Abstract
We have demonstrated that zinc exposure induces apoptosis in human prostate cancer cells (PC-3) and benign hyperplasia cells (BPH), but not in normal prostate cells (HPR-1). However, the mechanisms underlying the effects of zinc on prostate cancer cell growth and zinc homeostasis remain unclear.
To explore the zinc effect on gene expression profiles in normal (HPR-1) and malignant prostate cells (PC-3), we conducted a time course study of Zn treatment with microarray analysis. Microarray data were evaluated and profiled using computational approach for the primary and secondary data analyses. Final analyses were focused on the genes: 1. highly sensitive to zinc, 2. associated with zinc homeostasis, i.e. metallothioneins (MTs), solute zinc carriers (ZIPs) and zinc exporters (ZnTs), 3. relevant to several oncogenic pathways. Zinc-mediated mRNA levels of MT isotypes were further validated by semi-quantitative RT-PCR.
Results showed that zinc effect on genome-wide expression patterns was cell type specific, and zinc appeared to have mainly down-regulatory effects on thousands of genes (1,953 in HPR-1; 3,534 in PC-3) with a threshold of ±2.5-fold, while fewer genes were up-regulated (872 in HPR-1; 571 in PC-3). The patterns of zinc effect on functional MT genes’ expression provided evidence for the cell-type dependent zinc accumulation and zinc-induced apoptosis in prostate cells. In PC-3 cells, zinc significantly up-regulated the expression of MT-1 isotypes -J and -M, denoted previously as “non-functional” MT genes, and now a depictive molecular structure of MT-1J was proposed. Examination of genes involved in oncogenic pathways indicated that certain genes, e.g. Fos, Akt1, Jak3 and PI3K were highly regulated by zinc with cell type specificity.
This work provided an extensive database on zinc related prostate cancer research. The strategy of data analysis was devoted to find genes highly sensitive to Zn, and the genes associated with zinc accumulation and zinc-induced apoptosis. The results indicate that zinc regulation of gene expression is cell-type specific, and MT genes play important roles in prostate malignancy.
Keywords: microarray, zinc, prostate malignant cells, gene isoform, metallothionein, zinc transporter
Introduction
Zinc, as a structural, catalytic, and regulatory ion, is essential for the proper function of numerous enzymes and proteins in all cells [1]. This trace element plays a role in a wide variety of physiological and pathological processes involved in cell proliferation, differentiation and oncogenesis, in prevention of oxidative stress, and in immune status and aging [2,3]. In the human prostate, the high level of zinc, up to 3- to 7-fold higher than that in other organs, has been well documented [4]. On the other hand, the loss of zinc accumulation, 60–70% lower than that of normal prostate epithelial cells, is the most consistent characteristic of prostate malignancy [4]. Previous studies have demonstrated that accumulation of a high level of zinc in the prostate is associated with two major functions: citrate production and normal growth of prostate tissue [4]. However, to date, whether the malignancy causes the disruption of zinc accumulation or vice versa remains unknown. It has been demonstrated that zinc treatment at physiologic levels (15 μM) inhibits the growth of prostatic cancer cells via cell cycle arrest and induction of cell apoptosis both in vitro and in vivo [5–7]. Also, zinc-induced apoptosis in human prostate malignant PC-3 cells occurs via mitochondrial pathways through Bax-involved pore formation in mitochondrial membranes [5,8–10]. In contrast, no apoptotic effect of zinc was detected in the normal human prostate cell line HPR-1 [8]. However, the genes involved in the pathways of zinc-induced prostate cancer cell apoptosis have yet to be revealed. At the present time, there is a lack of information regarding the molecular mechanisms for: a) the lost capacity to accumulate zinc, and b) zinc-induced apoptosis in malignant prostate cells, which represent the major challenges for future study.
Regulation of zinc accumulation in cells involves several central components, e.g. metallothionein (MT) and zinc transporters (ZIP and ZnT), functionally associated with cellular zinc storage, uptake and efflux, respectively. MT is a family of low molecular weight (6–7 Kd), cysteine-rich, metal-binding proteins [11–13]. In humans, four MT isoforms ( MT-1, 2, 3 and 4) and 17 isotypes (13 isotypes for MT-1; 2 isotypes for MT-2 and 1 isotype each for MT-3 and 4) have been identified [14]. Among these isotypes, the biological functions of MT-1 C, D, I, J, K, L and MT-2B have not been identified and have been currently named as pseudo-genes. MTs, as receptor/donors of cellular zinc, are considered to be the major regulators for zinc accumulation, distribution and bioactivity. MTs are also involved in the homeostasis of other metals, including cadmium and copper, and protect cells against damage induced by oxidative stress and detoxify harmful metals [2,15]. In addition to MTs, zinc transporters, which all have transmembrane domains encoded by two solute-linked carrier gene families, ZIP (SLC39) and ZnT (SLC30), are also known to be major factors associated with cellular zinc homeostasis in many different types of cells [16,17]. The ZIP family that imports zinc is composed of 15 members, and the ZnT family that functions in releasing zinc or sequestering zinc internally is comprised of 10 integral membrane proteins. With opposite roles, they provide high-affinity systems for a dynamic maintenance of cellular zinc homeostasis. Previously, researchers determined zinc regulation of MT and zinc transporter expression in human prostate cancer cells and tissues [18–20]. The suppressed endogenous levels of MT-1/MT-2 were detected in prostate cancer cell lines of PC-3, DU145 and LNCaP [21]. An inhibitory effect of zinc on ZIP1 expression in prostate cancer cells was demonstrated [22]. However, due to the complexity of isoforms in the MT, ZIP and ZnT families, information about the differential effect of zinc on the expression of these genes in normal and malignant prostate was limited.
Advances in microarray technology, which provides a broad view of gene expression, have allowed us to discriminate possible biomarkers linked specifically to zinc accumulation and zinc-induced apoptosis in prostate cells. In this study, we employed microarray analysis and semi-quantitative RT-PCR to display the expression patterns of genes highly affected by zinc in human prostatic normal, hyperplastic and malignant cells. The results may lead to future elucidation of zinc-responsive genes in activation of the pathways involved in prostate malignancy.
Methods and Materials
Human Prostate Cell Lines and Zinc Treatment
The human prostatic cell line, HPR-1, derived from normal prostate epithelial cells (provided by Dr. C.K. Choo, University of Hong Kong), was cultured in Keratinocyte serum-free medium (Invitrogen) supplemented with EGF 0.005 mg/ml, bovine pituitary extract 0.05 mg/ml and 1U/ml penicillin and streptomycin sulfate (Invitrogen). The human benign prostatic hyperplasia cell line (BPH) (provided by Dr. S. Haywood, UCSF), human prostate malignant androgen-dependent LNCaP and androgen-independent PC-3 cell lines (ATCC, Rockville, MD) were maintained in RPMI 1640 medium (GIBCO, BRL) with 10% fetal bovine serum (5% for BPH) and 1U/ml penicillin-streptomycin. Cells were maintained in a humidified incubator with 5% CO2 atmosphere at 37°C. Time course studies of zinc effect were initiated with the cells at 70–80% confluence and the cells were synchronized by hormone/serum-depletion for 24 hrs before zinc treatment. Zinc (20 μM) was added with fresh hormone/serum-free media, and the cells were then collected at 1, 3, and 6 hours, respectively. Cells without zinc treatment were collected as the control. Cell pellets were immediately frozen and stored at −80 °C for RNA preparation. Total RNA was extracted using Qiagen (Valencia, CA) RNeasy Mini kit according to the manufacturer’s protocol. The quality of RNA was monitored by 28S/18S ribosomal RNA ratios and 260/280-nm absorbance ratios. The time course studies were conducted in each aforementioned cell line and repeated three times, respectively. RNA samples prepared from these bioassays were used for cDNA microarray and semi-quantitative RT-PCR analyses.
cDNA Microarray Expression Analysis
Zinc effects on gene expression of HPR-1 and PC-3 cells were evaluated using Applied Biosystems (Foster City, CA) human genome-wide microarray chips V2.0. The ABI human genome survey microarray chip contained 32,878 of 60-mer oligonucleotide probes for interrogation of 29,098 known genes. For each chip, 2 μg total RNA from each sample was used for generation of cRNA. The synthesis of cRNA and a subsequent hybridization were completed by the Core Facility at UMBI (University of Maryland Biotechnology Institute, Baltimore, MD) according to the ABI standard protocol. For each cell line, four groups of samples were employed including a control group and groups treated with zinc for 1 hr, 3 hrs, or 6 hrs. The management of microarray images, algorithm raw data, and primary data analysis including quantification and normalization was completed with the Applied Biosystems 1700 Chemiluminescent Microarray Analyzer and 1700 Gene Expression Microarray System. The primary data analysis is performed using the software provided with the Applied Biosystems Expression Array System. For secondary analysis, Spotfire (Somerville, MA) was used to export microarray data in a flat-file format and to determine the differentially expressed genes. Each array is normalized by the median chemiluminescent signal. The fold change of zinc treated sample vs. the control sample was analyzed by filtering the dataset using a threshold of ±2.5-fold and a signal-to-noise (background) ratio > 3 and without flags for determining a meaningful alteration. Final profile data management including sorting data and data mining procedures was conducted using Microsoft Office Excel (Redmond, WA). The experimental procedures and data are available at http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE5590 according to the Minimum Information About a Microarray Experiment standards.
