Abstract
Previous studies indicated increased levels of protein oxidation in brain from subjects with Alzheimer's disease (AD), raising the question of whether oxidative damage is a late effect of neurodegeneration or precedes and contributes to the pathogenesis of AD. Hence, in the present study we used a parallel proteomic approach to identify oxidatively modified proteins in inferior parietal lobule (IPL) from subjects with mild cognitive impairment (MCI) and early stage-AD (EAD). By comparing to age-matched controls, we reasoned that such analysis could help in understanding potential mechanisms involved in upstream processes in AD pathogenesis. We have identified four proteins that showed elevated levels of protein carbonyls: carbonic anhydrase II (CA II), heat shock protein 70 (Hsp70), mitogen-activated protein kinase I (MAPKI), and syntaxin binding protein I (SBP1) in MCI IPL. In EAD IPL we identified three proteins: phosphoglycerate mutase 1 (PM1), glial fibrillary acidic protein, and fructose bisphospate aldolase C (FBA-C). Our results imply that some of the common targets of protein carbonylation correlated with AD neuropathology and suggest a possible involvement of protein modifications in the AD progression. Antioxid. Redox Signal. 12, 327–336.
Introduction
Alzheimer's disease (AD) is a progressive neurodegenerative disorder characterized by memory loss and cognitive decline. Histopathologically, AD is characterized by the presence of senile plaques, neurofibrillary tangles, and synapse loss (37). Current diagnosis of AD is based on National Institute of Neurological and Communicative Disorders and Stroke and the Alzheimer's Disease and Related Disorders Association (NINCDS–ADRDA) Workgroup criteria for the clinical diagnosis of probable AD that involves mini-mental state examination (MMSE) scores (4). Based on MMSE scores and other clinical observations, two pre-AD stages have been determined, termed mild cognitive impairment (MCI) and early AD (EAD). Mild cognitive impairment is considered as the first stage of AD with limited symptomology and no dementia. These individuals are classified into two types, amnestic MCI and nonanmenstic MCI, based on the presence or absence of memory complaints, respectively. The yearly conversion rate of patients from MCI to AD to 10–15% (34), although reversion back to normal is possible. Most the cases of conversion to AD were reported from the individuals with amnestic MCI. Many MCI patients present with significant medial temporal lobe atrophy, while others have high cerebrospinal fluid Tau and/or low CSF-β amyloid (1–42) concentrations, factors that are associated with the senile plaques found in AD brain. There are also genetic similarities between the conditions. The strongest physiologic predictor of familial AD, for example, may be the presence of apolipoprotein E gene (ApoE) allele 4, which is over-represented in both AD and MCI patients (24). These characteristics, in combination with the fact that the onset of AD is insidious and has a course that is gradually progressive, suggest that neuropathology exists many years before any symptoms occur. These considerations are consonant with the concept that in many cases MCI is an early sign of AD. This boundary line is becoming the focus of much research with stress on approaches to slow or prevent development of AD.
Early Alzheimer's disease (EAD) is considered as an intermediate state between mild cognitive impairment (MCI) and Alzheimer's disease (AD) (29). EAD brain showed frontal lobe atrophy (14) and ventricular widening in magnetic resonance imaging (MRI). Histopathologically, EAD brains also showed increase in the number of neurofibrillary tangles compared to MCI patients in the frontal and temporal lobes (29) and also demonstrated synapse loss (36). Synapse loss has been previously reported in AD brain (36). One of the synaptic proteins, synaptophysin, that plays an important role in synapse formation and exocytosis has been reported to be present in low levels in AD hippocampus (44) that may consequently lead to a reduced number of synapses and also suggest its possible involvement in altered neurotransmission and learning and memory processes observed in AD.
A number of mechanisms have been proposed to explain the pathogenesis of AD including: amyloid cascade, excitoxicity, oxidative stress, and inflammation (5, 20, 21, 27, 45). The oxidative stress hypothesis is one of the well-accepted hypotheses of AD pathogenesis; however, it is difficult at this point to suggest whether oxidative stress is the primary contributor or the secondary effect of the disease (22). The oxidative stress hypothesis of AD suggests that there is an imbalance between the level of oxidants and antioxidants, favoring oxidants that will result in the increased production of reactive oxygen species (ROS) and reactive nitrogen species (RNS) (8). ROS and RNS can damage virtually all biological molecules: proteins, lipids, carbohydrates, DNA, and RNA. Oxidative damage of proteins is one of the modifications leading to a severe failure of biological functions and cell death. Free radicals may directly oxidize amino acid residue side-chains and also lead to damage of proteins by an adduction of products of glycoxidation and/or lipid peroxidation. Therefore, accumulation of oxidatively modified proteins disrupts cellular functions either by a loss of catalytic ability or by an interruption of regulatory pathways. Oxidative stress may cause reversible and/or irreversible modifications on sensitive proteins, leading to structural, functional, and stability modulations. Protein modifications such as carbonylation, nitration, and protein–protein cross-linking are generally associated with loss of function and may lead to either the unfolding and degradation of the damaged proteins, or aggregation leading to accumulation as cytoplasmic inclusions, as observed in age-related neurodegenerative disorders (18, 39).
In AD brain, an increase in protein oxidation (protein carbonyl and 3-nitrotyrosine), lipid peroxidation, DNA oxidation, advanced glycation end products, and reactive oxygen species (ROS) formation has been widely demonstrated (6, 26, 27, 38, 40). In both MCI and AD patients, plasma mean levels of nonenzymatic antioxidants and activity of antioxidant enzymes appear to be lower than in controls, with no parallel induction of antioxidant enzymes (8, 41). Individuals with MCI, and subsequently with AD, are likely to have inadequate antioxidant enzymatic activity, unable to counteract the increased production of free radicals during the pathogenesis of the disease. Very recent studies reported increased oxidative damage in nuclear and mitochondrial DNA, lipid peroxidation, and protein oxidation in MCI compared to age-matched controls (2, 7, 9, 16, 28, 48). Individuals with EAD also show an increase in the levels of oxidative stress as indicated by protein HNE and 3-NT, RNA oxidation in EAD compared to age-matched controls (49). Studies from our laboratory are actively in progress to elucidate the role of oxidative stress in the progression of AD, with particular relevance to oxidative damage occurring at protein level. In the present study, we used a redox proteomic approach to identify specifically carbonylated proteins in inferior parietal lobule (IPL) samples obtained from subjects with MCI and EAD to provide insight into the role of oxidative stress in the pathogenesis and progression of AD.
