Abstract
Substitution of NH3 by a range of amines in trans-[PtCl2(NH3)2] produces compounds with cytotoxicity significantly improved over the parent transplatin and in many cases equivalent to that of cisplatin. This microreview summarizes the chemistry and biology of trans-platinum compounds containing principally planar amines and succinctly reviews the current status of anticancer relevance of the trans-platinum geometry. The nature of bifunctional DNA adducts (intrastrand, interstrand) is remarkably dependent on the nature of the amine. Further, the stability of monofunctional adducts allows for competitive production of DNA-protein crosslinks and overall the results suggest that the trans-platinum chemotype may offer significant potential for design of selective DNA-protein crosslinking agents. A subset of proteins known to bind to DNA modified by trans-platinum is that comprised of zinc fingers – model studies show the potential for formation of heteronuclear thiolate-bridged species as precedent for zinc displacement from the biomolecule.
1 Introduction
Cisplatin and its analogs oxaliplatin and carboplatin are effective anticancer agents in the treatment of a variety of tumors. Platinum-based combinatorial chemotherapy has also been used with agents such as bleomycin, paclitaxel and 5-fluorouracil, with improved outcomes for patients. 1 The profound success of cisplatin cannot overshadow its clinical shortcomings such as toxic side effects and the development of resistance. The effectiveness of cisplatin lies in its ability to induce DNA damage. The subsequent recognition of the platinum-DNA adducts by cellular proteins may shield the lesion from repair resulting in a definitive cytotoxic insult to the cell. Carboplatin and oxaliplatin are direct structural analogues of cisplatin and similar DNA binding is expected. Studies have shown that cellular sensitivity to cisplatin, as well as clinical resistance to cisplatin is multifactorial, with a major mechanism being the ability of cells to repair DNA damage by nucleotide excision repair and mismatch repair. 1, 2 Resistance mechanisms also include decreased drug accumulation and/or cellular detoxification by thiol containing compounds such as plasma proteins (extracellular) and glutathione (intracellular).
The motivation for study of new compounds comes from the desire to expand the number of tumors susceptible to platinum drug treatment and to overcome clinical resistance to the currently used drugs. The lack of chemotherapeutic activity of the trans isomer trans-[PtCl2(NH3)2] has long been noted and this distinct difference remains the enduring structure-activity relationship for development of platinum antitumor compounds. Other, early, empirical rules such as the need for charge neutrality or a bifunctional compound (two leaving groups) have been defied. 3, 4 It is now clear that substitution of NH3 by a range of amines actually produces compounds with cytotoxicity significantly improved over the parent transplatin and in many cases equivalent to that of cisplatin. This microreview summarizes the chemistry and biology of trans-platinum compounds containing principally planar amines and succinctly reviews the current status of anticancer relevance of the trans-platinum geometry.
2 Cytotoxicity of trans-platinum Compounds
Substitution of the carrier ligand NH3 by planar amines such as pyridine was first reported to activate the trans geometry producing cytotoxicity equivalent to that of cisplatin.5-7 In general, the trans platinum aromatic heterocycles (TPA) complexes have cytotoxicities similar to that of their cis isomers and cisplatin while being markedly more effective than transplatin, Table 1. In a panel of human ovarian carcinoma (HOC) cell lines it is also clear that trans-[PtCl2(py)2] displays collateral sensitivity (is non-cross-resistant), retaining activity in tumor cell lines resistant to cisplatin.6 The pattern of cytotoxicity, even in this limited panel, is also different with cell lines such as HX/62 exhibiting higher sensitivity to trans-[PtCl2(py)2] than to cisplatin. The activation using planar amines is general and is also seen for the series trans-[PtCl2(NH3)(L)] (L = pyridine, isoquinoline, quinoline, thiazole etc.), Figure 1.8
Table 1.
In vitro cytotoxicity (IC50, μM) of transplanaramine (TPA) platinum compounds in a human ovarian cell line panel. 6-9
| Compound | HX/62 | SKOV-3 | PXN94 | 41M | 41McisR | CH1 | CH1cisR |
|---|---|---|---|---|---|---|---|
| trans-[PtCl2(py)2] | 4.5 | 5.9 | 7.7 | 2.2 | 2.0 (0.91) | 1.6 | 1.7 (1.1) |
| trans-[PtCl2(tz)2] | 4.1 | 6.2 | 9.6 | 2.2 | 1.6 (0.73) | 1.3 | 1.4 (1.1) |
| Transplatin | 245 | 255 | 222 | 57 | 69 (1.2) | 30 | 68.5 (2.3) |
| Cisplatin | 12.6 | 4.4 | 3.0 | 0.23 | 1.4 (6.1) | 0.1 | 0.67 (6.7) |
| trans-[Pt(O2CCH3)2(py)2] | 31.0 | 14.0 | 4.5 (0.32) | 19.0 | 4.1 (0.22) | ||
|
trans- [Pt(O2CCH3)2(NH3)(tz)] |
96.0 | 21.5 | 7.2 (0.33) | 24.0 | 8.2 (0.34) | ||
|
trans- Pt(O2CCH3)2(NH3)(iquin)] |
40.0 | 22.0 | 5.8 (0.26) | 20.0 | 7.4 (0.37) |
tz=thiazole; py=pyridine; iquin=isoquinoline; quin=quinoline; bztz=benzothiazole (see Figure1)
Figure 1.
Cytotoxic transplanar amine platinum compounds (TPAs)
An important criterion for any potential new platinum drug is to display a different profile of antitumor activity in comparison to the clinically used agents. Analysis of cytotoxicity data of 107 platinum compounds from the NCI human tumor panel using clustered image maps, the COMPARE algorithm and other numerical methods identified 12 groups, 11 of which showed distinctive activity profiles. One such group (Pyridine Group) consisted of only trans-platinum complexes with planar amines, including trans-[PtCl2(py2)10]. The group was identified as having a novel cytotoxicity profile and activity in cisplatin and oxaliplatin-resistant lines. Further, the activity profile of the Pyridine Group is poorly correlated with that of a representative set of alkylating agents, by Pearson correlations. The COMPARE activity profile is also poorly correlated with cisplatin and carboplatin.11 The overall analysis of the compounds in the NCI database clearly support the view that structurally different sets of platinum compounds, including the TPA series, display pre-clinical properties distinct from agents such as cisplatin and oxaliplatin, with implications also for potential differences in clinical activity.
2.1 Trans-platinum compounds with N-donor ligands other than aromatic heterocycles
Use of other carrier ligands besides planar amines subsequently showed that enhancement of cytotoxicity by replacement of NH3 was a general phenomenon. Notably iminoethers, aliphatic amines and heterocyclic aliphatic amines have all been used to generate a broad array of potentially useful compounds, Figure 2. The cytotoxicity data for the various series has been very well summarized recently.12-15 In general it is reasonable to say that most compounds display cytotoxicity in the micromolar (1-20 μM) range and the compounds consistently display cytotoxicity in cisplatin-resistant cells.
Figure 2.