Semi-Quantitative RT-PCR for MT-Isotypes
Semi-quantitative RT-PCR analyses were conducted to validate the alterations of gene expression induced by zinc obtained from microarray and also to further investigate zinc effects on the mRNA level of MT isotypes. cDNA was synthesized using 2 μg total RNA with reverse transcriptase (ABI, Foster City, CA) for a total 20 μl reaction following the manufacturer’s protocol. The sequences of primers for GAPDH and MT isotypes including MT-1A, MT-1E, MT-1F, MT-1G, MT-1J, MT-1K, MT-1X, MT-2A are listed in Table 6. A cDNA template (0.6 μl) was used for a 25 μl reaction mixed with 0.5 μl of 10 mM dNTP, 1 μl of 50mM MgCl2, 2.5 μl 10X Taq reaction buffer, and 0.1 μl of 5U platinum Taq (Invitrogen). Thirty (25 for MT-1J) cycles at 94°C for 30 sec, 55 °C (62 °C for MT-1J, 50 °C for MT-1M) for 30 sec and 72 °C for 30 sec were performed using a Perkin Elmer thermocycler (GeneAmp PCR System 9600, Shelton, CT). GAPDH (Glyceraldehydes-3-phosphate dehydrogenase) used as a control to normalize the MT transcript level was subjected to the same thermal cycle conditions as the MT isotypes except with an annealing temperature of 55 °C. PCR products were analyzed in 1.5% agarose gels, stained with ethidium bromide, and visualized by UV fluorescence. Densities of bands were scanned and quantitated with an LKB Ultra Scan XL laser densitometer (Image Quant, Molecular Dynamics, Sunnyvale, CA). The ratio of each target to its corresponding GAPDH control represents the normalized transcript abundance. The ratio of transcript abundance in zinc-treated samples vs. the corresponding untreated control represents the zinc effect on the transcripts of MT isotypes, which were statistically analyzed.
Table 6.
The sequences of primers for MT-isotypes
| Gene Name | Forward Primer Sequences | Reverse Primer Sequences | Expected Size (bp) |
|---|---|---|---|
| GAPDH | CCACCCATGGCAAATTCCATGGCA | TCTAGACGGCAGGTCAGGTCCACC | 598 |
| MT-1A | CTCGAAATGGACCCCAACT | ATATCTTCGAGCAGGGCTGTC | 219 |
| MT-1E | GCTTGTTCGTCTCACTGGTG | CAGGTTGTGCAGGTTGTCTA | 284 |
| MT-1F | AGTCTCTCCTCGGCTTGC | ACATCTGGGAGAAAGGTTGTC | 232 |
| MT-1G | CTTCTCGCTTGGGAACTCTA | AGGGGTCAAGATTGTAGCAAA | 303 |
| MT-1J | ACTGGTGGCTCCTGCACGTGCGCC | CCCACATCAGGCACAGCAG | 168 |
| MT-1M* | CTAGCAGTCGCTCCATTTATCG | CAGCTGCAGTTCTCCAACGT | 228 |
| MT-1X | TCTCCTTGCCTCGAAATGGAC | GGGCACACTTGGCACAGC | 151 |
| MT-2A | CCATGGATCCCAACTGCTC | TGGAAGTCGCGTTCTTTACA | 232 |
Source of MT-1M primer sequences: [42]
Statistical Analysis
Differences among subject groups were assessed by analysis of variance (ANOVA) and mean separations by least significant difference (LSD) analyses (SAS Institute, Cary, NC) performed using the Mixed procedure of the SAS 9.1 program package for semi-quantitative RT-PCR analysis. A 95% confidence interval (P ≤0.05) was considered significant. Each time point of zinc treatment was considered as an independent variable.
Results
Zinc Regulation of Gene Expression in HPR-1 & PC-3 Cells Determined by Microarray Analysis
A total of 6,959 genes were identified to exhibit altered expression patterns with at least a 2.5-fold change in response to zinc (Table 1). Among these genes, only 597 were common to both cell lines, including 538 and 59 genes that were consistently down- or up-regulated by zinc, respectively. From the differentially expressed genes induced by zinc, we selected 20 genes in each cell line with the most pronounced alterations in response to zinc, i.e. the top 10 list for up-regulated and down-regulated genes, which are presented in Table 2 (HPR-1) and Table 3 (PC-3). The genes associated with protein phosphorylation, cell differentiation, transcription factor and signal transduction, such as MAP kinases, zinc finger protein 223, TNF receptor-associated factor 6, and Bcl-2like 11, were markedly induced by zinc in HPR-1, but not in PC-3 cells (Table 2). In contrast, the expression of candidate genes for cell growth and apoptosis, such as heat shock 70 kDa protein 6 (HSP70B), heat shock 70 kDa protein 1-like, and Fos, was dramatically increased by zinc as much as 4- to 20-fold in PC-3 cells. Yet no significant zinc effect on these genes was observed in HPR-1 cells. MGC17301, encoding for methyltransferase, and several candidate genes for transcriptional DNA binding proteins were down-regulated in PC-3 cells (Table 3).
Table 1.
Summary of the number of genes differentially regulated by zinc detected by microarray
| Expression Pattern | Prostate cell lines |
||
|---|---|---|---|
| Normal (HPR-1) | Malignant (PC-3) | HPR-1 & PC-3 | |
| Increased | 872 | 571 | 59 |
| Decreased | 1,953 | 3,534 | 538 |
| Varied in Time Course | 13 | 16 | 0 |
| Total number | 2,838 | 4,121 | 597 |
Table 2.
Genes with the most zinc-altered expression levels in HPR-1 cells
| GenBank ID | Gene Name | Ratio of Zinc Treated vs Control in Gene Expression Level |
Major Biological Function | |||||
|---|---|---|---|---|---|---|---|---|
| HPR-1 |
PC-3 |
|||||||
| 1hr | 3hr | 6hr | 1hr | 3hr | 6hr | |||
|
Increased gene expression | ||||||||
| NM_006301 | Mitogen-activated protein kinase kinase kinase 12 (MAP3K12) | 12.14 | 8.56 | 2.62 | 1.09 | 1.30 | −1.51 | Protein phosphorylation; Embryogenesis |
| NM_148172 | Phosphatidylethanolamine N-methyltransferase (PEMT) | 9.45 | 4.25 | 5.00 | −1.29 | 1.12 | 1.22 | Phosphatidylcholine biosynthesis; lipid metabolism |
| NM_173621 | Chromosome 17 open reading frame 44 (C17orf44) | 8.42 | 2.62 | 1.54 | -* | -* | -* | Unclassified |
| NM_013361 | Zinc finger protein 223 (ZNF223) | 8.14 | 5.18 | 1.32 | -1.96 | -5.00 | -2.25 | Transcription factor; KRAB box transcription factor |
| NM_013313 | Yippee-like 1 (Drosophila) (YPEL1) | 6.39 | 5.18 | 5.42 | 1.76 | −1.03 | −4.17 | Regulation of cellular morphology |
| NM_138334 | Josephin domain containing 2 (JOSD2) | 4.37 | 5.92 | 2.79 | −1.20 | 1.59 | 1.11 | Unclassified |
| NM_032520 | N-acetylglucosamine-1-phosphate transferase, gamma subunit (GNPTG) | 3.92 | 6.93 | 3.26 | 1.51 | 1.17 | 1.05 | Transferase |
| NM_145803 | TNF receptor-associated factor 6 (TRAF6) | 3.83 | 5.29 | 4.60 | −2.37 | −1.59 | 1.09 | Signal transduction |
| NM_006538 | BCL2-like 11 (apoptosis facilitator) (BCL2L11) | 2.88 | 5.01 | 4.49 | 1.54 | 1.58 | 2.00 | Oncogenesis; Induction of apoptosis |
| NM_000076 | CDK inhibitor 1C (p57, Kip2) (CDKN1C) | 1.99 | 3.13 | 6.18 | 2.55 | 1.61 | 1.77 | Kinase modulator; Select regulatory molecule |
|
Decreased gene expression | ||||||||
| NM_152558 | IQ motif containing E (IQCE) | −12.5 | −4.55 | −3.03 | 1.03 | −1.42 | −1.85 | Unclassified |
| NM_145288 | Zinc finger protein 342 (ZNF342) | −8.33 | −2.70 | −3.45 | −1.22 | −2.88 | −2.34 | mRNA transcription regulation; Oncogenesis |
| NM_001452 | Forkhead box F2 (FOXF2) | −6.67 | −3.85 | −8.33 | -* | −1.13 | 2.82 | Developmental processes; Nucleic acid metabolism; mRNA transcription and its regulation |
| NM_172239 | RNA exonuclease 1 homolog (S. cerevisiae)-like 2 (REXO1L2) | −6.07 | −4.63 | −13.78 | −3.38 | −4.40 | −3.71 | Nucleic acid binding; Oxidoreductase |
| NM_152540 | Sec1 family domain containing 2 (SCFD2) | −5.84 | −7.93 | −4.42 | 1.10 | −1.71 | 12.14 | Molecular function unclassified |
| NM_002548 | Olfactory receptor, family 1, subfamily D, member 2 (OR1D2) | −5.77 | −5.26 | −8.36 | −1.99 | −2.42 | −7.27 | Chemosensory perception; Signal transduction |
| NM_002429 | Matrix metallopeptidase 19 (MMP19) | −5.54 | −24.01 | −6.47 | −1.09 | −1.64 | −1.08 | Extracellular matrix; Protease |
| XM_211291 | Chromosome 17 open reading frame 52 (C17orf52) | −5.03 | −13.16 | −9.11 | −1.76 | −1.10 | −1.28 | Unclassified |
| NM_173590 | Chromosome 11 open reading frame 36 (C11orf36) | −4.08 | −6.43 | −5.91 | −1.11 | −1.20 | −1.92 | Unclassified |
| NM_019013 | Family with sequence similarity 64, member A (FAM64A) | −2.75 | −5.38 | −20.98 | −1.91 | −1.14 | −2.60 | Unclassified |
NOTE: The genes were listed in a descending order according to the ratio of 1-hour zinc induced expression vs. the control in HPR-1 cells.
Corresponding results in PC-3 cells were listed aside for the comparison.
Signal intensities were flagged as unavailable.
Table 3.
Genes with the most zinc-altered expression levels in PC-3 cells
| GenBank ID | Gene Name | Ratio of Zinc Treated vs Control in Gene Expression Level |
Major Biological Function | |||||
|---|---|---|---|---|---|---|---|---|
| PC-3 |
HPR-1 |
|||||||
| 1hr | 3hr | 6hr | 1hr | 3hr | 6hr | |||
|
Increased gene expression | ||||||||
| NM_152857 | Wilms tumor 1 associated protein (WTAP) | 37.14 | 29.41 | 31.21 | −1.29 | −1.05 | −1.14 | Transcriptional and posttranscriptional regulationof certain cellular genes |
| NM_002155 | Heat shock 70kDa protein 6 (HSP70B) | 6.18 | 11.92 | 14.50 | 1.10 | −1.19 | −1.06 | Ubiquitous protein; Chaperon |
| NM_175622 | Metallothionein 1J (MT1J) | 4.44 | 5.74 | 4.21 | -* | -* | -* | Unclassified |
| NM_145276 | Zinc finger protein 563 (ZNF563) | 3.72 | 6.35 | 4.90 | -* | -* | -* | Transcription factor; KRAB box transcription factor |
| NM_005252 | V-fos FBJ murine osteosarcoma viral oncogene homolog (FOS) | 3.10 | 19.70 | 13.59 | 2.06 | −1.08 | −1.35 | Nucleic acid binding; Transcription factor |
| NM_005527 | Heat shock 70kDa protein 1-like (HSPA1L) | 1.18 | 6.15 | 5.33 | -* | -* | -* | Hsp 70 family protein; Chaperon |
| NM_172365 | Chromosome 14 open reading frame 50 (C14orf50) | −1.13 | 3.42 | 11.17 | 1.12 | 1.08 | −2.65 | Unclassified |
| NM_018602 | DnaJ (Hsp40) homolog, subfamily A, member 4 (DNAJA4) | 0.74 | 9.56 | 18.51 | 3.96 | 1.42 | −1.14 | Chaperone |
| NM_001885 | Crystallin, alpha B (CRYAB) | 0.74 | 7.30 | 9.19 | 1.64 | −1.18 | 1.05 | Structural protein; Miscellaneous function |
| NM_032961 | Protocadherin 10 (PCDH10) | 0.70 | 4.67 | 14.05 | −1.02 | 1.06 | −1.72 | Cadherin; Cell adhesion molecule |
|
Decreased gene expression | ||||||||
| NM_014212 | Homeobox C11 (HOXC11) | −14.25 | −9.38 | −7.25 | -* | -* | -* | DNA-binding protein; Homeobox transcription factor |
| NM_152637 | MGC17301 (MGC17301) | −10.73 | −3.78 | −4.44 | -* | 2.18 | 1.53 | Methyltransferase |
| NM_023070 | Zinc finger protein 643 (ZNF643) | −10.19 | −6.32 | −5.16 | −1.09 | −1.08 | −1.73 | Transcription factor; KRAB box transcription factor |
| NM_014562 | Orthodenticle homolog 1 (Drosophila) (OTX1) | −9.18 | −8.59 | −3.30 | −1.93 | −1.73 | −1.98 | Nucleic acid –binding protein; Transcription factor |
| NM_032208 | Anthrax toxin receptor 1 (ANTXR) | −8.43 | −5.22 | −3.71 | −1.19 | −1.19 | 1.05 | Receptor |
| NM_032563 | Late cornified envelope 3D (LCE3D) | −6.62 | −6.84 | −3.66 | −1.21 | 1.20 | −1.25 | Structural protein; Miscellaneous function |
| NM_002498 | NIMA-related kinase 3 (NEK3) | −6.44 | −5.30 | −4.74 | 1.90 | 1.24 | −1.07 | Protein kinase; Transferase |
| NM_001818 | Aldo-keto reductase family 1, member C4 (AKR1C4) | −5.94 | −6.20 | −4.41 | −2.06 | −1.75 | −5.70 | Oxidoreductase |
| NM_015396 | Armadillo repeat containing 8 (ARMC8) | −4.53 | −3.54 | −11.53 | −1.84 | 1.13 | −1.40 | Unclassified |
| NM_005693 | Nuclear receptor subfamily 1, group H, member 3 (NR1H3) | −4.30 | −3.58 | −10.83 | 2.43 | 1.31 | 1.25 | Unclassified |
NOTE: The genes were listed in a descending order according to the ratio of 1-hour zinc-induced expression vs. the control in PC-3 cells.
Corresponding results in HPR-1 cells were listed aside for the comparison.
Signal intensities were flagged as unavailable.
Zinc Regulation of Its Homeostasis Related Genes in HPR-1 and PC-3 Cells
To further understand the effect of zinc on genes associated with cellular zinc accumulation in normal and malignant prostate cells, the results of the expression of MTs and zinc transport genes (ZIPs and ZnT) are summarized in Table 4. Our microarray data showed a dramatic zinc induction of the expression of MT-1J and 1M genes in PC-3 and HPR-1 cells, respectively. In PC-3 cells, the increase in transcriptional levels of the MT-1J gene induced by zinc was approximately 4- to 5.7-fold, and was listed as one of the top three among all zinc up-regulated genes during the time course study (Table 3). Moreover, the zinc up-regulation of the MT-3 gene, recognized as a growth inhibitory factor [23], was also detected with a 2- to 5.6-fold increase in PC-3 cells. However, the level of MT-1J transcripts could not be detected validly by microarray analysis in HPR-1 cells, but was determined by RT-PCR as presented in Figure 1. The up-regulation of MT-1M transcripts with a greater than 2-fold increase was observed in both HPR-1 and PC-3 cells, and a remarkable enhancement of MT-1 M (13-fold) after longer durations of zinc exposure (6hrs) was found in HPR-1 cells. Transcriptional levels of other MT isotypes revealed unremarkable effects of zinc in both cell lines (Table 4).
Table 4.
Expression patterns for MT-isotypes and zinc transporters in response to zinc
| GenBank ID | Gene Name | Ratio of Zinc Treated vs Control in Gene Expression Level |
|||||
|---|---|---|---|---|---|---|---|
| HPR-1 |
PC-3 |
||||||
| 1hr | 3hr | 6hr | 1hr | 3hr | 6hr | ||
| BC029475 | Metallothionein 1A (MT1A) | 1.11 | −1.11 | 1.05 | −1.31 | −1.45 | −1.14 |
| NM_005947 | Metallothionein 1B (MT1B) | −1.25 | −1.03 | −1.40 | −1.08 | −1.01 | 1.13 |
| NM_175617 | Metallothionein 1E (MT1E) | −1.01 | −1.21 | −1.65 | −1.07 | 1.07 | 1.03 |
| NM_005949 | Metallothionein 1F (MT1F) | −1.34 | −1.40 | −1.24 | −1.14 | −1.12 | 1.08 |
| NM_005950 | Metallothionein 1G (MT1G) | 1.18 | −1.03 | 1.04 | −1.42 | −1.23 | −1.41 |
| NM_175622 | Metallothionein 1J (MT1J) | -* | -* | -* | 4.44 | 5.74 | 4.21 |
| BC070351 | Metallothionein 1L (MT1L) | −1.07 | −1.31 | −1.31 | −1.79 | −1.10 | −1.85 |
| BF973961 | Metallothionein 1M (MT1M) | 1.20 | 2.26 | 13.35 | −1.97 | 2.43 | 2.21 |
| NM_005952 | metallothionein 1X (MT1X) | 1.08 | −1.01 | -* | −1.06 | −1.37 | -* |
| NM_005954 | Metallothionein 3 (growth inhibitory factor (MT3) | -* | -* | -* | -2.26 | 2.35 | 5.62 |
| NM_032935 | Metallothionein IV (MT4) | 1.01 | −1.51 | 1.11 | 1.11 | 1.22 | 1.24 |
| BG181336 | MTM | 2.11 | 2.21 | 1.07 | 1.31 | 3.38 | 1.15 |
| NM_005955 | metal-regulatory transcription factor 1 (MTF1) | 1.01 | −1.09 | 1.03 | −2.57 | −1.82 | −1.24 |
| NM_021194 | zinc transporter, SLC30A1 (ZnT-1) | −1.07 | 1.15 | 1.27 | −1.11 | 1.16 | −1.07 |
| NM_003459 | zinc transporter, SLC30A3 (ZnT-3) | 1.10 | −2.78 | −1.73 | −2.30 | −1.97 | −2.35 |
| NM_022902 | zinc transporter, SLC30A5 (ZnT-5) | −1.19 | −1.21 | −1.05 | −1.21 | −1.52 | −1.65 |
| NM_024055 | zinc transporter, SLC30A5(ZnT-5) | 1.05 | 1.11 | 1.27 | −1.20 | −1.03 | −1.13 |
| NM_017964 | zinc transporter, SLC30A6(ZnT-6) | −1.94 | −1.32 | −1.76 | 1.21 | −1.08 | −2.03 |
| NM_133496 | zinc transporter, SLC30A7(ZnT-7) | −2.90 | −1.39 | −1.59 | −2.30 | −1.10 | −2.55 |
| NM_006345 | zinc transporter, SLC30A9 (ZnT-9) | −1.89 | −1.64 | −1.59 | −1.87 | −1.29 | −2.10 |
| NM_014437 | zinc transporter, SLC39A1 (ZIP1) | −1.14 | −1.17 | −1.40 | −1.07 | −1.12 | −1.08 |
| NM_014579 | zinc transporter, SLC39A2 (ZIP2) | -* | -* | -* | -* | -* | -* |
| NM_144564 | zinc transporter, SLC39A3 (ZIP3) | -* | -* | -* | -* | -* | -* |
| NM_130849 | zinc transporter, SLC39A4 (ZIP4) | −1.04 | 1.04 | −1.30 | −1.03 | −1.10 | −1.11 |
| NM_173596 | zinc transporter, SLC39A5 (ZIP5) | −1.22 | −2.53 | −2.05 | −2.22 | −1.43 | −3.57 |
| NM_012319 | zinc transporter, SLC39A6 (ZIP6) | −1.16 | −1.08 | −1.35 | 1.28 | 1.30 | 1.20 |
| NM_006979 | zinc transporter, SLC39A7 (ZIP7) | −1.59 | −2.35 | −1.51 | −1.68 | −1.29 | −2.65 |
| NM_022154 | zinc transporter, SLC39A8 (ZIP8) | −1.14 | −1.28 | −1.14 | −1.28 | −1.18 | −1.45 |
| NM_018375 | zinc transporter, SLC39A9 (ZIP9) | −3.10 | −2.31 | −3.41 | −1.81 | −1.26 | −1.12 |
| NM_020342 | zinc transporter, SLC39A10 (ZIP10) | −1.44 | 1.12 | −1.18 | −1.62 | −1.19 | −1.99 |
| NM_139177 | zinc transporter, SLC39A11 (ZIP11) | −1.21 | −1.06 | −1.16 | −1.06 | −1.14 | −1.67 |
| NM_152725 | zinc transporter, SLC39A12 (ZIP12) | -* | -* | -* | −3.04 | -* | −3.97 |
| NM_152264 | zinc transporter, SLC39A13 (ZIP13) | 1.12 | 1.28 | 1.30 | −1.15 | 1.13 | 1.14 |
| NM_015359 | zinc transporter, SLC39A14 (ZIP14) | -* | -* | -* | −4.09 | -* | −2.63 |
Signal intensities were flagged as unreliable.
FIGURE 1.
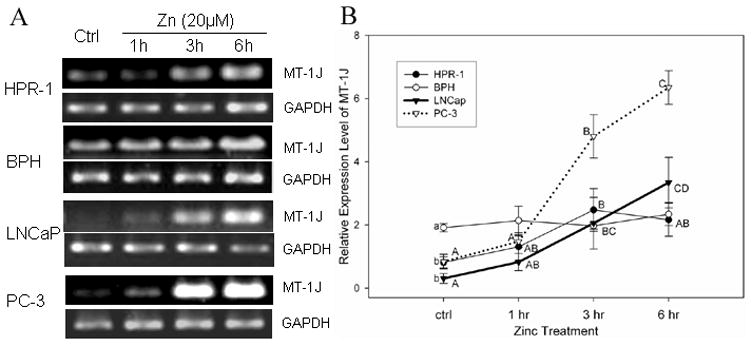
Time course study of zinc-induced MT-1J mRNA levels in prostate cells detected by semi-quantitative RT-PCR. Cells were incubated with or without zinc (20 μM) for different times, as indicated. A. RT- PCR products of MT-1J and GAPDH were analyzed by gel electrophoresis. B. MT-1J mRNA levels are expressed by the MT-1J relative optical densities normalized by related GAPDH signals. Each denoted point and bar represent the mean ± SE (n=3). A significant difference (p≤0.05, LSD) between samples from different time point in each cell line is indicated with different capitalized letters (A,B,C,D). A significant difference (p≤0.05, LSD) of the endogenous levels of MT-isotypes among four cell lines is denoted with different lowercase letters (a,b,c). The same illustration for the statistical significance analysis is used in Figure 2 and 3.
In addition to MTs, zinc transporters are known to play a pivotal role in maintenance of a dynamic homeostasis of cellular zinc. Correspondingly, we observed the inhibitory effect of zinc, in general, on the expression of the majority of zinc transporter genes (Table 4). The zinc down-regulation of ZnT expression with greater than 2-fold decrease, such as ZnT 3 and ZnT 7 genes was observed in both HPR-1 and PC-3 cells. Furthermore, zinc also appeared to inhibit most ZIP gene expression and zinc suppression with about a 3.5-fold reduction of ZIP5 gene expression observed in both cell lines. However, the inhibitory effect of zinc with an approximately 2 to 3.4-fold decrease in ZIP9 gene transcripts was only detected in HPR-1 cells.
Zinc Regulation of MT-1J and 1M Expression in HPR-1, BPH, LNCaP and PC-3 Cells Detected by Semi-Quantitative RT-PCR
Marked zinc-induced MT-1J gene expression observed in PC-3 cells via microarray analyses (Tables 3 and 4) was further validated using semi-quantitative RT-PCR with two additional human prostate cell lines, BPH and LNCaP cells (Fig. 1). In malignant PC-3 cells the significant zinc-induced MT-1J mRNA levels were detected at 3 hr and reached even higher at 6 hr by 5.7- and 7.6-fold increase compared to endogenous MT-1J mRNA level, respectively. A remarkable zinc induction of MT-1J transcripts was also observed in malignant LNCaP cells with about 6.6- and 10.8-fold increase at 3 hr and 6 hr, respectively. However, no zinc effect on MT-1J expression was observed either in BPH or HPR-1 cells. Our results indicate that zinc regulation of MT-1J gene expression is cell-type specific and time-dependent. Zinc appeared to be more effective on MT-1J gene transcription in malignant PC-3 and LNCaP cells than in normal and benign hyperplastic BPH cells.
In order to confirm the inducible effect of zinc on MT-1M gene expression obtained from microarray analysis (Table 4), and to further investigate whether the zinc regulation of MT-1M is cell-type specific, we conducted a time course study of zinc using RT-PCR with the primers targeting the MT-1M gene in four cell lines (Fig. 2). The results showed an extremely lower amount of endogenous MT-1M mRNA found in LNCaP and HPR-1 cells (p≤0.05). Except for LNCaP cells, a significant zinc induction of MT-1M gene expression was observed in the cell lines with 3- to 5-fold increases compared to their controls observed in the remaining three cell lines. Seemingly, the time course of MT-1M induction by zinc differs in the cells, being fast in PC-3 and delayed in HPR-1 and BPH cells. This early activation of gene transcription in PC-3 cells was not observed for MT-1J (Fig. 1).
FIGURE 2.
