Abstract
Mitochondrial dysfunction is a central feature of a number of acute and chronic neurodegenerative conditions, but clinically approved therapeutic interventions are only just emerging. Here we demonstrate the potential clinical utility of low molecular weight inhibitors of the hypoxia inducible factor prolyl-4-hydroxylases (HIF PHDs) in preventing mitochondrial toxin-induced cell death in mouse striatal neurons that express a “knock-in” mutant Huntingtin allele. Protection from 3-nitropropionic acid (3-NP, a complex II inhibitor)-induced toxicity by HIF PHD inhibition occurs without rescue of succinate dehydrogenase activity. Although HIF-1α mRNA is dramatically induced by mutant huntingtin, HIF-1α depletion by short interfering RNAs (siRNA) does not affect steady-state viability or protection from 3-NP-induced death by HIF PHD inhibitors in these cells. Moreover, 3-NP-induced complex II inhibition in control or mutant striatal neurons does not lead to activation of HIF-dependent transcription. HIF PHD inhibition also protects cortical neurons from 3-NP-induced cytotoxicity. Protection of cortical neurons by HIF PHD inhibition correlates with enhanced VEGF but not PGC-1α gene expression. Together, these findings suggest that HIF PHD inhibitors are promising candidates for preventing cell death in conditions such as Huntington's disease and Alzheimer's disease that are associated with metabolic stress in the central nervous system. Antioxid. Redox Signal. 12, 435–443.
Introduction
Mitochondrial dysfunction and aberrant energy metabolism appear to be a common upstream mediators of many acute and chronic neurodegenerative conditions. Of these, disordered energy metabolism is most closely linked with the pathophysiology of Huntington's disease (HD) (6, 10). HD is a movement disorder characterized by choreiform movements, cognitive dysfunction, and psychiatric manifestations. Two converging lines of inquiry support the hypothesis that mitochondrial energy metabolism may be the primary defect in HD. First, HD is attributable to an expanded glutamine repeat stretch in the protein huntingtin (mhtt). Among its many cellular manifestations, mhtt leads to transcriptional repression of many genes, including those controlling adaptation to low mitochondrial energy charge such as PPARγ coactivator 1α (PGC-1α) (7, 8, 35). Indeed, recent studies have shown that germline deletion of PGC-1α leads to striatal degeneration similar in localization and behavioral manifestations to HD (17); by contrast, PGC-1α overexpression via lentiviral delivery in vivo prevents striatal degeneration attributable to transgenic expression of mhtt (8). In this context, PGC-1α is believed to coactivate genes involved in mitochondrial proliferation and function, including a number of antioxidant enzymes localized to mitochondria (e.g., MnSOD and GPx) (32).
These molecular and genetic studies are amplified by a second line of investigation using 3-NP, a toxin that selectively poisons complex II (succinate dehydrogenase, SDH) of the mitochondrial electron transport chain. Intoxication with 3-NP can induce selective CNS damage in rodents that phenocopies human HD and rodent models of HD (4, 5). Moreover, agents that mitigate 3-NP-induced striatal degeneration in vivo can also attenuate disease onset or progression in rodent models of HD (3, 19).
Besides mitochondrial biogenesis and/or induction of mitochondrial proteins, an alternate strategy to compensate for mitochondrial energy deficit is to shift a cell's energy economy towards aerobic glycolysis and away from oxidative phosphorylation (14). Indeed, transcriptional upregulation of glycolytic enzymes is an essential feature of adaptation to hypoxia, a condition where oxygen is used inefficiently or is in short supply (30). Transcriptional induction of glycolytic enzymes in response to metabolic challenges such as hypoxia is mediated primarily via stabilization of the transcriptional activator HIF-1α and the consequent induction of >100 genes associated with adaptation to hypoxic stress (30). In addition to glycolytic enzymes, these genes include vascular endothelial growth factor (VEGF), erythropoietin, and p21waf1/cip1 (38).
Stabilization of HIF-1α in response to hypoxia is mediated via the inhibition of a family of dioxygenases known as the HIF prolyl hydroxylases (HIF PHDs) (12, 13). Prior studies from our laboratory and others have demonstrated a role for small molecule inhibitors of HIF PHDs in protecting neurons from ischemic or oxidative injury (2, 31, 38). Another study suggested that HIF PHD inhibition may prevent mitochondrial toxicity in C6 glioma cells (37). However, no studies to date have systematically evaluated the HIF pathway in disease models of mitochondrial dysfunction such as HD; moreover, the ability of HIF PHD inhibitors to prevent mitochondrial toxicity in normal or HD associated neurons has yet to be explored. Herein, we show that the HIF pathway is markedly induced in immortalized striatal cells bearing a full length huntingtin protein with a pathological number of repeats (111) but not in wild-type striatal neurons with 7 repeats. We further demonstrate that canonical low molecular weight HIF PHD inhibitors abrogate 3-NP-induced death in neurons. Unexpectedly, these inhibitors protect even with marked silencing of the HIF-1α message. Altogether, these studies add to the growing enthusiasm for HIF PHD inhibitors as neurological therapeutics and suggest that these agents may be appropriate for neurological conditions associated with metabolic dysfunction such as HD and stroke.
Material and Methods
Cell culture
Clonal striatal cell lines established from E14 striatal primordia of HdhQ111/Q111 (mutant) and HdhQ7/Q7 (wild-type) knock-in mouse littermates were generously provided by Dr. Marcy E. MacDonald (Massachusetts General Hospital). These cells were immortalized using a replication defective retrovirus transducing the tsA58/U19 large T-antigen (33). Striatal Q7 and Q111 cells were maintained in Dulbecco's modified Eagle medium (DMEM) containing 25 mM D-glucose, 1 mM L-glutamine, 10% fetal bovine serum (FBS), 1 mM sodium pyruvate, and 400 μg/mL Geneticin (Invitrogen, Carlsbad, CA) and were incubated at 33°C with 5% CO2.
Virus transduction
Cells were plated in 12-well plates at a density of 5 × 104 cells/ml for 16 h prior to viral transduction. Retroviruses carrying siRNAs against either HIF-1α or green fluorescent protein (GFP) were added at 5 MOI in the presence of hexadimethrine bromide (4 μg/ml; Sigma, St. Louis, MO) for 24 h. The cells were then subjected to selection by puromycin (4 μg/ml; Sigma) as previously described (1). Cell lines expressing the siRNAs were maintained as above.
Primary neurons
Primary cortical neurons were cultured from embryonic E17 Sprague–Dawley rats, as previously described with slight modification (22). Briefly, cortices were dissected, homogenized, and plated in minimum essential media containing 10% FBS, and 1% penicillin/streptomycin in 96-well plates or in 10 cm dishes. Neurons were maintained at 37°C and 5% CO2. All experiments were conducted 24 h after plating.
Western blot analysis
After treatment with PHD inhibitors, striatal cells were washed in cold PBS, scraped, and pelleted by centrifugation. Nuclear extracts were prepared from the resulting pellets using the NE-PER Nuclear and Cytoplasmic Extraction kits (Pierce, Rockford, IL) and protein concentrations were measured using the Bradford protein assay (Sigma). Nuclear proteins were separated by SDS-PAGE electrophoresis and transferred onto nitrocellulose membranes. The membranes were probed with a monoclonal antibody against HIF-1α (Upstate, Davers, MA), which was detected using infrared-fluorescent dye conjugated secondary antibodies (Li-cor, Lincoln, NE). Images were acquired using the Li-cor Odyssey quantitative Western blot system.
Luciferase assay
Q7 and Q111 cells were co-transfected with a plasmid expressing a firefly luciferase reporter gene driven by a hypoxia responsive element from the 5’ noncoding region of the enolase gene HRE promoter and a pTK-Renilla luciferase control plasmid (Promega, Madison, WI). Luciferase activity was assessed using a Promega dual luciferase assay and measured with an LMAXII384 luminometer (Molecular Devices, Sunnyvale, CA). Firefly luciferase luminescence was normalized to Renilla luciferase luminescence according to the manufacturer's protocol.
Cell viability
Q7, Q111, or primary neurons were plated in a 96-well black cell culture plates with clear bottoms designed for fluorescence imaging. Striatal cells were pretreated with 50 μM desferrioxamine (DFO), 50 μM 3,4-dihydroxybenzoic acid (DHB), or 2.5 μM ciclopirox (CPO) for 6 h and then exposed to 10 mM 3-NP in low glucose media for an additional 24 h. Primary neurons were treated concurrently with 100 μM DFO, 20 μM DHB, or 0.5 μM CPO and 10 mM 3-NP in low glucose media for 48 h. Fluorescence calcein and ethidium-homodimer (LIVE/DEAD) images of striatal cells were captured using a Zeiss Axiovert microscope (Carl Zeiss Microimaging, Thornwood, NY). For automated cell counting, a fluorescence image of the entire well of each well of striatal or primary neuron plates were captured using a flash cytometer (Trophos, France). Cell counts of viable cells (calcein-positive) were automatically generated from these pictures with the Trophos software.
Quantitative real time PCR
Two micrograms of total RNA extract from Q7, Q111, or primary cortical neurons were reverse transcribed using Superscript 3 (Invitrogen). The resulting cDNA was assayed by real-time PCR using TaqMan Gene Expression Assays for HIF-1α, HIF-2α, VEGF, p21, β-actin, and GAPDH (Applied Biosystems, Carlsbad, CA). PCR reactions were carried out using an Applied Biosystems 7500 Fast Real Time PCR System. Real time PCR data were normalized to β-actin levels and are expressed as a fraction of the mRNA expression level in untreated cells. Similar results were obtained when GAPDH was used as an internal loading control and are reported in Supplemental Fig. 1 (see www.liebertonline.com/ars).
SDH activity assay
Striatal cells were lysed with 0.1% vol/vol NP40 in 100 mM Tris-HCl (pH 7.4). Twenty microliters of the resulting lysate was added to an assay mixture containing 100 mM Tris-HCl (pH 8.3), 0.5 mM EDTA, 2 mM iodonitrotetrazolium chloride, 5 μM rotenone, 2 μM antimycin A, 10 mM sodium azide, and 20 mM succinate. The rate of succinate-dependent generation of the formazan product of iodonitrotetrazolium reduction was measured by absorbance at 500 nm at 37°C for 60 min and was taken as SDH activity as previously described (21).
Statistical analysis
All results are plotted as mean ± SEM using GraphPad Prism software. The statistical tests used for analysis are indicated in the figure legends. Statistical significance was taken as p < 0.05.
Results
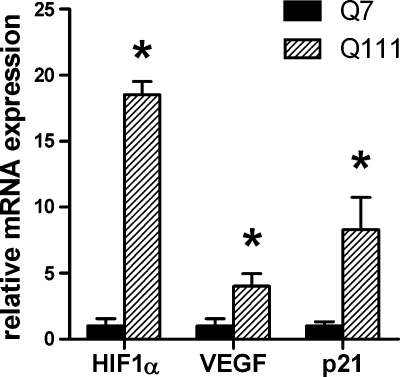
To begin to query the behavior of the HIF pathway in response to mitochondrial dysfunction in neurons, we compared RNA levels for HIF-1α and two of its target genes, VEGF and p21 in immortalized striatal neurons expressing normal huntingtin protein (7 repeats) and striatal neurons expressing a mutant huntingtin protein associated with juvenile onset HD (111 repeats). These cells were generated from the HdhQ111 knock-in mice (STHdhQ111, which bear full-length htt with an expanded polyglutamine (polyQ) tract of 111 CAG repeats; herein referred to as Q111) and control cells from the striatum of a knock-in mouse bearing a nonpathological polyQ expansion (STHdhQ7, herein referred to as Q7) (8, 33). Interestingly, we found that HIF-1α mRNA levels and levels of HIF target genes are significantly increased in Q111 cells as compared to Q7 cells, raising the possibility that HIF is induced to compensate for metabolic compromise in HD (Fig. 1). The change in HIF message has been observed in prior gene array studies of these cells (16) and in arrays from brain tissue homogenates of one rodent model of HD (analysis of R6/2 datasets) (15), but the biological role of this change has not been previously evaluated.
FIG. 1.
HIF-1α and HIF target gene mRNA are elevated in STHdhQ111 striatal cells. TaqMan real-time gene expression assays for HIF-1α, VEGF, and p21 show elevated basal expression of these genes in mutant huntingtin (Q111) cells compared to wild-type (Q7) striatal cells. Gene expression levels were normalized to β-actin levels and are expressed as fold increase over Q7 cells for the same gene. Data represent mean ± SEM of three independent experiments. *p < 0.05 compared to corresponding Q7, Student's t-test.
