Abstract
The purpose of the present study was to describe the bacterial diversity in the oral cavity of the elderly without root caries using bacterial microarrays, and to determine the site- and subject-specificity of bacterial colonization. Samples were collected from the tongue dorsum, mucosa of the buccal fold, hard palate, supragingival plaque from sound root surfaces, and subgingival plaque from the same roots. A new 16S rRNA gene based microarray method was used for the simultaneous detection of approximately 300 bacterial species. Overall, 175 species and clusters were detected, representing 8 phyla. Species belonging to the genera Streptococcus, Veillonella and Fusobacterium were common in all sites. The number of species per subject varied from 51 to 81. Statistical analyses revealed about 40 species or clusters with significant associations with at least one of the sites. The bacterial diversity was highest in the cheek and palate regions. Species typically associated with caries and periodontitis were detected rarely or not at all. The oral bacterial flora of the elderly appears to be diverse, and to a large extent site- rather than subject-specific.
Keywords: Oral microflora, bacteria, microbiology, aged 80 and over, microarray analysis
Introduction
The microbial flora of the human oral cavity can be highly diverse, encompassing approximately 700 bacterial species [1], of which about ½ has not yet been grown in vitro. It is therefore of interest to characterize the normal flora of the elderly present in different oral sites. Significant site- and subject-specificity has been shown in the microflora of the oral cavity of adults, but not of the elderly [2].
Various bodily functions, such as the physiologic defense mechanisms and the salivary gland secretions, are reduced with age [3, 4]. Colonization of the oral cavity in the elderly with bacteria that may be normally a small part of the oral resident flora has been described in the literature [5, 6]. Other age-related factors that can influence the composition of the oral microflora, are dentures, hormones, long-term medication, diet and reduced oral hygiene [7]. Together, these factors suggest that the oral microflora of the elderly may be different from that of younger individuals.
Most studies characterizing the oral microbiota have focused on young and middle-aged adults or on isolated sites [8, 9, 11]. Only limited information is available on the oral microflora of the elderly, and these studies used culture-based methods of single isolated sites or were limited to a few species [6, 8, 12–14]. The present study employed the Human Oral Microbial Identification Microarray (HOMIM) which targets about 300 predominant oral bacterial species, including cultivable and not-yet-cultivated phylotypes [15]. The method allowed the investigation of a statistically significant number of samples, i.e., from five different sites in each of 30 subjects. The aim of the study was to determine the site- and subject-specificity of the bacterial profiles in the oral cavity of the elderly.
Materials and Methods
Subjects
Thirty elderly subjects (24 females and 6 males) were included in the study. Eight of the subjects were residents of a long-term care facility (Cathinka Guldberg-Centre for elderly in Oslo, Norway), the other subjects were non-residents. Residence time varied from 17 to 44 months prior to examination. Mean age of the subjects was 83.9 years (range 73–93), mean number of teeth 23 (range 9–36). All the subjects were examined orally prior to sampling. The subjects showed no signs of oral mucosal disease, or root caries and had no antibiotic use up to 1 month prior to sampling. The subjects did not perform tooth cleaning the evening and the morning before plaque sampling. Eight of the subjects showed local or slight general redness and inflammation of the gingiva. Two of these subjects had periodontal pocket depths of the sampled tooth up to 4 mm, while the pockets were less than 3 mm in the other six. The recession (defined as the distance from the gingival level to the dento-enamel juncture) measured on the sampled site of the root varied from no recession to 7 mm. The study was approved by the local ethics committee.
Samples
Samples from the following five sites of the oral cavity were taken from each subject: tongue dorsum (area of tuberculum impar), buccal fold (mucosal area facing the first molar of the fourth quadrant), hard palate (cross area between the palatine raphe and the second plica palatina transversa), supra- and subgingival plaque from the same root surface. The plaque samples were collected using a Whatman sterile OmniSwab (Whatman, Community Drive Stanford, ME); the supragingival plaque samples were taken with a sterile Gracey curette, while the subgingival plaque was collected using sterile paper points (Roeko, Coltene/Whaledent, Langenau, Germany). The samples (n=150) were processed as previously described [16]. Sampling was performed by one examiner (D. P.).
DNA extraction and amplification of 16S rRNA genes
Bacterial DNA was extracted using the QIAamp® DNA Mini Kit (Qiagen, GmbH, Hilden, Germany) according to the instructions of the manufacturer. The extracts were stored at −20°C until use. The 16S rRNA genes were amplified as previously described [17]. Briefly, two separate PCR reactions were set up, using either forward primer 5′-CCA GAG TTT GAT YMT GGC-3′ with reverse primer 5′-GAA GGA GGT GWT CCA RCC GCA -3′ or forward primer 5′-GAC TAG AGT TTG ATY MTG GC-3′ with reverse primer 5′-GYT ACC TTG TTA CGA CTT-3′. Two μl of genomic DNA template were added to the reaction mixture (final volume 25 μl) containing 20 pmol of each primer, 40 nmol of deoxytriphosphates, 1.5 mM Mg2+ and 1 U of Platinum High Fidelity Taq polymerase (Invitrogen, San Diego, CA). The samples were preheated at 94°C for 2 min, followed by 32 cycles of amplification under the following conditions: 94°C for 30 s, 55°C for 30 s, and 68°C for 1.5 min, with an additional 1 s for each cycle. A final 10 min elongation step at 68°C was added. The results of the PCR amplification were examined by electrophoresis in a 1% agarose gel. The two parallel PCR reactions were combined and the products purified with QIAquick PCR Purification Kit (Qiagen).
Microarray procedure
The present work used the same microarrays (HOMIM), and the same labelling procedure and hybridization conditions as previously described [17]. Briefly, labelled nucleotide Cy3-dCTP was incorporated during a second, nested PCR reaction. Hybridization was performed overnight at 55oC; the arrays were washed at room temperature, spun dry and stored in a dark container until scanned by an Axon 4000B microarray scanner. Median pixel intensities for each individual spot were calculated using the analysis function of the GenePix Pro. The median background intensity for each individual feature was subtracted from the median feature intensity yielding a normalized “median intensity score” for each individual feature. The generated gpr files were exported to the HOMIM tool website for further analysis (http://bioinformatics.forsyth.org/homim/). This analysis allowed the determination of the presence or absence of a particular microorganism based on specific criteria set for that individual spot, and thus microbial profile maps for each sample, and subsequent cluster analyses. The cluster method used was UPGMA with a correlation distance function. The similarity measurement was calculated with the Person’s Correlation Coefficient with values from −1 to +1, when identical: 1, and completely opposition: −1. The signal intensities were categorized from 0 to 5. The oral taxon numbers of the bacterial species are provided at www.homd.org/.
Statistical analyses
When comparing the prevalence of a specific species in subjects, a Cochrane’s test was used to calculate p-values. Species showing p < 0.005 were selected for further analysis. In the selected species, when comparing p-values of bacterial species in two different sites in one subject, a Sign test was used. A significance level of 5% was chosen. H0 was defined as “the true prevalences of the two groups are identical”.
Results
General findings
Altogether, 156 species and 19 clusters (i.e., groups of species found by common probes) were detected. The species belonged to eight phylogenetic groups (Table 1). Firmicutes was the most common, and Synergistes the least prevalent phylum; TM7 and Spirochaetales were the only phyla that were not detected at all sites.
Table 1.
Distribution of phyla comprising the detected species and clusters in the different sample categories
| Phylogenetic group | Number of species per site |
Total | ||||
|---|---|---|---|---|---|---|
| Tongue dorsum | Buccal fold | Hard palate | Supragingival | Subgingival | ||
| Firmicutes | 45 | 58 | 58 | 52 | 55 | 72 |
| Bacteroidetes | 25 | 23 | 19 | 23 | 20 | 37 |
| Proteobacteria | 19 | 19 | 16 | 20 | 23 | 26 |
| Actinobacteria | 6 | 6 | 9 | 15 | 12 | 19 |
| Fusobacteria | 7 | 4 | 3 | 6 | 6 | 10 |
| Spirochaetales | 1 | 1 | - | 3 | 4 | 7 |
| TM7 | - | - | - | 3 | 1 | 3 |
| Synergistes | 1 | 1 | 1 | 1 | 1 | 1 |
| Total | 104 | 112 | 106 | 123 | 122 | 175 |
From 10–58 bacterial species and clusters were detected per site (Table 2). When combining the five sites for each subject, between 51 and 81 (mean 59) species were detected. The 20 species that were present in more than two thirds of the subjects are listed in Table 3. The most commonly-detected species were as follows: Streptococcus oralis oral taxon 707, Veillonella atypica oral taxon 524, Streptococcus parasanguinis oral taxon 721 and Fusobacterium nucleatum subsp. polymorphum oral taxon 202. A complete list of bacterial species detected at each site is supplied in Table S1 in the supplemental material.
Table 2.
Species diversity per site and per subject
| Tongue dorsum | Buccal fold | Hard palate | Supragingival | Subgingival | Total/subject | |
|---|---|---|---|---|---|---|
| Range | 13–42 | 10–58 | 11–51 | 10–42 | 10–42 | 51–81 |
| Mean | 27 | 27 | 26 | 22 | 26 | 59 |
Table 3.
Species with the highest prevalences in subjects regardless of sitea
| Species | No. of subjects (n=30) | No. of samples (n=150) |
|---|---|---|
| Streptococcus oralis oral taxon 707 | 30 | 122 |
| Veillonella atypica oral taxon 524 | 30 | 101 |
| Streptococcus parasanguinis oral taxon 721 | 30 | 97 |
| Campylobacter rectus/concisus oral taxon 748/575 | 30 | 96 |
| Streptoccous salivarius/FO042 oral taxon 755/067 | 30 | 88 |
| Gemella hemolysans/sanguinis oral taxon 626/575 | 30 | 82 |
| Fusobacterium nucleatum ss. polymorphum oral taxon 202 | 29 | 110 |
| Haemophilus parainfluenzae/A. aphrophilus oral taxon 718/545 | 27 | 84 |
| Parvimonas micra oral taxon 111 | 27 | 60 |
| Campylobacter gracilis oral taxon 623 | 27 | 49 |
| Veillonella parvula oral taxon 161 | 27 | 47 |
| Prevotella melaninogenica/BE073 oral taxon 469/298 | 25 | 29 |
| Gemella morbillorum oral taxon 046 | 24 | 64 |
| Fusobacterium periodonticum oral taxon 201 | 24 | 49 |
| Campylobacter concisus oral taxon 575 | 24 | 46 |
| Eubacterium saburreum/DO088 oral taxon 494/082 | 22 | 49 |
| Synergistes sp. clone W090 oral taxon 363 | 22 | 35 |
| Rothia mucilaginosa oral taxon 681 | 22 | 23 |
| Capnocytophaga sputigena oral taxon 775 | 21 | 44 |
| Kingella oralis oral taxon 706 | 21 | 35 |
Species found in more than two thirds of the subjects are arranged according to their overall prevalence.
Comparison between the five sites
The hierarchical order, as to the number of species belonging to different phyla, was the same regardless of site, with one minor exception (Proteobacteria were more common than Bacteroidetes in subgingival plaque) (Table 1). Although the total number of bacterial species detected was somewhat lower at the tongue dorsum and the hard palate, as compared to the other three sites, there were no appreciable differences as to overall bacterial diversity. The diversity within each subject ranged considerably at all sites. However, at species level, interesting differences were observed.
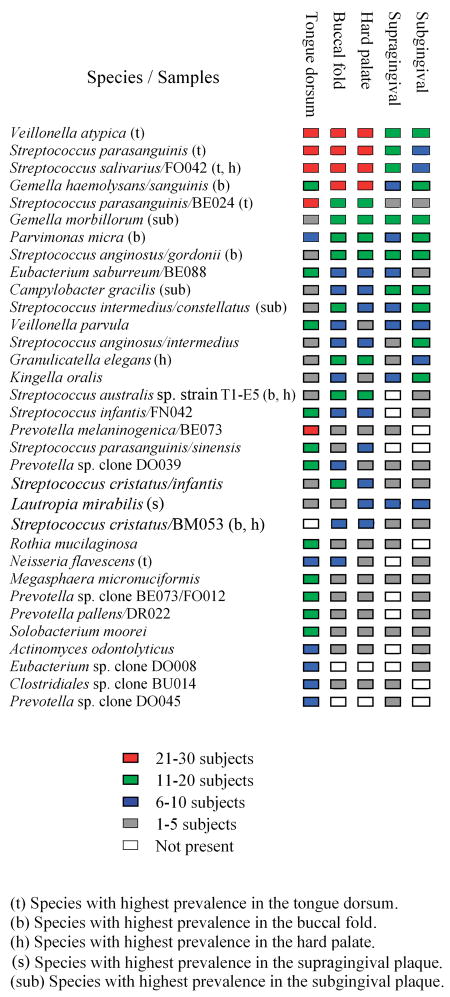
A statistical analysis (Cochrane test) was performed in order to determine if the prevalence of the 175 species or clusters differed at least in one of the sites. The 43 bacterial species with a p < 0.005 (see Table S2 in the supplemental material) were selected for further analysis with a Sign test. In this test, the 43 bacterial species were tested as significance of association with the five sites comparing pairwise, i.e., 430 significance tests were performed. The 33 species (clusters were not included) for which at least one of the comparisons had a p-value below 0.05 were chosen for further analyses (Fig. 1, 2).
Fig. 1.
Species with significant differences as to their prevalence at different sites. Species with p- value < 0.05 were included in the figure. The species are arranged according to their overall prevalence.
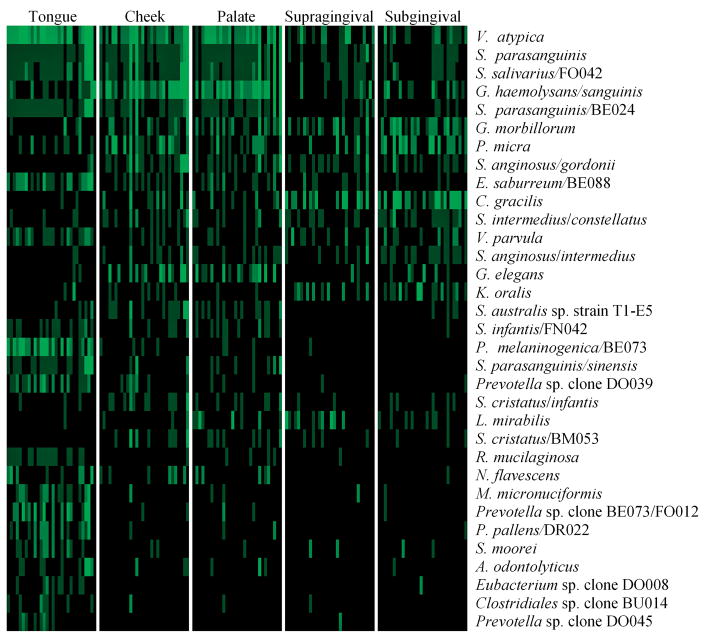
Fig. 2.
Bacterial profiles including only significant species between the different sites. The species are listed as in Fig. 1.
Based on the 5% rule of random finding, 23 false positive results were expected. The results showed 165 significant results, divided between 43 species and clusters, indicating that the majority of the results are true positives (Table S2). It is worth noting that most of these species showed p-values of 0.005 or less in many comparisons. The tongue had the highest number (16) of significant associations, while the supragingival plaque (1) had the lowest. Species most significantly associated with the tongue dorsum were Rothia mucilaginosa oral taxon 681, Megasphera micronuciformis oral taxon 122 and Prevotella melaninogenica oral taxon 469/P. sp. oral taxon 298, clone BE073 (Fig. 1). Overall, the genera Eubacterium and Prevotella showed a significant association with the tongue dorsum. Lautropia mirabilis oral taxon 022 was the only species significantly associated with the supragingival plaque, though present also in other sites. Actinomyces odontolyticus oral taxon 701, Streptococcus australis sp. oral taxon 073, strain T1-E5, Streptococcus infantis oral taxon 065/S. sp. oral taxon 638, clone FN042, S. parasanguinis oral taxon 721/S. sinensis oral taxon 411 and Prevotella pallens taxon 714/P. sp. taxon 310, clone DR022 were not found in the supragingival plaque, but were highly significant in at least one of the other sites. Treponema socranskii oral taxon 769/T. sp. oral taxon 268, clone IL034 was found only in the subgingival plaque, while P. melaninogenica oral taxon 469/P. sp. oral taxon 298, clone BE073, R. mucilaginosa oral taxon 681 and Clostridiales sp. oral taxon 085, clone BU014 were missing in this site. Significant species with the highest prevalence in the subgingival plaque were Kingella oralis oral taxon 706, S. intermedius oral taxon 576/S. constellatus oral taxon 644 and S. anginosus oral taxon 543/S. intermedius oral taxon 644. More detailed information on the species distribution within the different sites can be found in the supplemental material (Table S1).
Analyses of the microarray data were performed in an attempt to visualize the differences in relative proportions of bacterial profiles between the five sites. Only species with significant differences in prevalence were included (Fig. 2). The analyses revealed three distinct patterns of sites as follows: the tongue was dominated by species such as V. atypica oral taxon 524 and P. melaninogenica oral taxon 469/P. sp. oral taxon 298, clone BE073. P. melaninogenica oral taxon 469, Eubacterium sulci oral taxon 467, Lachnospiraceae sp. oral taxon 097, clone DO016 and Legionella pneumophila were exclusively associated with the tongue dorsum, while Actinomyces gerencseriae oral taxon 618 and Streptococcus cristatus oral taxon 578/S. sp. oral taxon 058, clone BM053 were found in all the sites except the tongue dorsum (Table S1). The buccal fold and the hard palate showed similarities, such as high prevalences of V. atypica oral taxon 524, S. parasanguinis oral taxon 721, S. salivarius oral taxon 755/S. sp. oral taxon 067, clone FO042, Gemella haemolysans oral taxon 626/G. sanguinis oral taxon 575, Granulicatella elegans oral taxon 596 and S. australis sp. oral taxon 073, strain T1-E5. In the supra- and subgingival plaque, on the other hand, species such as G. morbillorum oral taxon 046 and Campylobacter gracilis oral taxon 623 were more prevalent, while S. parasanguinis oral taxon 721/S. sinensis oral taxon 411 were not found at these two sites.
Individual differences
Although the number of species per sample differed considerably (from 10 to 58) (Table 2), overall the subjects could not easily be categorized as low or high in diversity. Individuals with a low diversity at one site tended to have a more diverse flora at other sites, thus the total number of species observed differed less (51–81). The diversity appeared to be independent of the number of teeth, signs of gingivitis, medicine use or place of residence. Moreover, cluster analyses, where the microarray results were divided as to persons (all sites combined), rather than to site, did not suggest any rational for dividing the subjects into subgroups. An attempt to look at differences in dominant species between subjects did not reveal any interesting variations. Some species were observed more often and together than others; Haemophilus parainfluenzae oral taxon 718/Aggregatibacter aphrophilus oral taxon 545 were found together with S. oralis oral taxon 707 in 13 out of 14 subjects, and always accompanied by either F. nucleatum. subsp. polymorphum oral taxon 202 or Veillonella cluster II (V. atypica/parvula/dispar/sp. clone BU083/sp. clone AA050), or both. This group of species was associated by either V. atypica oral taxon 524, S. parasanguinis oral taxon 721 (in one third of the subjects), C. rectus oral taxon 748/C. concisus oral taxon 575 (nine subjects), S. salivarius oral taxon 755/S. sp. oral taxon 067, clone FO042 (seven subjects) or G. morbillorum oral taxon 046 (six subjects).
Association with oral and non-oral diseases
All the subjects had reasonably healthy oral cavities, considering their age. Eight subjects had clinical signs of gingival inflammation. No obvious clustering was observed when comparing the supra- and subgingival microflora in subjects with redness and inflammation of the gums to those without signs of gingivitis. Species commonly associated with periodontitis (Porphyromonas gingivalis oral taxon 619, Treponema denticola oral taxon 584, Tannerella forsythia oral taxon 613) were not detected. Other Treponema species were found, but at very low prevalences, in both subjects with and without redness and inflammation of the gums. Similarly, putative cariogenic species were either absent (lactobacilli, Pseudoramibacter alactolyticus oral taxon 538, Atopobium and Olsenella species) or rarely observed (S. mutans oral taxon 686, Bifidobacterium species).
As to the presence of species typically associated with disease elsewhere in the body, some interesting observations were made. H. parainfluenzae oral taxon 718/A. aphrophilus oral taxon 545 were found in 90% and Cardiobacterium hominis oral taxon 633 in 20% of the subjects. Pseudomonas species (cluster I including Pseudomonas aeruginosa/pseudoalcaligenes/fluorescens/sp. clone AZ002) were significantly prevalent on the cheek and hard palate (Table S2).
Discussion
The purpose of the present study was to describe the microbial complexity of the oral cavity in the elderly, and to determine site- and subject-specificities of the detected bacterial species. A total of 175 species and clusters were detected.
Species of the genera Veillonella (particularly V. atypica oral taxon 524) and Streptococcus were ubiquitous (Table 3, Fig 1), as was the case in young adults [11]. S. oralis oral taxon 707 was by far the most prevalent species in the present study. It is recognized as a frequent cause of infections in immunocompromised patients; other streptococci are known to be associated with bacterial endocarditis [18, 19, 20]. High prevalences of F. nucleatum subsp. polymorphum oral taxon 202, H. parainfluenzae oral taxon 718/A. aphrophilus oral taxon 545 and Leptotrichia were also found. F. nucleatum subsp. polymorphum oral taxon 202 has previously been shown to be associated with healthy root surfaces in old age [16]. Species such as V. atypica oral taxon 524, S. parasanguinis oral taxon 721 and S. salivarius oral taxon 755/S. sp. oral taxon 067, clone FO042 were among the most prevalent species at all sites, but associated particularly with soft tissue samples. A high level of lactobacilli has been described as typical for the elderly [6], however, in the present study these bacteria were rarely detected. In the elderly, Lactobacillus species were commonly detected in carious root surfaces, but not in subjects without root caries [16].
When comparing the different sites sampled, three distinct bacterial patterns could be observed (Table 2, Fig 2): The tongue dorsum had the most distinct profile with the highest number of unique and significantly associated species. The papillary structure and the low redox potential of its surface may explain the significant site-specific bacterial association [2]. P. melaninogenica oral taxon 469 has long been associated with tongue flora [21]. R. mucilaginosa oral taxon 681 and E. sulcii oral taxon 467 have been most associated with the healthy tongue microflora, while Fusobacterium periodonticum oral taxon 201, Megasphera micronuciformis oral taxon 122 and Solobacterium moorei oral taxon 678 with halitosis [1]. These bacterial species were significantly associated with the tongue dorsum in the present study. The second bacterial pattern was observed on the cheek and hard palate having closely related bacterial profiles with the highest diversities per site per subject. The microflora of the hard palate has previously shown high variations in the number of species isolated, and the bacterial diversity on the cheek and palate epithelium have been reported to be similar [2]. The shedding of the epithelial cells and the shear forces from chewing in the buccal fold and the hard palate [2] may explain the high variation in the bacterial diversity in these sites. The third bacterial pattern was observed in the supra- and subgingival flora with closely related site diversity and number of phyla detected; however, at the species level, the two sites differed somewhat. Some species (T. socranskii oral taxon 769, S. intermedius oral taxon 576/S. constellatus oral taxon 644, K. oralis oral taxon 706, S. anginosus oral taxon 543/S. intermedius oral taxon 644) were significantly associated only with the subgingival plaque.
Though a direct comparison of studies is not possible due to different methodologies, the bacterial profiles in the elderly are more diverse (i.e., have more species) than those of the normal flora of healthy young and middle aged adults [11]. It has been suggested that age-related changes in the immune response may result in a higher oral bacterial diversity in the elderly [7].
Only subjects with a relatively good oral health, including absence of root caries, were included in the present study. It is known that root caries has a significant association with poor oral hygiene [22]. Root surfaces appear to be more susceptible to carious attack than enamel surfaces [23, 24]. The absence of root caries suggested, therefore, none or a low coronal caries activity. The bacterial profiles of the supragingival plaque showed a low prevalence, or absence, of species often associated with caries, such as lactobacilli, S. mutans oral taxon 686, Actinomyces, Pseudoramibacter alactolyticus oral taxon 538, Bifidobacterium, Atopobium and Olsenella species.
Eight of the subjects had signs of gingivitis. However, neither direct inspection of bacterial profiles, nor cluster analyses, revealed any distinct characteristics of the flora of these subjects (all sites) compared to the rest.
Actinomyces species were commonly found in the supragingival plaque samples of the healthy root surfaces. Members of this genus have been described to be associated both with healthy and carious root surfaces in the literature [10].
H. parainfluenzae oral taxon 718/A. aphrophilus oral taxon 545 were among the most prevalent species in the present study at all sites. Haemophilus species on the tongue surface have been associated with aspiration pneumonia in the elderly [14]. A compromised immune response may fail to eliminate these species, thus promoting infection. H. parainfluenzae oral taxon 718/A. aphrophilus oral taxon 545 and C. hominis oral taxon 633 are part of the so-called HACEK group (H. parainfluenzae, A. aphrophilus and A. paraphrophilus, Aggregatibacter actinomycetemcomitans, C. hominis, E. corrodens and Kingella kingae) because of their common microbiological (fastidious in their nutritional and atmospheric requirements, a long incubation time for growth) features. They may cause infective endocarditis involving native valves and are the most common Gram-negative bacterial causes of endocarditis among people who do not use intravenous drugs [18, 25, 26]. Pseudomonas species were significantly associated with the buccal fold and the hard palate in the present study. They have been repeatedly associated with tracheal and bronchial colonization, and nosocomial or aspiration pneumonia [27]. The presence of non-oral species such as L. pneumophila (tongue dorsum) was noteworthy because it is usually considered as an airway-pathogen and in rare cases as a cause of Legionella endocarditis [17].
The results presented in this report highlighted the diversity and complexity of the oral bacterial profiles in the elderly. Significant differences between sites were observed, but the subjects could not be grouped as to characteristics of their flora. There was no obvious set of bacterial species that tended to cluster together in comparing the different subjects. Some of the most prevalent site-specific species have been previously associated with systemic diseases.
Supplementary Material
Acknowledgments
We thank Sabah Tariq and particularly Marianne Wenaasen, the Cathinka Guldberg-Centre, Oslo, Norway, for patient management. We acknowledge Susan K. Boches and Sean J. Cotton, the Forsyth Institute, Boston, USA for excellent laboratory assistance. Professor Leiv Sandvik is kindly acknowledged for the statistical advice. The study was supported by the Faculty of Dentistry, University of Oslo, Oslo, Norway and NIH grant DE11443 (B.J.P.).
Contributor Information
Dorita Preza, Institute of Oral Biology, Faculty of Dentistry, University of Oslo, Postbox 1052 Blindern, 0316 Oslo, Norway.
Ingar Olsen, Institute of Oral Biology, Faculty of Dentistry, University of Oslo, Postbox 1052 Blindern, 0316 Oslo, Norway.
Tiril Willumsen, Department of Gerodontology Faculty of Dentistry, University of Oslo, Oslo, Norway.
Bjørn Grinde, Institute of Oral Biology, Faculty of Dentistry, University of Oslo, Postbox 1052 Blindern, 0316 Oslo, Norway. Norwegian Institute of Public Health, Oslo, Norway.
Bruce J. Paster, The Forsyth Institute, Department of Molecular Genetics and Department of Oral Medicine, Infection and Immunity, Harvard School of Dental Medicine, Boston, Massachusetts
References
- 1.Kazor CE, Mitchell PM, Lee AM, Stokes LN, Loesche WJ, Dewhirst FE, Paster BJ. Diversity of bacterial populations on the tongue dorsa of patients with halitosis and healthy patients. J Clin Microbiol. 2003;41:558–563. doi: 10.1128/JCM.41.2.558-563.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Marsh PD, Martin MV. Oral microbiology. 3. Chapman & Hall; London: 1992. [Google Scholar]
- 3.Amerongen AV, Veerman EC. Saliva--the defender of the oral cavity. Oral Dis. 2002;8:12–22. doi: 10.1034/j.1601-0825.2002.1o816.x. [DOI] [PubMed] [Google Scholar]
- 4.Saltzman RL, Peterson PK. Immunodeficiency of the elderly. Rev Infect Dis. 1987;9:1127–1139. doi: 10.1093/clinids/9.6.1127. [DOI] [PubMed] [Google Scholar]
- 5.Fure S. