Abstract
The expression of protein-coding genes is enhanced by the exquisite coupling of transcription by RNA polymerase II with pre-messenger RNA processing reactions, such as 5′-end capping, splicing and 3′-end formation. Integration between cotranscriptional processing events extends beyond the nucleus, as proteins that bind cotranscriptionally can affect the localization, translation and degradation of the mature messenger RNA. MicroRNAs are RNA polymerase II transcripts with crucial roles in the regulation of gene expression. Recent data demonstrate that processing of primary microRNA transcripts might be yet another cotranscriptional event that is woven into this elaborate nuclear network. This review discusses the extensive molecular interactions that couple the earliest steps in gene expression and therefore influence the final fate and function of the mature messenger RNA or microRNA produced.
Introduction
In vertebrates, RNA polymerase II (RNAPII) is responsible for the transcription of genes encoding proteins and many noncoding RNAs, including most microRNAs (miRNAs). For each of these gene classes, the initial primary transcript undergoes a number of processing reactions to produce the final RNA product or products. Most of these RNA remodeling steps occur in the cell nucleus.
In the case of protein-coding genes, the initial transcript, or pre-messenger (pre-mRNA), contains both coding sequences – exons – and intervening noncoding sequences – introns. Pre-mRNA processing involves 5′-end capping, the removal of introns by means of splicing, and 3′-end cleavage and polyadenylation (CPA) to produce the final messenger RNA (mRNA; Figure 1A). Many pre-mRNAs are subjected to additional processing steps, such as editing and surveillance, which can further influence the coding potential of transcripts [1,2]. Furthermore, several types of noncoding RNAs, such as small nucleolar RNAs (snoRNAs) and small Cajal body-associated RNAs (scaRNAs), are released from some introns of select pre-mRNA transcripts [3].
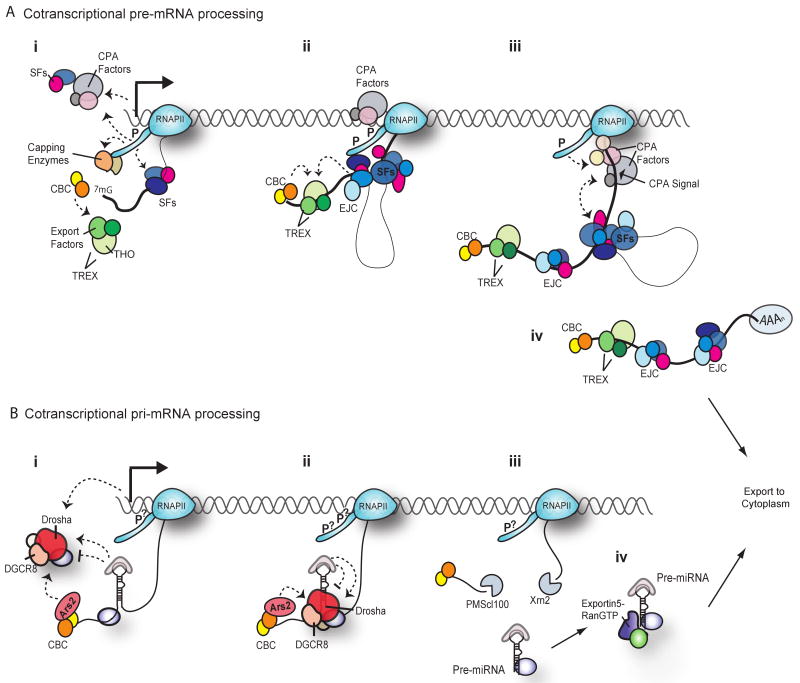
Figure 1.
Cotranscriptional RNA processing. (A) Cotranscriptional pre-mRNA processing. Exons are represented as thick solid lines and introns as thin lines. Dashed arrows depict interactions between proteins that might stabilize interactions or promote the corresponding reaction. (i) Capping. Near the transcription initiation site, RNAPII is phosphorylated (P) on Ser5, which results in recruitment and activation of the capping enzymes. The cap binding complex (CBC) subsequently binds to the 5′ cap. Splicing factors (SFs) and some components of the CPA machinery are recruited to gene promoters through interactions with RNAPII, as well as associated transcription factors. SFs also bind to exonic sequence enhancers, and interactions with the C-terminal domain (CTD) might stabilize complex formation. Export factors in conjunction with the THO complex form the transcription-export (TREX) complex, which is recruited to the nascent transcript through interactions with the CBC. (ii) Spliceosome assembly. Phosphorylation of RNAPII on Ser2 allows elongation into the gene body. The spliceosome assembles on the first intron; assembly is enhanced by binding of SR proteins and other splicing factors to both the nascent RNA and the CTD, serving to bring the first and second exons into close proximity. The exon-junction complex (EJC) is recruited by the splicing machinery and is deposited just upstream of the exon–exon junction. The TREX complex stably associates with nascent RNA owing to interactions with CBC, as well as SFs and/or the EJC. (iii) Splicing of the 3′-terminal exon and 3′-end formation. The final intron and the 3′-terminal exon have been transcribed, and splicing of the final intron is under way. The CPA signal has also been transcribed, and interactions with splicing factors bound to the terminal exon, as well as the CTD, enhance recruitment of additional components of the CPA machinery. The CPA machinery reciprocally stabilizes interaction of SFs with the 3′-terminal intron. The CPA machinery assembles on the CPA signal and will cleave the RNA to release the transcript from the DNA template after splicing of the last intron is complete. (iv) Export. The processed mRNA is exported to the cytoplasm. Note that many proteins deposited on the pre-mRNA during splicing remain associated with the mRNA in the cytoplasm and might affect downstream processes.
(B) Cotranscriptional pri-miRNA processing. (i) Microprocessor recruitment. The Microprocessor, consisting of Drosha, DGCR8 and additional accessory proteins, can be recruited to the pri-miRNA by means of the promoter, through interaction with Ars2 (which binds to CBC), through interaction with auxiliary proteins that bind to the pri-miRNA terminal loop (see Table 1 for examples) or through other, currently unknown, mechanisms. Auxiliary proteins might also inhibit Microprocessor interaction with the pri-miRNA. Note that the indicated interactions might affect pri-miRNA processing for only some miRNAs, as indicated by dashed arrows. (ii) Microprocessor assembly and cleavage. Drosha cleaves at the base of the hairpin to release the pre-miRNA. For some pri-miRNAs, Ars2 might facilitate Drosha cleavage. Proteins that bind to the pri-miRNA terminal loop could also facilitate or inhibit pri-miRNA cleavage. Note that, for intronic miRNAs, Drosha association with the pri-miRNA might be enhanced by interaction with splicing factors, and the bridging of exons mediated by binding of SR proteins to the flanking exons and to the CTD (as demonstrated in Figure 1Aii) would allow exons to be efficiently spliced despite prior cotranscriptional cleavage of the intronic miRNA. (iii) Pre-miRNA release and exonuclease recruitment. Exonucleases are recruited to the newly generated ends of the flanking pri-miRNA sequences. 5′–3′ degradation of the 3′ segment of the pri-miRNA might lead to premature termination of intergenic pri-miRNA transcription by RNAPII. (iv) Export. The pre-miRNA is exported to the cytoplasm by the Exportin5–RanGTP complex. Note that proteins that bind the pri-miRNA terminal loop, such as KSRP, might remain associated with the pre-miRNA and affect downstream processes in the cytoplasm such as Dicer cleavage. Hypothetical CTD phosphorylation states are indicated by question marks as the effect of CTD phosphorylation on pri-miRNA processing has not yet been determined.
The most recently discovered class of genes transcribed by RNAPII is the miRNA class. These tiny RNAs posttranscriptionally control expression of their target messenger RNAs and govern cellular functions ranging from cell growth and differentiation to apoptosis [4]. The biogenesis of miRNA involves cleavage of the primary miRNA (pri-miRNA) transcript in the nucleus by the Microprocessor complex [5], which contains the required RNase-III-like enzyme Drosha and its double-stranded RNA binding partner, DGCR8, to produce the hairpin-shaped precursor miRNA (pre-miRNA; Figure 1B). Further cleavage by another RNase-III-like enzyme, Dicer, in the cytoplasm produces the mature miRNA [6]. Approximately 80% of miRNAs are located within introns of either protein-coding or spliced, noncoding transcripts [7]. Thus, some RNAPII transcripts are simultaneously pre-mRNAs and pri-miRNAs.