Materials and Methods
Control, MCI, and EAD brains
For the present study, frozen inferior parietal lobule (IPL) samples were obtained from six MCI, six controls, and four EAD subjects. The Rapid Autopsy Program of the University of Kentucky Alzheimer's Disease Research Center (UK ADRC) provided autopsy samples with average postmortem intervals (PMIs) of 2.1 h for MCI patients and 2.9 h for age-matched control subjects (Table 1). The PMIs were 2.4 h and 2.9 h for EAD and age-matched controls, respectively (Table 1). Other neuropathological details of the subjects are provided in Table 2. All subjects came from the longitudinally followed normal control group who had annual neuropsychological testing and neurological and physical examinations every 2 years. Control subjects had no cognitive complaints, normal cognitive test scores, especially objective memory test scores, and normal neurological examinations. EAD patients met the criteria set by the NINCDS–ADRDA. These criteria include: progressive memory loss, two or more cognitive deficits, altered activities of daily living, onset of disease between age 40 and 90, Clinical Dementia Rating score of 0.5–1.0 (mild dementia), and a clinical evaluation. Patients with amnestic MCI met the criteria described by Petersen (34), which include the following: a memory complaint corroborated by an informant; objective memory test impairment (age and education adjusted); general normal global intellectual function; Clinical Dementia Rating score of 0.0–0.5 (no dementia), and a clinical evaluation that revealed no other cause for memory decline. Other characteristics of MCI, EAD, and age-matched control patients that were available from medical records are provided in Tables 1 and 2.
Table 1.
Profile of Subjects Used in this Study
| Samples | Age (yrs) | Gender (M/F) | Brain weight (g) (Mean ± SD) | Postmortem interval (h) | Mean Braak stage | MMSE score (Mean ± SD) | Lag (MMSE evaluation before death) | APOE genotype |
|---|---|---|---|---|---|---|---|---|
| Controls for EAD–IPL | 79 ± 2.4 | 2/2 | 1250 ± 57 | 2.9 ± 0.4 | 1.5 | 29 ± 0.8 | 189.7 ± 67.4 | 3/4 (2), 3/3 (2) |
| EAD | 86 ± 4.0 | 1/3 | 1220 ± 41 | 2.4 ± 0.3 | 5.0 | 24.3 ± 3.9 | 322 ± 178.62 | 3/4 (2), 2/3 (1), 4/4 (1) |
| Controls for MCI-IPL | 82 ± 2.6 | 2/4 | 1252.5 ± 44.3 | 2.9 ± 0.5 | 1.3 | 28.4 ± 1.3 | 415.4 ± 300.3 | 3/4 (1), 3/3 (4), 2/3 (1) |
| MCI-IPL | 88 ± 1.5 | 2/4 | 1121.7 ± 24.9 | 3.1 ± 0.4 | 3.8 | 25.2 ± 3.0 | 360 ± 159.5 | 3/4 (1), 3/3 (4), 2/3 (1) |
EAD, Early Alzheimer's disease; IPL, inferior parietal lobe; MCI, mild cognitive impairment.
Table 2.
Neuropathological Details of the Control, MCI, and EAD Subjects
| Subjects | DP | Neuritic plaque | Neurofibrillary tangles |
|---|---|---|---|
| Control for EAD IPL-1 | 3 | 7.59 | 0.4 |
| Control for EAD IPL-2 | 0 | 0 | 0 |
| Control for EAD IPL-3 | 0.2 | 0.2 | 0 |
| Control for EAD IPL-4 | 27.39 | 11 | 0 |
| EAD 1 | 48.4 | 12.39 | 1.39 |
| EAD 2 | 50 | 9.19 | 2.79 |
| EAD 3 | 20 | 9.19 | 4.59 |
| EAD 4 | 8.19 | 6.19 | 0.6 |
| Control for MCI-IPL-1 | 41.59 | 7.8 | 0 |
| Control for MCI-IPL-2 | 0 | 0 | 0 |
| Control for MCI-IPL-3 | 0 | 0 | 0 |
| Control for MCI-IPL-4 | 39.79 | 16.2 | 0 |
| Control for MCI-IPL-5 | 0 | 0 | 0 |
| Control for MCI-IPL-6 | 0 | 0 | 0 |
| MCI-IPL-1 | 4.4 | 11.39 | 0 |
| MCI-IPL-2 | 50 | 22 | 0 |
| MCI-IPL-3 | 0.2 | 5 | 0 |
| MCI-IPL-4 | 49 | 14.19 | 0 |
| MCI-IPL-5 | 50 | 16.2 | 0 |
| MCI-IPL-6 | 50 | 18.2 | 0 |
Sample preparation
Brain samples were minced and suspended in 10 mM HEPES buffer (pH 7.4) containing 137 mM NaCl, 4.6 mM KCl, 1.1 mM KH2PO4, 0.1 mM EDTA, and 0.6 mM MgSO4, as well as proteinase inhibitors: leupeptin (0.5 mg/ml), pepstatin (0.7 μg/ml), type II S soybean trypsin inhibitor (0.5 μg/ml), and PMSF (40 μg/ml). Homogenates were centrifuged at 14,000 g for 10 min to remove debris. Protein concentration in the supernatant was determined by the BCA method (Pierce, Rockford, IL).
Two-dimensional gel electrophoresis
Samples (150 μg) were incubated at room temperature for 30 min in four volumes of 10 mM 2,4-dinitrophenylhydrazine (DNPH) in either 2 M HCl for protein carbonyl derivatization/oxyblots or 2 M HCl for gel maps and mass spectrometry analysis, according to the method of Levine et al. (25). This was followed by precipitation of proteins by addition of ice-cold 100% trichloroacetic acid (TCA) to a final concentration of 15%, and samples were placed on ice for 10 min to allow precipitation of proteins. Precipitates were centrifuged at 15,800 g for 2 min. The pellets were washed with 0.5 ml of 1:1 (vol/vol) ethanol/ethyl acetate solution. After centrifugation and washing with ethanol/ethyl acetate solution three times, the samples were then dissolved with 185 μl of rehydration buffer (8 M urea, 20 mM dithiothreitol, 2.0% (wt/vol) CHAPS, 0.2% Biolytes, 2 M thiourea and bromophenol blue).
For the first-dimension electrophoresis, 200 μL of sample solution were applied to a ReadyStrip™ IPG strip (Bio-Rad, Hercules, CA). The strips were soaked in the sample solution for 1 ho to allow uptake of the proteins. The strip was then actively rehydrated in protean IEF cell (Bio-Rad) for 16 h at 50 V. The isoelectric focusing was performed at 300 V for 2 h linearly; 500 V for 2 h linearly; 1,000 V for 2 h linearly, 8,000 V for 8 h linearly, and 8,000 V for 10 h rapidly. All the processes above were carried out at 22°C. The focused IEF strip was stored at −80°C until second-dimension electrophoresis was performed.
For second-dimension electrophoresis, thawed IPG® Strips pH 3–10 were equilibrated for 10 min in 50 mM Tris-HCl (pH 6.8) containing 6 M urea, 1% (wt/vol) sodium dodecyl sulfate (SDS), 30% (vol/vol) glycerol, and 0.5% dithiothreitol, and then re-equilibrated for 15 min in the same buffer containing 4.5% iodacetamide in place of dithiothreitol. Linear Gradient (8–16%) Precast criterion Tris-HCl gels (Bio-Rad) were used to perform second-dimension electrophoresis. Precision Protein™ Standards (Bio-Rad) were run along with the sample at 200 V for 65 min.
After electrophoresis, the gels were incubated in fixing solution (7% acetic acid, 10% methanol) for 20 min. Approximately 40 ml of SYPRO Ruby Gel Stain (Bio-Rad) were used to stain the gels for 2 h, on a gently continuous rocker. The gels were placed in deionized water overnight for destaining.