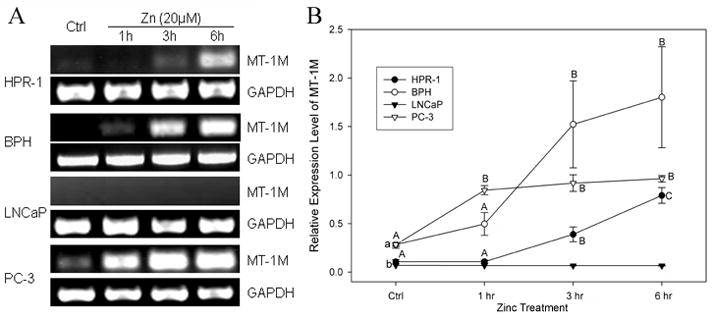
Zinc induced MT-1M mRNA levels in prostate cells detected by semi-quantitative RT-PCR. The experimental conditions are the same as in Figure 1. A. RT- PCR products of MT-1M and GAPDH were analyzed by gel electrophoresis. B. MT-1M mRNA levels are expressed as the MT-1J relative optical densities normalized by related GAPDH signals. The points and bars represent the mean ± SE (n=3).
Zinc Regulation of Functional MT-isotype expression in Prostate Cells
To further understand the roles of MTs in cellular zinc accumulation in prostate normal, BPH and malignant cells, we evaluated the zinc effect on functional MT gene expression, including five isotypes of MT-1 (1A, 1E, 1F, 1G, and 1X) and MT-2A by semi-quantitative RT-PCR with samples obtained from the time course studies (Fig. 3). Among these four cell lines, the significant higher endogenous mRNA levels for MT-1E and MT-2A were found in BPH and PC-3 cells, and higher endogenous MT-1G transcripts were detected in BPH and LNCaP cells. No difference of endogenous mRNA for MT-1X was detected among these cell lines. Within these functional MT-isotype genes, the endogenous mRNA levels for MT-1E, -1F, and -1G and their responses to zinc appeared to correspond to their cellular zinc levels reported previously [19]. The MT-isotype genes, which are highly sensitive to zinc induction, were determined in multiple cell lines with the probes targeting MT-1A and -1G (Fig. 3B). The distinct up-regulation of MT-1F and -1X gene expression was only recognized in PC-3 cells with a similar finding of MT-2A in LNCaP cells (Fig. 3B).
FIGURE 3.
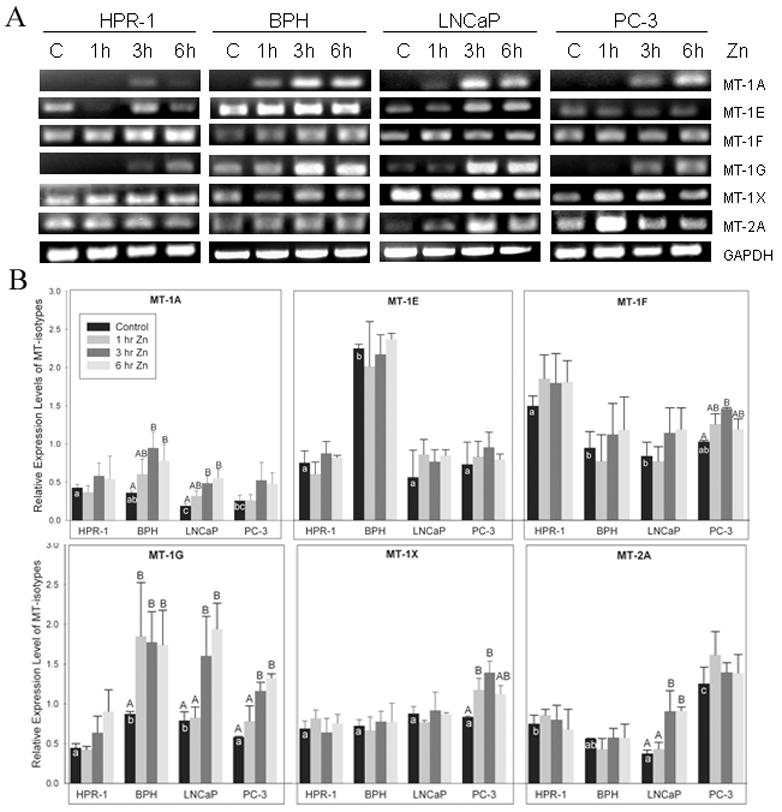
Time course studies of zinc effect on MT isotype mRNA levels in prostate cells by RT-PCR. A. Gel electrophoresis of MT isotype RT-PCR products in prostate cells. GAPDH signals were used as internal controls. B. The quantitative results of RT-PCR were plotted as the ratio of the densities from each MT isotype relative to optical densities normalized by related GAPDH signals. The columns and bars represent the mean ± SE (n=3).
Zinc Effect on the Genes Involved in Oncogenesis Pathways in HPR-1 & PC-3 Cells
The 40 genes listed in Tables 2 and 3, which we identified by microarray, were greatly altered at their transcriptional levels in response to zinc. Many of their associated proteins are involved in the critical biological processes of these cells. In order to further understand the mechanism of the inhibitory effect of zinc on prostate tumor cell growth, we examined expression patterns of the genes associated with oncogenesis. Oncogenes and tumor suppressor genes have been known to interact and most form a biological network as illustrated by the Pathway Architect (Cell Signaling Technology) (Table 5). Interestingly, many genes were either highly induced or suppressed by zinc or distinctly altered in the opposite direction in response to zinc depending on the cell type examined. Among these genes, Fos, which codes for a transcription factor associated with AP-1, was dramatically up-regulated by zinc in PC-3 cells. In HPR-1 cells, expression of the Akt1, JAK3 and PIK3 genes, associated with the Akt pathway, was dramatically induced with transcriptional levels up to 6-, 8- and 7-fold higher respectively, in response to zinc. In contrast, zinc had no significant effect on AKT1 and PIK3 mRNA levels and had an inhibitory effect (up to 75% lower) on JAK3 gene expression in PC-3 cells (Table 5). It has also been noticed that a tumor suppressor, TSC2, was induced up to 5-fold higher after 1hr exposure to zinc in HPR-1 cells, but no zinc effect on this gene was observed in PC-3 cells. Our data revealed that the genes involved in signaling pathways that are likely to be highly responsive to zinc are cell type-specific, which may lead to future studies on zinc targeting genes in prostate malignancy.
Table 5.
Summary of zinc altered oncogenes and tumor suppressor genes
| Pathway | Protein | GenBank ID | Gene symbol | Ratio of Gene Expression in the Cells of Zinc Treated vs. Control |
|||||
|---|---|---|---|---|---|---|---|---|---|
| HPR-1 |
PC-3 |
||||||||
| 1hr | 3hr | 6hr | 1hr | 3hr | 6hr | ||||
| TGF β | BMPR | NM_001204 | BMPR2 | −1.64 | 1.01 | −1.20 | −1.67 | −1.39 | −4.35 |
| Wnt | SOX | NM_005634 | SOX3 | 1.22 | 1.11 | 2.46 | −1.69 | −2.22 | −3.85 |
| NM_003107 | SOX4 | 1.35 | 1.15 | −1.82 | −1.30 | −1.43 | −2.50 | ||
| NM_017508 | SOX6 | 4.50 | 2.31 | 2.25 | 2.86 | -* | -* | ||
| NM_006943 | SOX12 | 5.34 | 1.88 | 3.00 | 2.57 | −1.37 | −2.13 | ||
| NM_004189 | SOX14 | −1.25 | 2.55 | 1.95 | −2.17 | −1.69 | −3.13 | ||
| NM_022454 | SOX17 | 1.38 | 1.53 | -* | −2.17 | −2.56 | −2.94 | ||
| APC | NM_000038 | APC | −1.16 | −1.39 | −1.72 | −1.72 | −3.45 | −8.33 | |
| Ras | Fos | NM_005252 | FOS | 2.06 | −1.09 | −1.35 | 3.10 | 19.70 | 13.59 |
| NM_006732 | FOSB | -* | -* | -* | 1.48 | 13.15 | 3.33 | ||
| Jun | NM_002228 | JUN | 1.47 | 1.00 | −1.30 | −1.37 | 1.08 | −1.20 | |
| ILK | NM_004517 | ILK | −1.69 | −1.61 | −2.63 | −1.32 | −1.06 | −1.09 | |
| Integrin | NM_003637 | ITGA10 | −1.27 | 1.06 | −1.43 | 1.30 | −1.43 | −2.70 | |
| NM_002203 | ITGA2 | −2.38 | −1.61 | −2.86 | −2.27 | −1.43 | −2.33 | ||
| NM_002204 | ITGA3 | −2.00 | −1.41 | 1.16 | −1.85 | −2.78 | −2.08 | ||
| NM_005353 | ITGAD | 1.31 | −2.38 | 1.65 | 1.39 | 3.09 | 2.31 | ||
| NM_000632 | ITGAM | 1.55 | 1.30 | 2.03 | −1.52 | −1.05 | 1.62 | ||
| NM_002210 | ITGAV | −2.00 | −2.33 | −3.85 | −1.25 | −1.96 | −1.30 | ||
| NM_000887 | ITGAX | -* | 3.44 | 2.40 | −2.38 | −1.05 | -* | ||
| NM_000211 | ITGB2 | −2.00 | −2.94 | −1.89 | −1.61 | −1.30 | −1.49 | ||
| NM_000888 | ITGB6 | -* | −5.00 | −2.38 | −2.13 | −1.04 | -* | ||
| NF1 | NM_000267 | NF1 | −1.79 | −1.96 | −2.56 | 1.22 | −2.78 | −3.23 | |
| VHL | NM_000551 | VHL | 1.08 | 1.80 | 1.58 | −1.35 | −2.27 | −1.49 | |
| Akt | Akt | NM_005163 | AKT1 | 6.38 | 2.73 | 5.00 | −1.05 | 2.02 | 1.46 |
| JAK | NM_000215 | JAK3 | 8.40 | 5.49 | 2.33 | -* | −2.50 | −4.00 | |
| PI3K | NM_002645 | PIK3C2A | −2.78 | −1.59 | −5.00 | −2.13 | −2.04 | −3.70 | |
| NM_002647 | PIK3C3 | 1.10 | −1.37 | −1.19 | −1.23 | −1.54 | −3.33 | ||
| NM_003629 | PIK3R3 | −2.94 | −2.70 | −2.17 | −2.08 | −1.35 | −1.82 | ||
| NM_014308 | PIK3R5 | 3.73 | -* | 7.18 | -* | −1.11 | 1.35 | ||
| TSC1/TSC2 | NM_000368 | TSC1 | 1.27 | −1.11 | −1.85 | −2.70 | −3.33 | −1.79 | |
| NM_000548 | TSC2 | 5.32 | 1.59 | 3.67 | −1.20 | −1.08 | −1.22 | ||
| Hedgehog | Smo | NM_005631 | SMO | 1.60 | 3.56 | 2.33 | −2.22 | 2.29 | −1.22 |
| Cyclin D | NM_053056 | CCND1 | 1.89 | 3.59 | −1.39 | −1.28 | −1.37 | −1.69 | |
| Cyclin E | NM_001238 | CCNE1 | 2.03 | −1.01 | −1.09 | −1.06 | −1.41 | 1.12 | |
| NM_057735 | CCNE2 | −2.17 | −1.96 | −3.23 | 1.07 | 1.18 | −1.61 | ||
| CDK2 | NM_001798 | CDK2 | -* | 1.02 | −1.27 | −3.57 | −1.08 | −2.08 | |
| Cell cycle | ARF | NM_001662 | ARF5 | −1.33 | −1.08 | −1.85 | −1.49 | −1.37 | −2.13 |
| NM_001663 | ARF6 | −1.00 | 1.25 | −2.22 | −1.03 | −2.63 | −2.63 | ||
| ATM | NM_000051 | ATM | −1.12 | −3.33 | −4.55 | −1.08 | −3.03 | -* | |
| ATR | NM_001184 | ATR | −1.03 | −1.18 | −2.00 | −2.22 | −1.39 | −1.69 | |
| BRCA1 | NM_007305 | BRCA1 | 1.33 | −1.41 | −1.33 | −1.28 | −1.03 | −2.50 | |
Signal intensities were flagged as unreliable.
Discussion
In this study, we surveyed the effect of zinc on genome-wide gene expression profiles using microarray assays in normal and cancer prostate cell lines with multiple exposure times. Microarray results indicated that zinc exposure appeared to have: 1) major down-regulatory effects on thousands of genes, while less than 900 genes were up-regulated in both prostate cell lines; and 2) cell-type specific effects on gene expression patterns. We previously reported that zinc treatment inhibits the growth of prostate malignant (PC-3) and benign hyperplasia (BPH) cells, mostly due to zinc-induced apoptosis through the mitochondrial pathway [5,8–10]. In contrast, no growth inhibitory effect of zinc was observed in normal prostate cells, which contain higher endogenous zinc levels and whose mitochondria do not respond to zinc with the release of cytochrome c [10]. The microarray assay provided us enormous information of zinc effect on the gene expression profiles which are involved in many biological and molecular events. The main challenge in this study is how to develop a strategy for sorting the microarray data. Considering our major goal in studying the mechanism of zinc effect on prostate cancer cell death, we developed a plan to determine and to sort the genes: 1) highly sensitive in response to zinc treatment, 2) relevant to zinc homeostasis, and 3) involved in cell growth and apoptosis, and related signaling pathways.
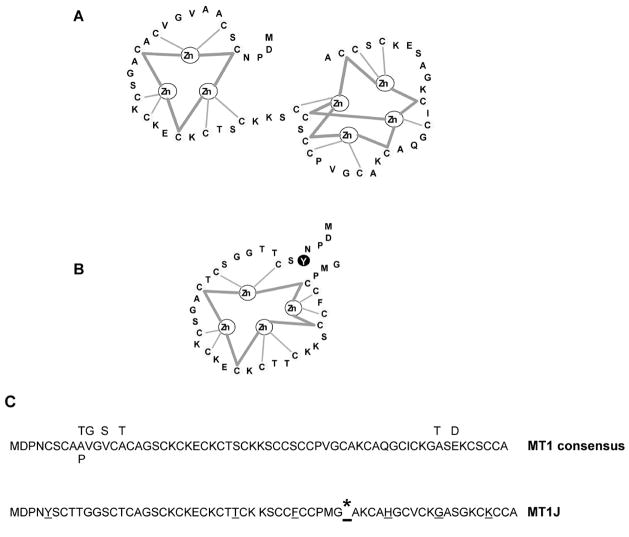
It is well recognized that MTs play major biological roles in the cells as metal binding/donors. The MT genes are highly inducible by heavy metals and zinc is the primary physiological inducer in mammalian cells [13]. Using human prostate cells characterized highly relevant to cellular zinc accumulation as a model system, we observed a specific zinc induction of MTs. Among 14 MT-1 isotypes, MT-1J was significantly induced by zinc detected via microarray analysis and verified by semi-quantitative RT-PCR, and this induction was observed specifically only in PC-3 and LNCaP, but not in HPR-1, BPH cells (Fig. 1). Schmidt et al. [24] indicated that MT-1J, previously regarded as a non-functional pseudogene, is a MT-related gene and has an exon/intron structure consistent with that of a functional MT gene. However, unlike the currently characterized functional MT genes, MT-1J has been predicted to be incapable of encoding a complete typical MT protein due to the presence of a truncated structure and atypical aromatic amino acid substitutions, which have never been found universally in any of the sequenced vertebrate MTs to date [24]. In addition, the overexpression of MT-1J ( hMT-I) in mouse cells failed to protect against cadmium (10 μM) toxicity [24]. With limited functional studies on MT-1J, it is still labeled as a “non-functional pseudogene” [24,25]. The MT genes are highly conserved in DNA sequences within both coding and regulatory regions [24]. In particular, the positions of the cysteine residues, which serve to chelate heavy metal ions via thiolate complexes, are invariant, and bind to three Zn ions in the N-terminal β domain and four Zn ions in the C-terminal a domain. Zn ions bound to thiolates (apo MT) of the β domain are more labile than those of the a domain. It has been suggested by Cousin et al. (2006) [26] that the β domain is physiologically relevant, while the a domain may be related to metal detoxification. Based on the predicted peptide sequence, MT-1J may encode a truncated protein which possesses a complete β domain with a tyrosine substitution of a conserved cysteine at position 5 and a coil region with four cysteine residues and one phenylalanine replacement at position 35 as proposed in Figure 4. With this molecular structure, it could be highly possible that MT-1J might carry a reduced capability to bind metals (up to four zinc ions instead of a total of seven zinc ions), but retain the function to release the zinc from the β domain in response to cellular demands. Interestingly, our results revealed that in PC-3 cells, MT-1J transcripts showed the highest sensitivity to zinc treatment with a time-dependent increase. Previously, endogenous levels of MT-1J were reported to be down-regulated by an average ratio of 0.22 in 19 pancreatic carcinoma cell lines in comparison to the immortalized human pancreatic ductal epithelial cell line (HPDE) [27]. In their study, the authors suggested that the biological function of MT-1J was to “modulate apoptosis,” which would be the first report regarding the biological functions of MT-1J other than “metal ion binding” or “unclassified.” However, at the present time, no detailed information is available regarding the biological function of MT-1J in apoptosis, which emphasizes the importance of zinc’s effect on prostate cancer for future studies.
FIGURE 4.
Predicted protein structure of MT-1J. According to the protein sequences, MT-1J only possesses a β domain of the MT-1 molecule with a tyrosine substitution at position 5 and a coil residue with four cysteine residues and one phenylalanine replacement at position 35. MT-1J might carry a reduced metal ion binding capability down to four zinc ions compared to seven zinc ions in a normal MT-1 molecule. * indicates the presence of a TGA stop codon.