Coupling of pre-mRNA processing and transcription
Each of the processing steps mentioned above is catalyzed by its own specific molecular machinery. Yet, a vast amount of data argues that, for pre-mRNAs, these reactions occur not as a series of independent stepwise events but, rather, in a complex and highly coordinated network [1,2,8,9]. This molecular coupling, or cross-talk, between multiple processes results in dramatically enhanced efficiency and accuracy of gene expression, as discussed below. Recent work suggests that the initial stages of miRNA biogenesis might also be interwoven into this remarkably sophisticated network.
Three mechanisms underlying the coupling of transcription and pre-mRNA processing have been described: (i) physical coupling, (ii) kinetic coupling and (iii) allosteric activation [2]. In physical coupling, interactions between the transcription machinery and pre-mRNA processing factors result in increased concentrations of factors adjacent to the pre-mRNA as it emerges from the exit channel of elongating RNAPII [9]. Thus, the correct processing enzymes bind to nascent pre-mRNAs before other abundant, nonspecific nuclear RNA-binding proteins gain access. Major players in physical coupling are RNAPII and its unique C-terminal domain (CTD; Box 1), as well as RNAPII-associated transcription factors [10]. In kinetic coupling, alterations in the rate of RNAPII elongation modulate the time available for splicing to occur [11]. Consequently, a slow RNAPII elongation rate allows inclusion of an exon with weak splice sites, whereas rapid elongation leads to exclusion because of more efficient splicing to a downstream exon with a stronger 3′ splice site [11]. In allosteric coupling, interactions between RNAPII and pre-mRNA processing factors result in activation of their catalytic activity [2]. In the following, we discuss how these mechanisms contribute to specific pre-mRNA processing steps.
Box 1. The RNAPII CTD.
Eukaryotic RNAPII is unlike all other DNA-dependent RNA polymerases in that it contains a unique appendage at the C terminus of its largest subunit, termed the C-terminal domain (CTD). The CTD comprises a number of heptapeptide repeats with the sequence Y1S2P3T4S5P6S7. The yeast CTD contains 26 conserved repeats, whereas the human CTD contains 52 repeats, 31 of which diverge from the consensus sequence in one or more residues [29]. Phosphorylation plays a crucial role in CTD function. The unphosphorylated form of RNAPII is recruited to preinitiation complexes at gene promoters, where it engages in nonprocessive transcription and generates short, abortive transcripts. Phosphorylation of the CTD on Ser5 of the repeat is required for initiation of processive transcription and escape of RNAPII from promoter-proximal pause sites; subsequent phosphorylation of Ser2 allows elongation into the gene body [10,86].
General transcription-factor-initiated RNA synthesis can proceed in vitro without the CTD, suggesting that the CTD has crucial functions aside from transcription [2,10]. Indeed, the CTD plays an essential role in facilitating and integrating a number of cotranscriptional events by simultaneously interacting with a wide range of factors, including pre-mRNA processing factors, DNA-remodeling complexes, chromatin and the nascent RNA [29]. Many proteins bind the CTD only when it is phosphorylated on specific residues or combinations of residues. Upon dephosphorylation by CTD phosphatases, the bound processing factors are released. In addition to phosphorylation, residues on the CTD can also be modified by glycosylation or proline isomerization. Because all seven residues of each heptapeptide can be modified, an extremely large number of combinations is possible. These different modification patterns contribute to a ‘CTD code’, whereby dynamic changes properly coordinate recruitment of the correct processing factors at specific times during pre-mRNA transcription and maturation [10,86]. Recent work has expanded our understanding of the CTD code by demonstrating that Ser7 phosphorylation is essential for transcription and 3′-end formation of snRNA genes, as well as of some protein-coding genes [10]. Different CTD phosphorylation states might also affect the rate of elongation of RNAPII, which in turn affects the timing of pre-mRNA processing events [11]. Thus, the CTD plays an integral role in the coordination of RNAPII transcription and pre-mRNA processing.
Coupling of 5′-end capping to transcription
The capping reaction, in which RNAPII transcripts receive an m7GpppN cap structure, is coupled to transcription through direct physical interactions of the two capping enzymes, HCE1 and HCM1, with phosphorylated forms of the RNAPII CTD (Figure 1Ai) [1,12]. Binding to the CTD of RNAPII phosphorylated on serine 5 (Ser5P) also allosterically activates the guanylyltransferase activity of HCE1 [2,12]. As RNAPII with Ser5P CTD is most abundant in promoter-proximal regions, capping enzymes are both concentrated and activated near transcription initiation sites, resulting in rapid capping of nascent transcripts that are only 20–40 nucleotides long. The 5′ cap is crucial for stability of the pre-mRNA, as well as for its recognition by the cap binding complex (CBC) in the nucleus and by eIF4E in the cytoplasm [12]. Thus, functional coupling of capping to transcription dramatically enhances the efficiency and fidelity of pre-mRNA production, stability and translation.
Coupling of transcription and pre-mRNA splicing
Multiple lines of evidence indicate that pre-mRNA splicing is both physically and kinetically coupled to transcription. Specific mutations within the CTD greatly reduce the efficiency of splicing without affecting transcription [2]. Some splicing factors directly interact with RNAPII through the CTD [13], as well as with RNAPII-associated transcription factors and coactivators [14]. The SR family of essential splicing factors plays a particularly important role in splicing by binding to specific sequence elements in exons termed exonic sequence enhancers (ESEs) and recruiting the rest of the splicing machinery to constitutive or alternative splice sites [15]. Some SR proteins can also bind indirectly to the CTD of RNAPII [13,16] or directly to histones [17] and thus serve as bridges that tether nascent exons to RNAPII or to the chromatin template (Figure 1Aii). This phenomenon of ‘exon tethering’ is thought to dramatically increase the fidelity of splicing by bringing short exons into close proximity, even when intervening introns are thousands of nucleotides long [16,18]. However, additional data suggest that SR proteins are recruited to chromatin primarily through interactions with the nascent transcript [19]. Therefore, it is possible that chromatin, RNAPII and the nascent RNA act cooperatively in the formation of stable nascent ribonucleoprotein (RNP) complexes [19,20].
In support of this hypothesis, in vitro coupled transcription–splicing systems have demonstrated that pre-mRNAs transcribed by RNAPII more efficiently recruit splicing factors and are more efficiently spliced than those transcribed by T7 phage polymerase or RNA polymerase III [13,21]. The CTD of RNAPII is necessary but not sufficient for this recruitment; thus, other RNAPII-specific factors are also likely to contribute [21]. Additional evidence for coupling between transcription and splicing includes observations that different promoters, enhancers, transcription elongation factors and transcriptional activators favor alternative splice-site choices [11,22] and that the rate of transcription elongation can dramatically affect the efficiency of splicing, as well as splice-site decisions for both constitutive and alternative splicing [2,11]. In many cases, promoter structure or gene context affects the specific repertoire of transcription and splicing factors recruited to gene promoters, as well as the rate of RNAPII elongation, which in turn affects the efficiency of splicing [11,23,24].
Conversely, splicing can affect transcription. The U2 snRNP directly interacts with TAT-SF1, a transcription elongation factor [1,8]. Recruitment of TAT-SF1 to spliced genes in turn stimulates elongation by binding to and activating the positive transcription elongation factor b (P-TEFb) complex [9], which contains the kinase responsible for phosphorylation of the RNAPII CTD on serine 2 (Ser2) [25]. The essential SR splicing factor SC35 also recruits P-TEFb to specific pre-mRNAs, an interaction that is vital to promote more efficient RNAPII escape from pause sites [26].
Coupling of transcription and 3′-end formation
During 3′-end formation, the nascent pre-mRNA is cleaved, released from the DNA template and, in most cases, polyadenylated and exported to the cytoplasm [27]. Components of the CPA machinery are recruited to promoters and transcription sites through direct interactions with multiple targets within the transcription machinery, including RNAPII, the RNAPII-associated transcription factor TFIID and transcriptional coactivators such as PC4 [8,9,27]. Pausing of RNAPII near the promoter and in the 3′-flanking region of the gene might enhance association of 3′-end processing factors with the RNAPII ternary complex [28]. The promoter structure likewise modulates the specificity of 3′-end processing factor recruitment by influencing the modification state of the RNAPII CTD [27]. Confirming the importance of RNAPII in coordinating the CPA reaction, purified RNAPII or the CTD can stimulate CPA in vitro [29]. Thus, the efficiency of 3′-end cleavage and polyadenylation is dramatically enhanced by coupling to transcription.