Western blot analysis
The same amount of protein samples (150 μg) was used for detecting specific protein carbonyl levels, and the electrophoresis was carried out in the same way as described above. 150 μg of proteins were incubated with 4 vol of 20 mM 2,4-dinitrophenylhydrazine (DNPH) at room temperature (25°C) for 20 min. The gels were prepared in the same manner as 2D-electrophoresis. The proteins from the second-dimension electrophoresis gels were transferred to nitrocellulose (Bio-Rad) using a Transblot-Blot® SD semi-Dry Transfer Cell (Bio-Rad) at 15 V for 2 h. The 2,4-dinitrophenyl hydrazone (DNP) adducts of the carbonyls of the proteins were detected on the nitrocellulose paper using a primary rabbit antibody (Millipore, Temecula, CA) specific for DNP–protein adduct (1:100), followed by a secondary goat anti-rabbit IgG (Sigma, St Louis, MO) antibody. The resultant stain was developed using 5-bromo-4-chloro-3-indolyl phosphate/nitro blue tetrazolium (BCIP/NBT) solution (SigmaFast tablets; Sigma).
Image analysis
The gels (n = 16) and nitrocellulose blots (n = 16) were scanned and saved in TIF format using a Scanjet 3300C (Hewlett Packard). PDQuest 2-D Analysis Software (Bio-Rad, Inc.) was used for matching and analysis of visualized protein spots among differential gels and membranes to compare protein and DNP immunoreactivity content between MCI/EAD IPL samples and age-matched controls. This sophisticated software offers powerful comparative analysis and is specifically designed to analyze many gels or blots at once. Powerful automatching algorithms quickly and accurately match gels or blots and sophisticated statistical analysis tools allow identification of experimentally significant spots. The principles of measuring intensity values by 2-D analysis software were similar to those of densitometric measurement. The average mode of background subtraction was used to normalize intensity values, which represents the amount of protein (total protein on gel and DNP-bound protein on the membrane) per spot. After completion of spot matching, the normalized intensity of each protein spot from individual gels (or membranes) was compared between groups using statistical analysis. Statistical significance was assessed by a two-tailed Student's t-test. p Values <0.05 were considered significant for comparison between control (age-matched subjects) and experimental data (MCI/EAD subjects).
Trypsin digestion
In-gel digestion on selected gel spots was performed according to Sultana et al. (40). The selected protein spots were excised with a clean blade and transferred into clean microcentrifuge tubes. The protein spots were then washed with 0.1 M ammonium bicarbonate (NH4HCO3) at room temperature for 15 min. Acetonitrile was added to the gel pieces and incubated at room temperature for 15 min. The solvent was removed, and the gel pieces were dried in a flow hood. The protein spots were incubated with 20 μl of 20 mM DTT in 0.1 M NH4HCO3 at 56°C for 45 min. The DTT solution was then removed and replaced with 20 μl of 55 mM iodoacetamide in 0.1 M NH4HCO3. The solution was incubated at room temperature in the dark for 30 min. The iodoacetamide was removed and replaced with 0.2 ml of 50 mM NH4HCO3 and incubated at room temperature for 15 min. 200 μl of acetonitrile was added. After a 15-min incubation, the solvent was removed, and the gel spots were dried in a flow hood for 30 min. The gel pieces were rehydrated with 20 ng/μl methylated trypsin (Promega, Madison, WI) in 50 mM NH4HCO3 with the minimal volume to cover the gel pieces. The gel pieces were chopped into smaller pieces and incubated at 37°C overnight in shaking incubator.
Mass spectrometry
All mass spectra reported in this study were acquired at the Department of Pharmacology in the University of Louisville School of Medicine and VAMC. A Spec 2E MALDI TOF (matrix assisted laser desorption ionization-time of flight) mass spectrometer operated in the reflectron mode was used to generate peptide mass fingerprints. Peptides resulting from in-gel digestion with trypsin were analyzed on a 384 position, 600 μm AnchorChip™ Target (Bruker Daltonics, Bremen, Germany) and prepared according to AnchorChip recommendations (AnchorChip Technology, Rev. 2, Bruker Daltonics). Briefly, 1 μl of digest was mixed with 1 μl of alpha-cyano-4-hydoxycinnamic acid (0.3 mg/ml in ethanol:acetone, 2:1 ratio) directly on the target and allowed to dry at room temperature. The sample spot was washed with 1 μl of a 1% TFA solution for ∼60 s. The TFA droplet was gently blown off the sample spot with compressed air. The resulting diffuse sample spot was recrystallized (refocused) using 1 μl of a solution of ethanol: acetone: 0.1% TFA (6:3:1 ratio). Reported spectra are a summation of 100 laser shots. External calibration of the mass axis was used for acquisition and internal calibration using either trypsin autolysis ions or matrix clusters was applied post acquisition for accurate mass determination.
The MALDI spectra used for protein identification from tryptic fragments were searched against the NCBI protein databases using the MASCOT search engine (http://www.matrixscience.com). Peptide mass fingerprinting used the assumption that peptides are monoisotopic, oxidized at methionine residues and carbamidomethylated at cysteine residues (6, 11). Up to one missed trypsin cleavage was allowed. Mass tolerance of 150 ppm was the window of error allowed for matching the peptide mass values. Probability based MOWSE scores were estimated by comparison of search results against estimated random match population and were reported as –10 *LOG10(p), where p is the absolute probability. MOWSE score greater than 61 were considered significant (P < 0.05). All the protein identifications were in the expected size range based on position in the gel.
Immunoprecipitation
To confirm the correct identification of the proteins identified by mass spectrometry, control or MCI samples (150 μg) were first precleared by incubation with protein A-agarose (Amersham Pharmacia Biotech AB, Uppsala, Sweden) for 1 h at 4°C. Samples were then incubated overnight with the relevant antibody followed by 1 h of incubation with protein A-Agarose, then washed three times with buffer B (50 mM Tris HCl pH 8.0, 150 mM NaCl, and 1% NP40). Proteins were resolved by SDS-PAGE, followed by immunoblotting on a nitrocellulose membrane (BioRad). Proteins were detected by the alkaline phosphate-linked secondary antibody using 5-bromo-4-chloro-3-indolyl phosphate/nitro blue tetrazolium (BCIP/NBT) solution (Sigma Fast tablets; Sigma).
Enzyme assay
Carbonic anhydrase II
Carbonic anhydrase activity was measured as described in (10) with minor modifications. For assay of carbonic anhydrase activity, a decrease in absorbance at 560 nm was recorded after the addition of 5 μl of the samples to CO2 saturated Tris buffer (pH 8.3, 0.2 M Tris-HCl, phenol red).
MAPK kinase I
MAPK kinase activity was measured with the MAPK kinase Assay Kit according to the manufacturer instructions (Chemicon, Inc.).
Statistics
The data of protein levels and protein-specific carbonyl levels were analyzed by two-tailed Student's t test. A value of p < 0.05 was considered statistically significant.
Results
Specific protein carbonyl level
Two-dimensional (2D) electrophoresis offers an efficient tool for screening of abundant protein changes in different disease states, as well as differences in metabolic pathways (5, 6). Western blot analysis and subsequent immunochemical detection of DNP-adducts allowed identification of specifically carbonylated proteins in the IPL tissue of MCI/EAD samples in comparison to their respective age-matched controls (Figs. 1–3). The specific carbonyl levels were obtained by dividing the carbonyl level of a protein spot on the nitrocellulose membrane by the protein level of its corresponding protein spot on the gel. Such numbers give the carbonyl level per unit of protein. Using this approach, we confirmed that not all of the protein spots with increased immunoreactivity are excessively modified proteins, since the normalization step also takes into account the protein levels to avoid the artifactual identification of a protein as oxidatively modified. This approach may limit the identification of some of the other proteins that are present below the threshold limit of detection. We used a parallel approach to quantify the protein levels by SYPRO Ruby staining and the extent of DNP-bound proteins by immunochemistry. Both the gels and blots used in this study showed reproducible patterns of protein separation. Based on the stringent conditions applied in the data analysis in this study, we found that in comparison with age-matched control samples, MCI samples have four proteins significantly more oxidized (Figs. 1 and 2). These proteins are: heat shock protein 70 (Hsp70), carbonic anhydrase II (CAII), syntaxin binding protein I (SBP1), and mitogen-activated protein kinase I (MAPKI). Further, in EAD IPL we identified three proteins: phosphoglycerate mutase 1 (PM 1), fructose bisphosphate aldolase C (FBA-C), and glial fibrillary acidic protein (GFAP) that show a significant increase in protein carbonyls compared to age-matched controls (Fig. 3). Table 3 shows the proteins that were successfully identified by mass spectrometry in IPL from both MCI and EAD subjects, along with the peptides matched, percentage coverage, pI, Mr values, and the increase of specific carbonyl levels, indexed as percentage of control.
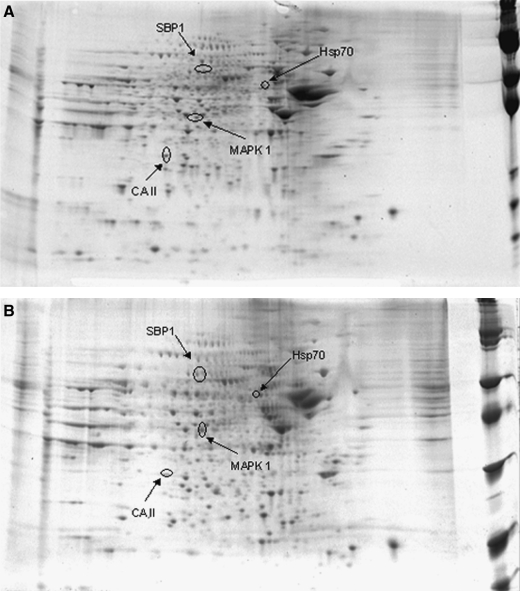
FIG. 1.
Representative SYPRO Ruby 2-D gels of the inferior parietal lobule from control (A) or MCI (B) brain. Protein (150 μg) was separated on immobilized pH 3–10 IPG strips, followed by separation on an 8–16% gradient SDS-PAGE gels.
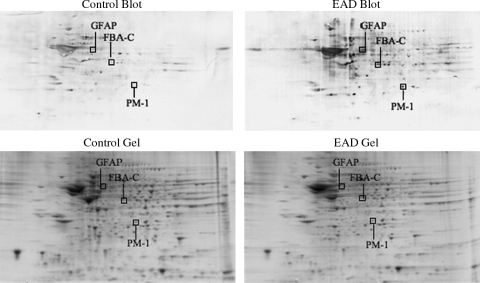
FIG. 3.
Two-dimensional carbonyl immunoblots from EAD and CTR subjects (IPL). Positions of the three identified proteins are shown on the blots.
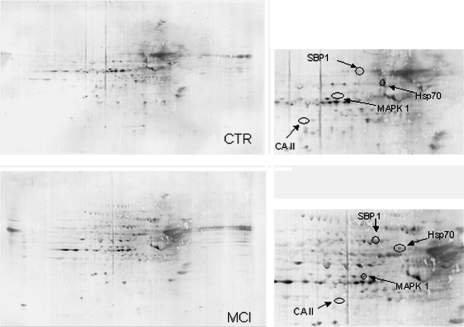
FIG. 2.
Two-dimensional carbonyl immunoblots from MCI and control (CTR) subjects (IPL). Positions of the four identified protein are shown on the blots. Expanded images of the selected proteins on the blots are also shown. Relative change in carbonyl immunoreactivity, after normalization of the immunostaining intensities to the protein content, was significant for four proteins. See text. Expanded images of the selected proteins on the blots are also shown.
Table 3.
Summary of Proteins Identified as Significantly Oxidized in MCI Patients Compared to Control Subjects
| Samples | Oxidatively modified protein | % Coverage of matched peptides | pI | Mr (kDa) | MOWSE score* | Protein oxidation | p value |
|---|---|---|---|---|---|---|---|
| MCI–IPL | CA II | 39 | 6.82 | 29.1 | 86 | 64 ± 12 | < 0.02 |
| MCI–IPL | Syntaxin binding protein I | 29 | 6.49 | 67.9 | 66 | 70 ± 15 | < 0.03 |
| MCI–IPL | Hsp70 | 25 | 5.37 | 71.1 | 95 | 71 ± 9 | < 0.03 |
| MCI–IPL | MAPK kinase I | 21 | 6.5 | 41.7 | 64 | 9 ± 2 | < 0.02 |
| EAD–IPL | FBA-C | 49 | 6.46 | 39.7 | 166 | 195 ± 76.16 | < 0.05 |
| EAD–IPL | PM-1 | 39 | 6.75 | 28.8 | 96 | 4.09 ± 1.13 | < 0.05 |
| EAD–IPL | GFAP | 41 | 5.42 | 49.9 | 120 | 444 ± 1.66 | < 0.05 |
CA II, carbonic anhydrase II; FBA-C, Fructose bisphosphate aldolase C; GFAP, glial fibrillary acidic protein; Hsp 70, Heat shock protein 70; PM-1, MAPK kinase I, mitogen activated protein kinase I; Mr, relative mobility Phosphoglycerate mutase 1; pI, isoelectric point.
Probability based MOWSE scores were estimated by comparison of search results against estimated random match population and were reported as –10 *LOG10(p), where p is the absolute probability.
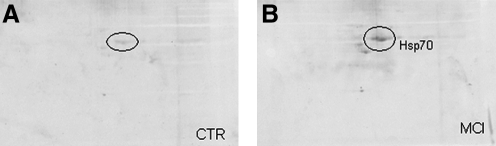
To validate the proteomics results, immunochemical selection of two proteins, Hsp70 and CA II, was undertaken. The blot probed with anti-Hsp70 antibody (Fig. 4) showed three spots among which the biggest corresponded to Hsp70 (Fig. 4). The position of HSP70 protein spot on the blot probed with anti-HSP70 antibody was the same as that observed on the carbonyl protein blot. In addition, we used anti-CA II antibody to immunoprecipitate the selected protein from brain homogenate, both from MCI and controls samples. Upon immunoprecipitation, the spot was absent in the 2-D gels (Fig. 5). Thus, the identification of the Hsp70 and CA II was validated by immunochemistry, indicating that identification of proteins by mass spectrometry is equivalent to those by immunochemistry.