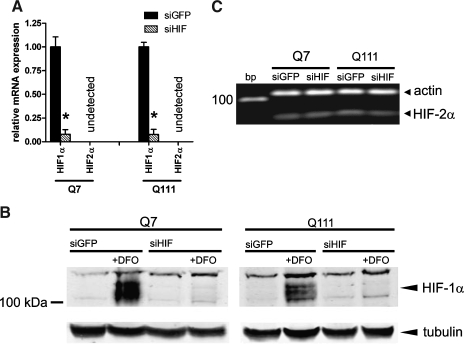
To explore whether the increase in HIF-1α allows Q111 cells to adapt to mitochondrial dysfunction, we utilized a well-characterized retrovirus that can transduce murine cells to express an siRNA to GFP or HIF-1α (1). As expected, we found that siRNA to HIF-1α significantly reduced message levels for HIF-1α by >80% in both cell lines (Fig. 2A). To examine whether reduction in HIF-1α message levels were associated with reductions in HIF-1α protein, we performed immunoblots from lysates of Q7 or Q111 lines treated with vehicle or the HIF PHD inhibitor desferrioxamine (DFO), which mimics hypoxia by stabilizing HIF-1α protein levels. As expected, in both Q7 and Q111 lines DFO induced HIF-1α protein, which was completely suppressed by the siRNA to HIF-1α but not the siRNA to GFP (Fig. 2B). As other HIFα isoforms may be induced to compensate for the loss of HIF-1α, we assessed HIF-2α mRNA levels following HIF-1α knockdown. We were unable to detect HIF-2α message in either cell line by real-time RT-PCR (Fig. 2A), though we were able to resolve a PCR product corresponding to HIF-2α by agarose gel electrophoresis, which was not changed by HIF-1α knockdown (Fig. 2C).
FIG. 2.
Stable silencing of HIF-1α in striatal cells with si-RNA against HIF-1α. Q7 and Q111 striatal cells were transduced with a retrovirus delivering hp/GFP (siGFP) or hp/HIF-1α (siHIF-1). (A) HIF-1α and HIF-2α expression levels were determined by RT-PCR. HIF-1α expression was significantly reduced in siHIF cells of either genotype whereas HIF-2α expression was undetectable by this method. (B) HIF-1α protein levels were determined by Western blot analysis. Under normoxic conditions, HIF-1α protein is rapidly degraded. DFO (200 μM), a pharmacologic stabilizer of HIF-1α protein, increased HIF-1α levels in both Q7 and Q111 cells expressing siGFP. In contrast, expression of siHIF prevented the DFO-mediated increase in HIF-1α protein. (C) The PCR products from the RT-PCR for HIF-2α expression assay were resolved by agarose gel electrophoresis and detected by ethidium bromide fluorescence. Though HIF-2α levels were undetectable by RT-PCR, bands corresponding to HIF-2α were present. Importantly, HIF-2α levels were not induced in cells expressing siHIF. Data represent mean ± SEM of three independent experiments. *p < 0.05 compared to corresponding siGFP expression levels, Student's t-test.
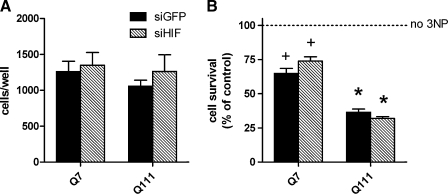
We next examined the role of HIF-1α in regulating viability in response to the selective complex II inhibitor, 3-NP. We first confirmed prior studies from our laboratory and others that showed that Q111 cells are more sensitive to 3-NP as compared to Q7 cells (Fig. 3B) (28). Despite these differences, we found that HIF-1α deletion had no effect on basal viability or sensitivity to 3-NP (10 mM) in control Q7 or disease associated Q111 cells (Fig. 3). Altogether, these findings suggest that HIF-1α is not necessary to maintain viability of striatal neurons in response to an expanded polyQ repeat in the huntingtin protein or in response to the selective complex II inhibitor 3-NP. Of note, our results also do not support a prodeath role for HIF-1α in striatal neurons (1).
FIG. 3.
Knockdown of HIF-1α does not affect viability or vulnerability to 3-NP toxicity in striatal cells. (A) Basal cell counts 36 h following an initial plating of 500 cells/well in a 96-well plate were unchanged by HIF-1α knockdown. (B) Cell viability was determined in siGFP and siHIF following 3-NP (10 mM) exposure for 24 h. While Q111 cells were more sensitive to 3-NP than Q7 cells, there was no effect of HIF-1α knockdown on vulnerability to 3-NP. Data represent mean ± SEM of 3–5 independent experiments. +p < 0.05 compared to no 3-NP, *p < 0.05 compared to no 3-NP and Q7/3-NP, two-way ANOVA with Bonferroni post-test.
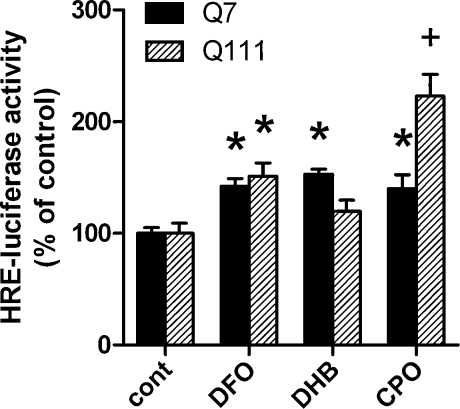
To investigate whether HIF activation in Q111 cells is sufficient to protect striatal neurons, we examined the effect of pharmacological inhibition of HIF PHD inhibitors on 3-NP induced death. HIF PHDs negatively regulate the stability of HIF family transcription factors, and thus inhibitors of HIF PHDs stabilize HIF-1α protein. Consistent with Fig. 2B, we found that structurally diverse inhibitors of the HIF PHDs that target the iron (DFO, CPO) or 2-oxoglutarate (3,4-DHB) cofactors significantly increase HIF-1α protein (Supplemental Fig. 2; see www.liebertonline.com/ars) and drive the expression of luciferase reporter under the control of the hypoxia response element from the 5’ noncoding region of the enolase gene (Fig. 4). We then examined the effect of 6 h pretreatment with each of the inhibitors on 24 h of 3-NP exposure in Q111 cells. Although there was a reduction in cell proliferation and a small increase in cell death, each of the inhibitors provided a significant level of neuroprotection as measured by the LIVE/DEAD assay (Fig. 5), which was corroborated by qualitative, visual observations using phase contrast microscopy (Supplemental Fig. 3; see www.liebertonline.com/ars).
FIG. 4.
Structurally diverse, low molecular weight prolyl 4-hydroxylase inhibitors enhance the activity of a hypoxia-response element-driven reporter in Q7 and Q111 striatal cells. Treatment with 50 μM desferrioxamine (DFO), 50 μM 3, 4-dihydroxybenzoic acid (DHB), or 2.5 μM ciclopirox (CPO) for 6 h increases the activity of a HRE-driven firefly luciferase reporter in Q7 and Q111 striatal cells. Data represent mean ± SEM of 3–12 independent experiments. *p < 0.05 comppared to corresponding genotype untreated control, +p < 0.05 compared to untreated control and corresponding Q7 cells, two-way ANOVA with Bonferroni post-test.
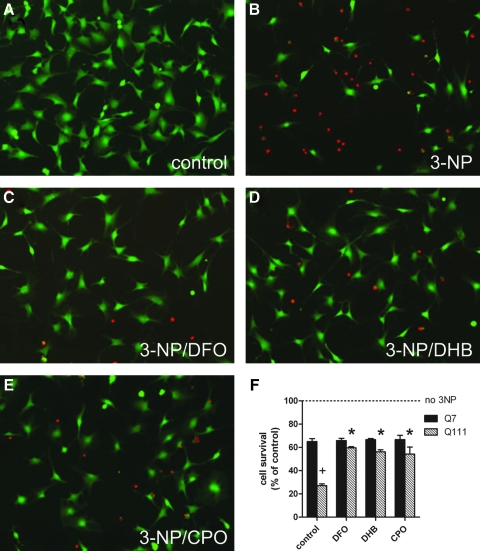
FIG. 5.
Prolyl 4-hydroxylase inhibitors abrogate 3-NP toxicity in Q111 striatal cells. Q7 and Q111 Striatal cells were pretreated with the indicated PHD inhibitors for 6 h and then exposed to 3-NP (10 mM) for an additional 24 h, after which cell survival was assessed by LIVE/DEAD assay. (A–E) Images of Q111 cells following viability assay: green fluorescence (calcein) indicates live cells while red fluorescence (ethidium homodimer) labels the nuclei of dead cells. (A) untreated control cells, (B) 3-NP (10 mM), (C) 50 μM desferrioxamine (DFO) + 3-NP, (D) 50 μM 3,4-dihydroxybenzoic acid (DHB) + 3-NP, (E) 2.5 μM ciclopirox (CPO) + 3-NP, (F) quantitation of cell survival following pretreatment and 3-NP exposure. Data represent mean ± SEM of four independent experiments. All cells treated with 3-NP were significantly different than no 3-NP treatment, *p < 0.01 compared to control Q111 cells treated with 3-NP only, +p < 0.001 compared to corresponding Q7 cells, two-way ANOVA with Bonferroni post-test.
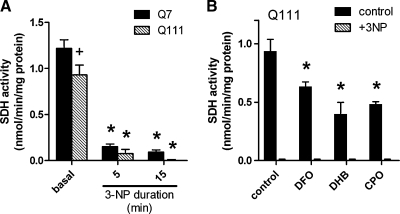
To establish whether HIF PHD inhibitors act upstream or downstream of SDH inhibition, we examined the effects of HIF PHD inhibitors on the suppression of SDH activity by 3-NP. We found small but significant reductions in basal SDH activity in Q111 cells compared to Q7 cells and observed that SDH activity is rapidly depleted after treatment with 3-NP in both cell types (Fig. 6A). This reduction in SDH activity was not associated with inhibition of the HIF PHDs as monitored by HRE driven luciferase activity as others have reported (not shown). Interestingly, exogenous addition of HIF PHD inhibitors led to a reduction in steady state SDH activity in Q7 cells, while SDH activity remained completely lost following 3-NP inhibition in Q111 cells treated with DFO, DHB, or CPO (Fig. 6B), suggesting that the observed protection was not simply through preserving SDH activity.
FIG. 6.
SDH activity following 3-NP and PHD inhibitors. Striatal cells were treated with 3-NP or 3-NP + PHD inhibitors for the indicated times, lysed, and assayed for SDH activity. (A) Q111 cells exhibited lower basal SDH activity, which was rapidly abolished by 3-NP treatment in both cell types. SDH activity was undetectable in Q111 cells by 15 min of exposure. (B) Q111 cells were co-treated with 3-NP and PHD inhibitors for 15 min and then assayed for SDH activity. While PHD inhibitors decreased basal SDH activity, they did not prevent the complete loss of SDH activity caused by 3-NP exposure. Data represent mean ± SEM of three independent experiments. *p < 0.01 compared to corresponding untreated control cells, +p < 0.05 compared to corresponding Q7 cells, two-way ANOVA with Bonferroni post-test.
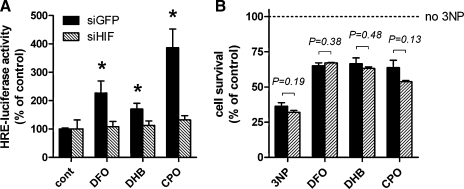
To examine whether HIF-1α is necessary for protection by HIF PHD inhibitors in striatal Q111 cells, we again utilized a retrovirus expressing an siRNA to GFP or HIF-1α. As expected, siHIF reduced the activation of a luciferase reporter driven by the hypoxia response element in Q111 cells whereas a retrovirus expressing siGFP had no effect (Fig. 7A). However, siHIF did not affect the protection induced by HIF PHD inhibitors (Fig. 7B). Together these studies demonstrate that HIF PHD inhibitors abrogate 3-NP toxicity independent of HIF-1α and downstream of SDH inhibition.
FIG. 7.
Knockdown of HIF-1α inhibits PHD inhibitor-mediated induction of HRE-reporter activity but does not abrogate their protection against 3-NP toxicity. HRE-luciferase activity was measured in Q111 cells following treatment with PHD inhibitors for 6 h. (A) PHD inhibitors do not induce HRE-luciferase activity in Q111 striatal cells expressing siHIF, indicating significant attenuation of the HIF pathway. (B) PHD inhibitors confer similar protection to Q111 with diminished HIF activity compared to cells expressing siGFP. Data represent mean ± SEM of three independent experiments. *p < 0.05 compared to corresponding untreated control cells, two-way ANOVA with Bonferroni post-test. In (B), all PHD inhibitor-treated cells are significantly different from corresponding 3-NP only cells, two-way ANOVA with Bonferoni post-test. Student's t-test p values are shown comparing survival between siGFP and siHIF cells.