RNA editing
The most common type of RNA editing is conversion of adenosine to inosine, which is catalyzed by adenosine deaminases acting on RNA (ADARs). As inosine is read as guanosine during translation, editing of a single nucleotide can introduce or remove a stop codon or splice site, as well as alter single codons [30]. Because intronic sequences are frequently required for editing, editing must occur before splicing. Transcriptional coupling seems to be crucial for coordinating these events. For example, for both the glutamate receptor B and ADAR2 pre-mRNAs, the RNAPII CTD is required to delay splicing until after ADAR editing has occurred [31–33].
RNA export
The coupling of mRNA export to transcription was initially suggested by the observation that spliced mRNAs exit the nucleus more efficiently than unspliced mRNAs or RNAs derived from cDNAs [9]. In vertebrates, this increase in export efficiency is mediated by the transcription-export complex (TREX) complex, which consists of two export factors, Aly and UAP56, in conjunction with the THO complex, which is involved in transcription elongation [34]. The TREX complex is recruited to the 5′ ends of pre-mRNAs through a direct interaction between Aly and CBP80 of the CBC (Figure 1Ai) [34]. This cap-dependent recruitment of the TREX complex might be stabilized by interactions between TREX and the exon-junction complex (EJC), which is deposited during splicing just upstream of exon–exon junctions (Figure 1Aii) [8,34]. Splicing-induced structural changes within the mRNP can promote the CBP80–Aly/TREX interaction as well [34]. Splicing-dependent recruitment and stabilization of TREX explain why mRNAs produced from cDNAs are not efficiently exported, even though these transcripts acquire a 5′ cap.
Surveillance
Defective pre-mRNAs and mRNAs arising from improper processing or mutation are recognized and targeted for destruction by multiple surveillance systems or checkpoints, most of which are coupled to transcription [16]. Soon after processive transcription begins, the transcription complex is stalled by binding of NELF (negative elongation factor) and DSIF (DRB-sensitivity inducing factor) to RNAPII, thereby facilitating 5′-end capping and recruitment of 3′-end processing factors [28]. Subsequent binding of capping enzymes to Ser5P CTD disables NELF- and DSIF-mediated repression of transcription and allows RNAPII to pass through this checkpoint [16]. Another checkpoint occurs during 3′-end processing, as transcripts that are not properly cleaved and polyadenylated are retained at transcription sites through a mechanism that involves the CTD [35]. The exosome, which is a complex of several 3′–5′ exoribonucleases, also participates in cotranscriptional surveillance of pre-mRNAs and ensures that aberrantly processed transcripts are degraded [36]. A component of the nuclear exosome, Rrp6, is recruited to elongating RNAPII through interactions with transcription elongation factor Spt6, which binds Ser2P CTD [37]. Depletion of the Spt6 binding partner Iws1 reduces recruitment of Rrp6 to nascent transcripts, resulting in mRNA processing and export defects [37]. Nonsense-mediated decay (NMD) is yet another surveillance system; it subjects mRNAs with premature translation termination codons to degradation [38]. Certain components of the NMD machinery are deposited on spliced mRNAs cotranscriptionally with the EJC. If a stop codon stalls the ribosome greater than 50 nucleotides upstream of an exon–exon junction, then the mRNA is degraded primarily in the cytoplasm during the initial (‘pioneer’) round of translation [38]. Coupling of these multiple surveillance pathways to transcription greatly increases the fidelity of gene expression by rapidly targeting aberrant transcripts for decay, in many cases even before they are released from transcription sites.
Chromatin modification and pre-mRNA processing
During transcription, the DNA template, which is wrapped around histone octamers to form nucleosomes, is rearranged and made accessible to the transcription apparatus. Interestingly, histone remodeling and modification complexes affect not only the DNA structure, but also splicing [39]. For example, the SWI/SNF complex uses the energy of ATP hydrolysis to fashion an altered chromatin state at promoters, allowing transcription factors to bind [39]. For some genes, this complex is also detected within the coding regions [40]. The SWI/SNF catalytic subunit, Brm, promotes exon inclusion both by recruiting spliceosomal components to alternative exons and by inducing selective accumulation of a hyperphosphorylated form of RNAPII with a decreased rate of elongation over alternative exons [40]. Thus, splicing is enhanced through both physical and kinetic coupling to transcription by means of a protein involved in nucleosome remodeling.
Specific chromatin modifications can also directly recruit spliceosomal proteins to nascent pre-mRNAs. An example is the chromodomain-containing protein CHD1, which binds to the active chromatin mark of trimethylated histone H3 lysine 4 (H3K4me3) and interacts with the U2 snRNP, thereby tethering a core spliceosomal component to active DNA [41].
Coupling between pre-mRNA processing events
In addition to coupling to transcription, specific molecular interactions link the various pre-mRNA processing reactions to each other (Figure 1A). The CBC not only recruits export factors to nascent pre-mRNAs [34], but also interacts directly with splicing factors to stimulate recognition of splice sites in the cap-proximal intron [9]. Similarly, splicing factors recruited to the 3′ splice site of the terminal intron or to specific sequence elements in the 3′ end of the transcript bind to polyadenylation factors, thus increasing the efficiency of 3′-end CPA (Figure 1Aiii) [2,27]. Polyadenylation complexes can reciprocally stimulate splicing of the 3′-most intron [27]. An additional level of coupling exists in that 3′-end CPA proceeds only after splicing of the 3′-terminal exon [42]. Ultimately, transcription termination is tightly coupled to 3′-end formation and requires an intact CPA signal [43].
The production of noncoding snoRNAs is also coupled to transcription and to splicing. Proteins involved in the processing of intronic box C/D snoRNAs are recruited to spliceosomal complexes specifically in the C1 stage, linking box C/D snoRNA release to splicing [44]. Similarly, box H/ACA snoRNA biogenesis is coupled to RNAPII transcription and requires RNAPII-associated factors for efficient assembly of the snoRNA processing machinery [45].
Regulation of pre-mRNA processing
Alternative splicing and alternative 3′-end formation allow the production of distinct mRNAs from the same pre-mRNA and thus play crucial roles in expanding the coding potential of the human genome. Alternative pre-mRNA processing must be tightly regulated because alternative splice isoforms can perform antagonistic functions. Although some pre-mRNA processing factors are tissue-specific, other mechanisms can impart regulation of alternative pre-mRNA processing. Many of these involve coupling to transcription. For example, promoter structure dramatically affects alternative splice-site choice by recruiting different transcription complexes, which in turn can engage various combinations of splicing factors or lead to differences in RNAPII elongation rates [11]. Kinetic coupling is also influenced by RNAPII pause sites within the DNA template, which affect splice-site choices [11]. Cotranscriptional regulation of pre-mRNA processing can be responsive to external stimuli and cell signaling cascades, allowing splicing of some transcripts to be rapidly modulated in response to changes in the environment [46]. Signaling cascades can affect alternative splicing by either physical or kinetic coupling to transcription. For example, stimulation of mitogen-activated protein (MAP) kinase leads to phosphorylation of splicing factor Sam68, allowing it to associate with the SWI/SNF chromatin remodeling component Brm, which binds to regulatory sequences in a normally excluded variant exon of the CD44 pre-mRNA [40]. In this case, physical coupling to a chromatin-associated protein leads to inclusion of the CD44 variant exon [40,47]. Alternatively, signaling cascades can alter the elongation rate of RNAPII, in turn affecting alternative splice-site choices by kinetic coupling to transcription. For instance, DNA damaging agents such as ultraviolet (UV) radiation or neural cell depolarization in response to high levels of extracellular potassium can induce either hyperphosphorylation of RNAPII or histone modifications in the vicinity of variant exons, leading to altered rates of transcriptional elongation [48,49]. These changes lead in turn to inclusion or exclusion of variant exons during splicing and thus affect the protein isoforms produced [48,49]. Such splicing alterations are reversed following removal of the extracellular signal, demonstrating that specific and reversible epigenetic changes can have dramatic effects on alternative splicing decisions through either physical or kinetic coupling to transcription.
Pri-miRNA processing: cotranscriptional, but is it coupled to transcription?
A recent addition to the list of RNA processing events that occur cotranscriptionally is pri-miRNA cleavage by Drosha (Figure 1B). Hints initially came from observations that miRNA-containing introns are spliced more slowly than neighboring introns, and even more slowly when Drosha levels are reduced [7]. Direct evidence then emerged demonstrating that nascent RNAPII transcripts are indeed substrates for Drosha and that Drosha cleavage of intronic miRNAs occurs before splicing [50]. Consistently, chromatin immunoprecipitation (ChIP) experiments revealed that the presence of Drosha correlates with that of RNAPII at pri-miRNA genomic loci, suggesting that Drosha recruitment is linked to transcription [50]. In HeLa cells, Drosha interaction with miRNA genomic loci seems to be both restricted to and dependent on the pre-miRNA hairpin [50]. By contrast, in cell lines expressing the leukemogenic fusion protein All1/Af4, Drosha is detected on genomic miRNA loci at distances up to 3.5 kb away from a pre-miRNA hairpin; even at these loci, Drosha presence is correlated with increased pri-miRNA processing efficiency [51]. The association of Drosha with miRNA genomic loci is also dependent on its interaction with DGCR8 [50], implying that DGCR8 will be found at miRNA genomic loci as well.
Evidence for coupling of pri-miRNA processing and transcription
The finding that pri-miRNA processing occurs cotranscriptionally does not necessarily mean that pri-miRNA processing and transcription are functionally coupled. However, recent observations that pri-miRNAs are processed with enhanced efficiency at their transcription sites provide indirect support for coupling. Transiently expressed pri-miRNAs that lack a CPA signal are retained on the DNA template and are processed to pre- and mature miRNA with 3- to 4-fold greater efficiency than pri-miRNAs that undergo CPA [52]. By contrast, pri-miRNAs containing either a viral RNA stabilizing element or a ribozyme near their 3′ end accumulate to high nuclear levels after release from transcription sites, but are not efficiently processed to pre- or mature miRNA [52,53]. For endogenous pri-miRNAs, those that undergo efficient processing are enriched in chromatin-associated nuclear fractions [50,52]. These findings raise the questions of how coupling of transcription and pri-miRNA processing might be achieved and whether the main mechanisms of physical, kinetic and allosteric coupling contribute. Recruitment of pre-mRNA processing factors through the CTD or other RNAPII-specific transcription factors is clearly important for enhancing the efficiency of capping, splicing and 3′-end formation. Although a direct interaction between the CTD and Drosha or DGCR8 has not yet been demonstrated, many proteins that interact with Drosha also interact with the CTD (Table 1), suggesting that Drosha recruitment might be indirectly mediated by the CTD. In addition, sequences or structural features within the nascent pri-miRNA could act cooperatively with the CTD or associated factors to promote the formation of stable complexes involving RNAPII and Microprocessor components. Kinetic coupling could explain the finding that increased retention of the nascent pri-miRNA at the site of transcription by means of deletion of 3′-end processing signals or use of a miRNA promoter leads to an increase in pri-miRNA processing efficiency [52,54]. Here, kinetic competition between transcription and release of the pri-miRNA from the DNA template seem strikingly similar to the situation whereby stalling of RNAPII induced by the topoisomerase I inhibitor camptothecin results in enhanced accumulation of splicing factors on the constitutively spliced FOS pre-mRNA [20], leading to an increase in cotranscriptional FOS splicing. Thus, in the case of both constitutive splicing at strong splice sites [20] and pri-miRNA cleavage [52,54], processing is limited by kinetic competition between elongation and release of the completed transcript from the template.
Table 1. Regulators of pri-miRNA processing.
| Protein | Major function | Role in pri-miRNA processing | Link to transcription | Refs |
|---|---|---|---|---|
| ADAR1 & 2 | RNA editing (A→I) | Blocks Drosha cleavage or Dicer cleavage | Coordinated with splicing by the CTD | [30–33,73] |
| ALL/AF4 ALL/AF9 | Histone methyl-transferase fusion protein | Recruits Drosha to DNA resulting in enhanced pri-miRNA processing | Regulates transcription through chromatin remodeling | [51,87] |
| Ars2 | Essential for maintenance of cell proliferation | Interacts with Drosha, enhances processing of specific pri-miRNAs | Interacts with CBP80 and CBP20 | [59] |
| hnRNPA1 | Splicing regulation | Binds pri-miRNA terminal loop, promotes processing to pre-miRNA | Interacts with nascent pre-mRNA, regulates alternative splicing | [65,80,88] |
| KSRP | Splicing regulation mRNA decay | Interacts with Drosha and Dicer, binds to pri-miRNA terminal loop, promotes processing to pre- and mature miRNA | Regulates alternative splicing cotranscriptionally | [78,79,89,90] |
| Lin-28 | Transcription factor Translation regulation | Binds to pri-miRNA terminal loop, inhibits processing to pre-miRNA | Binds to DNA, activates transcription | [66-68,91,92] |
| Nanog | Transcription factor | Interacts with Drosha and p68, promotes processing to pre- and mature miRNA | Activates or represses transcription | [72,93,94] |
| NF90/NF45 | Transcription factor | Interacts with Drosha, inhibits Drosha cleavage of some pri-miRNAs | Facilitates pre-mRNA splicing, activates transcription | [5,69,95] |
| p53 | Transcription factor Tumor suppressor | Interacts with Drosha and p68, promotes processing to pre- and mature miRNA | Regulates transcription in response to DNA damage | [71,96] |
| p68/p72 | RNA helicase | Interacts with Drosha, enhances Drosha cleavage of some pri-miRNAs | Interacts with CTD, transcriptional coactivators | [64,97] |
| SMAD1/SMAD5 | Signal transduction Transcription regulation | Recruits Drosha and p68 to pri-miRNAs after TGF-β stimulation to miRNAs after TGF-β stimulation to enhance processing to pre-miRNA | Interacts with p68, activates transcription | [70,98] |
| SNIP1 | Transcription regulation | Interacts with Drosha, enhances processing of specific pri-miRNAs | Interacts with transcription factors including SMADs, recruits RNA processing factors to DNA | [57,58,99] |
Proteins with roles in pri-miRNA processing are listed and their known associations with transcription are described. Note that several other proteins with roles in transcription or cotranscriptional processes also interact with Drosha [5], including EWS, FUS/TLS, TAF15, TDP-43 and SRPK1a; these proteins might play a role in cotranscriptional pri-miRNA processing as well (see also [53]).
Promoter structure and gene context might also influence the coupling of pri-miRNA processing to transcription by enhancing the recruitment of Microprocessor components. For instance, transient expression of pri-miRNAs from an endogenous miRNA promoter leads to higher levels of Drosha recruitment, increased retention of the pri-miRNA on the DNA template and enhanced pri-miRNA processing in comparison with pri-miRNAs expressed from mRNA or snRNA RNAPII promoters [54]. Interestingly, some intronic pri-miRNAs have recently been shown to contain their own promoter elements, which do not correspond to the host protein gene promoter [55,56].
Chromatin structure or chromatin remodeling complexes could likewise affect Drosha association with active pri-miRNA genomic loci. Notably, the active chromatin mark of H3K4me3 binds to CHD1, which in turn binds to NF90 [41], a component of the Microprocessor complex that copurifies with Drosha [5]. Similarly, Drosha interacts with SNIP1 [57], a protein that controls transcription through interaction with several transcription factors and with Brg1, a component of the SWI/SNF chromatin remodeling complex [58]. Pri-miRNA processing is also linked to another cotranscriptional process: 5′-end capping. The CBC is important for recruiting Ars2 (arsenite resistance protein 2), a protein that has an important role in maintenance of proliferative expansion in mammalian cells [59]. Ars2 interacts with the CBC, as well as with Drosha, and is necessary for both the stability and processing of a subset of pri-miRNAs [59]. These data suggest that Drosha recruitment to nascent pri-miRNAs could be mediated by Ars2 interaction with the CBC (Figure 1B). However, as Ars2 depletion does not affect all miRNAs, and Ars2 is expressed only in proliferating cells, it is likely that other mechanisms contribute to the recruitment of the Microprocessor complex to nascent pri-miRNAs.
Interestingly, pri-miRNA processing might reciprocally affect other cotranscriptional processes, such as splicing and transcription termination. For instance, Drosha and DGCR8 associate with components of the spliceosome [60]. Cotranscriptional cleavage of nascent pri-miRNAs by Drosha results in recruitment of the 5′–3′ and 3′–5′ exonucleases XRN2 and PMScl100, respectively, to the newly generated ends of the cleaved transcript (Figure 1Biii) [50,54]. After Drosha cleavage of intergenic pri-miRNAs, recruitment of XRN2 might result in termination of transcription through a hypothesized torpedo-like mechanism in which its 5′–3′ exonuclease activity allows XRN2 to catch up with RNAPII and ‘torpedo’ it off the DNA template [54]. For intronic miRNAs, the degradation of intronic sequences might enhance splicing efficiency by clearing away sequences that interfere with the splicing apparatus, thereby allowing more efficient exon ligation [50]. Such coordination would enable a single primary transcript to be efficiently and accurately processed to produce both a mature miRNA and a protein-coding mRNA. Together, these findings argue for intricate coordination of pri-miRNA processing with transcription, splicing, exonucleolytic degradation and transcription termination.
Regulation of miRNA expression
The expression of miRNA is extensively regulated by developmental and tissue-specific signaling. Control of miRNA expression can occur at the level of transcription, as many pri-miRNA promoters contain binding sites for RNAPII-associated transcription factors, such as p53, Myc and muscle-specific myogenin, resulting in expression of these miRNAs only under specific conditions [6]. MiRNA expression can be regulated at several posttranscriptional steps as well, including pri-miRNA cleavage by the Microprocessor complex, pre-miRNA export, Dicer cleavage and miRNA stability [6].
Regulation of pri-miRNA processing plays a crucial role during development, and a block in Microprocessor activity contributes to aberrant miRNA expression in numerous diseases, including cancer and neurological disorders [61–63]. Several proteins interact with Drosha and can either promote or inhibit cleavage of specific pri-miRNAs (Table 1). For example, the DEAD-box helicases p68 and p72 interact with Drosha to increase the processing efficiency of a subset of miRNAs [64]. The multifunctional protein hnRNP A1 is necessary for processing of miR-18a from its primary transcript [65]. The RNA binding protein Lin-28 plays a crucial role in inhibiting Drosha cleavage of pri-let-7 family members during embryonic development [66–68]. Transcription factors NF90 and NF45 also interact with Drosha and affect the processing efficiency of some pri-miRNAs [69]. Interestingly, terminal loop nucleotides within the pri-miRNA hairpin play essential roles in regulation by several of these auxiliary proteins (Box 2). As most of these Drosha-associated proteins function in both transcription and regulation of alternative splicing (Table 1) and are known to bind to nascent transcripts, future research might well uncover their importance for cotranscriptional regulation of pri-miRNA processing.
Box 2. The importance of loop nucleotides in pri-miRNA processing.
Several recent studies have demonstrated that nucleotides within the terminal loop of some pri-miRNA hairpins play an important role in regulation of miRNA biogenesis, as well as in the function of the miRNA itself [85]. These terminal loops have been demonstrated to be the major binding sites for regulatory proteins such as hnRNP A1, Lin-28 and KSRP [65,68,78,80]. Protein binding is crucial for regulation of processing, as hnRNPA1 and KSRP are required for Drosha cleavage of specific pri-miRNAs, whereas Lin-28 binding inhibits cleavage (see the text).
The majority of pri-miRNA terminal loops are not well conserved across mammalian species. However, a subset of miRNA precursors (14%) exhibit higher conservation in the loop sequence than expected [80]. Consistently, transfection of 2′-O-methyl-modified oligonucleotides complementary to terminal loop sequences inhibits cleavage of pri-miRNAs with conserved loops, but has no effect on pri-miRNAs with nonconserved loops [80]. Thus, terminal loop binding sites for auxiliary proteins seem to be crucial to either promote or inhibit Drosha cleavage of select pri-miRNAs.
Further studies suggest that the terminal loop might play additional roles in miRNA function. The sequences of mature miR-181a-1 and miR-181c differ in only one nucleotide, but these miRNAs have dramatically different functions in thymocyte T cells [85]. Surprisingly, the terminal loop sequences of the corresponding pri-miRNAs dictate final function, as demonstrated by terminal loop swapping experiments [85]. It has been postulated that the terminal loop affects Drosha cleavage, binding of export factors, subcellular localization, Dicer processing or loading into the RISC complex [85]. As many terminal loop binding proteins, such as hnRNPA1 and KSRP, interact with Drosha and might therefore bind cotranscriptionally, these findings raise the possibility that the cotranscriptional binding of multifunctional proteins to terminal loops influences the final fate and function of the mature miRNA.
External signaling pathways can also stimulate pri-miRNA processing. Induction of transforming growth factor TGF-β or BMP (bone morphogenetic protein) signaling in smooth muscle cells activates SMAD proteins, which then interact with p68 and Drosha to enhance Drosha cleavage of pri-miR-21 [70]. In response to DNA damage, activated p53 binds to Drosha complexed to p68 and enhances the processing efficiency of several pri-miRNAs, indicating that regulation of pri-miRNA cleavage by Drosha is a component of the p53-induced DNA damage response [71]. Similarly, activation of protein kinase Cε (PKCε), through events such as hyaluronan binding to the CD44 receptor, results in phosphorylation of the transcription factor Nanog, its translocation to the nucleus, and its subsequent interaction with the Drosha–p68 complex to promote processing of pri-miR-21 [72]. It will be of great interest to determine whether physical or kinetic coupling to transcription contributes to regulation of pri-miRNA processing in response to external signaling cascades, promoter structure, chromatin structure, chromatin remodeling complexes or transcription factors. The biogenesis of miRNAs can be regulated through ADAR editing as well. Several pri-miRNAs undergo editing at multiple sites, resulting in an altered structure of the miRNA-containing hairpin, thus preventing cleavage by Drosha or Dicer [73]. Because the CTD coordinates cotranscriptional splicing and pre-mRNA editing [31–33], it is possible that editing of pri-miRNAs occurs cotranscriptionally, orchestrated by the CTD. Observations that miRNA-containing introns are spliced more slowly than adjacent introns [7] support the idea that splicing of miRNA-containing introns is delayed until after Drosha cleavage. In this scenario, the CTD could act to ensure that the miRNA-harboring intron is not excised prematurely. Conversely, rapid splicing promoted by the CTD or other interacting factors could render mRNA and miRNA expression from the same transcript mutually exclusive. Interestingly, Microprocessor components Drosha and DGCR8 regulate each other posttranscriptionally. A pre-miRNA-like hairpin in the 5′ untranslated region (UTR) of DGCR8 mRNA is cleaved by the Microprocessor complex, downregulating DGCR8 expression [74,75]. In Drosophila, several protein-coding pre-mRNAs that do not contain annotated miRNAs also contain pre-miRNA-like hairpins in their 5′ UTRs and are probably substrates for cleavage by the Microprocessor complex [76]. Thus, Microprocessor cleavage might impact transcripts other than pri-miRNAs and could involve cotranscriptional binding of Drosha.
In summary, many proteins involved in regulation of pri-miRNA processing by Drosha (Table 1) play roles in transcription, in the cotranscriptional regulation of pre-mRNA processing and/or in the coordination of cotranscriptional events. It will therefore be of significant interest to establish whether the regulation of pri-miRNA processing also occurs cotranscriptionally, and whether any of the mechanisms of coupling described for pre-mRNAs above have a role in this coordination (Box 3).
Box 3. Outstanding questions.
How are Drosha and DGCR8 recruited to sites of pri-miRNA transcription?
Is DGCR8 recruited cotranscriptionally, and, if so, does its recruitment depend on the nascent pri-miRNA and on RNAPII?
Does regulation of pri-miRNA processing occur cotranscriptionally?
Does promoter identity influence recruitment of the Microprocessor and/or pri-miRNA processing efficiency?
Does RNAPII or the CTD directly or indirectly recruit the Microprocessor complex?
Does CTD phosphorylation affect Microprocessor recruitment?
Does the rate of transcription elongation affect pri-miRNA processing?
Do chromatin modifications or chromatin remodeling complexes affect recruitment or activity of the Microprocessor complex?
Does DNA damage result in changes in pri-miRNA processing?
Does pri-miRNA processing have reciprocal effects on transcription elongation and/or other pre-mRNA processing events?
Are factors that bind to pri-miRNAs cotranscriptionally carried with the pre-miRNA into the cytoplasm and do these proteins participate in the final functions of the mature miRNA?
Do RNA helicases such as p68/p72 have roles in disassembly of protein complexes and remodeling of miRNPs?
Do components of the Microprocessor complex that are known to have roles in transcription and regulation of pre-mRNA processing events also have roles in miRNA biogenesis (i.e. EWS, TLS, FUS)?