FIG. 4.
Two-dimensional carbonyl immunoblots of CTR (A) and MCI (B) samples after immunoprecipitation with specific anti-Hsp70 antibody. To confirm the correct identification of Hsp70 by mass spectrometry, control or MCI samples (150 μg) were immunoprecipitated with anti-Hsp70 antibody and protein resolved by SDS-PAGE, followed by immunoblotting on nitrocellulose.
FIG. 5.
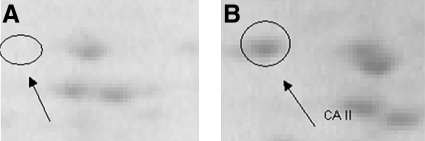
(A, left) 2D electrophoresis gel from supernatant of MCI IPL homogenate described in this study after immunoprecipitation by anti-CA II antibody. (B, right) 2D electrophoresis gel from the same sample without immunoprecipitation. The arrowhead indicates that CA II disappears after immunoprecipitation, confirming its identity.
CA II and MAPKI enzyme activity
In order to investigate the role of oxidative modifications on enzyme activity, we measured the catalytic activity of CA II and MAPKI on brain tissue homogenates. We found that the activity of CA II was significantly lower in MCI samples (50% decrease) compared with their age-matched controls (Table 4). This result showed that the drop in functional activity matched the increase in oxidative modifications of the enzyme. Conversely, we observed that MAPKI activity was higher in MCI samples compared with control samples (Table 4). Both the MAPK1 and CAII did not show any significant difference in the activities compared to respective controls. We suggest that dysregulation of the MAPK pathways might play a role in the intracellular mechanisms of neurodegeneration.
Table 4.
Carbonic Anhydrase and MAP Kinase Activity in Control, MCI, and EAD Inferior Parietal Lobule
| Enzymes activities | Control for MCI (units per mg of protein, Mean ± SD) | MCI | Control for EAD | EAD |
|---|---|---|---|---|
| Carbonic anhydrase | 37.9 ± 5.09 | 19.5 ± 4.80 | 40 ± 3.82 | 35.525 ± 8.47 |
| MAP kinase* | 462.2 ± 73.87 | 591.7 ± 52.22 | 481.3 ± 184.54 | 630.82 ± 108.95 |
One unit is the amount of ERK2 kinase that will incorporate 1 nmol phosphate onto myelin basic protein substrate per minute at 30°C.
Discussion
Previous studies have reported the identification of specifically carbonylated proteins in both the brain and plasma of AD subjects (8, 12, 40). In the present study, we used a redox proteomics approach to identify specific targets of proteins carbonylation in AD and MCI brain. The IPL regions of the brain was specifically selected in this study as this region of the brain has been shown to be one of the severely affected region in AD brain, and also a recent study using MRI showed that the entorhinal cortex and IPL regions are first to be affected in AD pathogenesis (13, 31). We have identified four proteins that showed elevated levels of protein carbonyls in the inferior parietal lobule of MCI patients compared with their age-matched controls: carbonic anhydrase II (CA II), syntaxin binding protein I (SBP1), heat shock protein 70 (Hsp70), and mitogen-activated protein kinase I (MAPKI). These proteins are involved in different biological functions; hence, the impairment of their activity might contribute to neuropathological events leading to neurodegeneration.
Carbonic anhydrases represent a group of isozymes that catalyze the reversible hydration of carbon dioxide to form H2CO3 that in turn decomposes to H+ and HCO3−. CA II is the main isozyme of the CA family in the human brain. It is located mainly in oligodendroglia and less in astrocytes (10). CA II has multiple functions in the brain and its deficiency is associated with pathological consequences such as mental retardation and brain calcification (47). CA II leads to HCO3−, which regulates membrane transport of Na+/water and contributes to cerebrospinal fluid formation. CA II has been proposed to participate in the processes of myelination and also is involved in pH regulation, HCO3− reabsorption, and CO2 expiration. In addition, it plays a crucial role in signal processing, long-term synaptic transformation, and attentional gating of memory storage (42).
Memory abnormalities that characterize the early stages of AD involve multiple neurotransmitter deficits in hippocampal formation. It is known that alterations in synaptic spines and loss of dendrites during aging are associated with a significant decline in CA in the brain, and that this decline is even more dramatic in brains of AD subjects (36). Previous studies reported a decreased activity and oxidative modification of CA II in AD brain compared with their age-matched controls (8, 30, 40). In the present work, we found that CA II showed increased carbonyl levels in MCI subjects compared to control subjects, with a parallel enzyme activity decline. Based on our findings, we suggest that dysfunction of CA II, due to its oxidatively modifications, impairs cognition and might be associated with decreased cognition in AD. The second protein found to be excessively carbonylated in MCI brain is syntaxin binding protein 1. Syntaxin binding protein 1 (MUNC18-1, MUNC18a) is a 67-kDa hydrophilic, neuron-specific protein that binds strongly to syntaxin 1 and is required for synaptic vesicle exocytosis and neurotransmitter release (19). Loss of function of syntaxin binding protein 1 as a result of protein oxidation could lead to loss of fusion of synaptic vesicles, release of neurotransmitters, and subsequently, loss of synaptic transmission and neuronal function. All these events might strongly contribute to exacerbate loss of memory and cognition and neurodegenerative processes involved in progression of MCI to AD.
A number of neurodegenerative diseases, including AD, are associated with protein misfolding and aggregation of protein outside and within the cells. Misfolded proteins and protein aggregation are controlled by molecular chaperones such as heat shock proteins (HSPs) that are constitutively and inducibly expressed in the nervous system. Among the proteins chaperoned are Aβ and tau in AD. Here, we found increased carbonyl levels of Hsp70 in IPL from MCI subjects compared with control subjects. Based on this finding, we propose that inactivation of Hsp70 by oxidative modification makes this protein unable to facilitate misfolded protein degradation by the proteasome, which would contribute to the accumulation of toxic, oligomeric assemblies of Aβ peptides. It is interesting to note that previous studies from our laboratory found Hsc71, the constitutive isoform of Hsp70, oxidatively modified in the inferior parietal region of AD brain (11). This overlapping result suggests that dysfunction of chaperones occurs as early as the MCI state and may contribute to neuronal death associated with AD. Hence, HSPs are expected to be critically involved in the progression of AD, making them potential therapeutic targets.
The mitogen-activated protein kinases (MAPKs) have recently emerged as possible key regulators of the formation of plaques and tau hyperphosphorylation occurring in AD (15, 50). In this study, we found that MAPK kinase I showed increased carbonyl levels in IPL from MCI subjects compared with controls. Oxidative modification of the protein kinase was accompanied by increased enzyme activity, leading to the activation of the MEK/MAPK cascade. Since all MAPKs pathways transduce intracellular signalling to increase expression of different proteins, dysregulation of MAPK-dependent pathways suggests a systematic disorder of protein translation regulation in MCI brains. ERK activation is present in early AD astroglia, while in more advanced AD it is associated with neuronal cell bodies and dystrophic neurites around plaques, suggesting that ERK activation in astroglia may be an important early response to the onset of AD pathology (1). This study suggested that MAPK cascade could be a molecular link between the increased activity of MAP kinases and tau phosphorylation. More recently, abnormal phosphorylation of tau was reported to correlate with increase activity of ERK1/2 in postmortem AD brains (43). The possibility exists that increased phosphorylation of cytoskeletal proteins by various MAP kinases found in AD could reflect part of the pathological outcome of oxidative stress. We suggest that oxidative modifications of MAPKs might make them more prone to phosphorylation or may be an alternative mechanism of their activation, thus initiating signaling cascades ultimately leading to hyperphosphorylation of tau. Inhibiting MAPKs in the early phase of AD could have two valuable effects. First, it putatively would lower phosphorylation of tau, and second, would prevent cell death by obstructing the cascade that leads to premature cell death.
In EAD, three proteins were found to be specifically carbonylated compared to age-matched controls. Two of the identified carbonylated proteins, PM1 and FBA-C, belong to the glycolytic pathway. PM1 catalyzes the interconversion of 3-phosphoglycerate to 2-phosphoglycerate, and in this process it results in the production of a second equivalent of ATP in glycolysis. FBA-C catalyzes the conversion of hexoses to trioses, for example, fructose 1,6 -bisphosphate into dihydroxyacetone phosphate (DHAP) and glyceraldehyde-3-phosphate (G3P). Previous studies showed that the levels of PM1 and FBA-C are decreased in AD brain (23, 40). PM1 is oxidatively modified in AD hippocampus and in an in vivo model of Aβ (3, 8, 40). Further, aldolase has been reported to be oxdatively modified in MCI and AD brain (33, 35), and FBA-C has been shown to be oxidatively modified in the olfactory bulb of aged mice (46). The oxidation and decrease activity of FBA-C may lead to increase levels of fructose 1,6- bisphosphate that may inhibit glycolysis and favors the glycogenesis process. Oxidative modifications generally inhibit enzyme activity, hence oxidative modification of FBA-C and PM1 may increase glycolytic intermediates, decrease pyruvate production, eventually leading to inhibition of complete glycolysis and glucose metabolism and eventually to ATP depletion that further support the PET studies of MCI, EAD, and AD.
GFAP is exclusively found in astrocytes and has been shown to undergo activation in AD (32). The oxidation of GFAP were consistent with the previous study that show GFAP undergoing oxidation early in the progression of AD, consistent with an on-going inflammatory process and consequently with increase oxidative stress (17).
In conclusion, our results imply that some of the common targets of protein carbonylation in comparison to LAD suggest their possible involvement in AD progression. MCI is considered to be relatively early in the progression of disease and the identification of a number of overlapping proteins between LAD and MCI suggest that they are key in the process of disease progression. In the current study, we did not observe any common target of oxidation in IPL between MCI and EAD. We have no definitive explanation for this lack of overlap, but do note that due to the rarity of obtaining EAD specimens, we had only four independent samples of EAD with which to investigate. It is noteworthy that most of the commonly oxidized proteins from MCI, EAD, and LAD belong to the glucose metabolic pathway (8), suggesting that alteration in the energy metabolism pathway is critical in the progression of the disease. Further, our laboratory is investigating why certain proteins are modified at certain stages of disease to provide insights into potential therapeutic options to treat or delay the progression of AD.
Abbreviations Used
- AD
Alzheimer's disease
- ALS
amyotrophic lateral sclerosis
- ApoE
apolipoprotein E
- BCIP
5-bromo-4-chloro-3-indolyl phosphate
- CA-II
carbonic anhydrase II
- DHAP
dihydroxyacetone phosphate
- DHPH-2,4
dinitrophenylhydrazine
- DTT
dithiothreitol
- EAD
early AD
- FBA-C
fructose bisphosphate aldolase C
- GFAP
glial fibrillary acidic protein
- G3P
glyceraldehyde-3-phosphate
- HNE
4-hydroxy 2-trans nonenal
- Hsc71
heat shock cognate 71
- Hsp70
heat shock protein 70
- IEF
isoelectrofocusing
- IPL
inferior parietal lobe
- LAD
late stage Alzheimers disease
- MALDI TOF
matrix assisted laser desorption ionization-time of flight
- MAPK kinase I
mitogen activated protein kinase
- MCI
mild cognitive impairment
- NBT
nitro blue tetrazolium
- 3-NT
3-nitrotyrosine
- PD
Parkinson's disease
- PM1
phosphoglycerate mutase 1
- PMI
postmortem intervals
- RNS
reactive nitrogen speies
- ROS
reactive oxugen speices
- SBP1
synthaxin binding protein I (SBP1)
- SDS
sodium dodecyl sulfate
- SDS-PAGE
sodium dodecyl sulfate-polyacrylamide gel electrophoresis
- SOD
superoxide dismutase
- TCA
trichloroacetic acid
- TFA
trifluoroaceticacid
Acknowledgment
This work was supported in part by grants from the National Institutes of Health AG-10836; AG-05119 (DAB). The authors also wish to thank Drs. Fred Schmitt, Richard Kryscio, Charles Smith, and Greg Cooper for clinical data, diagnosis information, and tissue procurement.
Author Disclosure Statement
No competing financial interests exist.