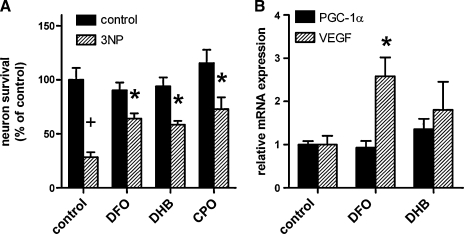
It is formally possible that the protection we observe in Q111 cells is facilitated by the immortalization of these cells by the expression of a temperature sensitive T-antigen. To exclude this possibility, we examined the ability of DFO, DHB, and CPO to prevent 3-NP toxicity in postmitotic cortical neurons (1–2 days in vitro) cultured from E17 rat embryos. As expected, all three compounds showed neuroprotection as measured by calcein uptake and automated cell counting (Fig. 8A); and unlike dividing striatal cells, no reduction in cell number was evident in primary cortical neurons (Fig. 8A). Further, these protective concentrations of HIF PHD inhibitors induced VEGF expression, a HIF-1α target gene, in these neurons (Fig. 8B). These data also suggest that HIF PHD inhibitors may be generally protective to neurons under conditions of metabolic stress independent of mhtt expression. Recent studies have suggested a connection between HIF-1α and PGC-1α, and given the role of PGC-1α as a target for mhtt toxicity, we determined whether HIF PHD inhibitors induce PGC-1α message. Indeed, while DFO and DHB induced the expected increases in VEGF message in wild-type cortical neurons, there was no effect on PGC-1α message in these cells (Fig. 8B), suggesting that HIF PHD inhibitors act independently of PGC-1α rescue to prevent 3-NP toxicity.
FIG. 8.
PHD inhibitors abrogate 3-NP toxicity and do not induce PGC-1α in embryonic cortical neuronal cultures. (A) Cortical neuron cultures from E17 rats were co-treated with PHD inhibitors and/or 3-NP (10 mM) for 48 h and viability was assessed by LIVE/DEAD assay. DFO (100 μM), DHB (20 μM), and CPO (0.5 μM) protected primary neurons against 3-NP toxicity. (B) Cortical neuron cultures were treated with PHD inhibitors for 24 h and assayed the expression of PGC-1α and VEGF mRNA by real-time PCR. PGC-1α levels were unchanged by PHD inhibitors while VEGF, a HIF target gene, was significantly induced with these agents. Data represent mean ± SEM of 3–6 independent experiments. *P < 0.05 compared to corresponding control cells treated with 3-NP only (A) or untreated control cells (B), +P < 0.05 compared to corresponding untreated control cells, two-way ANOVA with Bonferroni post-test.
Discussion
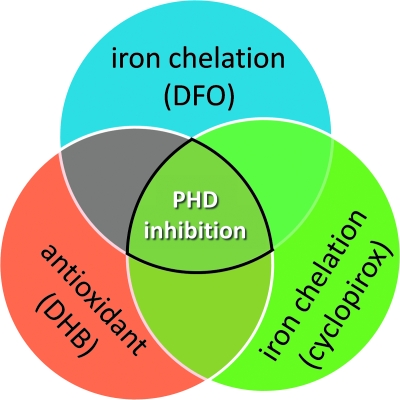
Despite its prominent role in metabolic adaptation, no studies have formally evaluated the role of hypoxia inducible factors and their upstream regulators, the HIF PHDs, in compensating for mitochondrial dysfunction in Huntington's disease and other neurological conditions. Here we demonstrate that three structurally diverse, low molecular weight inhibitors of the HIF PHDs prevent toxicity from 3-NP in striatal neuronal cells that accurately express a mutant huntingtin allele with 111 polyQ repeats that leads to abnormalities in ATP/ADP ratios (Fig. 9) (20). DFO and CPO bind iron with high affinity and are predicted to inhibit HIF PHDs by binding to iron in the active sites of these 2-oxoglutarate and oxygen-dependent dioxgenases (9, 34). By contrast, DHB appears to inhibit the HIF PHDs, stabilize HIF-1α, and drive HIF-dependent expression by iron-independent mechanisms (18). Indeed, prior studies from our laboratory showed that 20–50 μM DHB (unlike DFO and CPO) does not affect intracellular calcein fluorescence, IRP binding in an RNA gel shift assay, or total iron levels as measured by inductively coupled mass spectrometry, in neurons (31). Thus, HIF PHDs, and not iron, are the common established target for these compounds (Fig. 9). Metal chelators have been shown to prolong survival and diminish pathology in an R6/2 model of HD but the mechanism of protection in this model remains obscure (25). Our data argue that metal chelators may have benefit in HD by inhibiting one or all of the HIF PHDs (three HIF PHD isoforms are expressed in brain) (36). Of course, our data do not allow us to exclude the possibility that HIF-independent, 2-oxoglutarate-dependent dioxygenases may be relevant for protection in HD or in response to mitochondrial toxins. Indeed, the fact that HIF-1α is not required for protection by DFO, DHB, or CPO is congruent with this possibility. Future studies using siRNAs to each isoform or conditional deletion of each HIF PHD isoform (1–3) will clarify the role of HIF PHD inhibition in protection from mitochondrial dysfunction in HD and other disorders.
FIG. 9.
Structurally diverse compounds inhibit proyl-4-hydroxylases. A series of structurally diverse proyl-4-hydroxylase inhibitors target distinct cofactors necessary for enzyme function. These compounds are known to chelate iron (DFO and CPO) or block 2-oxoglutarate binding (DHB) in the active site of the enzyme and are protective under conditions of oxidative and metabolic stress. Further, the neuroprotective properties of these compounds may be independent of their ability to stabilize HIF-1α. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article at www.liebertonline.com/ars).