In addition to MT-1J, MT-1M, also labeled as a pseudogene and previously denoted as MT-1K, has been reported to be suppressed in pancreatic carcinoma cell lines in comparison to the immortalized HPDE cell line [27], and to be up-regulated by zinc with an 81-fold change in bronchial epithelial cells [28]. In this study, a significantly higher level of endogenous MT-1M transcripts, which was further rapidly induced by zinc with a 3- to 6-fold increase, was detected in PC-3 and BPH cells. In contrast, normal HPR-1 cells contain lower endogenous MT-1M mRNA and only a late induction by zinc was detected. In fact, the zinc-induced expression of MT-1J and -1M was detected in both PC-3 and BPH cell lines, which were demonstrated to be sensitive to zinc-induced apoptosis [8,10]. This correlation appears to lead to the apoptotic function of MT-1J and -1M, which was illustrated by Missiaglia et al. [27]. As to the gene for MT-1M, Stennard et al. [25] noted that the presence of a histidine residue and the loss of a lysine residue, a highly conserved residue among mammalian MTs. Thus, whether these genes and their protein products serve as natural detoxifiers or as acceptors/donors for zinc [29] in prostate cancer cells requires further study, including: 1) the characterization of MT-1J and -1M translated products, 2) the identification of the capability and capacity of zinc binding in MT-1J and/or -1M, and 3) the relationship of MT-1J and -1M expression and zinc-induced cell apoptosis.
The expression of MT-1/2 isotypes was reported to occur in a tissue-specific pattern [15], and their protein products may exert specific functions in human normal and/or malignant cells and tissues [30]. Previously, MT-2A gene expression had been connected with cell proliferation [31] and tumor invasiveness [32] in breast cancers. Our results showed that the higher abundance of MT-2A transcripts was only detected in PC-3 cells (Fig. 3), and this analogous expression pattern across different cancer types (37, 38) suggested a possible candidacy of MT-2A as a biomarker for potentially malignant cells and tumor tissues. Moreover, our data also showed that among six MT-1/2 isotypes (Fig. 3), the higher endogenous mRNA levels of MT-1A and -1F were detected in normal HPR-1 cells; likewise, those of MT-1E and -1G were found in BPH cells. The cell-type specific expression of MTs highly corresponds to the levels of their protein products which were determined by Western blot analysis [33], and also to the capacity of cellular zinc accumulation in different prostate tissues described previously (1).
Our data indicate that zinc highly induced the expression of a group of genes functionally related to cell proliferation and apoptosis, including HSP70B (14-fold increase), heat shock 70kDa protein 1-like (6-fold increase), Hsp40 homolog (18-fold increase) as well as MT-1J (5.7-fold increase) in the PC-3 cell line, while no or lesser expression alteration by zinc was detected in HPR-1. This finding in PC-3 cells corresponds to the reports of Koizumi’s group [34,35]. They demonstrated that human genes coding for HSPs (40, 60, 70, 90) and MTs (1E, 1B, 3) were up-regulated by cadmium in HeLa cancer cells. The expression of HSP70 is also induced by a variety of heavy metals including Zn, Cu, Hg and Ag [34,36]. The regulation of MT and HSP gene expression is mediated by metal-activated transcription factors, MTF-1 (MRE (metal-responsive element)-binding transcription factor-1) and HSF1 (heat shock factor1), respectively. Up-regulation of HSP70 inhibits cell apoptosis induced by a wide range of insults [37], and does so through mechanisms involving the interactions with Apaf-1, thereby preventing the recruitment of procaspase-9 to the apoptosome [38]. Altogether, zinc-induced HSP70 over-expression in prostate cancer cells can be considered as a cell self-defensive reaction. Moreover, the biological effect of zinc-induced HSP70 on prostate cancer cell growth should be further studied.
Another notable gene whose expression is regulated by zinc is the fos gene, which encodes the transcriptional factor Fos, which dimerizes with members of Jun and CREB/ATF families, forming the AP-1 complex [39]. AP-1 is activated in the regulation of genes involved in a variety of cellular events including normal development, neoplastic transformation, and apoptosis [40]. Fos has been shown to be associated with apoptosis in photoreceptor cells and hepatocytes [41]. Fos-related molecular events in prostate malignancy have not yet been elucidated. Our microarray assays revealed a dramatic zinc-induction of fos mRNA expression in PC-3 cells, but not in HPR-1. It would be important to further elucidate whether this zinc induction of fos gene expression is involved in regulation of zinc-induced apoptosis pathways in PC-3 cells. If such roles are identified as cell type-specific, fos could be a potential candidate used as a target gene for oncogenetic screening. This is the first report to address the zinc effect on increased fos transcripts in prostate cells. Further studies focused on the relationship of zinc-induced fos and zinc-induced tumor cell apoptosis will provide crucial information for the implementation of zinc in the clinical treatment of prostate malignancy.
Conclusions
This is the first report on the roles of zinc in differential regulation of gene expression profile in human prostate normal and malignant cell lines. A strategy for microarray data analysis was developed and led to distinguish the genes which are: a) highly sensitive to zinc, b) associated with zinc homeostasis, c) relevant to several oncogenic pathways. Our microarray data indeed provide evidence on zinc-altered gene transcripts which are involved in cellular zinc accumulation and cell apoptosis with oncogenic cell type specificity. Among MT-1 isotypes, MT-1J and -1M, previously considered as pseudogenes, were found to be significantly up-regulated at the transcriptional level by zinc in malignant PC-3 and LNCaP cells only. We believe that this study delivered important insight to a new field of research on zinc and its roles in the prevention and intervention of prostate cancer.
Acknowledgments
This study was supported by funding from NIH/National Cancer Institute grant (R01-116815) to P. Feng.
Footnotes
Competing interests
No competing interests are declared for this work.
Authors’ contributions
Shu-fei Lin performed the microarray study, RT-PCR, data analysis, related statistical analyses and participated in manuscript preparation. Hua Wei performed RT-PCR, contributed and participated in producing the tables and figures. Dennis Maeder predicted the protein structure for MT-1J. Renty B. Franklin participated in the discussions and manuscript preparation. Pei Feng conceived the study, responded to its design, coordination, data analysis and the completion of the manuscript. All authors read and approved the final manuscript.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Shu-fei Lin, Email: linshufei@gmail.com.
Hua Wei, Email: dotdotwei@gmail.com.
Dennis Maeder, Email: maeder@umbi.umd.edu.
Renty B. Franklin, Email: rfranklin@umaryland.edu.
Pei Feng, Email: pfeng@umaryland.edu.
Reference List
- 1.Tapiero H, Tew KD. Trace elements in human physiology and pathology: zinc and metallothioneins. Biomed Pharmacother. 2003;57:399–411. doi: 10.1016/s0753-3322(03)00081-7. [DOI] [PubMed] [Google Scholar]
- 2.Stefanidou M, Maravelias C, Dona A, Spiliopoulou C. Zinc: a multipurpose trace element. Arch Toxicol. 2006;80:1–9. doi: 10.1007/s00204-005-0009-5. [DOI] [PubMed] [Google Scholar]
- 3.Sensi SL, Ton-That D, Sullivan PG, Jonas EA, Gee KR, Kaczmarek LK, et al. Modulation of mitochondrial function by endogenous Zn2+ pools. Proc Natl Acad Sci U S A. 2003;100:6157–6162. doi: 10.1073/pnas.1031598100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Costello LC, Franklin RB. Novel role of zinc in the regulation of prostate citrate metabolism and its implications in prostate cancer. Prostate. 1998;35:285–296. doi: 10.1002/(sici)1097-0045(19980601)35:4<285::aid-pros8>3.0.co;2-f. [DOI] [PubMed] [Google Scholar]
- 5.Liang JY, Liu YY, Zou J, Franklin RB, Costello LC, Feng P. Inhibitory effect of zinc on human prostatic carcinoma cell growth. Prostate. 1999;40:200–207. doi: 10.1002/(sici)1097-0045(19990801)40:3<200::aid-pros8>3.0.co;2-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Feng P, Li TL, Guan ZX, Franklin RB, Costello LC. Effect of zinc on prostatic tumorigenicity in nude mice. Ann N Y Acad Sci. 2003;1010:316–320. doi: 10.1196/annals.1299.056. [DOI] [PubMed] [Google Scholar]
- 7.Iguchi K, Hamatake M, Ishida R, Usami Y, Adachi T, Yamamoto H, et al. Induction of necrosis by zinc in prostate carcinoma cells and identification of proteins increased in association with this induction. Eur J Biochem. 1998;253:766–770. doi: 10.1046/j.1432-1327.1998.2530766.x. [DOI] [PubMed] [Google Scholar]
- 8.Feng P, Liang JY, Li TL, Guan ZX, Zou J, Franklin R, et al. Zinc induces mitochondria apoptogenesis in prostate cells. Mol Urol. 2000;4:31–36. [PubMed] [Google Scholar]
- 9.Untergasser G, Rumpold H, Plas E, Witkowski M, Pfister G, Berger P. High levels of zinc ions induce loss of mitochondrial potential and degradation of antiapoptotic Bcl-2 protein in in vitro cultivated human prostate epithelial cells. Biochem Biophys Res Commun. 2000;279:607–614. doi: 10.1006/bbrc.2000.3975. [DOI] [PubMed] [Google Scholar]
- 10.Feng P, Li TL, Guan ZX, Franklin RB, Costello LC. Direct effect of zinc on mitochondrial apoptogenesis in prostate cells. Prostate. 2002;52:311–318. doi: 10.1002/pros.10128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vallee BL. The function of metallothionein. Neurochem Int. 1995;27:23–33. doi: 10.1016/0197-0186(94)00165-q. [DOI] [PubMed] [Google Scholar]
- 12.Davis SR, Cousins RJ. Metallothionein expression in animals: a physiological perspective on function. J Nutr. 2000;130:1085–1088. doi: 10.1093/jn/130.5.1085. [DOI] [PubMed] [Google Scholar]
- 13.Andrews GK. Regulation of metallothionein gene expression by oxidative stress and metal ions. Biochem Pharmacol. 2000;59:95–104. doi: 10.1016/s0006-2952(99)00301-9. [DOI] [PubMed] [Google Scholar]
- 14.Theocharis S, Karkantaris C, Philipides T, Agapitos E, Gika A, Margeli A, et al. Expression of metallothionein in lung carcinoma: correlation with histological type and. Histopathology. 2002;40:143–151. doi: 10.1046/j.1365-2559.2002.01325.x. [DOI] [PubMed] [Google Scholar]
- 15.Cherian MG, Jayasurya A, Bay BH. Metallothioneins in human tumors and potential roles in carcinogenesis. Mutat Res. 2003;533:201–209. doi: 10.1016/j.mrfmmm.2003.07.013. [DOI] [PubMed] [Google Scholar]
- 16.Liuzzi JP, Cousins RJ. Mammalian zinc transporters. Annu Rev Nutr. 2004;24:151–172. doi: 10.1146/annurev.nutr.24.012003.132402. [DOI] [PubMed] [Google Scholar]
- 17.Colvin RA, Fontaine CP, Laskowski M, Thomas D. Zn2+ transporters and Zn2+ homeostasis in neurons. Eur J Pharmacol. 2003;479:171–185. doi: 10.1016/j.ejphar.2003.08.067. [DOI] [PubMed] [Google Scholar]
- 18.Hasumi M, Suzuki K, Matsui H, Koike H, Ito K, Yamanaka H. Regulation of metallothionein and zinc transporter expression in human prostate cancer cells and tissues. Cancer Lett. 2003;200:187–195. doi: 10.1016/s0304-3835(03)00441-5. [DOI] [PubMed] [Google Scholar]
- 19.Garrett SH, Sens MA, Shukla D, Flores L, Somji S, Todd JH, et al. Metallothionein isoform 1 and 2 gene expression in the human prostate: Downregulation of MT-1X in advanced prostate cancer. Prostate. 2000;43:125–135. doi: 10.1002/(sici)1097-0045(20000501)43:2<125::aid-pros7>3.0.co;2-s. [DOI] [PubMed] [Google Scholar]
- 20.Sens MA, Somji S, Garrett SH, Beall CL, Sens DA. Metallothionein isoform 3 overexpression is associated with breast cancers having a poor prognosis. American Journal of Pathology. 2001;159:21–26. doi: 10.1016/S0002-9440(10)61668-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jacob ST, Majumder S, Ghoshal K. Suppression of metallothionein-I/II expression and its probable molecular mechanisms. Environ Health Perspect. 2002;110 (Suppl 5):827–830. doi: 10.1289/ehp.02110s5827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Costello LC, Liu Y, Zou J, Franklin RB. Evidence for a zinc uptake transporter in human prostate cancer cells which is regulated by prolactin and testosterone. J Biol Chem. 1999;274:17499–17504. doi: 10.1074/jbc.274.25.17499. [DOI] [PubMed] [Google Scholar]
- 23.Palmiter RD, Findley SD, Whitmore TE, Durnam DM. MT-III, a brain-specific member of the metallothionein gene family. Proc Natl Acad Sci U S A. 1992;89:6333–6337. doi: 10.1073/pnas.89.14.6333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schmidt CJ, Jubier MF, Hamer DH. Structure and expression of two human metallothionein-I isoform genes and a related pseudogene. J Biol Chem. 1985;260:7731–7737. [PubMed] [Google Scholar]
- 25.Stennard FA, Holloway AF, Hamilton J, West AK. Characterisation of six additional human metallothionein genes. Biochim Biophys Acta. 1994;1218:357–365. doi: 10.1016/0167-4781(94)90189-9. [DOI] [PubMed] [Google Scholar]
- 26.Cousins RJ, Liuzzi JP, Lichten LA. Mammalian zinc transport, trafficking, and signals. J Biol Chem. 2006 doi: 10.1074/jbc.R600011200. [DOI] [PubMed] [Google Scholar]
- 27.Missiaglia E, Blaveri E, Terris B, Wang YH, Costello E, Neoptolemos JP, et al. Analysis of gene expression in cancer cell lines identifies candidate markers for pancreatic tumorigenesis and metastasis. Int J Cancer. 2004;112:100–112. doi: 10.1002/ijc.20376. [DOI] [PubMed] [Google Scholar]
- 28.Li Z, Stonehuerner J, Devlin RB, Huang YC. Discrimination of vanadium from zinc using gene profiling in human bronchial epithelial cells. Environ Health Perspect. 2005;113:1747–1754. doi: 10.1289/ehp.7947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Magda D, Lecane P, Miller RA, Lepp C, Miles D, Mesfin M, et al. Motexafin gadolinium disrupts zinc metabolism in human cancer cell lines. Cancer Res. 2005;65:3837–3845. doi: 10.1158/0008-5472.CAN-04-4099. [DOI] [PubMed] [Google Scholar]
- 30.Jasani B, Schmid KW. Significance of metallothionein overexpression in human. Histopathology. 1997;31:211–214. doi: 10.1046/j.1365-2559.1997.2140848.x. [DOI] [PubMed] [Google Scholar]
- 31.Jin R, Chow VT, Tan PH, Dheen ST, Duan W, Bay BH. Metallothionein 2A expression is associated with cell proliferation in breast cancer. Carcinogenesis. 2002;23:81–86. doi: 10.1093/carcin/23.1.81. [DOI] [PubMed] [Google Scholar]
- 32.Tai SK, Tan OJ, Chow VT, Jin R, Jones JL, Tan PH, et al. Differential expression of metallothionein 1 and 2 isoforms in breast cancer lines with different invasive potential: identification of a novel nonsilent metallothionein-1H mutant variant. Am J Pathol. 2003;163:2009–2019. doi: 10.1016/S0002-9440(10)63559-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wei H, Hong Y, Lin S, Clark A, Feng P. Regulation of metallothionein expression by zinc involved in oncogenesis of human prostate cells. Proc Amer Assoc Cancer Res. 412006;47 [Google Scholar]
- 34.Yamada H, Koizumi S. DNA Microarray analysis of human gene expression induced by a non-lethal dose of cadmium. Industrial Health. 2002;40:159–166. doi: 10.2486/indhealth.40.159. [DOI] [PubMed] [Google Scholar]
- 35.Uenishi R, Gong PF, Suzuki K, Koizumi S. Cross talk of heat shock and heavy metal regulatory pathways. Biochemical and Biophysical Research Communications. 2006;341:1072–1077. doi: 10.1016/j.bbrc.2006.01.066. [DOI] [PubMed] [Google Scholar]
- 36.Murata M, Gong P, Suzuki K, Koizumi S. Differential metal response and regulation of human heavy metal-inducible genes. J Cell Physiol. 1999;180:105–113. doi: 10.1002/(SICI)1097-4652(199907)180:1<105::AID-JCP12>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- 37.Seo JS, Park YM, Kim JI, Shim EH, Kim CW, Jang JJ, et al. T cell lymphoma in transgenic mice expressing the human Hsp70 gene. Biochem Biophys Res Commun. 1996;218:582–587. doi: 10.1006/bbrc.1996.0103. [DOI] [PubMed] [Google Scholar]
- 38.Beere HM, Wolf BB, Cain K, Mosser DD, Mahboubi A, Kuwana T, et al. Heat-shock protein 70 inhibits apoptosis by preventing recruitment of procaspase-9 to the Apaf-1 apoptosome. Nat Cell Biol. 2000;2:469–475. doi: 10.1038/35019501. [DOI] [PubMed] [Google Scholar]
- 39.Hess J, Angel P, Schorpp-Kistner M. AP-1 subunits: quarrel and harmony among siblings. Journal of Cell Science. 2004;117:5965–5973. doi: 10.1242/jcs.01589. [DOI] [PubMed] [Google Scholar]
- 40.Milde-Langosch K. The Fos family of transcription factors and their role in tumourigenesis. Eur J Cancer. 2005;41:2449–2461. doi: 10.1016/j.ejca.2005.08.008. [DOI] [PubMed] [Google Scholar]
- 41.Yu RA, Yang CF, Chen XM. DNA damage, apoptosis and C-myc, C-fos, and C-jun overexpression induced by selenium in rat hepatocytes. Biomed Environ Sci. 2006;19:197–204. [PubMed] [Google Scholar]
- 42.Rozsa FW, Reed DM, Scott KM, Pawar H, Moroi SE, Kijek TG, et al. Gene expression profile of human trabecular meshwork cells in response to long-term dexamethasone exposure. Mol Vis. 2006;12:125–141. [PubMed] [Google Scholar]