References
- 1.Arendt T. Disturbance of neuronal plasticity is a critical pathogenetic event in Alzheimer's disease. Int J Dev Neurosci. 2001;19:231–245. doi: 10.1016/s0736-5748(01)00007-7. [DOI] [PubMed] [Google Scholar]
- 2.Beal MF. Oxidative damage as an early marker of Alzheimer's disease and mild cognitive impairment. Neurobiol Aging. 2005;26:585–586. doi: 10.1016/j.neurobiolaging.2004.09.022. [DOI] [PubMed] [Google Scholar]
- 3.Bigl M. Bruckner MK. Arendt T. Bigl V. Eschrich K. Activities of key glycolytic enzymes in the brains of patients with Alzheimer's disease. J Neural Transm. 1999;106:499–511. doi: 10.1007/s007020050174. [DOI] [PubMed] [Google Scholar]
- 4.Braak H. Braak E. Demonstration of amyloid deposits and neurofibrillary changes in whole brain sections. Brain Pathol. 1991;1:213–216. doi: 10.1111/j.1750-3639.1991.tb00661.x. [DOI] [PubMed] [Google Scholar]
- 5.Butterfield DA. Amyloid beta-peptide (1–42)-induced oxidative stress and neurotoxicity: Implications for neurodegeneration in Alzheimer's disease brain. A review. Free Radic Res. 2002;36:1307–1313. doi: 10.1080/1071576021000049890. [DOI] [PubMed] [Google Scholar]
- 6.Butterfield DA. Boyd-Kimball D. Castegna A. Proteomics in Alzheimer's disease: Insights into potential mechanisms of neurodegeneration. J Neurochem. 2003;86:1313–1327. doi: 10.1046/j.1471-4159.2003.01948.x. [DOI] [PubMed] [Google Scholar]
- 7.Butterfield DA. Poon HF. St Clair D. Keller JN. Pierce WM. Klein JB. Markesbery WR. Redox proteomics identification of oxidatively modified hippocampal proteins in mild cognitive impairment: Insights into the development of Alzheimer's disease. Neurobiol Dis. 2006;22:223–232. doi: 10.1016/j.nbd.2005.11.002. [DOI] [PubMed] [Google Scholar]
- 8.Butterfield DA. Reed T. Newman SF. Sultana R. Roles of amyloid beta-peptide-associated oxidative stress and brain protein modifications in the pathogenesis of Alzheimer's disease and mild cognitive impairment. Free Radic Biol Med. 2007;43:658–677. doi: 10.1016/j.freeradbiomed.2007.05.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Butterfield DA. Reed T. Perluigi M. De Marco C. Coccia R. Cini C. Sultana R. Elevated protein-bound levels of the lipid peroxidation product, 4-hydroxy-2-nonenal, in brain from persons with mild cognitive impairment. Neurosci Lett. 2006;397:170–173. doi: 10.1016/j.neulet.2005.12.017. [DOI] [PubMed] [Google Scholar]
- 10.Cammer W. Bieler L. Fredman T. Norton WT. Quantitation of myelin carbonic anhydrase-development and subfractionation of rat brain myelin and comparison with myelin from other species. Brain Res. 1977;138:17–28. doi: 10.1016/0006-8993(77)90781-8. [DOI] [PubMed] [Google Scholar]
- 11.Castegna A. Aksenov M. Aksenova M. Thongboonkerd V. Klein JB. Pierce WM. Booze R. Markesbery WR. Butterfield DA. Proteomic identification of oxidatively modified proteins in Alzheimer's disease brain. Part I: Creatine kinase BB, glutamine synthase, and ubiquitin carboxy-terminal hydrolase L-1. Free Radic Biol Med. 2002;33:562–571. doi: 10.1016/s0891-5849(02)00914-0. [DOI] [PubMed] [Google Scholar]
- 12.Dalle-Donne I. Scaloni A. Giustarini D. Cavarra E. Tell G. Lungarella G. Colombo R. Rossi R. Milzani A. Proteins as biomarkers of oxidative/nitrosative stress in diseases: The contribution of redox proteomics. Mass Spectrom Rev. 2005;24:55–99. doi: 10.1002/mas.20006. [DOI] [PubMed] [Google Scholar]
- 13.Desikan RS. Cabral HJ. Hess CP. Dillon WP. Glastonbury CM. Weiner MW. Schmansky NJ. Greve DN. Salat DH. Buckner RL. Fischl B. Automated MRI measures identify individuals with mild cognitive impairment and Alzheimer's disease. Brain. 2009;132:2048–2057. doi: 10.1093/brain/awp123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Farrow TF. Thiyagesh SN. Wilkinson ID. Parks RW. Ingram L. Woodruff PW. Fronto-temporal-lobe atrophy in early-stage Alzheimer's disease identified using an improved detection methodology. Psychiatry Res. 2007;155:11–19. doi: 10.1016/j.pscychresns.2006.12.013. [DOI] [PubMed] [Google Scholar]
- 15.Ferrer I. Gomez-Isla T. Puig B. Freixes M. Ribe E. Dalfo E. Avila J. Current advances on different kinases involved in tau phosphorylation, and implications in Alzheimer's disease and tauopathies. Curr Alzheimer Res. 2005;2:3–18. doi: 10.2174/1567205052772713. [DOI] [PubMed] [Google Scholar]
- 16.Greilberger J. Koidl C. Greilberger M. Lamprecht M. Schroecksnadel K. Leblhuber F. Fuchs D. Oettl K. Malondialdehyde, carbonyl proteins and albumin-disulphide as useful oxidative markers in mild cognitive impairment and Alzheimer's disease. Free Radic Res. 2008;42:633–638. doi: 10.1080/10715760802255764. [DOI] [PubMed] [Google Scholar]
- 17.Gu F. Zhu M. Shi J. Hu Y. Zhao Z. Enhanced oxidative stress is an early event during development of Alzheimer-like pathologies in presenilin conditional knock-out mice. Neurosci Lett. 2008;44:44–48. doi: 10.1016/j.neulet.2008.05.050. [DOI] [PubMed] [Google Scholar]
- 18.Halliwell B. Oxidative stress and neurodegeneration: Where are we now? J Neurochem. 2006;97:1634–1658. doi: 10.1111/j.1471-4159.2006.03907.x. [DOI] [PubMed] [Google Scholar]
- 19.Hamos JE. DeGennaro LJ. Drachman DA. Synaptic loss in Alzheimer's disease and other dementias. Neurology. 1989;39:355–361. doi: 10.1212/wnl.39.3.355. [DOI] [PubMed] [Google Scholar]
- 20.Ho GJ. Drego R. Hakimian E. Masliah E. Mechanisms of cell signaling and inflammation in Alzheimer's disease. Curr Drug Targets Inflamm Allergy. 2005;4:247–256. doi: 10.2174/1568010053586237. [DOI] [PubMed] [Google Scholar]
- 21.Hynd MR. Scott HL. Dodd PR. Glutamate-mediated excitotoxicity and neurodegeneration in Alzheimer's disease. Neurochem Int. 2004;45:583–595. doi: 10.1016/j.neuint.2004.03.007. [DOI] [PubMed] [Google Scholar]
- 22.Ischiropoulos H. Beckman JS. Oxidative stress and nitration in neurodegeneration: Cause, effect, or association? J Clin Invest. 2003;111:163–169. doi: 10.1172/JCI17638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Iwangoff P. Armbruster R. Enz A. Meier-Ruge W. Glycolytic enzymes from human autoptic brain cortex: normal aged and demented cases. Mech Ageing Dev. 1980;14:203–209. doi: 10.1016/0047-6374(80)90120-7. [DOI] [PubMed] [Google Scholar]
- 24.Kryscio RJ. Schmitt FA. Salazar JC. Mendiondo MS. Markesbery WR. Risk factors for transitions from normal to mild cognitive impairment and dementia. Neurology. 2006;66:828–832. doi: 10.1212/01.wnl.0000203264.71880.45. [DOI] [PubMed] [Google Scholar]
- 25.Levine RL. Williams JA. Stadtman ER. Shacter E. Carbonyl assays for determination of oxidatively modified proteins. Methods Enzymol. 1994;233:346–357. doi: 10.1016/s0076-6879(94)33040-9. [DOI] [PubMed] [Google Scholar]
- 26.Lovell MA. Markesbery WR. Ratio of 8-hydroxyguanine in intact DNA to free 8-hydroxyguanine is increased in Alzheimer disease ventricular cerebrospinal fluid. Arch Neurol. 2001;58:392–396. doi: 10.1001/archneur.58.3.392. [DOI] [PubMed] [Google Scholar]
- 27.Markesbery WR. Oxidative stress hypothesis in Alzheimer's disease. Free Radic Biol Med. 1997;23:134–147. doi: 10.1016/s0891-5849(96)00629-6. [DOI] [PubMed] [Google Scholar]
- 28.Markesbery WR. Kryscio RJ. Lovell MA. Morrow JD. Lipid peroxidation is an early event in the brain in amnestic mild cognitive impairment. Ann Neurol. 2005;58:730–735. doi: 10.1002/ana.20629. [DOI] [PubMed] [Google Scholar]
- 29.Markesbery WR. Schmitt FA. Kryscio RJ. Davis DG. Smith CD. Wekstein DR. Neuropathologic substrate of mild cognitive impairment. Arch Neurol. 2006;63:38–46. doi: 10.1001/archneur.63.1.38. [DOI] [PubMed] [Google Scholar]
- 30.Meier-Ruge W. Iwangoff P. Reichlmeier K. Neurochemical enzyme changes in Alzheimer's and Pick's disease. Arch Gerontol Geriatr. 1984;3:161–165. doi: 10.1016/0167-4943(84)90007-4. [DOI] [PubMed] [Google Scholar]
- 31.Modrego PJ. Fayed N. Pina MA. Conversion from mild cognitive impairment to probable Alzheimer's disease predicted by brain magnetic resonance spectroscopy. Am J Psychiatry. 2005;162:667–675. doi: 10.1176/appi.ajp.162.4.667. [DOI] [PubMed] [Google Scholar]
- 32.Mouser PE. Head E. Ha KH. Rohn TT. Caspase-mediated cleavage of glial fibrillary acidic protein within degenerating astrocytes of the Alzheimer's disease brain. Am J Pathol. 2006;168:936–946. doi: 10.2353/ajpath.2006.050798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Perluigi M. Sultana R. Cenini G. Di Demenico F. Memo M. Pierce WM. Coccia R. Butterfield DA. Redox proteomics identification of HNE-modified brain proteins in Alzheimers disease: Role of lipid peroxidation in AD pathogenesis. Proteomics Clin Appli. 2009;3:682–693. doi: 10.1002/prca.200800161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Petersen RC. Mild cognitive impairment: Transition between aging and Alzheimer's disease. Neurologia. 2000;15:93–101. [PubMed] [Google Scholar]
- 35.Reed T. Perluigi M. Sultana R. Pierce WM. Klein JB. Turner DM. Coccia R. Markesbery WR. Butterfield DA. Redox proteomic identification of 4-hydroxy-2-nonenal-modified brain proteins in amnestic mild cognitive impairment: Insight into the role of lipid peroxidation in the progression and pathogenesis of Alzheimer's disease. Neurobiol Dis. 2008;30:107–120. doi: 10.1016/j.nbd.2007.12.007. [DOI] [PubMed] [Google Scholar]
- 36.Scheff SW. Price DA. Schmitt FA. Mufson EJ. Hippocampal synaptic loss in early Alzheimer's disease and mild cognitive impairment. Neurobiol Aging. 2006;27:1372–1384. doi: 10.1016/j.neurobiolaging.2005.09.012. [DOI] [PubMed] [Google Scholar]
- 37.Selkoe DJ. Alzheimer's disease: Genes, proteins, and therapy. Physiol Rev. 2001;81:741–766. doi: 10.1152/physrev.2001.81.2.741. [DOI] [PubMed] [Google Scholar]
- 38.Smith MA. Richey Harris PL. Sayre LM. Beckman JS. Perry G. Widespread peroxynitrite-mediated damage in Alzheimer's disease. J Neurosci. 1997;17:2653–2657. doi: 10.1523/JNEUROSCI.17-08-02653.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stadtman ER. Berlett BS. Reactive oxygen-mediated protein oxidation in aging and disease. Chem Res Toxicol. 1997;10:485–494. doi: 10.1021/tx960133r. [DOI] [PubMed] [Google Scholar]
- 40.Sultana R. Boyd-Kimball D. Poon HF. Cai J. Pierce WM. Klein JB. Merchant M. Markesbery WR. Butterfield DA. Redox proteomics identification of oxidized proteins in Alzheimer's disease hippocampus and cerebellum: An approach to understand pathological and biochemical alterations in AD. Neurobiol Aging. 2006;27:1564–1576. doi: 10.1016/j.neurobiolaging.2005.09.021. [DOI] [PubMed] [Google Scholar]
- 41.Sultana R. Piroddi M. Galli F. Butterfield DA. Protein levels and activity of some antioxidant enzymes in hippocampus of subjects with amnestic mild cognitive impairment. Neurochem Res. 2008;33:2540–2546. doi: 10.1007/s11064-008-9593-0. [DOI] [PubMed] [Google Scholar]
- 42.Sun MK. Alkon DL. Carbonic anhydrase gating of attention: Memory therapy and enhancement. Trends Pharmacol Sci. 2002;23:83–89. doi: 10.1016/s0165-6147(02)01899-0. [DOI] [PubMed] [Google Scholar]
- 43.Swatton JE. Sellers LA. Faull RL. Holland A. Iritani S. Bahn S. Increased MAP kinase activity in Alzheimer's and Down syndrome but not in schizophrenia human brain. Eur J Neurosci. 2004;19:2711–2719. doi: 10.1111/j.0953-816X.2004.03365.x. [DOI] [PubMed] [Google Scholar]
- 44.Sze CI. Troncoso JC. Kawas C. Mouton P. Price DL. Martin LJ. Loss of the presynaptic vesicle protein synaptophysin in hippocampus correlates with cognitive decline in Alzheimer disease. J Neuropathol Exp Neurol. 1997;56:933–944. doi: 10.1097/00005072-199708000-00011. [DOI] [PubMed] [Google Scholar]
- 45.Tuppo EE. Arias HR. The role of inflammation in Alzheimer's disease. Int J Biochem Cell Biol. 2005;37:289–305. doi: 10.1016/j.biocel.2004.07.009. [DOI] [PubMed] [Google Scholar]
- 46.Vaishnav RA. Getchell ML. Poon HF. Barnett KR. Hunter SA. Pierce WM. Klein JB. Butterfield DA. Getchell TV. Oxidative stress in the aging murine olfactory bulb: redox proteomics and cellular localization. J Neurosci Res. 2007;85:373–385. doi: 10.1002/jnr.21130. [DOI] [PubMed] [Google Scholar]
- 47.Vlkolinsky R. Cairns N. Fountoulakis M. Lubec G. Decreased brain levels of 2′,3′-cyclic nucleotide-3′-phosphodiesterase in Down syndrome and Alzheimer's disease. Neurobiol Aging. 2001;22:547–553. doi: 10.1016/s0197-4580(01)00218-4. [DOI] [PubMed] [Google Scholar]
- 48.Wang J. Markesbery WR. Lovell MA. Increased oxidative damage in nuclear and mitochondrial DNA in mild cognitive impairment. J Neurochem. 2006;96:825–832. doi: 10.1111/j.1471-4159.2005.03615.x. [DOI] [PubMed] [Google Scholar]
- 49.Williams TI. Lynn BC. Markesbery WR. Lovell MA. Increased levels of 4-hydroxynonenal and acrolein, neurotoxic markers of lipid peroxidation, in the brain in mild cognitive impairment and early Alzheimer's disease. Neurobiol Aging. 2006;27:1094–1099. doi: 10.1016/j.neurobiolaging.2005.06.004. [DOI] [PubMed] [Google Scholar]
- 50.Zhu X. Lee HG. Raina AK. Perry G. Smith MA. The role of mitogen-activated protein kinase pathways in Alzheimer's disease. Neurosignals. 2002;11:270–281. doi: 10.1159/000067426. [DOI] [PubMed] [Google Scholar]