An interesting and surprising finding is the dramatic elevation in HIF-1α message in Q111 versus Q7 cells. This change in message does not appear to be associated with an increase in HIF-1α protein (Fig. 2B) or HIF-dependent reporter expression (data not shown), although we do see associated elevation of two known HIF-dependent genes, VEGF and p21 (Fig. 1). Most attention has focused on HIF-1α protein stability or increased message translation as a mechanism for enhancing nuclear HIF activity. Recent studies have shown that the versatile, nuclear threonine/serine kinase homeodomain interacting protein kinase 2 (HIPK2) can negatively regulate transcription of HIF-1α (23, 24). These studies raise the interesting possibility that mutant huntingtin negatively regulates HIPK2. Under basal conditions, HIPK2 co-represses a host of genes including HIF-1α; thus loss of HIPK2 activity related to mutant huntingtin could lead to de-repression of HIF-1α, leading to the observed changes in message in Q111 cells. Despite the dramatic increase in HIF-1α in Q111 cells, HIF-1α does not appear to be necessary for steady-state viability or HIF PHD inhibitor-induced neuroprotection. Both HIF-1α and HIF-2α are hypoxia-responsive HIFα subunit isoforms encoded by distinct gene loci and show some overlap in tissue localization and target gene expression, though both subunits may also have distinct transcriptional targets (27). Notably, the expression of glycolysis-related genes seems to be regulated primarily by HIF-1α (11). Thus along with nearly undetectable HIF-2α levels, it is unlikely that the dissociation of silencing HIF-1α and protection is related to redundancy provided by HIF-2α (Fig. 2A and C).
Another unexpected finding of the current study was our inability to see an induction in HIF protein stabilization or HIF reporter activity in response to 3-NP treatment. Prior studies have shown that pharmacological or molecular suppression of SDH by 3-NP or siRNA to SDH subunits leads to a cytosolic buildup of succinate and competitive inhibition of HIF PHDs (29). These events would result in HIF stabilization and induction of genes that compensate for hypoxia. The model has been confirmed for tumor cells, but little data support the notion that SDH inhibition leads to stabilization of HIF or activation of HIF-dependent gene expression in neurons either via succinate or alternatively, reactive oxygen species. Future studies that utilize siRNAs to SDH subunits in postmitotic neurons should address these open questions. At this time we cannot exclude the possibility that off-target effects of 3-NP mask the succinate-induced inhibition of HIF PHDs and activation of the HIF pathway.
Our findings set the table for a study of PHD inhibitors in fly or rodent models of HD to determine whether these drugs will also be effective in vivo. In our laboratory, HIF PHD inhibition in vitro is a good predictor of neuroprotection in animal models of stroke (26, 31), suggesting that these drugs may be beneficial for many neurological disorders that involve metabolic stress, including stroke and Alzheimer's disease. However, much work is yet to be done to fully understand the target and mechanism of action of these drugs. Additionally, our findings suggest that at the very least, HIF PHD inhibition is a good screen to identify agents that could prevent mitochondrial dysfunction and oxidative stress in these diseases.
Supplementary Material
Abbreviations Used
- CPO
ciclopirox
- DFO
desferrioxamine
- DHB
3,4-dihydrobenzoic acid
- FBS
fetal bovine serum
- GFP
green fluorescent protein
- HD
Huntington's disease
- HIF
hypoxia inducible factor
- HIPK2
nuclear threonine/serine kinase homeodomain interacting protein kinase 2
- HRE
hypoxia responsive element
- htt
wild-type huntingtin protein
- mhtt
mutant huntingtin protein
- 3-NP
3-nitropropionic acid
- PGC-1α
PPARγ coactivator 1α
- PHD
proyl-4-hydroxylase
- PolyQ
poly glutamine
- Q7
STHdhQ7 striatal cell line
- Q111
STHdhQ111 striatal cell line
- SDH
succinate dehydrogenase
- siRNA
short interfering RNA
- VEGF
vascular endothelial growth factor
Acknowledgments
This work was supported by a National Institutes of Health PO1 AG014930 grant and New York State Center of Research Excellence Grant to RRR (CO19772).
Author Disclosure Statement
No completing financial interests exist.
References
- 1.Aminova LR. Chavez JC. Lee J. Ryu H. Kung A. Lamanna JC. Ratan RR. Prosurvival and prodeath effects of hypoxia-inducible factor-1alpha stabilization in a murine hippocampal cell line. J Biol Chem. 2005;280:3996–4003. doi: 10.1074/jbc.M409223200. [DOI] [PubMed] [Google Scholar]
- 2.Aminova LR. Siddiq A. Ratan RR. Antioxidants, HIF prolyl hydroxylase inhibitors or short interfering RNAs to BNIP3 or PUMA, can prevent prodeath effects of the transcriptional activator, HIF-1alpha, in a mouse hippocampal neuronal line. Antioxid Redox Signal. 2008;10:1989–1998. doi: 10.1089/ars.2008.2039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Andreassen OA. Dedeoglu A. Ferrante RJ. Jenkins BG. Ferrante KL. Thomas M. Friedlich A. Browne SE. Schilling G. Borchelt DR. Hersch SM. Ross CA. Beal MF. Creatine increase survival and delays motor symptoms in a transgenic animal model of Huntington's disease. Neurobiol Dis. 2001;8:479–491. doi: 10.1006/nbdi.2001.0406. [DOI] [PubMed] [Google Scholar]
- 4.Beal MF. Brouillet E. Jenkins BG. Ferrante RJ. Kowall NW. Miller JM. Storey E. Srivastava R. Rosen BR. Hyman BT. Neurochemical and histologic characterization of striatal excitotoxic lesions produced by the mitochondrial toxin 3-nitropropionic acid. J Neurosci. 1993;13:4181–4192. doi: 10.1523/JNEUROSCI.13-10-04181.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brouillet E. Jacquard C. Bizat N. Blum D. 3-Nitropropionic acid: A mitochondrial toxin to uncover physiopathological mechanisms underlying striatal degeneration in Huntington's disease. J Neurochem. 2005;95:1521–1540. doi: 10.1111/j.1471-4159.2005.03515.x. [DOI] [PubMed] [Google Scholar]
- 6.Browne SE. Mitochondria and Huntington's disease pathogenesis: Insight from genetic and chemical models. Ann NY Acad Sci. 2008;1147:358–382. doi: 10.1196/annals.1427.018. [DOI] [PubMed] [Google Scholar]
- 7.Cha JH. Transcriptional signatures in Huntington's disease. Prog Neurobiol. 2007;83:228–248. doi: 10.1016/j.pneurobio.2007.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cui L. Jeong H. Borovecki F. Parkhurst CN. Tanese N. Krainc D. Transcriptional repression of PGC-1alpha by mutant huntingtin leads to mitochondrial dysfunction and neurodegeneration. Cell. 2006;127:59–69. doi: 10.1016/j.cell.2006.09.015. [DOI] [PubMed] [Google Scholar]
- 9.Epstein AC. Gleadle JM. McNeill LA. Hewitson KS. O'Rourke J. Mole DR. Mukherji M. Metzen E. Wilson MI. Dhanda A. Tian YM. Masson N. Hamilton DL. Jaakkola P. Barstead R. Hodgkin J. Maxwell PH. Pugh CW. Schofield CJ. Ratcliffe PJ. C. elegans EGL-9 and mammalian homologs define a family of dioxygenases that regulate HIF by prolyl hydroxylation. Cell. 2001;107:43–54. doi: 10.1016/s0092-8674(01)00507-4. [DOI] [PubMed] [Google Scholar]
- 10.Gil JM. Rego AC. Mechanisms of neurodegeneration in Huntington's disease. Eur J Neurosci. 2008;27:2803–2820. doi: 10.1111/j.1460-9568.2008.06310.x. [DOI] [PubMed] [Google Scholar]
- 11.Hu CJ. Wang LY. Chodosh LA. Keith B. Simon MC. Differential roles of hypoxia-inducible factor 1alpha (HIF-1alpha) and HIF-2alpha in hypoxic gene regulation. Mol Cell Biol. 2003;23:9361–9374. doi: 10.1128/MCB.23.24.9361-9374.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ivan M. Kondo K. Yang H. Kim W. Valiando J. Ohh M. Salic A. Asara JM. Lane WS. Kaelin WG., Jr HIFalpha targeted for VHL-mediated destruction by proline hydroxylation: Implications for O2 sensing. Science. 2001;292:464–468. doi: 10.1126/science.1059817. [DOI] [PubMed] [Google Scholar]
- 13.Jaakkola P. Mole DR. Tian YM. Wilson MI. Gielbert J. Gaskell SJ. Kriegsheim A. Hebestreit HF. Mukherji M. Schofield CJ. Maxwell PH. Pugh CW. Ratcliffe PJ. Targeting of HIF-alpha to the von Hippel–Lindau ubiquitylation complex by O2-regulated prolyl hydroxylation. Science. 2001;292:468–472. doi: 10.1126/science.1059796. [DOI] [PubMed] [Google Scholar]
- 14.Kim JW. Tchernyshyov I. Semenza GL. Dang CV. HIF-1-mediated expression of pyruvate dehydrogenase kinase: A metabolic switch required for cellular adaptation to hypoxia. Cell Metab. 2006;3:177–185. doi: 10.1016/j.cmet.2006.02.002. [DOI] [PubMed] [Google Scholar]
- 15.Kuhn A. Goldstein DR. Hodges A. Strand AD. Sengstag T. Kooperberg C. Becanovic K. Pouladi MA. Sathasivam K. Cha JH. Hannan AJ. Hayden MR. Leavitt BR. Dunnett SB. Ferrante RJ. Albin R. Shelbourne P. Delorenzi M. Augood SJ. Faull RL. Olson JM. Bates GP. Jones L. Luthi-Carter R. Mutant huntingtin's effects on striatal gene expression in mice recapitulate changes observed in human Huntington's disease brain and do not differ with mutant huntingtin length or wild-type huntingtin dosage. Hum Mol Genet. 2007;16:1845–1861. doi: 10.1093/hmg/ddm133. [DOI] [PubMed] [Google Scholar]
- 16.Lee JM. Ivanova EV. Seong IS. Cashorali T. Kohane I. Gusella JF. MacDonald ME. Unbiased gene expression analysis implicates the huntingtin polyglutamine tract in extra-mitochondrial energy metabolism. PLoS Genet. 2007;3:e135. doi: 10.1371/journal.pgen.0030135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lin J. Wu PH. Tarr PT. Lindenberg KS. St-Pierre J. Zhang CY. Mootha VK. Jager S. Vianna CR. Reznick RM. Cui L. Manieri M. Donovan MX. Wu Z. Cooper MP. Fan MC. Rohas LM. Zavacki AM. Cinti S. Shulman GI. Lowell BB. Krainc D. Spiegelman BM. Defects in adaptive energy metabolism with CNS-linked hyperactivity in PGC-1alpha null mice. Cell. 2004;119:121–135. doi: 10.1016/j.cell.2004.09.013. [DOI] [PubMed] [Google Scholar]
- 18.Majamaa K. Gunzler V. Hanauske-Abel HM. Myllyla R. Kivirikko KI. Partial identity of the 2-oxoglutarate and ascorbate binding sites of prolyl 4-hydroxylase. J Biol Chem. 1986;261:7819–7823. [PubMed] [Google Scholar]
- 19.Matthews RT. Yang L. Jenkins BG. Ferrante RJ. Rosen BR. Kaddurah-Daouk R. Beal MF. Neuroprotective effects of creatine and cyclocreatine in animal models of Huntington's disease. J Neurosci. 1998;18:156–163. doi: 10.1523/JNEUROSCI.18-01-00156.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Milakovic T. Johnson GV. Mitochondrial respiration and ATP production are significantly impaired in striatal cells expressing mutant huntingtin. J Biol Chem. 2005;280:30773–30782. doi: 10.1074/jbc.M504749200. [DOI] [PubMed] [Google Scholar]
- 21.Munujos P. Coll-Canti J. Gonzalez-Sastre F. Gella FJ. Assay of succinate dehydrogenase activity by a colorimetric-continuous method using iodonitrotetrazolium chloride as electron acceptor. Anal Biochem. 1993;212:506–509. doi: 10.1006/abio.1993.1360. [DOI] [PubMed] [Google Scholar]
- 22.Murphy TH. Schnaar RL. Coyle JT. Immature cortical neurons are uniquely sensitive to glutamate toxicity by inhibition of cystine uptake. FASEB J. 1990;4:1624–1633. [PubMed] [Google Scholar]
- 23.Nardinocchi L. Puca R. Guidolin D. Belloni AS. Bossi G. Michiels C. Sacchi A. Onisto M. D'Orazi G. Transcriptional regulation of hypoxia-inducible factor 1alpha by HIPK2 suggests a novel mechanism to restrain tumor growth. Biochim Biophys Acta. 2009;1793:368–377. doi: 10.1016/j.bbamcr.2008.10.013. [DOI] [PubMed] [Google Scholar]
- 24.Nardinocchi L. Puca R. Sacchi A. D'Orazi G. Inhibition of HIF-1alpha activity by homeodomain-interacting protein kinase-2 correlates with sensitization of chemoresistant cells to undergo apoptosis. Mol Cancer. 2009;8:1. doi: 10.1186/1476-4598-8-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nguyen T. Hamby A. Massa SM. Clioquinol down-regulates mutant huntingtin expression in vitro and mitigates pathology in a Huntington's disease mouse model. Proc Natl Acad Sci USA. 2005;102:11840–11845. doi: 10.1073/pnas.0502177102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ratan RR. Siddiq A. Aminova L. Langley B. McConoughey S. Karpisheva K. Lee HH. Carmichael T. Kornblum H. Coppola G. Geschwind DH. Hoke A. Smirnova N. Rink C. Roy S. Sen C. Beattie MS. Hart RP. Grumet M. Sun D. Freeman RS. Semenza GL. Gazaryan I. Small molecule activation of adaptive gene expression: Tilorone or its analogs are novel potent activators of hypoxia inducible factor-1 that provide prophylaxis against stroke and spinal cord injury. Ann NY Acad Sci. 2008;1147:383–394. doi: 10.1196/annals.1427.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ratcliffe PJ. HIF-1 and HIF-2: Working alone or together in hypoxia? J Clin Invest. 2007;117:862–865. doi: 10.1172/JCI31750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ruan Q. Lesort M. MacDonald ME. Johnson GV. Striatal cells from mutant huntingtin knock-in mice are selectively vulnerable to mitochondrial complex II inhibitor-induced cell death through a non-apoptotic pathway. Hum Mol Genet. 2004;13:669–681. doi: 10.1093/hmg/ddh082. [DOI] [PubMed] [Google Scholar]
- 29.Selak MA. Armour SM. MacKenzie ED. Boulahbel H. Watson DG. Mansfield KD. Pan Y. Simon MC. Thompson CB. Gottlieb E. Succinate links TCA cycle dysfunction to oncogenesis by inhibiting HIF-alpha prolyl hydroxylase. Cancer Cell. 2005;7:77–85. doi: 10.1016/j.ccr.2004.11.022. [DOI] [PubMed] [Google Scholar]
- 30.Semenza GL. Oxygen-dependent regulation of mitochondrial respiration by hypoxia-inducible factor 1. Biochem J. 2007;405:1–9. doi: 10.1042/BJ20070389. [DOI] [PubMed] [Google Scholar]
- 31.Siddiq A. Ayoub IA. Chavez JC. Aminova L. Shah S. LaManna JC. Patton SM. Connor JR. Cherny RA. Volitakis I. Bush AI. Langsetmo I. Seeley T. Gunzler V. Ratan RR. Hypoxia-inducible factor prolyl 4-hydroxylase inhibition. A target for neuroprotection in the central nervous system. J Biol Chem. 2005;280:41732–41743. doi: 10.1074/jbc.M504963200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.St-Pierre J. Drori S. Uldry M. Silvaggi JM. Rhee J. Jager S. Handschin C. Zheng K. Lin J. Yang W. Simon DK. Bachoo R. Spiegelman BM. Suppression of reactive oxygen species and neurodegeneration by the PGC-1 transcriptional coactivators. Cell. 2006;127:397–408. doi: 10.1016/j.cell.2006.09.024. [DOI] [PubMed] [Google Scholar]
- 33.Trettel F. Rigamonti D. Hilditch-Maguire P. Wheeler VC. Sharp AH. Persichetti F. Cattaneo E. MacDonald ME. Dominant phenotypes produced by the HD mutation in STHdh(Q111) striatal cells. Hum Mol Genet. 2000;9:2799–2809. doi: 10.1093/hmg/9.19.2799. [DOI] [PubMed] [Google Scholar]
- 34.Wanner RM. Spielmann P. Stroka DM. Camenisch G. Camenisch I. Scheid A. Houck DR. Bauer C. Gassmann M. Wenger RH. Epolones induce erythropoietin expression via hypoxia-inducible factor-1 alpha activation. Blood. 2000;96:1558–1565. [PubMed] [Google Scholar]
- 35.Weydt P. Pineda VV. Torrence AE. Libby RT. Satterfield TF. Lazarowski ER. Gilbert ML. Morton GJ. Bammler TK. Strand AD. Cui L. Beyer RP. Easley CN. Smith AC. Krainc D. Luquet S. Sweet IR. Schwartz MW. La Spada AR. Thermoregulatory and metabolic defects in Huntington's disease transgenic mice implicate PGC-1alpha in Huntington's disease neurodegeneration. Cell Metab. 2006;4:349–362. doi: 10.1016/j.cmet.2006.10.004. [DOI] [PubMed] [Google Scholar]
- 36.Willam C. Maxwell PH. Nichols L. Lygate C. Tian YM. Bernhardt W. Wiesener M. Ratcliffe PJ. Eckardt KU. Pugh CW. HIF prolyl hydroxylases in the rat; Organ distribution and changes in expression following hypoxia and coronary artery ligation. J Mol Cell Cardiol. 2006;41:68–77. doi: 10.1016/j.yjmcc.2006.04.009. [DOI] [PubMed] [Google Scholar]
- 37.Yang YT. Ju TC. Yang DI. Induction of hypoxia inducible factor-1 attenuates metabolic insults induced by 3-nitropropionic acid in rat C6 glioma cells. J Neurochem. 2005;93:513–525. doi: 10.1111/j.1471-4159.2005.03032.x. [DOI] [PubMed] [Google Scholar]
- 38.Zaman K. Ryu H. Hall D. O'Donovan K. Lin KI. Miller MP. Marquis JC. Baraban JM. Semenza GL. Ratan RR. Protection from oxidative stress-induced apoptosis in cortical neuronal cultures by iron chelators is associated with enhanced DNA binding of hypoxia-inducible factor-1 and ATF-1/CREB and increased expression of glycolytic enzymes, p21(waf1/cip1), and erythropoietin. J Neurosci. 1999;19:9821–9830. doi: 10.1523/JNEUROSCI.19-22-09821.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.