Structures of selected trans-platinum compounds with carrier ligand other than planar amines
2.2. Cytotoxic trans-platinum compounds with N2O2 Donor Sets
In conjunction with the carrier ligand modification, optimization of the trans geometry can be attained by modification of leaving group. Use of organic amines reduces aqueous solubility and the compounds of Figure 2 still retain the Cl-Pt-Cl axis, expected to be more reactive than a N-Pt-Cl axis due to the trans influence. To address the poor aqueous solubility of the [PtN2Cl2] chemotype, the use of carboxylates as leaving groups was introduced. Complexes trans-[Pt(O2CR)2(L)(L’)] are very water-soluble and surprisingly stable toward hydrolysis, resembling carboplatin in their chemical reactivity (See below). Carboxylate derivatives have shown similar cytotoxicities to their parent chlorides, Table 1.9, 16 The use of carboxylate ligands also affords these trans-platinum compounds increased cellular accumulation even in cisplatin and oxaliplatin-resistant cell lines.9, 16
The carboxylate strategy is a general one – as evidenced by the solution properties of trans-[Pt(O2CCH3)2(isopropylamine)(N-methylimidazole)]17. The cytotoxicity may be further modulated by changing the nature of the carboxylate leaving group.18, 19 In this respect most notable is the formate derivative, trans-[Pt(O2CH)2(NH3)(iquin)], which has an IC50 of 7.2 μM in HCT116 WT colon carcinoma cells compared to 5.4 μM for cisplatin; while in A2780 human ovarian cells, trans-[Pt(O2CH)2(NH3)(4-pic)] has a IC50 of 6.2 μM compared to that of cisplatin at 3.0 μM. These compounds have further been shown to activate p53 and PARP cleavage with higher levels of apoptosis than cisplatin at early time-points, pathways once thought to be restricted to cisplatin and its congeners.20
2.3 Platinum IV compounds
Although the anticancer activity of Pt(IV) complexes has been noted since 1967, their development has not been explored to a similar level as that of Pt(II) compounds. In theory, Pt(IV) compounds can serve as prodrugs, being reduced to the Pt(II) analogues under physiological conditions.21 JM335, (trans,trans,trans-[PtCl2(OH)2(NH3)(cyclohexylamine)]), is one of the most potent Pt(IV) compounds developed as part of a collaborative effort by Johnson Matthey and the Institute of Cancer Research along with over 300 other compounds including picoplatin and satraplatin.22 Other Pt(IV) compounds studied include trans,trans,trans-[PtCl2(OH)2(dimethylamine)(isopropylamine)].23
3 Chemistry
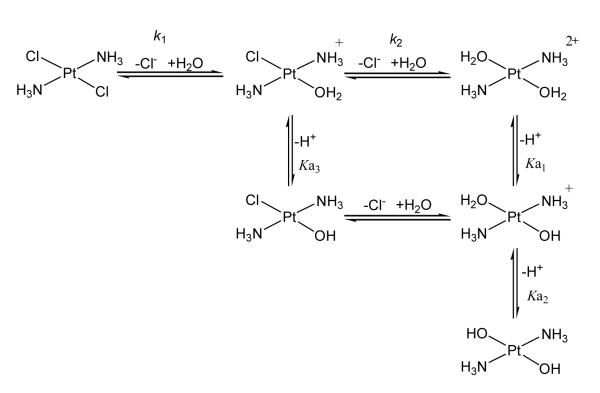
3.1. Aquated Trans-platinum species. Hydrolysis of trans-platinum
Similar to cisplatin, the neutral trans-platinum compound needs to be activated to the monoaqua or diaqua species to react with DNA (Scheme 1). Abstraction of the first chloride from trans-[PtCl2(NH3)2] by silver salts is relatively easy due to the mutual trans effect of Cl–. Removal of the second chloride is not so easy as it is now trans to the weaker H2O ligand.24 An interesting study by McGowan et al. measured the first hydrolysis step of trans-[PtCl2(NH3)(pic)] to be 2.6 × 10−5 ; 12.7 × 10−5 and 5.2 × 10−5 s −1 for the 2-picoline, 3-picoline and 4-picoline compounds respectively, Table 2.25 Under the conditions of the experiment, little hydrolysis of the second chloride was observed.
Scheme 1.
Proposed scheme for hydrolytic activation of transplatin
Table 2.
Effect of carrier ligand on pKa values of the diaqua species and rate constant for the first hydrolysis step of various trans-(diam(m)ine)platinum(II) complexes and cisplatin.
| Compound | pKa1 | pKa2 | k1 (s−1) | Reference |
|---|---|---|---|---|
| cis-[PtCl2(NH3)] | 5.37 | 7.21 | 2.5 × 10−5 | 25, 28 |
| trans-[PtCl2(NH3)2] | 4.35 | 7.40 | 9.8 × 10−5 | 25, 28 |
| trans-[PtCl2(NH3)(mba)] | 4.16 | 7.17 | -- | 25 |
| trans-[PtCl2(NH3)(ipa)((S)-mba)] | 4.21 | 7.33 | -- | 25 |
| trans-[PtCl2(NH3)(2-pic)] | 4.03 | 7.01 | 2.6 × 10−5 | 25 |
| trans-[PtCl2(NH3)(3-pic)] | 3.97 | 6.78 | 12.7 × 10−5 | 25 |
| trans-[PtCl2(NH3)(4-pic)] | 3.94 | 6.88 | 5.2 × 10−5 | 25 |
| trans-[Pt(O2CCH3)2(py)2] | 3.87 | 6.70 | 3.2 × 10−7 | 9 |
| trans-[Pt(O2CCH3)2(NH3)(quin)] | 3.89 | 7.01 | 3.7 × 10−7 | 9 |
| trans-[Pt(O2CCH3)2(NH3)(iquin)] | 3.78 | 6.92 | 7.4 × 10−7 | 9 |
The equilibrium constants for the formation of the monoaqua species are also approximately an order of magnitude lower than those reported for cisplatin, but similar to those for transplatin.25 The pKa values for major types of trans-platinum compounds are also shown in Table 2. The values are similar for most trans-platinum compounds and note that the first pKa of the diaqua species is uniformly lower than for the cis-isomer by an order of magnitude. The nature of the amine does have some effect on the pKa1 – coordinated water is more acidic in the presence of a planar amine than NH3, agreeing with earlier results on the [Pt(bipy)(H2O)2]2+ ion.26 Under physiological conditions (pH 7.4), the monoaqua TPA species is therefore likely to exist as the hydroxo species. Interestingly, trans-[PtCl(OH)(NH3)2]·H2O may be isolated as a stable solid.27 In diaqua species, the pKa2 values indicate that an equilibrium should exist between aqua and hydroxo species, making it possible for reaction between N-donor ligands such as that of the N7 of guanine.
Use of mutually trans carboxylates reduces the rate of hydrolysis by approximately two orders of magnitude in comparison to chlorides, Table 2. Indeed, observed rate constants for carboxylate trans-platinum compounds are similar to that of carboplatin. The compounds are significantly more inert than one might expect from two mutually trans ligands with weak trans effect and trans influence. Thus, it is notable that the cytotoxicity of TPA compounds with carboxylate ligands is very close to the chlorides, despite their “carboplatin-like” properties, that very little hydrolyzed species is apparent at biological pH– production of aquated species of carboplatin and oxaliplatin is pH dependent 29-31. The measured IC50 values could represent a balance between lesser deactivation and increased intracellular platinum accumulation.9, 16
3.2 Reactions with sulfur residues of proteins and peptide models
Deactivation of transplatin by thiol containing compounds such as human serum albumin (HSA, plasma) and glutathione (GSH, intracellular) may contribute to the observed differences in antitumor activity of cis and trans-DDP. Incubation of trans-[PtCl2(OH)2(dimethylamine)(isoproylamine)] and its Pt(II) analogue, cisplatin and transplatin with HSA over a 24 day time period showed both trans-[PtCl2(OH)2(dimethylamine)(isoproylamine)] and cisplatin have similar profiles with substantially less binding compared to transplatin and the Pt(II) analogue.23 The effects of sterically bulky ligands in the trans geometry on GSH and methionine reactivity have not been systematically studied or compared with transplatin.32-34 However, monofunctional adducts of both trans-[PtCl2(isopropylamine)(3-hydroxymethyl-pyridine)] and trans-[PtCl2(isopropylamine)(4-hydroxymethyl-pyridine)] were shown to react with GSH by HPLC analysis, albeit to a lesser extent than transplatin.23
4 DNA binding and adduct formation
4.1 DNA binding
DNA is the established primary target of platinum compounds in cells, making DNA adduct formation one of the key determinants of platinum-mediated cytotoxicity. The extent and nature of these DNA adducts has been used to explain the differences in biological activity of cis and transplatin. Indeed, the hypothesis that altered modes of DNA binding, and the subsequent differences in protein recognition and downstream processing, may eventually translate into a different profile of antitumor activity has been a driving force for development and design of potential platinum-based clinical agents. Bifunctional compounds may produce interstrand (IXL) and intrastrand crosslinks and monofunctional adducts. Cisplatin forms a variety of adducts of which the 1,2-GG intrastrand predominates, with other lesions including 1,2-AG intrastrand, 1,3-GNG intrastrand and to a lesser extent the 1,2-GG interstrand crosslink. In contrast, geometric constraints prevent the formation of 1,2 cross-links by transplatin – instead 1,3-intrastrand cross-links are formed.35 Interstrand crosslinks of transplatin are formed between G and C residues of the same base pair, Figure 3. Monofunctional (and also bifunctional intrastrand) adducts may convert to interstrand crosslinks. The slow rate of conversion of monofunctional to bifunctional interstrand may account for the fact that transplatin forms very few IXL in cells.35 Replacement of NH3 by an amine as carrier ligand affects both the structural nature of adducts as well as the kinetics of formation.36, 37 A summary of the effect of amine ligand on DNA adduct formation is shown in Table 3.
Figure 3.
Interstrand cross-link for cisplatin 45 873, 46 (left) and transplatin47 (right) highlighting the markedly different DNA bending induced by platinum compounds.
Table 3.
Effect of carrier ligand on distribution of DNA adducts for transplatinum complexes
| [PtCl2(L)(L’)] | interstrand CLs/adduct (%)a |
monofunctional lesions/adduct (%)a |
intrastrand CLs/adduct (%)a |
Reference |
|---|---|---|---|---|
| cisplatin | 6 | ~1-4 | ~90 | 38 |
| transplatin | 12 | ~60 | ~28 | 38 |
| NH3,thiazole | 30-40 | 33 | 27-37 | 36, 39 |
| NH3,piperidine | 26 | 15 | 59 | 40 |
| NH3,piperazine | 18 | 22 | 60 | 40 |
| NH3, 4-picoline | 40 | 34 | 26 | 40 |
| NH3, 2-methylbutyl- amine |
40 | 36 | 24 | 41 |
| (E-iminoether)2 | 10 | 90 | 0 | 42, 43 |
Adapted from ref. 44
4.2 Effect of carrier ligand on DNA adduct formation
The presence of a planar ligand results in formation of significantly more interstrand crosslinks in comparison to parent trans-[PtCl2(NH3)2]. In the specific case of thiazole, interstrand, intrastrand and monofunctional adducts are almost equally distributed as adducts.36 Further, the interstrand crosslinks are formed between the adjacent guanines of GC base pairs, similar to that of cisplatin, Figure 4.48 Thus, the planar amine forces the crosslink to be more “cisplatin-like” and is an example of using steric effects to turn a presumably non-toxic into a toxic lesion. The cation [Pt(9-EtGua)(5′-GMP)(NH3)(quin)]2+ gives conformers for both guanine residues and quinoline at room temperature due to restricted rotation.49
Figure 4.
Schematic representation of the modes of DNA interstrand cross-linking by transplatin (left), cisplatin (center), and trans-[PtCl2(NH3)(iquin)]. The latter is a representative example of the effect of a planar amine on DNA adduct structure. 48, 49
In contrast, bulky piperidine and piperazine ligands produce more intrastrand crosslinks whereas, in the presence of two iminoether ligands very few bifunctional crosslinks are formed at all. This latter fact may be attributed to steric reasons as the trans-[PtCl2(NH3)(E-iminoether)], compound shows significantly more interstrand cross-linking in plasmid DNA than either its cis isomer or cisplatin.50 However steric effects with two bulky carrier groups are not universal – trans-[PtCl2(py)2] is as efficient as trans-[PtCl2(NH3)(planar ligand)] in forming interstrand crosslinks in plasmid DNA.51 When L = 2-Me-butylamine or sec-butylamine mainly interstrand cross-links are formed in ~40-50% proportion.41
There are interesting differences between cis and trans isomers containing planar ligands. In general cis-[PtCl2(py)2] was 2-3 times more efficient at binding calf thymus (CT) and pUC19 DNA than trans-[PtCl2(py)2]51. This was also the case for poly (dG)-poly(dC) DNA, albeit to a lesser extent. In contrast, in poly(dG-dC)-poly(dG-dC) DNA the trans compound was the more effective at binding.52 While cis-[PtCl2(py)2] was able to bind DNA more efficiently than its trans isomer, trans-[PtCl2(py)2] had higher unwinding angles (ϕ=17°) than cis-[PtCl2(py)2] (ϕ=4°) 51, actually reversing the trend seen for PtCl2(NH3)2 isomers where the cis isomer exhibited higher unwinding angles than the trans, Table 4. However, it should be noted that trans-[PtCl2(NH3)(quin)] was more efficient at binding CT DNA than its cis isomer.39, 53 This variation in the trend could be due to steric effects of the larger quinoline group.
Table 4.
Summary and comparison of structural characteristics of DNA cross-links of trans-[PtCl2(NH3)(tz)] (t-PtTz), transplatin and cisplatin.54
| 1,3-intra- strand XL of t-PtTz |
Interstrand XL of t- PtTz |
Monofunctional adduct of t-PtTz |
Interstrand XL of transplatin |
Interstrand XLof cisplatin |
1,2-intra- strand XL of cisplatin |
|
|---|---|---|---|---|---|---|
| Frequency | 20-40% | 30-40% | 30-40% | ~12% | ~6% | ~90% |
| DNA bending | 40° toward minor groove |
22° toward minor groove |
34° toward major groove |
~20° toward minor groove |
40-45° toward minor groove |
32-34° toward major groove |
| DNA unwinding | 15° | 20° | 12° | ~12° | 76-79° | 13° |
| HMGB1a affinity | > 1.5 5μM | > 1.5 μM | 38.5 nM | ND | > 1.5 μM | 30.8 nM |
| HMGB1b affinity | > 30 μM | 13.40 μM | ND | |||
| NER by eukaryotic excinuclease (relative excision) |
1.0 | no | 1.4 | no | no | 1.5 |
4.3 DNA binding of N2O2 donor sets
In comparison to the chloride species, the carboxylates show less DNA binding and consequently interstrand cross-linking over time. This can be attributed to the observed lower rate constants for hydrolysis. The general trend showed that the more reactive formate derivatives exhibited significantly more DNA damage than the acetate or hydroxyacetate compounds, and in cells produced similar levels of DNA damage as trans-[PtCl2(NH3)(tz)]19. The influence of the carrier ligand is exemplified by the higher rate of interstrand cross-links seen for the trans-[Pt(O2CH)2(NH3)(iquin)] as compared to the trans-[Pt(O2CH)2(NH3)(4-pic)]20. Interestingly, similar steric effects were also observed in binding to ubiquitin.55
4.4 Conversion of monofunctional to bifunctional adducts
A key determinant of the formation of cross-links, inter or intra, is the stability of the monofunctional adduct formed by these trans-platinum compounds. The monofunctional adducts formed by trans-[PtCl2(NH3)(tz)] lead to bending and unwinding angles similar to that of cisplatin (See below); however, monofunctional adducts are very slowly converted to a bifunctional mode.54 trans-[PtCl2(NH3)(quin)] adducts persist for 48h in the monofunctional state; similarly the trans-[PtCl2(NH3)(tz)] compound shows only a 2-5% conversion rate to interstrand cross-links after 24h, increasing to 40% within 48h.54 These results are markedly different from transplatin, which has a 70% conversion rate.
5 Structural consequences of DNA adduct formation. Protein recognition of trans-platinum-DNA adducts
A multitude of proteins recognize platinated DNA adducts.56 In cells the tumor suppressor protein p53 is activated in response to damage induced by trans-platinum compounds.19, 57 This activation confirms that trans-platinum-treated cells process and respond to the damage, at least initially, in a similar manner to that of cisplatin. Interestingly, in a cell-free assay, DNA modified by trans-[PtCl2(NH3)(4-hydroxymethylpyridine)] reduces the affinity of p53 protein to bind to its consensus sequence. The diminution is comparable to that of cisplatin.58
One well characterized system is that of recognition of Pt-DNA adducts by high mobility group (HMG) proteins. The crystal and molecular structure of HMG recognition to duplex DNA containing a cisplatin 1,2-GG intrastrand crosslink has allowed the molecular details of this interaction to be studied.59, 60 The duplex bending caused by the adduct is a critical feature in protein recognition. Further, specific proteins like HMGB1 may protect the 1,2 GG cross-link from repair by excinuclease activity thus augmenting the cytotoxicity of the drug.61 Transplatin modified DNA and the monofunctional platinated adducts of compounds such as [PtCl(NH3)3]Cl are not recognized by this group of proteins.
A detailed analysis of the structures of DNA adducts of trans-[PtCl2(NH3)(tz)] allows examination of factors affecting protein recognition. The conformational changes of the monofunctional adduct are most similar to that of cisplatin (Table 4) and therefore it is not surprising that the lesion is recognized with high affinity by HMG-family proteins. Furthermore, the interstrand cross-link bends DNA with an angle of distortion, 22°, toward the minor groove but may be a substrate for some HMG family proteins. The details, and the consequences for NER (Nucleotide Excision Repair), are summarized in Table 4. Unlike trans-[PtCl2(NH3)(tz)], trans-[PtCl2(NH3)(quin)] adducts were not recognized by HMGB1.62 Unlike cisplatin, trans-[PtCl2(E-iminoether)2] adducts are not recognized by either HMGB1a or HMGB1b.37 Similarly, there was no recognition of DNA cross-linked by trans-[PtCl2(NH3)(pip)] by HMGB1, which is not surprising as DNA bends towards the minor groove.40
Since the monofunctional adduct contains a further substitution-labile chloride ligand, one might assume that ternary DNA-protein crosslinks could be formed with HMG proteins. Control of conditions allows observation of crosslinking of HMG group proteins 1 and 2 to platinated DNA in micrococcal nuclease accessible regions of chromatin.63 DNA modified by cis-DDP may also undergo photoinduced cross-linking to HMG1 proteins upon irradiation.64 No ternary DNA-Pt-protein crosslinking is observed between monofunctional TPA adducts and HMG proteins.54 An explanation may be found by examining the molecular model of the monofunctional adduct, Figure 5. In this case monofunctional platination in the major groove results in the Pt-Cl bond pointing into the major groove, rather than the minor groove where HMG associates. The stability of the monofunctional adduct with respect to bifunctional adduct conversion may also be explained by noting the stacking interactions of thiazole with the neighbouring bases.
Figure 5.

Molecular model of the monofunctional DNA adduct of trans-[PtCl2(NH3)(tz)].
6 Novel cellular protein targets for TPA compounds. Induction of topoisomerase-DNA compounds
The most striking feature of the biological activity of transplanar amine (TPA) compounds is the production of single-stranded protein-associated DNA strand breaks in cells. The pharmacology of these strand breaks was suggested to be remarkably similar to those produced by topoisomerase poisons.65 This is a unique result in platinum chemistry. At 24 and 48h exposure of breast cancer MCF-7 cells to trans-[PtCl2(NH3)(thiazole)], the formation of DNA-topoisomerase I (Topo I) complexes is detected, coincident with a high degree of apoptosis. Trace levels of Topo I-DNA complexes were observed at 10μM dose but at 50μM there was substantial increase in the level of the protein-DNA complex. Similar effects were seen for trans-[PtCl2(py)2]65. In contrast, no trapping of Topoisomerase II-DNA complexes was seen. The ability to produce Topo I-DNA adducts follows the same order as that of the production of strand breaks: trans-[PtCl2(py)2] > trans-[PtCl2(NH3)(tz)] >> trans-[PtCl2(NH3)(quin)]. The cis isomers produce very little protein-associated strand breaks and, as a corollary, do not form ternary protein-DNA adducts. Furthermore the effect appears restricted to those trans compounds containing planar rings. Treatment with cisplatin at the same concentrations also did not result in any detectable Topo I-DNA complexes. Topoisomerases – enzymes that relieve torsional strain in DNA and which play important roles in replication and transcription – have been recognized in cancer therapy as an attractive target for inhibition since the 1970’s.66 The structural resolution of Topo-DNA complexes has added to the attraction of this area.67 The fact that the induction of Topoisomerase I-DNA complexes is part of the biological response to TPA compounds, and is a property not shared by cis-DDP and congeners, suggests that cytotoxic trans-platinum complexes with planar amines have a different cellular injury response in comparison to that of cisplatin.
A further unusual target for TPA compounds has been mentioned as telomerase. Treatment of MCF-7 cells with varying concentrations of trans-[PtCl2(py)2] for 24h induced a small and rapid decrease of telomerase activity.68 Telomerase activity was restored within 5 days after continuous exposure to drug. The time course of telomerase inhibition and telomere dynamics did not correlate with observed cell death but the observed initial reduction could be explained by rapid distribution of drug into cellular compartments. In a cell-free assay the trans-pyridine compound showed a dose dependent inhibition of semi-purified telomerase and a telomerase inhibitory effect.68 It is of interest that stabilization of human telomeric quadruplex DNA and inhibition of telomerase has also been shown by platinum-phenanthroline and platinum-acridine complexes, suggesting that the structural role of planar ligands may have some effect.69, 70 Many telomerase-inhibitory compounds are large delocalized planar aromatic systems.
7 Ternary DNA-protein crosslink formation
Topoisomerases relax supercoiled DNA by formation of transient phosphodiester linkages between the backbone phosphate and a tyrosine residue on the protein. Ternary adduct formation by platinum would prevent the reannealing step and release of the bound protein, resulting in the observed strand breaks. By analogy with known mechanisms of Topo inhibition, the limiting mechanisms for formation of ternary DNA-protein adducts involve interactions between a mono- (or bifunctional) Pt-DNA adduct and a protein site on topoisomerase, Figure 6.
Figure 6.
Postulated competition between DNA-DNA and DNA-protein crosslink formation in trans-Pt adducts
There is precedence for DNA-protein formation mediated by trans-platinum. The long-lived nature of the monofunctional adduct of trans-DDP has been exploited previously for DNA/RNA-protein cross-linking. This approach, allowing contact points between RNA and protein, was demonstrated with the ribosome from E. coli and an aminoacyl-tRNA synthetase/tRNA complex.71 Likewise, trans-DDP has been used to cross-link the zinc-finger nucleocapsid protein to HIV-1 RNA.72 However, ternary DNA-Pt-protein crosslink formation is not a general feature of trans-platinum compounds with sterically hindered amines. As noted, no ternary DNA-HMG protein formation was noted for the monofunctional adduct of trans-[PtCl2(NH3)(tz)]54. Monofunctional adducts of transplatin itself on 44-mer DNA templates are stable in the presence of a variety of DNA-binding proteins – KF−, histone H1 and NF-κB.73 In contrast, oligodeoxyribonucleotide platinated by trans-[PtCl2(E-iminoether)2] cross-links histone H1 and the subsequent DNA-protein crosslinks inhibit DNA replication and repair.37 Overall the results suggest that the trans-platinum chemotype may offer significant potential for design of selective DNA-protein crosslinking agents.
7.1 Models for ternary DNA-protein formation
Nucleobase cations of general formula [PtCl(L)(L’)(Nucleobase)]+ represent convenient structures for studying competitive reactions of model DNA (e.g. N-donor 5′-guanosine monophosphate, 5′-GMP) or protein (e.g. S-donor N-acetylmethionine, N-AcMet) residues. The effects of complex geometry and the nature of L and L’ on the kinetic differences between S and N-donors gives a measure of the possible preferences for protein over DNA interaction.74-76 The 1:1 reactions with 5′-guanosine monophosphate (5′-GMP) or N-AcMet of (SP-4-2)-[PtCl(9-EtGua)(NH3)(L)]+ [L = NH3 or quinoline], showed that displacement of Cl− showed clear kinetic preference for the sulfur in both cases, with the sterically more hindered quinoline compound reacting more slowly (~t½’s = 1.5 and 3.5 h with N-AcMet against 7 and 17 h for 5′-GMP for L = NH3 or quinoline respectively). The S/N ratio (estimated by t½ (5′-GMP)/t½(N-Ac-L-Met)) was even greater for trans-[PtCl(9-EtGua)(py)2]+. This sulfur preference also extends to the cis isomer of the latter compound. A unique feature of the trans-platinum-mononucleobase compounds is that the Pt-Cl bond undergoes extremely slow hydrolysis. A major difference between the requirements for nucleotide and sulfur binding to Pt(II) is that the latter substitution proceeds via direct substitution of chloride whereas N7 binding may require prior hydrolysis of the chloride.77
7.2 Interactions of trans-platinum mononucleobase compounds with Zinc Fingers and Zinc Finger models
The high sulfur affinity of trans-platinum-mononucleobase compounds has been exploited in reactions with proteins containing zinc-finger motifs with cysteine and methionine residues. This important family of DNA-regulating proteins is widespread and a large number of zinc-finger proteins is recognized as encoded by the human genome78, 79 Zinc fingers have become important cellular targets for potential anti-cancer drugs as well as HIV inactivation. Interaction of the C-terminal finger of HIV NCp7 protein with trans-[PtCl(9-EtGua)(py)2]+ results in Zn ejection from the peptide accompanied by loss of tertiary structure.80 Targeting the NCp7-DNA interaction for drug design represents a conceptual advance over electrophiles designed for chemical attack on the zinc finger alone. The results demonstrate examples of a new platinum structural class targeting specific biological processes, distinct from the bifunctional DNA-DNA binding of cytotoxic agents like cisplatin. The results confirm the validity of a chemical biological approach for metallodrug design for selective ternary DNA(RNA)-protein interactions.
In order to provide precedents for the possible interactions of platinum DNA adducts with zinc finger proteins, complexes such as trans-[PtCl(9-EtGua)(py)2], and [Pt(dien)Cl]Cl (dien = diethylenetriamine) and [Pt(terpy)Cl]Cl (terpy = 2,2′: 6′,2″-terpyridine) were exposed to the Zn dithiolate, N, N’-bis(2-mercaptoethyl)-1,4-diazacycloheptanezinc(II) dimer, [Zn(bme-dach)]2.81, 82 The products defined by Electrospray Ionization Mass Spectrometry (ESI-MS), x-ray crystallography and 195Pt-NMR spectroscopy yielded evidence for Zn-(μ-SR)-Pt bridges followed by zinc ejection from the N2S2 coordination sphere and subsequent formation of a trimetallic Zn-(μ-SR)2-Pt-(μ-SR)2-Zn bridged species.
8 Cytotoxicity and in vivo efficacy of trans-platinum drugs
Space precludes a detailed analysis of cellular effects by trans-platinum compounds. As stated, there is an extensive literature on cytotoxicity and also DNA modifications by trans-platinum compounds. 11-15 There is somewhat less information on in vivo antitumor activity. Nevertheless some evidence of in vivo activity has been reported. 8, 11-15, 44 The trans-[PtCl2(NH3)(tz)] shows moderate activity against P388 leukemia. 8 In Lewis lung carcinoma xenografts, trans-[PtCl2(E-iminoether)2] decreased the level of lung metastases over that of the vehicle control but levels were still higher than that of cisplatin. Furthermore, there was no significant enhancement in survival in mice treated with trans-EE (33 days) over vehicle control (30 days).83 The complex also showed significant in P388 leukemia, but less than cisplatin.84 The therapeutic efficacy of the trans-[PtCl2(NH3)(pip-pip)] and trans-[PtCl2(NBA)(pip-pip)] was evaluated in Balb/c mice with either A2780 or A2780cisR. Though exhibiting similar cytotoxicities, cisplatin was able to extend the life expectancy by 160% while trans-[PtCl2(NH3)(pip-pip)] and trans-[PtCl2(NBA)(pip-pip)] only 79% to 46% respectively. Furthermore, though both compounds have similar cytotoxicities, trans-[PtCl2(NH3)(pip-pip)] showed no improvement in outcome of Balb/c/A2780cisR mice whereas both cisplatin and trans-[PtCl2(NBA)(pip-pip)] did.85
JM335 was one of the most active Pt(IV) compounds that showed activity against ADJ/PC6 tumor, a cisplatin resistant murine plasmacytoma xenograft.86 JM355 was also the most effective at delaying tumor growth, as much as 2 months in the case of PXN/100.87 trans-[PtCl2(dimethylamine)(isoproylamine)] at 30mg/kg showed no antitumor effects compared to untreated control in the human ovarian CH1 xenograft, was attributed to its’ inactivation by plasma proteins. However, the Pt(IV) analog trans,trans,trans-[PtCl2(OH)2(dimethylamine)(isopropylamine)] was effective at delaying the growth progression of CH1 xenografts at 15mg/kg for 15 days, similar to that of cisplatin.23
9 Summary
The use of sterically hindering amines in the trans-platinum geometry affords many complexes with cytotoxicity equivalent to that of cisplatin. Indeed, examining the diversity of amine used (Fig. 2) it appears that transplatin itself is the exception ! The chemistry and DNA binding modes of cytotoxic trans-platinum agents shows a rich carrier ligand-dependent diversity, both with respect to the structural consequences of the adducts produced and their stability and reactivity. Mechanistic studies also indicate that the trans-platinum scaffold may be especially useful for design of selective ternary DNA-protein crosslinking agents. Noteworthy in this respect is the ability to crosslink zinc finger proteins with subsequent zinc ejection from the protein. Finally, although some in vivo antitumor activity has been reported there is not such a wide variety of data available as there is for the cis chemotype. This may be due to the expense of in vivo studies but also to as yet unstudied issues of drug metabolism and biodistribution. While the greatest emphasis has been placed on comparing the target (DNA) interactions of cytotoxic trans-platinum compounds with those of cisplatin and congeners, the ultimate determinant of drug efficacy probably lies more in the pharmacokinetic and pharmacodynamic realm than with specific drug-DNA interactions. Nevertheless, modification of the “parent’ trans-[PtCl2(NH3)2] molecule by substitution of NH3 with a host of amines has produced a rich chemistry and biology of the trans geometry, which may eventually be translated to the development of clinically useful drugs.
10 Acknowledgements
This work was supported by grants from The National Institutes of Health and The National Science Foundation. It is a pleasure to acknowledge the collaborations of Viktor Brabec and his group as well as the contributions of Uli Bierbach to the DNA-binding studies of TPA complexes.
11 Dedication This review is dedicated to the memory of Lloyd R. Kelland a friend and collaborator, and a great advocate for platinum. Ar dheis Dé go raibh a hanam.
Definitions
- ipa
isopropylamine
- pz
piperazine
- pip
piperadine
- mba
2-methylbutylamine
- dma
dimethylamine
- sba
sec-butaneamine
References
- 1.Kelland L. Nat Rev Cancer. 2007;7:573–584. doi: 10.1038/nrc2167. [DOI] [PubMed] [Google Scholar]
- 2.Jung YW, Lippard SJ. Chemical Reviews. 2007;107:1387–1407. doi: 10.1021/cr068207j. [DOI] [PubMed] [Google Scholar]
- 3.Farrell N. Metal Ions in Biological Systems. 2004;42:251–296. [PubMed] [Google Scholar]
- 4.Hollis LS, Sundquist WI, Burstyn JN, Heiger-Bernays WJ, Bellon SF, Ahmed KJ, Amundsen AR, Stern EW, Lippard SJ. Cancer Research. 1991;51:1866–1875. [PubMed] [Google Scholar]
- 5.Farrell N, Ha TTB, Souchard JP, Wimmer FL, Cros S, Johnson NP. Journal of Medicinal Chemistry. 1989;32:2240–2241. doi: 10.1021/jm00130a002. [DOI] [PubMed] [Google Scholar]
- 6.Farrell N, Kelland LR, Roberts JD, Van Beusichem M. Cancer Research. 1992;52:5065–5072. [PubMed] [Google Scholar]
- 7.Van Beusichem M, Farrell N. Inorganic Chemistry. 1992;31:634–639. [Google Scholar]
- 8.Farrell N. Metal Ions in Biological Systems. 1996;32:603–639. [PubMed] [Google Scholar]
- 9.Ma ESF, Bates WD, Edmunds A, Kelland LR, Fojo T, Farrell N. Journal of Medicinal Chemistry. 2005;48:5651–5654. doi: 10.1021/jm050539d. [DOI] [PubMed] [Google Scholar]
- 10.Fojo T, Farrell N, Ortuzar W, Tanimura H, Weinstein J, Timothy G. Myers. Critical Reviews in Oncology/Hematology. 2005;53:25–34. doi: 10.1016/j.critrevonc.2004.09.008. [DOI] [PubMed] [Google Scholar]
- 11.Farrell N. Cancer Investigation. 1993;11:578–589. doi: 10.3109/07357909309011676. [DOI] [PubMed] [Google Scholar]
- 12.Coluccia M, Natile G. Anti-Cancer Agents in Medicinal Chemistry. 2007;7:111–123. doi: 10.2174/187152007779314080. [DOI] [PubMed] [Google Scholar]
- 13.Natile G, Coluccia M. Metal Ions in Biolgical Systems, Vol 42: Metal Complexes in Tumor Diagnosis and as Anticancer Agents. vol. 42. 2004. pp. 209–250. Editon edn. [PubMed] [Google Scholar]
- 14.Radulovic S, Tesic Z, Manic S. Current Medicinal Chemistry. 2002;9:1611–1618. doi: 10.2174/0929867023369376. [DOI] [PubMed] [Google Scholar]
- 15.Kalinowska-Lis U, Ochocki J, Matlawska-Wasowska K. Coordination Chemistry Reviews. 2008;252:1328–1345. [Google Scholar]
- 16.Quiroga AG, Perez JM, Alonso C, Navarro-Ranninger C, Farrell N. Journal of Medicinal Chemistry. 2006;49:224–231. doi: 10.1021/jm050804v. [DOI] [PubMed] [Google Scholar]
- 17.van Zutphen S, Pantoja E, Soriano R, Soro C, Tooke DM, Spek AL, den Dulk H, Brouwer J, Reedijk J. Dalton Transactions. 2006:1020–1023. doi: 10.1039/b512357g. [DOI] [PubMed] [Google Scholar]
- 18.Bulluss GH, Knott KM, Ma ESF, Aris SM, Alvarado E, Farrell N. Inorganic Chemistry. 2006;45:5733–5735. doi: 10.1021/ic060741m. [DOI] [PubMed] [Google Scholar]
- 19.Aris SM, Gewirtz DA, Ryan JJ, Knott KM, Farrell NP. Biochemical Pharmacology. 2007;73:1749–1757. doi: 10.1016/j.bcp.2007.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Aris SM, Knott KM, Yang X, Gewirtz DA, Farrell NP. Inorganica Chimica Acta. doi:10.1016/j.ica.2008.01.025. [Google Scholar]
- 21.Hall MD, Dolman RC, Hambley TW. Metal Ions in Biological Systems. 2004;42:297–322. [PubMed] [Google Scholar]
- 22.Kelland LR, Barnard CFJ, Mellish KJ, Jones M, Goddard PM, Valenti M, Bryant A, Murrer BA, Harrap KR. Cancer Research. 1994;54:5618–5622. [PubMed] [Google Scholar]
- 23.Perez JM, Kelland LR, Montero EI, Boxall FE, Fuertes MA, Alonso C, Navarro-Ranninger C. Molecular Pharmacology. 2003;63:933–944. doi: 10.1124/mol.63.4.933. [DOI] [PubMed] [Google Scholar]
- 24.Appleton TG, Bailey AJ, Barnham KJ, Hall JR. Inorganic Chemistry. 1992;31:3077–3082. [Google Scholar]
- 25.McGowan G, Parsons S, Sadler PJ. Inorganic Chemistry. 2005;44:7459–7467. doi: 10.1021/ic050763t. [DOI] [PubMed] [Google Scholar]
- 26.Wimmer S, Castan P, Wimmer FL, Johnson NP. Journal of the Chemical Society-Dalton Transactions. 1989:403–412. [Google Scholar]
- 27.Arpalahti J, Sillanpaa R, Mikola M. Journal of the Chemical Society-Dalton Transactions. 1994:1499–1500. [Google Scholar]
- 28.Miller SE, Gerard KJ, House DA. Inorganica Chimica Acta. 1991;190:135–144. [Google Scholar]
- 29.Frey U, Ranford JD, Sadler PJ. Inorganic Chemistry. 1993;32:1333–1340. [Google Scholar]
- 30.Canovese L, Cattalini L, Chessa G, Tobe ML. Journal of the Chemical Society-Dalton Transactions. 1988:2135–2140. [Google Scholar]
- 31.Jerremalm E, Videhult P, Alvelius G, Griffiths WJ, Bergman T, Eksborg S, Ehrsson H. Journal of Pharmaceutical Sciences. 2002;91:2116–2121. doi: 10.1002/jps.10201. [DOI] [PubMed] [Google Scholar]
- 32.Oehlsen ME, Qu Y, Farrell N. Inorganic Chemistry. 2003;42:5498–5506. doi: 10.1021/ic030045b. [DOI] [PubMed] [Google Scholar]
- 33.Berners-Price SJ, Kuchel PW. Journal of Inorganic Biochemistry. 1990;38:327–345. doi: 10.1016/0162-0134(90)80006-j. [DOI] [PubMed] [Google Scholar]
- 34.Berners-Price SJ, Kuchel PW. Journal of Inorganic Biochemistry. 1990;38:305–326. doi: 10.1016/0162-0134(90)80006-j. [DOI] [PubMed] [Google Scholar]
- 35.BernalMendez E, Boudvillain M, GonzalezVilchez F, Leng M. Biochemistry. 1997;36:7281–7287. doi: 10.1021/bi9703148. [DOI] [PubMed] [Google Scholar]
- 36.Kasparkova J, Novakova O, Farrell N, Brabec V. Biochemistry. 2003;42:792–800. doi: 10.1021/bi026614t. [DOI] [PubMed] [Google Scholar]
- 37.Novakova O, Kasparkova J, Malina J, Natile G, Brabec V. Nucleic Acids Research. 2003;31:6450–6460. doi: 10.1093/nar/gkg863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Brabec V, Leng M. Proceedings of the National Academy of Sciences of the United States of America. 1993;90:5345–5349. doi: 10.1073/pnas.90.11.5345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zakovska A, Novakova O, Balcarova Z, Bierbach U, Farrell N, Brabec V. European journal of biochemistry / FEBS. 1998;254:547–557. doi: 10.1046/j.1432-1327.1998.2540547.x. [DOI] [PubMed] [Google Scholar]
- 40.Kasparkova J, Marini V, Najajreh Y, Gibson D, Brabec V. Biochemistry. 2003;42:6321–6332. doi: 10.1021/bi0342315. [DOI] [PubMed] [Google Scholar]
- 41.Prokop R, Kasparkova J, Novakova O, Marini V, Pizarro A, Navarro-Ranninger C, Brabec V. Biochemical Pharmacology. 2004;67:1097–1109. doi: 10.1016/j.bcp.2003.11.001. [DOI] [PubMed] [Google Scholar]
- 42.Brabec V, Vrana O, Novakova O, Kleinwachter V, Intini FP, Coluccia M, Natile G. Nucleic Acids Research. 1996;24:336–341. doi: 10.1093/nar/24.2.336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zaludova R, Zakovska A, Kasparkova J, Balcarova Z, Kleinwachter V, Vrana O, Farrell N, Brabec V. European journal of biochemistry / FEBS. 1997;246:508–517. doi: 10.1111/j.1432-1033.1997.00508.x. [DOI] [PubMed] [Google Scholar]
- 44.Brabec V. Progress in Nucleic Acid Research and Molecular Biology. 2002;71:1–68. doi: 10.1016/s0079-6603(02)71040-4. [DOI] [PubMed] [Google Scholar]
- 45.Takahara PM, Rosenzweig AC, Frederick CA, Lippard SJ. Nature (London) 1995;377:649–652. doi: 10.1038/377649a0. [DOI] [PubMed] [Google Scholar]
- 46.Coste F, Malinge JM, Serre L, Shepard W, Roth M, Leng M, Zelwer C. Nucleic Acids Res. 1999;27:1837–1846. doi: 10.1093/nar/27.8.1837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Leng M, Schwartz A, Giraud-Panis M-J. In: Platinum-Based Drugs in Cancer Therapy. Kelland LR, Farrell NP, editors. Humana Press; Totowa, New Jersey: 2000. pp. 63–85. Editon edn. [Google Scholar]
- 48.Brabec V, Neplechova K, Kasparkova J, Farrell N. Journal of Biological Inorganic Chemistry. 2000;5:364–368. doi: 10.1007/pl00010665. [DOI] [PubMed] [Google Scholar]
- 49.Bierbach U, Farrell N. Inorganic Chemistry. 1997;36:3657–3665. doi: 10.1021/ic970154o. [DOI] [PubMed] [Google Scholar]
- 50.Intini FP, Boccarelli A, Francia VC, Pacifico C, Sivo MF, Natile G, Giordano D, De Rinaldis P, Coluccia M. Journal of Biological Inorganic Chemistry. 2004;9:768–780. doi: 10.1007/s00775-004-0572-x. [DOI] [PubMed] [Google Scholar]
- 51.Zou Y, Van Houten B, Farrell N. Biochemistry. 1993;32:9632–9638. doi: 10.1021/bi00088a015. [DOI] [PubMed] [Google Scholar]
- 52.Bierbach U, Qu Y, Hambley TW, Peroutka J, Nguyen HL, Doedee M, Farrell N. Inorganic Chemistry. 1999;38:3535–3542. doi: 10.1021/ic981181x. [DOI] [PubMed] [Google Scholar]
- 53.Kharatishvili M, Mathieson M, Farrell N. Inorganica Chimica Acta. 1997;255:1–6. [Google Scholar]
- 54.Marini V, Christofis P, Novakova O, Kasparkova J, Farrell N, Brabec V. Nucleic Acids Research. 2005;33:5819–5828. doi: 10.1093/nar/gki884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Najajreh Y, Peleg-Shulman T, Moshel O, Farrell N, Gibson D. Journal of Biological Inorganic Chemistry. 2003;8:167–175. doi: 10.1007/s00775-002-0402-y. [DOI] [PubMed] [Google Scholar]
- 56.Kartalou M, Essigmann JM. Mutat Res. 2001;478:1–21. doi: 10.1016/s0027-5107(01)00142-7. [DOI] [PubMed] [Google Scholar]
- 57.O’Neill CF, Hunakova L, Kelland LR. Chemico-Biological Interactions. 1999;123:11–29. doi: 10.1016/s0009-2797(99)00115-5. [DOI] [PubMed] [Google Scholar]
- 58.Stehlikova K, Kasparkova J, Novakova O, Martinez A, Moreno V, Brabec V. FEBS J. 2006;273:301–314. doi: 10.1111/j.1742-4658.2005.05061.x. [DOI] [PubMed] [Google Scholar]
- 59.Ohndorf U-M, Rould MA, He Q, Pabo CO, Lippard SJ. Nature (London) 1999;399:708–712. doi: 10.1038/21460. [DOI] [PubMed] [Google Scholar]
- 60.Ohndorf UM, Whitehead JP, Raju NL, Lippard SJ. Biochemistry. 1997;36:14807–14815. doi: 10.1021/bi9717643. [DOI] [PubMed] [Google Scholar]
- 61.Zamble DB, Mu D, Reardon JT, Sancar A, Lippard SJ. Biochemistry. 1996;35:10004–10013. doi: 10.1021/bi960453+. [DOI] [PubMed] [Google Scholar]
- 62.Marples B, Adomat H, Billings PC, Farrell NP, Koch CJ, Skov KA. Anti-Cancer Drug Design. 1994;9:389–399. [PubMed] [Google Scholar]
- 63.Scovell WM, Muirhead N, Kroos LR. Biochemical and Biophysical Research Communications. 1987;142:826–835. doi: 10.1016/0006-291x(87)91488-4. [DOI] [PubMed] [Google Scholar]
- 64.Kane SA, Lippard SJ. Biochemistry. 1996;35:2180–2188. doi: 10.1021/bi952240a. [DOI] [PubMed] [Google Scholar]
- 65.Farrell N, Povirk LF, Dange Y, DeMasters G, Gupta MS, Kohlhagen G, Khan QA, Pommier Y, Gewirtz DA. Biochemical Pharmacology. 2004;68:857–866. doi: 10.1016/j.bcp.2004.05.023. [DOI] [PubMed] [Google Scholar]
- 66.Pommier Y, Kohlhagen G, Kohn KW, Leteurtre F, Wani MC, Wall ME. Proceedings of the National Academy of Sciences of the United States of America. 1995;92:8861–8865. doi: 10.1073/pnas.92.19.8861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Stewart L, Redinbo MR, Qiu XY, Hol WGJ, Champoux JJ. Science. 1998;279:1534–1541. doi: 10.1126/science.279.5356.1534. [DOI] [PubMed] [Google Scholar]
- 68.Colangelo D, Ghiglia A, Viano I, Mahboobi H, Ghezzi A, Cassino C, Osella D. Journal of Inorganic Biochemistry. 2004;98:61–67. doi: 10.1016/j.jinorgbio.2003.08.009. [DOI] [PubMed] [Google Scholar]
- 69.Reed JE, Neidle S, Vilar R. Chemical Communications. 2007:4366–4368. doi: 10.1039/b709898g. [DOI] [PubMed] [Google Scholar]
- 70.Rao L, Bierbach U. Journal of the American Chemical Society. 2007;129:15764–15765. doi: 10.1021/ja077390a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Tukalo MA, Kubler MD, Kern D, Mougel M, Ehresmann C, Ebel JP, Ehresmann B, Giege R. Biochemistry. 1987;26:5200–5208. doi: 10.1021/bi00390a045. [DOI] [PubMed] [Google Scholar]
- 72.Darlix JL, Gabus C, Nugeyre MT, Clavel F, Barresinoussi F. Journal of Molecular Biology. 1990;216:689–699. doi: 10.1016/0022-2836(90)90392-Y. [DOI] [PubMed] [Google Scholar]
- 73.Chvalova K, Brabec V, Kasparkova J. Nucleic Acids Research. 2007;35:1812–1821. doi: 10.1093/nar/gkm032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Bierbach U, Farrell N. Journal of Biological Inorganic Chemistry. 1998;3:570–580. [Google Scholar]
- 75.Anzellotti A, Stefan S, Gibson D, Farrell N. Inorganica Chimica Acta. 2006;359:3014–3019. [Google Scholar]
- 76.Strukl JV, de Paula QA, Yang XH, Qu Y, Farrell NP. Australian Journal of Chemistry. 2008;61:694–699. [Google Scholar]
- 77.Djuran MI, Lempers ELM, Reedijk J. Inorganic Chemistry. 1991;30:2648–2652. [Google Scholar]
- 78.Venter JC, Adams MD, Myers EW, Li PW, Mural RJ, Sutton GG, Smith HO, Yandell M, Evans CA, Holt RA, Gocayne JD, Amanatides P, Ballew RM, Huson DH, Wortman JR, et al. Science. 2001;291:1304–1351. doi: 10.1126/science.1058040. [DOI] [PubMed] [Google Scholar]
- 79.Schwabe JWR, Klug A. Nature structural biology. 1994;1:345–349. doi: 10.1038/nsb0694-345. [DOI] [PubMed] [Google Scholar]
- 80.Anzellotti AI, Liu Q, Bloemink MJ, Scarsdale JN, Farrell N. Chemistry & Biology (Cambridge, MA, United States) 2006;13:539–548. doi: 10.1016/j.chembiol.2006.04.004. [DOI] [PubMed] [Google Scholar]
- 81.Almaraz E, de Paula QA, Liu Q, Reibenspies JH, Darensbourg MY, Farrell NP. Journal of the American Chemical Society. 2008;130:6272–6280. doi: 10.1021/ja711254q. [DOI] [PubMed] [Google Scholar]
- 82.Liu Q, Golden M, Darensbourg MY, Farrell N. Chemical Communications. 2005:4360–4362. doi: 10.1039/b507751f. [DOI] [PubMed] [Google Scholar]
- 83.Coluccia M, Nassi A, Boccarelli A, Giordano D, Cardellicchio N, Locker D, Leng M, Sivo M, Intini FP, Natile G. Journal of Inorganic Biochemistry. 1999;77:31–35. doi: 10.1016/s0162-0134(99)00139-7. [DOI] [PubMed] [Google Scholar]
- 84.Coluccia M, Nassi A, Loseto F, Boccarelli A, Mariggio MA, Giordano D, Intini FP, Caputo P, Natile G. Journal of Medicinal Chemistry. 1993;36:510–512. doi: 10.1021/jm00056a012. [DOI] [PubMed] [Google Scholar]
- 85.Najajreh Y, Elena Khazanov, Seba Jawbry, Ardeli-Tzaraf, Perez Jose M., Jana Kasparkova, Viktor Brabec, Yechezkel Barenholz, Dan Gibson. Journal of Medicinal Chemistry. 2006;49:4665–4673. doi: 10.1021/jm060237r. [DOI] [PubMed] [Google Scholar]
- 86.Kelland LR, Sharp SY, O’Neill CF, Raynaud FI, Beale PJ, Judson IR. Journal of Inorganic Biochemistry. 1999;77:111–115. [PubMed] [Google Scholar]
- 87.Kelland LR, Barnard CF, Evans IG, Murrer BA, Theobald BR, Wyer SB, Goddard PM, Jones M, Valenti M, Bryant A. Journal of Medicinal Chemistry. 1995;38:3016–3024. doi: 10.1021/jm00016a004. [DOI] [PubMed] [Google Scholar]