How widespread is the role of Microprocessor cleavage of hairpins in protein-coding gene 5′ UTRs in mRNA stability control?
Coupling nuclear events to microRNA fate
Nuclear history can dramatically affect the final functions of pre-mRNA transcripts [8,77]. For example, some SR proteins, as well as components of the EJC, remain on the mRNA after splicing and exit to the cytoplasm, providing a ‘molecular memory’ of nuclear events that can modulate the localization, stability or translational efficiency of the transcript [8,19,77]. An important area of future research will be to determine whether the nuclear history of a pri-miRNA likewise affects the functioning of its mature miRNA product. Supporting evidence is provided by the multifunctional protein KSRP. KSRP is required for Drosha cleavage of a subset of pri-miRNAs but remains associated with the pre-miRNA during nuclear export by Exportin 5 and promotes Dicer processing [78]. As KSRP is also involved in recruiting the decay machinery to mRNAs containing AU-rich elements in their 3′ UTRs [79], it might remain associated with the mature miRNA in the RNA induced silencing complex (RISC) and contribute to miRNA-mediated mRNA decay. Several other multifunctional proteins that are mediators of pri-miRNA processing by Drosha or that bind to pri-miRNA terminal loops also shuttle between the nucleus and cytoplasm and have reported roles in the regulation of translation, including hnRNP A1, hnRNP L, PTB, p53, and lin-28 (Box 2) [71,80,81]. Drosha-interacting proteins that are found in cytoplasmic complexes containing Argonautes, Dicer and RISC activity include NF90, NF45, SRPK1 and hnRNP proteins [5,82]. RBM4, another multifunctional protein with roles in both splicing and translation [83,84], interacts with Argonaute 2 and is required for miRNA-mediated repression of translation [82]. RBM4 binds to several pre-mRNAs cotranscriptionally [84], raising the possibility that it could load onto pri-miRNAs cotranscriptionally as well. Thus, cotranscriptional binding of multifunctional regulatory proteins could dictate the final fate and function of the mature miRNA, consistent with the recent demonstration that pri-miRNA terminal loops play a role in determining mature miRNA function (Box 2) [85].
Concluding remarks
Future experiments should focus on elucidating the molecular mechanisms underlying pri-miRNA processing (Box 3) and will need to consider these events in the context of transcription, chromatin state and pre-mRNA processing. Such investigations promise to reveal novel connections that allow pri-miRNA processing to be interwoven into the elaborate nuclear network of gene expression.
Acknowledgments
We are grateful to Anita Nag, Kasandra Riley, Kazio Tycowski and Kristina Herbert for critical comments on this manuscript. We thank all members of the Steitz lab for stimulating discussions. This work was supported by grant R01GM026154 from the NIGMS. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIGMS or the NIH. J.A.S is an investigator of the Howard Hughes Medical Institute.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Neugebauer KM. On the importance of being co-transcriptional. J Cell Sci. 2002;115:3865–3871. doi: 10.1242/jcs.00073. [DOI] [PubMed] [Google Scholar]
- 2.Bentley DL. Rules of engagement: co-transcriptional recruitment of pre-mRNA processing factors. Curr Opin Cell Biol. 2005;17:251–256. doi: 10.1016/j.ceb.2005.04.006. [DOI] [PubMed] [Google Scholar]
- 3.Matera AG, et al. Non-coding RNAs: lessons from the small nuclear and small nucleolar RNAs. Nat Rev Mol Cell Biol. 2007;8:209–220. doi: 10.1038/nrm2124. [DOI] [PubMed] [Google Scholar]
- 4.Carthew RW, Sontheimer EJ. Origins and mechanisms of miRNAs and siRNAs. Cell. 2009;136:642–655. doi: 10.1016/j.cell.2009.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gregory RI, et al. The Microprocessor complex mediates the genesis of microRNAs. Nature. 2004;432:235–240. doi: 10.1038/nature03120. [DOI] [PubMed] [Google Scholar]
- 6.Kim VN, et al. Biogenesis of small RNAs in animals. Nat Rev Mol Cell Biol. 2009;10:126–139. doi: 10.1038/nrm2632. [DOI] [PubMed] [Google Scholar]
- 7.Kim YK, Kim VN. Processing of intronic microRNAs. EMBO J. 2007;26:775–783. doi: 10.1038/sj.emboj.7601512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Moore MJ, Proudfoot NJ. Pre-mRNA processing reaches back to transcription and ahead to translation. Cell. 2009;136:688–700. doi: 10.1016/j.cell.2009.02.001. [DOI] [PubMed] [Google Scholar]
- 9.Maniatis T, Reed R. An extensive network of coupling among gene expression machines. Nature. 2002;416:499–506. doi: 10.1038/416499a. [DOI] [PubMed] [Google Scholar]
- 10.Egloff S, Murphy S. Cracking the RNA polymerase II CTD code. Trends Genet. 2008;24:280–288. doi: 10.1016/j.tig.2008.03.008. [DOI] [PubMed] [Google Scholar]
- 11.Kornblihtt AR. Coupling transcription and alternative splicing. Adv Exp Med Biol. 2007;623:175–189. doi: 10.1007/978-0-387-77374-2_11. [DOI] [PubMed] [Google Scholar]
- 12.Shuman S. Structure, mechanism, and evolution of the mRNA capping apparatus. Prog Nucleic Acid Res Mol Biol. 2001;66:1–40. doi: 10.1016/s0079-6603(00)66025-7. [DOI] [PubMed] [Google Scholar]
- 13.Das R, et al. SR proteins function in coupling RNAP II transcription to pre-mRNA splicing. Mol Cell. 2007;26:867–881. doi: 10.1016/j.molcel.2007.05.036. [DOI] [PubMed] [Google Scholar]
- 14.Auboeuf D, et al. A subset of nuclear receptor coregulators act as coupling proteins during synthesis and maturation of RNA transcripts. Mol Cell Biol. 2005;25:5307–5316. doi: 10.1128/MCB.25.13.5307-5316.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Long JC, Caceres JF. The SR protein family of splicing factors: master regulators of gene expression. Biochem J. 2009;417:15–27. doi: 10.1042/BJ20081501. [DOI] [PubMed] [Google Scholar]
- 16.de Almeida SF, Carmo-Fonseca M. The CTD role in cotranscriptional RNA processing and surveillance. FEBS Lett. 2008;582:1971–1976. doi: 10.1016/j.febslet.2008.04.019. [DOI] [PubMed] [Google Scholar]
- 17.Loomis RJ, et al. Chromatin binding of SRp20 and ASF/SF2 and dissociation from mitotic chromosomes is modulated by histone H3 serine 10 phosphorylation. Mol Cell. 2009;33:450–461. doi: 10.1016/j.molcel.2009.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dye MJ, et al. Exon tethering in transcription by RNA polymerase II. Mol Cell. 2006;21:849–859. doi: 10.1016/j.molcel.2006.01.032. [DOI] [PubMed] [Google Scholar]
- 19.Sapra AK, et al. SR protein family members display diverse activities in the formation of nascent and mature mRNPs in vivo. Mol Cell. 2009;34:179–190. doi: 10.1016/j.molcel.2009.02.031. [DOI] [PubMed] [Google Scholar]
- 20.Listerman I, et al. Cotranscriptional coupling of splicing factor recruitment and precursor messenger RNA splicing in mammalian cells. Nat Struct Mol Biol. 2006;13:815–822. doi: 10.1038/nsmb1135. [DOI] [PubMed] [Google Scholar]
- 21.Natalizio BJ, et al. The carboxyl-terminal domain of RNA polymerase II is not sufficient to enhance the efficiency of pre-mRNA capping or splicing in the context of a different polymerase. J Biol Chem. 2009;284:8692–8702. doi: 10.1074/jbc.M806919200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Barboric M, et al. 7SK snRNP/P-TEFb couples transcription elongation with alternative splicing and is essential for vertebrate development. Proc Natl Acad Sci U S A. 2009;106:7798–7803. doi: 10.1073/pnas.0903188106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pleiss JA, et al. Rapid, transcript-specific changes in splicing in response to environmental stress. Mol Cell. 2007;27:928–937. doi: 10.1016/j.molcel.2007.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bittencourt D, et al. Cotranscriptional splicing potentiates the mRNA production from a subset of estradiol-stimulated genes. Mol Cell Biol. 2008;28:5811–5824. doi: 10.1128/MCB.02231-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bres V, et al. The multi-tasking P-TEFb complex. Curr Opin Cell Biol. 2008;20:334–340. doi: 10.1016/j.ceb.2008.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lin S, et al. The splicing factor SC35 has an active role in transcriptional elongation. Nat Struct Mol Biol. 2008;15:819–826. doi: 10.1038/nsmb.1461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Danckwardt S, et al. 3′ end mRNA processing: molecular mechanisms and implications for health and disease. EMBO J. 2008;27:482–498. doi: 10.1038/sj.emboj.7601932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Glover-Cutter K, et al. RNA polymerase II pauses and associates with pre-mRNA processing factors at both ends of genes. Nat Struct Mol Biol. 2008;15:71–78. doi: 10.1038/nsmb1352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hirose Y, Ohkuma Y. Phosphorylation of the C-terminal domain of RNA polymerase II plays central roles in the integrated events of eucaryotic gene expression. J Biochem. 2007;141:601–608. doi: 10.1093/jb/mvm090. [DOI] [PubMed] [Google Scholar]
- 30.Bass BL. RNA editing by adenosine deaminases that act on RNA. Annu Rev Biochem. 2002;71:817–846. doi: 10.1146/annurev.biochem.71.110601.135501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Riedmann EM, Jantsch MF. An editor controlled by transcription. EMBO Rep. 2006;7:269–270. doi: 10.1038/sj.embor.7400650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ryman K, et al. The C-terminal domain of RNA Pol II helps ensure that editing precedes splicing of the GluR-B transcript. RNA. 2007;13:1071–1078. doi: 10.1261/rna.404407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schoft VK, et al. Regulation of glutamate receptor B pre-mRNA splicing by RNA editing. Nucleic Acids Res. 2007;35:3723–3732. doi: 10.1093/nar/gkm314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cheng H, et al. Human mRNA export machinery recruited to the 5′ end of mRNA. Cell. 2006;127:1389–1400. doi: 10.1016/j.cell.2006.10.044. [DOI] [PubMed] [Google Scholar]
- 35.Custodio N, et al. Splicing- and cleavage-independent requirement of RNA polymerase II CTD for mRNA release from the transcription site. J Cell Biol. 2007;179:199–207. doi: 10.1083/jcb.200612109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schmid M, Jensen TH. The exosome: a multipurpose RNA-decay machine. Trends Biochem Sci. 2008;33:501–510. doi: 10.1016/j.tibs.2008.07.003. [DOI] [PubMed] [Google Scholar]
- 37.Yoh SM, et al. The Spt6 SH2 domain binds Ser2-P RNAPII to direct Iws1-dependent mRNA splicing and export. Genes Dev. 2007;21:160–174. doi: 10.1101/gad.1503107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Isken O, Maquat LE. Quality control of eukaryotic mRNA: safeguarding cells from abnormal mRNA function. Genes Dev. 2007;21:1833–1856. doi: 10.1101/gad.1566807. [DOI] [PubMed] [Google Scholar]
- 39.Allemand E, et al. Splicing, transcription, and chromatin: a menage a trois. Curr Opin Genet Dev. 2008;18:145–151. doi: 10.1016/j.gde.2008.01.006. [DOI] [PubMed] [Google Scholar]
- 40.Batsche E, et al. The human SWI/SNF subunit Brm is a regulator of alternative splicing. Nat Struct Mol Biol. 2006;13:22–29. doi: 10.1038/nsmb1030. [DOI] [PubMed] [Google Scholar]
- 41.Sims RJ, 3rd, et al. Recognition of trimethylated histone H3 lysine 4 facilitates the recruitment of transcription postinitiation factors and pre-mRNA splicing. Mol Cell. 2007;28:665–676. doi: 10.1016/j.molcel.2007.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rigo F, Martinson HG. Polyadenylation releases mRNA from RNA polymerase II in a process that is licensed by splicing. RNA. 2009;15:823–836. doi: 10.1261/rna.1409209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Richard P, Manley JL. Transcription termination by nuclear RNA polymerases. Genes Dev. 2009;23:1247–1269. doi: 10.1101/gad.1792809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hirose T, et al. Splicing-dependent and -independent modes of assembly for intron-encoded box C/D snoRNPs in mammalian cells. Mol Cell. 2003;12:113–123. doi: 10.1016/s1097-2765(03)00267-3. [DOI] [PubMed] [Google Scholar]
- 45.Richard P, et al. Cotranscriptional recognition of human intronic box H/ACA snoRNAs occurs in a splicing-independent manner. Mol Cell Biol. 2006;26:2540–2549. doi: 10.1128/MCB.26.7.2540-2549.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shin C, Manley JL. Cell signalling and the control of pre-mRNA splicing. Nat Rev Mol Cell Biol. 2004;5:727–738. doi: 10.1038/nrm1467. [DOI] [PubMed] [Google Scholar]
- 47.Matter N, et al. Signal-dependent regulation of splicing via phosphorylation of Sam68. Nature. 2002;420:691–695. doi: 10.1038/nature01153. [DOI] [PubMed] [Google Scholar]
- 48.Munoz MJ, et al. DNA damage regulates alternative splicing through inhibition of RNA polymerase II elongation. Cell. 2009;137:708–720. doi: 10.1016/j.cell.2009.03.010. [DOI] [PubMed] [Google Scholar]
- 49.Schor IE, et al. Neuronal cell depolarization induces intragenic chromatin modifications affecting NCAM alternative splicing. Proc Natl Acad Sci U S A. 2009;106:4325–4330. doi: 10.1073/pnas.0810666106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Morlando M, et al. Primary microRNA transcripts are processed co-transcriptionally. Nat Struct Mol Biol. 2008;15:902–909. doi: 10.1038/nsmb.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nakamura T, et al. Oncogenic All1 fusion proteins target Drosha-mediated microRNA processing. Proc Natl Acad Sci U S A. 2007;104:10980–10985. doi: 10.1073/pnas.0704559104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pawlicki JM, Steitz JA. Primary microRNA transcript retention at sites of transcription leads to enhanced microRNA production. J Cell Biol. 2008;182:61–76. doi: 10.1083/jcb.200803111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pawlicki JM, Steitz JA. Subnuclear compartmentalization of transiently expressed polyadenylated pri-microRNAs: processing at transcription sites or accumulation in SC35 foci. Cell Cycle. 2009;8:345–356. doi: 10.4161/cc.8.3.7494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ballarino M, et al. Coupled RNA processing and transcription of intergenic primary miRNAs. Mol Cell Biol. 2009;29:5632–5638. doi: 10.1128/MCB.00664-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ozsolak F, et al. Chromatin structure analyses identify miRNA promoters. Genes Dev. 2008;22:3172–3183. doi: 10.1101/gad.1706508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Corcoran DL, et al. Features of mammalian microRNA promoters emerge from polymerase II chromatin immunoprecipitation data. PLoS One. 2009;4:e5279. doi: 10.1371/journal.pone.0005279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yu B, et al. The FHA domain proteins DAWDLE in Arabidopsis and SNIP1 in humans act in small RNA biogenesis. Proc Natl Acad Sci U S A. 2008;105:10073–10078. doi: 10.1073/pnas.0804218105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Roche KC, et al. The FHA domain protein SNIP1 is a regulator of the cell cycle and cyclin D1 expression. Oncogene. 2004;23:8185–8195. doi: 10.1038/sj.onc.1208025. [DOI] [PubMed] [Google Scholar]
- 59.Gruber JJ, et al. Ars2 links the nuclear cap-binding complex to RNA interference and cell proliferation. Cell. 2009;138:328–339. doi: 10.1016/j.cell.2009.04.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kataoka N, et al. Functional association of the Microprocessor complex with the spliceosome. Mol Cell Biol. 2009;29:3243–3254. doi: 10.1128/MCB.00360-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Thomson JM, et al. Extensive post-transcriptional regulation of microRNAs and its implications for cancer. Genes Dev. 2006;20:2202–2207. doi: 10.1101/gad.1444406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Perkins DO, et al. microRNA expression in the prefrontal cortex of individuals with schizophrenia and schizoaffective disorder. Genome Biol. 2007;8:R27. doi: 10.1186/gb-2007-8-2-r27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kluiver J, et al. Regulation of pri-microRNA BIC transcription and processing in Burkitt lymphoma. Oncogene. 2007;26:3769–3776. doi: 10.1038/sj.onc.1210147. [DOI] [PubMed] [Google Scholar]
- 64.Fukuda T, et al. DEAD-box RNA helicase subunits of the Drosha complex are required for processing of rRNA and a subset of microRNAs. Nat Cell Biol. 2007;9:604–611. doi: 10.1038/ncb1577. [DOI] [PubMed] [Google Scholar]
- 65.Guil S, Caceres JF. The multifunctional RNA-binding protein hnRNP A1 is required for processing of miR-18a. Nat Struct Mol Biol. 2007;14:591–596. doi: 10.1038/nsmb1250. [DOI] [PubMed] [Google Scholar]
- 66.Piskounova E, et al. Determinants of microRNA processing inhibition by the developmentally regulated RNA-binding protein Lin28. J Biol Chem. 2008;283:21310–21314. doi: 10.1074/jbc.C800108200. [DOI] [PubMed] [Google Scholar]
- 67.Viswanathan SR, et al. Selective blockade of microRNA processing by Lin28. Science. 2008;320:97–100. doi: 10.1126/science.1154040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Newman MA, et al. Lin-28 interaction with the Let-7 precursor loop mediates regulated microRNA processing. RNA. 2008;14:1539–1549. doi: 10.1261/rna.1155108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sakamoto S, et al. The NF90–NF45 complex functions as a negative regulator in the microRNA processing pathway. Mol Cell Biol. 2009;29:3754–3769. doi: 10.1128/MCB.01836-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Davis BN, et al. SMAD proteins control DROSHA-mediated microRNA maturation. Nature. 2008;454:56–61. doi: 10.1038/nature07086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Suzuki HI, et al. Modulation of microRNA processing by p53. Nature. 2009;460:529–533. doi: 10.1038/nature08199. [DOI] [PubMed] [Google Scholar]
- 72.Bourguignon LY, et al. Hyaluronan–CD44 interaction with PKC-epsilon promotes oncogenic signaling by the stem cell marker, Nanog and the production of microRNA-21 leading to downregulation of the tumor suppressor protein, PDCD4, anti-apoptosis and chemotherapy resistance in breast tumor cells. J Biol Chem. 2009;284:26533–26546. doi: 10.1074/jbc.M109.027466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ohman M. A-to-I editing challenger or ally to the microRNA process. Biochimie. 2007;89:1171–1176. doi: 10.1016/j.biochi.2007.06.002. [DOI] [PubMed] [Google Scholar]
- 74.Han J, et al. Posttranscriptional crossregulation between Drosha and DGCR8. Cell. 2009;136:75–84. doi: 10.1016/j.cell.2008.10.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Triboulet R, et al. Post-transcriptional control of DGCR8 expression by the Microprocessor. RNA. 2009;15:1005–1011. doi: 10.1261/rna.1591709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kadener S, et al. Genome-wide identification of targets of the drosha-pasha/DGCR8 complex. RNA. 2009;15:537–545. doi: 10.1261/rna.1319309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Giorgi C, Moore MJ. The nuclear nurture and cytoplasmic nature of localized mRNPs. Semin Cell Dev Biol. 2007;18:186–193. doi: 10.1016/j.semcdb.2007.01.002. [DOI] [PubMed] [Google Scholar]
- 78.Trabucchi M, et al. The RNA-binding protein KSRP promotes the biogenesis of a subset of microRNAs. Nature. 2009;459:1010–1014. doi: 10.1038/nature08025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Gherzi R, et al. A KH domain RNA binding protein, KSRP, promotes ARE-directed mRNA turnover by recruiting the degradation machinery. Mol Cell. 2004;14:571–583. doi: 10.1016/j.molcel.2004.05.002. [DOI] [PubMed] [Google Scholar]
- 80.Michlewski G, et al. Posttranscriptional regulation of miRNAs harboring conserved terminal loops. Mol Cell. 2008;32:383–393. doi: 10.1016/j.molcel.2008.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Polesskaya A, et al. Lin-28 binds IGF-2 mRNA and participates in skeletal myogenesis by increasing translation efficiency. Genes Dev. 2007;21:1125–1138. doi: 10.1101/gad.415007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Hock J, et al. Proteomic and functional analysis of Argonaute-containing mRNA-protein complexes in human cells. EMBO Rep. 2007;8:1052–1060. doi: 10.1038/sj.embor.7401088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Lin JC, et al. Cell stress modulates the function of splicing regulatory protein RBM4 in translation control. Proc Natl Acad Sci U S A. 2007;104:2235–2240. doi: 10.1073/pnas.0611015104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Lin JC, Tarn WY. Exon selection in alpha-tropomyosin mRNA is regulated by the antagonistic action of RBM4 and PTB. Mol Cell Biol. 2005;25:10111–10121. doi: 10.1128/MCB.25.22.10111-10121.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Liu G, et al. Pre-miRNA loop nucleotides control the distinct activities of mir-181a-1 and mir-181c in early T cell development. PLoS One. 2008;3:e3592. doi: 10.1371/journal.pone.0003592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Buratowski S. The CTD code. Nat Struct Biol. 2003;10:679–680. doi: 10.1038/nsb0903-679. [DOI] [PubMed] [Google Scholar]
- 87.Krivtsov AV, Armstrong SA. MLL translocations, histone modifications and leukaemia stem-cell development. Nat Rev Cancer. 2007;7:823–833. doi: 10.1038/nrc2253. [DOI] [PubMed] [Google Scholar]
- 88.He Y, Smith R. Nuclear functions of heterogeneous nuclear ribonucleoproteins A/B. Cell Mol Life Sci. 2009;66:1239–1256. doi: 10.1007/s00018-008-8532-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Ruggiero T, et al. LPS induces KH-type splicing regulatory protein-dependent processing of microRNA-155 precursors in macrophages. FASEB J. 2009;23:2898–2908. doi: 10.1096/fj.09-131342. [DOI] [PubMed] [Google Scholar]
- 90.Min H, et al. A new regulatory protein, KSRP, mediates exon inclusion through an intronic splicing enhancer. Genes Dev. 1997;11:1023–1036. doi: 10.1101/gad.11.8.1023. [DOI] [PubMed] [Google Scholar]
- 91.Rybak A, et al. A feedback loop comprising lin-28 and let-7 controls pre-let-7 maturation during neural stem-cell commitment. Nat Cell Biol. 2008;10:987–993. doi: 10.1038/ncb1759. [DOI] [PubMed] [Google Scholar]
- 92.Heo I, et al. Lin28 mediates the terminal uridylation of let-7 precursor microRNA. Mol Cell. 2008;32:276–284. doi: 10.1016/j.molcel.2008.09.014. [DOI] [PubMed] [Google Scholar]
- 93.Chambers I, et al. Functional expression cloning of Nanog, a pluripotency sustaining factor in embryonic stem cells. Cell. 2003;113:643–655. doi: 10.1016/s0092-8674(03)00392-1. [DOI] [PubMed] [Google Scholar]
- 94.Pan G, Pei D. The stem cell pluripotency factor NANOG activates transcription with two unusually potent subdomains at its C terminus. J Biol Chem. 2005;280:1401–1407. doi: 10.1074/jbc.M407847200. [DOI] [PubMed] [Google Scholar]
- 95.Saunders LR, et al. Characterization of two evolutionarily conserved, alternatively spliced nuclear phosphoproteins, NFAR-1 and -2, that function in mRNA processing and interact with the double-stranded RNA-dependent protein kinase, PKR. J Biol Chem. 2001;276:32300–32312. doi: 10.1074/jbc.M104207200. [DOI] [PubMed] [Google Scholar]
- 96.Vogelstein B, et al. Surfing the p53 network. Nature. 2000;408:307–310. doi: 10.1038/35042675. [DOI] [PubMed] [Google Scholar]
- 97.Fuller-Pace FV, Ali S. The DEAD box RNA helicases p68 (Ddx5) and p72 (Ddx17): novel transcriptional co-regulators. Biochem Soc Trans. 2008;36:609–612. doi: 10.1042/BST0360609. [DOI] [PubMed] [Google Scholar]
- 98.Schmierer B, Hill CS. TGFbeta-SMAD signal transduction: molecular specificity and functional flexibility. Nat Rev Mol Cell Biol. 2007;8:970–982. doi: 10.1038/nrm2297. [DOI] [PubMed] [Google Scholar]
- 99.Bracken CP, et al. Regulation of cyclin D1 RNA stability by SNIP1. Cancer Res. 2008;68:7621–7628. doi: 10.1158/0008-5472.CAN-08-1217. [DOI] [PMC free article] [PubMed] [Google Scholar]