification and can thus be used as a filter to reduce the false discovery rate (FDR). In addition to this, the enzymatic deamidation step that is used to remove the N-glycan structure and make the peptides amenable to LC-MS, results in conversion of the formerly glycosylated asparagine to aspartic acid. This generates a small mass shift for the resultant peptide, easily measurable in higher accuracy mass spectrometers. This serves as yet another confirmation of peptide identity, and further provides positive identification of the N-glycosylation site on the protein, information that may turn out to also be biologically relevant down the road. Finally, with the availability of sequence databases and the existence of a N-glycosylation motif, the finite number of possible N-glycosylated peptides, along with their expected sequences, is known. This has allowed for the building of N-glycosylation databases, such as UniPep [21], that can be a useful resource in a candidate-based biomarker discovery workflow.
For relevant biomarker discovery research, one also needs to perform quantitative measurements on populations of samples, and identify discriminating peptides for which the abundance segregates the populations into disease states. Traditional quantification methods for proteomics involve stable isotope tags of distinct masses, which enable multiplexing of samples in one LC-MS measurement [22–27]. However, there is a strict limit to the number of samples that can be multiplexed, restricting their use in population studies. Multiplexing also reduces the available dynamic range, since the amount of sample loaded into the LC-MS system is a finite quantity. Multiplexing 5 samples is equivalent to loading only 1/5 of the original material on a per sample basis.
Traditional shotgun proteomic workflows have significant limitations for biomarker discovery. Automatic precursor selection introduces a bias towards high intensity precursor ions, leaving the lower intensity ones that frequently correspond to lower abundance peptides unidentified. Typically, the majority of the reliably detected peptides at the MS1 level do not get identified by automatic precursor selection [28]. In order to concentrate our efforts on the specific and discriminating features, our approach compares LC-MS profiles first, to find statistically significant changes in peptide abundance between different populations. Once those peptides are found, their m/z and retention times are placed in an inclusion list to be sequenced and identified, on reanalysis of the same samples by tandem mass spectrometry (MS/MS) [29].
In label-free LC-MS quantification, only one sample is analyzed at a time, which requires a highly reproducible analytical platform. To infer relative peptide abundance, peak areas from different LC-MS experiments analyzed under similar conditions are compared. The first step in label-free LC-MS quantification is to find all the peaks detected in a sample, a process called peak picking. The next step is peak alignment, where the peaks found in some samples are tentatively mapped to other samples, compensating for small changes in m/z and retention times. This type of analysis assumes that the samples have a majority of peaks in common and that only a small fraction of those will change between subjects. This approach has been formulated in various software packages [30–33]. However, it was found that most tools were designed with a particular kind of data in mind and did not perform well with data acquired with another type of mass spectrometer. Factors such as mass accuracy, noise level and resolving power, which are specific not only to a particular instrument, but to how that instrument is operated, can limit the scope of such tools. The Corra framework (M. Brusniak et al., manuscript submitted) was designed to facilitate the use of those tools and combine them with statistical methods for data analysis within a common interface where we can mix and match different software tools and instruments.
The goal of this study was thus to demonstrate the applicability of a highly integrated label-free LC-MS proteomics workflow to a disease biomarker discovery effort. We used a p19ARF mouse model for skin cancer [34] to perform a proof-of-concept experiment to show that we were able to identify peptides and proteins that were differentially abundant between plasma samples from skin cancer bearing mice and normal controls. We utilized N-glycopeptide enrichment for plasma protein complexity reduction, followed by LC-MS and Corra data analysis to identify differentially abundant peptides. We then used inclusion list tandem mass spectrometry to identify the peptides and the proteins they originated from. Finally, we used protein interaction and functional data to identify protein networks and molecular functions that were enriched in the regulated proteins. This confirmed that cancer-related proteins and processes were indeed detectable and quantifiable in plasma by this method, thus demonstrating its utility as part of any larger biomarker discovery project.
MATERIALS AND METHODS
Mouse model
The p19ARF mouse model is a well-characterized in vivo model of epithelial neoplasia, as deletion of the p19ARF tumor suppressor makes them highly susceptible to tumor formation upon topical application of carcinogenic agents. The two stage chemical protocol involves treatment of mice with a carcinogen, 7,12-dimethylbenz[a]anthracene (DMBA) followed by multiple applications of the promoting agent, 12-O-tetradecanoylphorbol-13-acetate (TPA). This treatment induces benign squamous cell papillomas, with sustained activating mutations in the H-ras oncogene. Early papillomas consist of folded epidermal or follicular hyperplasias that begin to protrude from the skin surface. In wild type mice, a small fraction of these benign papillomas will progress over time to malignant carcinomas as further genetic mutations occur. Mice heterozygous for the tumor suppressor p19ARF were used for the skin tumor model in this study due to their reduced latency in developing papillomas and carcinomas compared to wild type mice [34].
Breeder pairs of genotype p19ARF+/− X p19ARF +/+ on a NIH01a background strain, were set up to generate a cohort of 10 experimental and 10 control mice of genotype p19ARF +/−. Each experimental mouse was paired with a control mouse from the same litter and sex, and housed in the same cage during the course of the experiment. Care was taken to ensure that no systematic biases were introduced between cases and controls. All mice were maintained on a 12 hour light dark cycle and had access to autoclaved food and water ad libitum. For the experimental mice, the backs of 8 week old male and female mice were shaved and treated with a single application of DMBA (Sigma, Saint-Louis, MO) 25μg in 200μl acetone, followed a week later by twice weekly applications of TPA (Sigma) (200μl of 10-4M solution in acetone), for 15 weeks. Control mice were treated with TPA alone for 15 weeks. Benign squamous cell papillomas appeared by 8 weeks post-DMBA treatment that progressed to malignant squamous cell carcinomas beginning at 24 weeks. The experimental mice had an average of 5 papillomas and 1 carcinoma. No tumors were detected on the backs of control mice. Mice were monitored daily and any abnormal health or behavior noted. Benign to malignant conversion was accompanied by a dramatic change in the appearance of the tumor and its invasion and growth into the underlying dermis. Conversion was easily quantified by visual inspection and confirmed using histological analysis of tumor sections. Carcinoma-bearing experimental mice and the matched control were sacrificed within 1–2 weeks following first visual detection of carcinoma. Mice were euthanized by CO2 inhalation and whole blood was collected by cardiac puncture. Plasma was purified from the whole blood by K3EDTA addition and centrifugation. Plasma was stored in conical cryovials in 100 μl aliquots and stored in a liquid nitrogen tank.
Sample preparation
The isolation of N-linked glycopeptides from total mouse plasma samples (40 μl total from each mouse) was performed essentially as described elsewhere [35, 36]. Unless otherwise noted, all chemicals and reagents were from Sigma. Individual plasma samples were diluted with 40 μl trypsin buffer (0.1% Rapigest (Waters, Milford, MA) in 25 mM KHPO4, pH 8) and desalted using spin columns (SNS P060, The Nest Group, Southborough, MA) pre-equilibrated in trypsin buffer, eluting in a final volume of 80 μl. Proteins were denatured with the addition of 75 μl trifluoroethanol for 1 h, 60°C. Proteins were then reduced with the addition of 10 μl of 80 mM Tris (2-carboxyethyl) phosphine (Pierce, Rockford, IL) for 30 min, 60°C, followed by alkylation with 20 μl of 100 mM iodoacetamide for 30 min, room temperature, in the dark. Proteins were next proteolyzed with the addition of 550 μl trypsin buffer and 40 μl 0.5 mg/ml trypsin (Promega, Madison, WI) for 2 h, 37°C. Glycopeptides were next oxidized with the addition of 100 μl coupling buffer (1 M sodium acetate, 2.5 M NaCl, pH 5.5) and 100 μl of 100 mM NaIO4 (Sigma) for 1 h, 4°C, in the dark and then captured to a solid-phase with the addition 100 μl of a 50% slurry of hydrazide resin (Bio-Rad, Hercules, CA) pre-washed with coupling buffer, 3 h at room temperature with gentle mixing. Beads were washed 3x each with 1.5 M NaCl, H2O and freshly made bicarbonate buffer (100 mM NH4HCO3, pH 8.3) and the N-linked glycopeptides released by the addition to the beads of 25 μl bicarbonate buffer and 3 μl protein N-glycosidase F (PNGaseF, New England Biolabs, Ipswich, MA) and incubation overnight at 37°C with gentle mixing. Released peptides were then recovered with 2 additional washes with 100 μl bicarbonate buffer, and desalted and cleaned up on Sep-Pak C18 cartridges (Waters, Milford, MA) eluting with 50% acetonitrile, 0.1% trifluoroacetic acid and finally evaporated to dryness under vacuum in clean glass vials.
Differential mapping
Peptides were separated on an 1100 Series HPLC system (Agilent, Santa Clara, CA) equipped with a nanoflow pump, operating at a flow rate of 1 μL/min. Mobile phase A was a 0.1% formic acid in water and mobile phase B was 0.1% formic acid, 5% water and 95% acetonitrile. A binary gradient from 5% to 35% B was used to separate the peptides on a monolithic C18 10 cm long × 100 μm inner diameter column (Merck KGaA, Germany). A self-packed integraFrit column (New Objective, Woburn, MA) with a bed of Magic C18 5 μm particles (Michrom Bioressources, Auburn, CA) 2 cm × 100 μm was used as a pre-column. Sample volumes of 5 μL were injected by the autosampler and every batch of replicates was randomized prior to injection.
Mass analysis was performed on a MicrOTOF electrospray time-of-flight mass spectrometer (Bruker Daltonics, Billerica, MA) with a mass accuracy of 5 ppm and a resolving power of 9000 or better. The mass scale was calibrated using glu1-fibrinopeptide B (Sigma) and mass spectra were acquired at 1 spectra/s over the range of 300–1600 m/z. High mass accuracy for the differential mapping on the MicrOTOF was maintained by automatic instrument recalibration between every sample. This was achieved by injecting 320 fmol of glu1-fibrinopeptide B (Sigma) with a 15 minutes gradient and increasing the cone voltage to 220 V, inducing in-source CID. The fragments were used by the visual basic script to recalibrate on-the-fly the mass spectrometer, insuring the same mass accuracy from the first to the last sample. This measure also had the benefit of preventing carry over between samples, and provided a way to monitor the sensitivity and reproducibility of the system during large batch analyses.
Corra statistical analysis
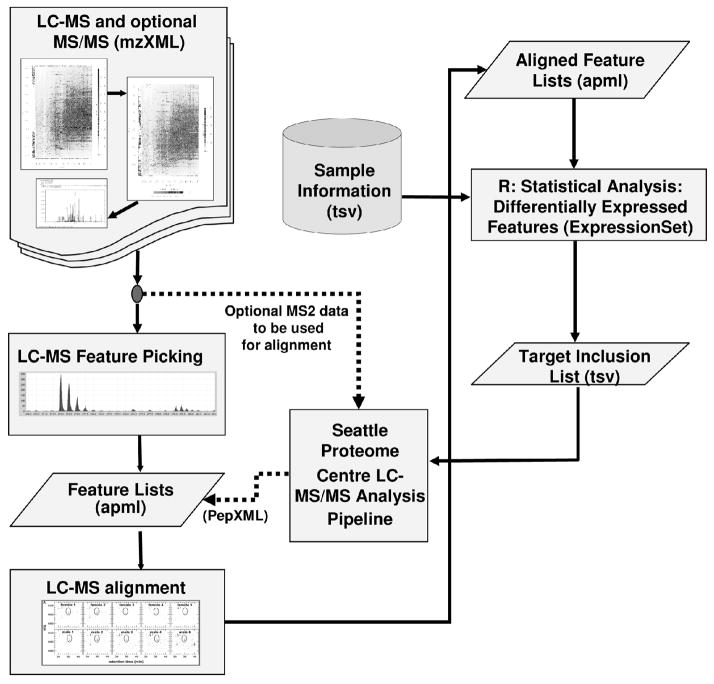
The mzXML data was imported into the Corra framework (M. Brusniak et al., manuscript submitted) to perform statistical analysis to reveal peptides that are differentially abundant between the two sample populations. Peak detection and alignment were performed within Corra using the SpecArray [30] algorithm. The alignment was performed such that only features that were detected in half of the samples made it to the aligned feature table. Figure 1 represents schematically the Corra framework. Corra was running on a six dual core, dual processor AMD Opteron 275, 2.2GHz, 1MB level 2 cache hardware configuration.
Figure 1.
Workflow of the Corra framework. The pipeline accepts mzXML files as input, then it performs peak picking and alignment. The statistics module is used to find differentially abundant features that are significant, and generates an inclusion list from them. Identified peptides are then annotated back to the feature list.
Targeted sequencing of discriminatory peptides by LC-MS/MS
The top 300 discriminatory peptides of charge 2 and 3 were made into an inclusion list for sequencing on an LTQ-FT mass spectrometer (ThermoFisher, San Jose, CA), as described elsewhere [29]. A mass tolerance of 25 ppm was specified and ion accumulation time was set at 500 ms for both IT-MS and FT-MS scans. The scan rate for the FT-MS scans was set to 0.89 seconds over the range of 300–1600 at a resolving power of 100000. MS/MS scan rate was set at 0.2 seconds. The LTQ-FT connected to an 1100 Series HPLC system, but used a self-packed, 15 cm capillary column with a 150 μm inner diameter packed with a bed of Magic C18 5 μm particles (Michrom Bioresource), without a pre-column. The flow rate was 1.2 μL/min and the gradient was the same as described for the differential mapping. The identified peptides were mapped back to the aligned features, keeping the same mass accuracy and retention time tolerances as were used for the inclusion list.
Protein identification and function
The proteins from the inclusion lists were identified using the Trans Proteomic Pipeline [37] using SEQUEST version 27 and the mouse IPI database version 3.5. The searches were performed using a mass tolerance of 0.1 Da on the precursor ion and of 3 Da on the fragment ions. Variable modifications were included for oxidized methionine (16.0 Da) and conversion of asparagine to aspartic acid due to the deglycosylation (0.98 Da) were added to the search, as well as a static modification for cysteine carbamidomethylation (57.02 Da). Trypsin was selected as the proteolytic enzyme. Peptide and Protein FDRs are automatically calculated in a data set-dependant manner by the PeptideProphet and ProteinProphet components of the Trans Proteomic Pipeline [37].
Because biological function is more commonly annotated to genes than proteins, the first step of the protein data analysis was to find the Entrez GeneIDs that coded for the identified proteins. These GeneIDs allowed us to identify enriched biological processes and molecular functions, protein-protein interactions, and protein associations. The data mining was performed using Cytoscape [38] and Gaggle [39], through the Protein Function Exploration WorkBench [40] using the following data sources: Entrez GeneID identifications were obtained primarily from the mouse database version 3.5 (ftp://ftp.ebi.ac.uk) and then supplemented (for IPIs that had no Entrez GeneID) by searching the IPI protein sequence against the NCBI ‘nt’ database, using tblastn (http://blast.wustl.edu/) dynamically translating in all six reading frames, selecting only high scoring, complete matches with Entrez GeneIDs. Enriched Gene Ontology biological processes and molecular functions were calculated using the Bioconductor GOstats package (http://bioconductor.org) on annotations provided by the Affymetrix Mouse Genome 430 2.0 Array annotation data. Protein-protein interactions were inferred from HPRD, the Human Protein Reference Database (http://www.hprd.org) via homology, using NCBI’s Homologene (http://www.ncbi.nlm.nih.gov). Other gene associations were provided by EMBL STRING (http://string.embl.de).
RESULTS AND DISCUSSION
LC-MS Analysis of N-glycosylated tryptic peptides by LC-MS
For this experiment, we enriched formerly N-glycosylated tryptic peptides from the blood plasma of 10 cancer-bearing mice and 10 control mice. All 20 samples were initially analyzed by LC-MS on an ESI-TOF system. Sample run order was randomized for all 20 samples. This randomization was to both average out any cross contamination that might occur due to sample carryover, and to ensure no statistical bias for run order was introduced for subsequent data analysis. To further minimize sample carryover, a standard injection of 320 fmol of the calibration standard peptide glu1-fibrinopeptide B was inserted between each of the 20 LC-MS injections. An advantage of doing this between each LC-MS run was that this calibration standard could be used to recalibrate the machine between each run in an automated fashion. This gave us great confidence in, and knowledge of, the mass resolution and accuracy from run to run. This, in trun, gave us increased confidence in the LC-MS data alignments, which is done on the basis of accurate mass and LC retention time.
Feature Selection and Clustering
All 20 LC-MS maps were processed with Corra. The number of detected features per LC-MS injection varied from 1096 to 1407, with a median of 1354 features on the ESI-TOF platform. In order to increase our chances of identifying cancer-specific regulated proteins, we only made use of features that aligned across, minimally, half of the LC-MS runs. This approach would, in theory, find any differentially abundant feature observed in all of the cancer mice and not the controls, and reduce the processing time to a manageable amount, since it grows exponentially with the number of features to compare. In using this filter, we would expect to loose some genuine cancer marker candidates. However, this was done to help control the false discovery rate. While we would expect that some N-glycoproteins that might be up-regulated and detectable in cancer and below the level of detection in controls, they may not be detected in all the cancer mouse LC-MS runs. This likelihood increases as their relative abundance decreases. However, for the purposes of this study, it was important to have a low false-discovery rate in terms of the alignments, so that we could have high confidence in the downstream protein function analyses. The next step in the data analysis was to superimpose observed signal intensities for the aligned peptide features to determine which showed differential abundance between case and control mice. This information was then used to see whether the aligned LC-MS data alone could be used to distinguish between the two disease populations in a blind analysis.
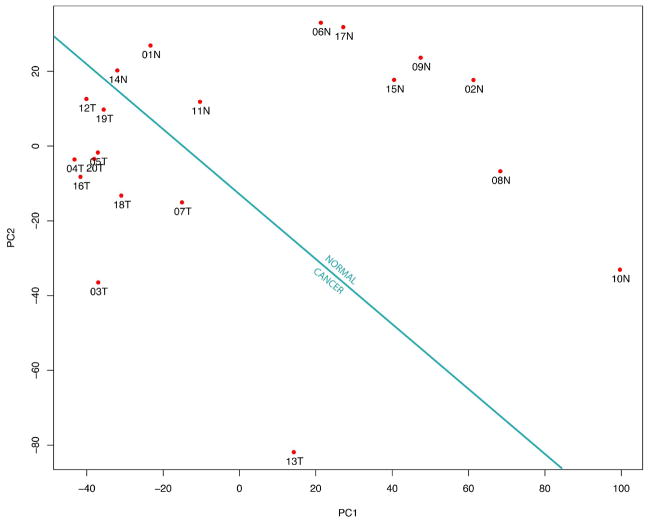
Figure 2 shows how the use of Principal Components Analysis (PCA) allowed us to successfully differentiate between the healthy and disease states on the basis of the LC-MS profiles alone. The samples from the tumor bearing mice clustered in the bottom left corner of the PCA space, while the normal mice samples clustered in the top right corner. In Figure 2, each data point represents a single sample, on which a series of measurements have been made, in this case of peak intensities at aligned retention times and m/z. All the measurements are projected in orthogonal space, on a reduced number of axes, typically two or three. This dimensionality reduction facilitates the visualization of trends in the data. Since PCA is an unsupervised mode of analysis it reveals clustering in an unbiased way, without risk of over fitting by using a priori knowledge of class membership (in this case, those from cancer and those from control mice). More importantly with this plot, that we observed two linearly separable clusters tells us that there were differences at the protein level between the normal and the tumor-bearing mice. It also tells us that those differences could be measured, as well as quantified, and that they contain information of sufficient power to correctly classify all 20 LC-MS data files on the basis of disease state. This result further goes to illustrate the potential applicability of such an LC-MS workflow to a larger biomarker discovery effort.
Figure 2.
PCA plot of the clustering of the 20 LC-MS in two different disease states are linearly separated. Each data point is annotated with a number representing the order in which the samples were analyzed, followed by a letter, N or T, which indicate the normal and tumor-bearing mice, respectively. In this plot, the first principal component represents 25% and the second principal component represents 17% of the total variance.
Targeted sequencing by LC-MS/MS for peptide and protein identification
With the aligned peptide feature signal intensities for the cancer versus normal mice, the next step was to identify the peptides and proteins of most interest, according to apparent differential abundance, via inclusion list-based targeted LC-MS/MS. To do this, we selected 300 aligned features (based on raking according to Corra-generated p-values) to form the inclusion list. In turn, this was used for LC-MS/MS sequencing using and LTQ-FT MS system, and was performed on 6 randomly chosen plasma N-glycopeptide isolates, 3 each from the cancer and normal sample sets.
Since the mass spectrometer was set to monitor selected ion traces of all the targeted features throughout the chromatographic gradient. There were more identified peptides than targeted features as the mass (i.e. m/z) of a feature can correspond to multiple peptides, especially for a complex sample such as isolates from plasma, within the resolving power of the LTQ portion of the mass spectrometer. Corra then determined the correct assignment of identified peptides to a particular feature, based not just on observed mass, but also on expected retention times. The MS/MS spectra obtained from the inclusion lists were then searched against a sequence database, so that the corresponding peptides sequences could be mapped back to their respective LC-MS features. In this way, we identified 80 peptides out of the 300 differentially abundant features with a PeptideProphet score of ≥0.9. In addition, we identified another 8 glycopeptides that had a score lower than 0.9, but were manually validated to be correct (shown in Supplemental Figure S1). The PeptideProphet-calculated FDR for a cut-off of ≥0.9, for this data set, was ~2 %.
Variability and reproducibility of the analytical platform
In order for an LC-MS platform such as this to be useful for studies such as biomarker discovery, it is important that the reproducibility of the experimental platform as a whole is not so great as to confound subsequent data analyses. We thus performed some additional analyses to look at the reproducibility of the platform used in this study.
As was discussed in the Methods section above, the use of an internal MS calibration standard in the wash cycles between analytical runs effectively removed variation in the accuracy of the MS measurements themselves. Much more variation, however, could be expected to come from the LC-MS system. We had previously determined that the normalization process and alignment tools of Corra adequately control for inherent variation in global signal intensity and LC retentions times for the LC systems used in this study (Brusniak et al., manuscript submitted). However, since we were also mapping back subsequent MS/MS identifications onto the original, aligned LC-MS data, we looked to see how well these data correlated.
Figure 3 shows a plot of observed retention times for the LC-MS data (from an ESI-TOF) versus the subsequent LC-MS/MS data (from an LTQ-FT) for the 48 positively identified aligned features/peptides (from the inclusion list experiment above) for one of the serum samples analyzed on both machines. Here, the identified peptides from the targeted MS/MS experiment were mapped back manually to the original aligned LC-MS features, using Corra. This plot shows that, with the exception of the beginning of the chromatography run, where the peptides eluted earlier from the LC system on the LTQ-FT platform, due to the absence of a trapping column, the retention times coincide well, within five minutes of each other. This observation reconfirmed the reproducibility of the LC system and Corra alignments. Also based on this observation, we used a LC elution time window of 5 minutes for the software-based annotation of the LC-MS features by Corra. It additionnaly demonstrates the portability of the features form one instrument to the other.
Figure 3.
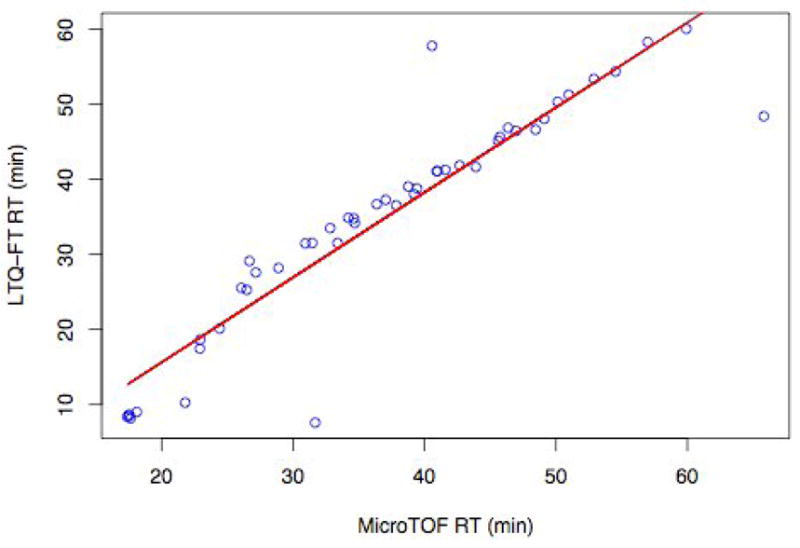
Correlation between LC elution times for MS/MS identified peptides using an LTQ-FT platform versus their respective LC-MS aligned features from an ESI-TOF platform. Shown are 48 MS/MS identified peptides from the analysis of one mouse plasma glycopeptide isolate that could be mapped back onto the LC-MS data. Both instruments use a similar HPLC hardware set-up, and all 48 features were validated by visual examination of the original MS data files.
It is generally assumed that the largest form of variation in a LC-MS based disease biomarker study is likely to be between the individuals (patients, mice etc.) being used for the study. While this variation is expected to be high in human studies, transgenic mice, as used in this study, might generate less variation due to their common genetic background and the use of littermates (i.e. siblings) as controls. In order to be able to accurately identify differences in the LC-MS profiles of the individuals in a given study, it is important that the observed variation between the individuals is less that the variation observed for the analytical platform as a whole. To investigate whether this was the case for the experimental platform being used for this study, we thus performed additional LC-MS profiling of the 20 samples, however in triplicate this time (60 runs in all) to better examine the variation introduced into the data via the experimental platform.
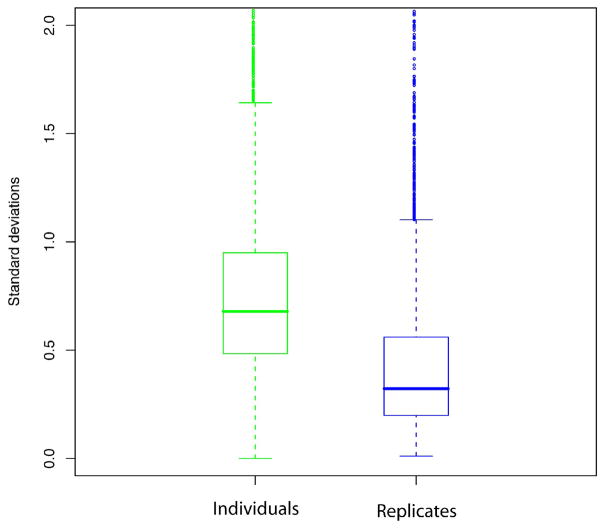
The plot shown in Figure 4 displays the distribution of calculated standard deviations for the observed signal intensities observed for all features that aligned across at last 3 of the 60 LC-MS runs (i.e. to be observed in at least one sample, in all 3 replicates of that sample). On the left side, we show the standard deviation distribution observed for the same features aligned across multiple individuals in the same disease class, i.e. for cancer and normal, separately. On the right side, we show the standard deviation distribution observed for the same features, but instead aligned across the replicate analyses of the same sample/individual. As expected, desired for a successful outcome, we found that the variation between individual mice was, indeed, significantly higher than the variation introduced by the analytical platform itself, again highlighting the good reproducibility of the LC-MS based approach.
Figure 4.
Distribution of standard deviation for the intensity of the aligned features across individuals in the same group (i.e. cancer and normal) (green) and across technical replicates (blue), for 3 replicate LC-MS runs of each 20 samples. The horizontal line represents the median value, and the upper and lower edges of the boxes represent the 75 and 25 percentile data points, respectively.
Biological inference via protein network and functional analyses
The final step in the data analysis process was to take the peptides/proteins identified from the targeted MS/MS, and use protein network and functional analyses, to see if we can identify and interesting proteins or protein families enriched in the data set. In so doing, by using network inference, we might also expect to identify additional proteins, not actually identified in the MS/MS data, but that might also be of interest with respect to disease, on account of being linked to other proteins in a network or functional group that were identified. To increase our confidence in the protein networks, we restricted these analyses to proteins identified by at least one identified glycosylated peptide (i.e. containing the N-glycosylation consensus motif of -N-X-S/T-, where X is any amino acid except proline, and S/T is serine or threonine). Using these criteria, a total of 21 proteins, shown in Table 1, were used for the functional protein network analysis.
Table 1.
Gene name, description, IPI number, log2 ratio (CAN WE CHANGE THESE TO DIRECT RATIO?), PeptideProphet and ProteinProphet probabilities for the differentially abundant glycoproteins that were identified by targeted MS/MS analysis.
| Gene | Description | IPI | log2 Ratio | PeptideProphet | ProteinProphet |
|---|---|---|---|---|---|
| Ahsg | alpha-2-HS-glycoprotein | IPI001 28249 |
−0.39 | 0.62 | 0.6 |
| Apob | apolipoprotein B | IPI003 50772 |
0.53 | 1.00 | 1 |
| C9 | complement component 9 | IPI002 30718 |
−0.35 | 0.59 | 1 |
| Cfh | complement component factor h | IPI001 30010 |
0.44 | 1.00 | 1 |
| Cfi | complement component factor i | IPI003 20675 |
0.27 | 0.93 | 0 |
| Hp | haptoglobin | IPI004 09148 |
0.04 | 1.00 | 1 |
| Hpx | hemopexin | IPI001 28484 |
0.70 | 0.74 | 1 |
| Igh1a | immunoglobulin heavy chain 1a (serum IgG2a) | IPI001 11657 |
−0.44 | 0.96 | 0 |
| Ighg1 | immunoglobulin heavy constant gamma 1 (G1m marker) | IPI003 08213 |
0.59 | 0.83 | 0.98 |
| Itih3 | inter-alpha trypsin inhibitor, heavy chain 3 | IPI001 24725 |
0.50 | 1.00 | 1 |
| Itih4 | inter alpha-trypsin inhibitor, heavy chain 4 | IPI001 19818 |
0.74 | 0.99 | 1 |
| Klkb1 | kallikrein B, plasma 1 | IPI001 13057 |
−0.81 | 1.00 | 1 |
| Lcat | lecithin cholesterol acyltransferase | IPI001 33500 |
0.22 | 0.60 | 1 |
| Lifr | leukemia inhibitory factor receptor | IPI001 19299 |
0.64 | 1.00 | 1 |
| LOC640207 | similar to Ig heavy chain V region 102 precursor | IPI001 25877 |
0.38 | 1.00 | 1 |
| Lrg1 | leucine-rich alpha-2-glycoprotein 1 | IPI001 29250 |
0.52 | 0.99 | 0.96 |
| Pzp | pregnancy zone protein | IPI001 26194 |
0.27 | 0.92 | 1 |
| Serpina10 | serine (or cysteine) peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 10 | IPI001 53258 |
−0.03 | 0.85 | 1 |
| Serpina6 | serine (or cysteine) peptidase inhibitor, clade A, member 6 | IPI001 16105 |
−0.58 | 0.99 | 1 |
| Serpind1 | serine (or cysteine) peptidase inhibitor, clade D, member 1 | IPI001 13227 |
0.28 | 1.00 | 0.88 |
| Spil-6 | serine protease inhibitor 1–6 | IPI001 17857 |
0.46 | 0.71 | 1 |
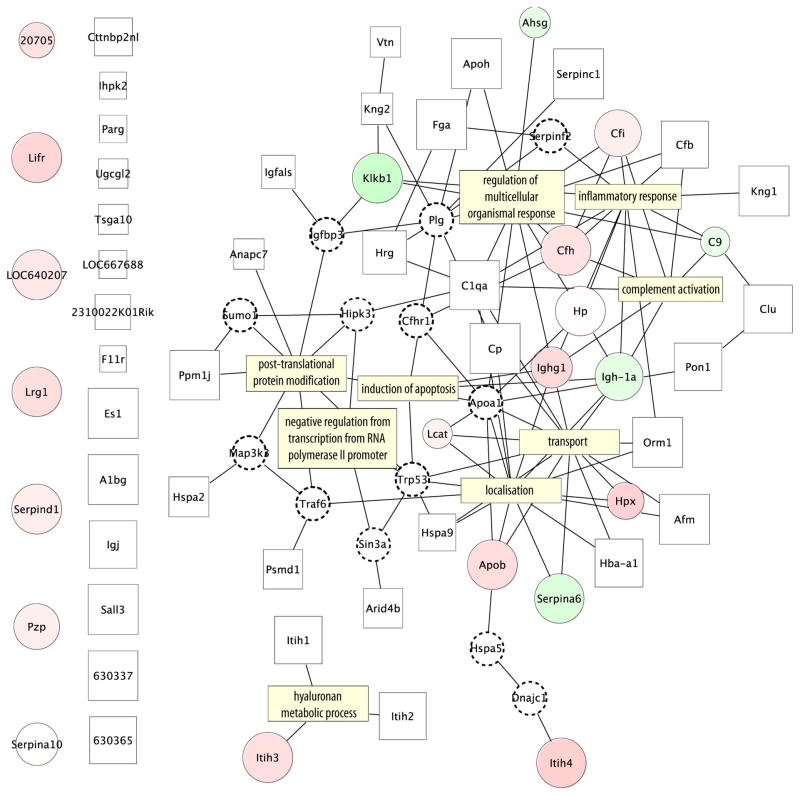
Figure 5 shows a protein-protein interaction network, generated using human orthologs of the identified mouse proteins, and the human protein reference database (HPRD) (www.hprd.org). Proteins that were found to be more abundant in cancer are shown as red circles, and those less abundant in cancer as green circles. The white rectangles represent the proteins that were identified by MS2 peptide sequencing at high confidence, but not assigned to a LC-MS feature. All identified glycoproteins were used, not just the differentially abundant, in order give the network more context around the differentially abundant ones. To ensure that they better pertain to the disease being studied, we also only included those proteins with MS/MS evidence that they were present in one or more the cancer samples analyzed by MS/MS. To enhance the information density of the network, and thereby connect more proteins, some proteins were inferred (i.e. had no observational evidence for their presence in this experiment) and are depicted in Figure 5 by the small white circles with dotted borders.
Figure 5.
Protein-protein interaction map constructed from the proteins identified by targeted MS/MS, using inferred human protein orthologs and the protein interaction database HRPD. Proteins that were observed with increased abundance in cancer are represented by red circles, those observed to decrease in abundance by green circles. Increased circle size represents higher observed ProteinProphet probability, and increased fold-change observed with Corra is represented by a darker hue. White squares represent proteins identified in only cancer mice, with increased box size again indicative of higher ProteinProphet probability. Inferred proteins, observed to interact with identified proteins according to the HRPD database, but not observed in the MS/MS data, are represented by smaller dotted circles, and were added to the network to connect more nodes together. The GO biological processes for which those proteins are involved are indicated by yellow rectangles.
Another way to add information to the network is to look for enrichment of biological process from the Gene Ontology (GO). The GO terms are a controlled vocabulary that describe gene and gene product attributes according to their molecular functions, cellular components or biological process. The p-values, probabilities that a certain term is over-represented in a gene list are used as a measure of likelihood of these processes have been activated in the mouse cancer model studied. Filtering for low p-values will highlight the processes that are unlikely to appear within a group of randomly selected genes or proteins. The yellow rectangles represent those GO biological processes that were prevalent in this data set. As it is shown in Figure 5, that major differences between the two populations are the result of the cancer bearing mice being seriously sick, due to the advanced state of the cancer. Hence we observed proteins whose function is related to the host response to disease, such as inflammatory response, regulation of multicellular organism processes and complement activation. For example, we observed proteins such as C9, Cfi, Cfh and C1qa, which are complement factors related to innate immunity, a general defense mechanism against pathogens and infections. Likewise, the increased abundance of proteins involved in blood coagulation, such as Klkb1, Fga, Apoh, Serpind1 and Serpinc1 is likely an indication of wounding, if we view the morphology of the carcinoma as akin to an open wound. The functional enrichment for these pathways confirmed our ability to detect significant changes in protein abundances caused by disease. Considering the advanced state of the cancer in this study, it was not unexpected that it would generate a very strong host response, and that Corra would detect those proteins in our data.
Another protein cluster can be seen that includes Lcat, Hpx, Serpina6, Apob, which are clustered together around the transport and localization GO categories. A post-translational protein modification enrichment region has been identified featuring where Anapc7 has been identified. Anapc7 is regulated by Trp53 [41], which is a tumor repressor protein. Proteins related to cell death and cell cycle include complement component 9 (C9) and Ighg1. C9 protein is a membrane attack protein and can form large pores that will lyse a cell. Ighg1 appears to be more abundant in the cancer samples and it has been shown to be produced by cancer cells [42]. Hyaluronan metabolic process was also represented by the differentially abundant Itih3. Hyaluronan contributes significantly to cell proliferation and migration, and may also be involved in the progression of some malignant tumors [42, 43]. Using a different mouse skin cancer model, but the same glycopeptide enrichment technique, Zhang et al. previously found carboxipeptidase N, hemopexin and complement component factor h to be differentially regulated [44].
Taken together, while the proteins identified in this study and discussed here are insufficient to formulate a strong cancer hypothesis, they can be a starting point for a more targeted type of analysis such as high sensitivity multiple reaction monitoring experiments [45, 46]. Additionally, the inferred proteins from the protein-protein interaction network would also make prime candidates for follow up analysis. It is important to note that proteins that were not identified by MS/MS but present in this network were either not differentially abundant or were present below the detection limit of the technique.
CONCLUDING REMARKS
This study demonstrated that it is possible to start from a population of complex plasma samples and perform label-free LC-MS quantitative proteomics, to identify a list of differentially abundant features that segregate two or more sample populations and link those features to proteins present in serum. We also showed that the changes detected do reflect actual biological processes that are related to the presence of disease, such as host response, cell cycle, cell death, cell adhesion and DNA damage.
We demonstrated that a modern LC-MS platform has sufficient reproducibility and to perform label free protein quantification of small sample populations, which is challenging for most other techniques used for biomarker discovery. We also confirmed that run-to-run variations were smaller than individual variation, an important prerequisite for this mode of analysis. It was possible to map the retention times from one instrument to another to perform targeted peptide sequencing. Simpler and affordable LC-ESI-TOF platforms can thus be used to analyze the bulk of the data and keep the smaller number of sequencing experiments on the more costly tandem mass spectrometer.
Measuring the difference in protein abundance is the first step of biomarker discovery. The proteins identified were clearly not all specific markers for skin cancer. Many were related to the host immune responses that are expected during tumor growth. However, some proteins seem to be involved in biological processes that are critical to cancer. In order to detect early disease biomarkers, it would be interesting to repeat the experiment with earlier stages of cancer. This data may still be of value to inform follow-up targeted MRM experiments for the cancer-related proteins that were identified, as well as those that were not identified, but inferred from the building of the protein interaction network.
Finally, this methodology has no definite limit in the number of samples that can be compared, making it suitable for large-scale studies. The principle was demonstrated with a small number of cancer mouse model samples, but can also be directly applied to other organisms and other diseases.
Supplementary Material
Acknowledgments
This work was supported with federal funds from the National Cancer Institute, National Institutes of Health, under contract No. N01-CO-12400 (to J.W.), the National Heart, Lung, and Blood Institute, National Institutes of Health, under contract N01-HV-28179 (to R.A.), the National Cancer Institute by Grants R21-CA-114852 (to H.Z.), and by Grant No. 31000-107-67 by the Swiss National Science Foundation (to R.A.).
References
- 1.Jemal A, Siegel R, Ward E, Murray T, et al. Cancer statistics. CA Cancer J Clin. 2007;57:43–66. doi: 10.3322/canjclin.57.1.43. [DOI] [PubMed] [Google Scholar]
- 2.van’t Veer LJ, Bernards R. Enabling personalized cancer medicine through analysis of gene-expression patterns. Nature. 2008;452:564–570. doi: 10.1038/nature06915. [DOI] [PubMed] [Google Scholar]
- 3.Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100:57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 4.Srivastava S, Srivastava RG. Proteomics in the Forefront of Cancer Biomarker Discovery. J Proteome Res. 2005;4:1098–1103. doi: 10.1021/pr050016u. [DOI] [PubMed] [Google Scholar]
- 5.Anderson KS, LaBaer J. The sentinel within: exploiting the immune system for cancer biomarkers. J Proteome Res. 2005;4:1123–1133. doi: 10.1021/pr0500814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sawyers CL. The cancer biomarker problem. Nature. 2008;452:548–552. doi: 10.1038/nature06913. [DOI] [PubMed] [Google Scholar]
- 7.Anderson NL, Anderson NG. The human plasma proteome: history, character, and diagnostic prospects. Mol Cell Proteomics. 2002;1:845–867. doi: 10.1074/mcp.r200007-mcp200. [DOI] [PubMed] [Google Scholar]
- 8.Wang H, Hanash S. Intact-protein based sample preparation strategies for proteome analysis in combination with mass spectrometry. Mass Spectrom Rev. 2005;24:413–426. doi: 10.1002/mas.20018. [DOI] [PubMed] [Google Scholar]
- 9.Fortier MH, Bonneil E, Goodley P, Thibault P. Integrated Microfluidic Device for Mass Spectrometry-Based Proteomics and Its Application to Biomarker Discovery Programs. Anal Chem. 2005;77:1631–1640. doi: 10.1021/ac048506d. [DOI] [PubMed] [Google Scholar]
- 10.Ranish JA, Yi EC, Leslie DM, Purvine SO, et al. The study of macromolecular complexes by quantitative proteomics. Nature genetics. 2003;33:349–355. doi: 10.1038/ng1101. [DOI] [PubMed] [Google Scholar]
- 11.Chelius D, Zhang T, Wang G, Shen RF. Global protein identification and quantification technology using two-dimensional liquid chromatography nanospray mass spectrometry. Anal Chem. 2003;75:6648–6657. doi: 10.1021/ac034607k. [DOI] [PubMed] [Google Scholar]
- 12.Stuart LM, Boulais J, Charriere GM, Hennessy EJ, et al. A systems biology analysis of the Drosophila phagosome. Nature. 2007;445:95–101. doi: 10.1038/nature05380. [DOI] [PubMed] [Google Scholar]
- 13.Gilchrist A, Au CE, Hiding J, Bell AW, et al. Quantitative proteomics analysis of the secretory pathway. Cell. 2006;127:1265–1281. doi: 10.1016/j.cell.2006.10.036. [DOI] [PubMed] [Google Scholar]
- 14.Sitnikov D, Chan D, Thibaudeau E, Pinard M, Hunter JM. Protein depletion from blood plasma using a volatile buffer. J Chromatogr B Analyt Technol Biomed Life Sci. 2006;832:41–46. doi: 10.1016/j.jchromb.2005.12.013. [DOI] [PubMed] [Google Scholar]
- 15.Liu T, Qian WJ, Mottaz HM, Gritsenko MA, et al. Evaluation of multi-protein immunoaffinity subtraction for plasma proteomics and candidate biomarker discovery using mass spectrometry. Mol Cell Proteomics. 2006;5:2167–2174. doi: 10.1074/mcp.T600039-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu T, Qian WJ, Strittmatter EF, Camp DG, 2nd, et al. High-throughput comparative proteome analysis using a quantitative cysteinyl-peptide enrichment technology. Anal Chem. 2004;76:5345–5353. doi: 10.1021/ac049485q. [DOI] [PubMed] [Google Scholar]
- 17.Trinidad JC, Specht CG, Thalhammer A, Schoepfer R, Burlingame AL. Comprehensive Identification of Phosphorylation Sites in Postsynaptic Density Preparations. Mol Cell Proteomics. 2006;5:914–922. doi: 10.1074/mcp.T500041-MCP200. [DOI] [PubMed] [Google Scholar]
- 18.Zhang H, Li XJ, Martin DB, Aebersold R. Identification and quantification of N-linked glycoproteins using hydrazide chemistry, stable isotope labeling and mass spectrometry. Nat Biotechnol. 2003;21:660–666. doi: 10.1038/nbt827. [DOI] [PubMed] [Google Scholar]
- 19.Kakugawa Y, Wada T, Yamaguchi K, Yamanami H, et al. Up-regulation of plasma membrane-associated ganglioside sialidase (Neu3) in human colon cancer and its involvement in apoptosis suppression. Proc Natl Acad Sci U S A. 2002;99:10718–10723. doi: 10.1073/pnas.152597199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Orntoft TF, Vestergaard EM. Clinical aspects of altered glycosylation of glycoproteins in cancer. Electrophoresis. 1999;20:362–371. doi: 10.1002/(SICI)1522-2683(19990201)20:2<362::AID-ELPS362>3.0.CO;2-V. [DOI] [PubMed] [Google Scholar]
- 21.Zhang H, Loriaux P, Eng J, Campbell D, et al. UniPep--a database for human N-linked glycosites: a resource for biomarker discovery. Genome Biol. 2006;7:R73. doi: 10.1186/gb-2006-7-8-r73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gygi SP, Rist B, Gerber SA, Turecek F, et al. Quantitative analysis of complex protein mixtures using isotope-coded affinity tags. Nature Biotechnology. 1999;17:994–999. doi: 10.1038/13690. [DOI] [PubMed] [Google Scholar]
- 23.Ong S-E, Blagoev B, Kratchmarova I, Kristensen DB, et al. Stable Isotope Labeling by Amino Acids in Cell Culture, SILAC, as a Simple and Accurate Approach to Expression Proteomics. Mol Cell Proteomics. 2002:1. doi: 10.1074/mcp.m200025-mcp200. [DOI] [PubMed] [Google Scholar]
- 24.Ross PL, Huang YN, Marchese JN, Williamson B, et al. Multiplexed Protein Quantitation in Saccharomyces cerevisiae Using Amine-reactive Isobaric Tagging Reagents. Mol Cell Proteomics. 2004;2:1154–1169. doi: 10.1074/mcp.M400129-MCP200. [DOI] [PubMed] [Google Scholar]
- 25.Conrads TP, Alving K, Veenstra TD, Belov ME, et al. Quantitative analysis of bacterial and mammalian proteomes using a combination of cysteine affinity tags and 15N metabolic labeling. Anal Chem. 2001;73:2132–2139. doi: 10.1021/ac001487x. [DOI] [PubMed] [Google Scholar]
- 26.Veenstra TD, Martinovic S, Anderson GA, Pasa-Tolic L, Smith RD. Proteome analysis using selective incorporation of isotopically labeled amino acids. J Am Soc Mass Spectrom. 2000;11:78–82. doi: 10.1016/S1044-0305(99)00120-8. [DOI] [PubMed] [Google Scholar]
- 27.Zhou H, Ranish JA, Watts JD. Quantitative proteome analysis by solid-phase isotope tagging and mass spectrometry. Nature biotechnol. 2002;19:512–515. doi: 10.1038/nbt0502-512. [DOI] [PubMed] [Google Scholar]
- 28.Finney GL, Blackler AR, Hoopmann MR, Canterbury JD, et al. Label-Free Comparative Analysis of Proteomics Mixtures Using Chromatographic Alignment of High-Resolution muLC-MS Data. Anal Chem. 2008;80:961–971. doi: 10.1021/ac701649e. [DOI] [PubMed] [Google Scholar]
- 29.Schmidt A, Gehlenborg N, Bodenmiller B, Mueller L, et al. An integrated, directed mass spectrometric approach for in-depth characterization of complex peptide mixtures. Mol Cell Proteomics. doi: 10.1074/mcp.M700498-MCP200. in press 10.1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li X-j, Yi EC, Kemp CJ, Zhang H, Aebersold R. A Software Suite for the Generation and Comparison of Peptide Arrays from Sets of Data Collected by Liquid Chromatography-Mass Spectrometry. Mol Cell Proteomics. 2005;4:1328–1340. doi: 10.1074/mcp.M500141-MCP200. [DOI] [PubMed] [Google Scholar]
- 31.Bellew M, Coram M, Fitzgibbon M, Igra M, et al. A suite of algorithms for the comprehensive analysis of complex protein mixtures using high-resolution LC-MS. Bioinformatics. 2006;22:1902–1909. doi: 10.1093/bioinformatics/btl276. [DOI] [PubMed] [Google Scholar]
- 32.Jaffe JD, Mani DR, Leptos KC, Church GM, et al. PEPPeR: A platform for experimental proteomic pattern recognition. Mol Cell Proteomics. 2006;5:1927–1941. doi: 10.1074/mcp.M600222-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mueller L, Rinner O, Schmidt A, Letarte S, et al. Superhirn - A Novel Tool For High Resolution LC-MS Based Peptide/Protein Profiling. Journal of Proteome Research. 2007;19:3470–3480. doi: 10.1002/pmic.200700057. [DOI] [PubMed] [Google Scholar]
- 34.Kelly-Spratt KS, Gurley KE, Yasui Y, Kemp CJ. p19Arf suppresses growth, progression, and metastasis of Hras-driven carcinomas through p53-dependent and -independent pathways. PLoS Biol. 2004;2:E242. doi: 10.1371/journal.pbio.0020242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tian Y, Zhou Y, Elliott S, Aebersold R, Zhang H. Solid-phase extraction of N-linked glycopeptides. Nat Protoc. 2007;2:334–339. doi: 10.1038/nprot.2007.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhou Y, Aebersold R, Zhang H. Isolation of N-linked glycopeptides from plasma. Anal Chem. 2007;79:5826–5837. doi: 10.1021/ac0623181. [DOI] [PubMed] [Google Scholar]
- 37.Keller A, Eng J, Zhang N, Li XJ, Aebersold R. A uniform proteomics MS/MS analysis platform utilizing open XML file formats. Mol Syst Biol. 2005;1 doi: 10.1038/msb4100024. 2005 0017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shannon P, Markiel A, Ozier O, Baliga NS, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shannon PT, Reiss DJ, Bonneau R, Baliga NS. The Gaggle: an open-source software system for integrating bioinformatics software and data sources. BMC Bioinformatics. 2006;7:176. doi: 10.1186/1471-2105-7-176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ramos H, Shannon P, Aebersold R. The Protein Information and Property Explorer: an easy-to-use, rich-client web application for the management and functional analysis of proteomic data. Bioinformatics. 2008 doi: 10.1093/bioinformatics/btn363. 10.1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hearnes JM, Mays DJ, Schavolt KL, Tang L, et al. Chromatin immunoprecipitation-based screen to identify functional genomic binding sites for sequence-specific transactivators. Mol Cell Biol. 2005;25:10148–10158. doi: 10.1128/MCB.25.22.10148-10158.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chen Z, Gu J. Immunoglobulin G expression in carcinomas and cancer cell lines. Faseb J. 2007;21:2931–2938. doi: 10.1096/fj.07-8073com. [DOI] [PubMed] [Google Scholar]
- 43.Heldin P, Karousou E, Bernert B, Porsch H, et al. Importance of hyaluronan-CD44 interactions in inflammation and tumorigenesis. Connect Tissue Res. 2008;49:215–218. doi: 10.1080/03008200802143323. [DOI] [PubMed] [Google Scholar]
- 44.Zhang H, Yi EC, Li X-j, Mallick P, et al. High Throughput Quantitative Analysis of Serum Proteins Using Glycopeptide Capture and Liquid Chromatography Mass Spectrometry. Mol Cell Proteomics. 2005;4:144–155. doi: 10.1074/mcp.M400090-MCP200. [DOI] [PubMed] [Google Scholar]
- 45.Anderson L, Hunter CL. Quantitative mass spectrometric multiple reaction monitoring assays for major plasma proteins. Mol Cell Proteomics. 2006;5:573–588. doi: 10.1074/mcp.M500331-MCP200. [DOI] [PubMed] [Google Scholar]
- 46.Stahl-Zeng J, Lange V, Ossola R, Eckhardt K, et al. High sensitivity detection of plasma proteins by multiple reaction monitoring of N-glycosites. Mol Cell Proteomics. 2007;6:1809–1817. doi: 10.1074/mcp.M700132-MCP200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.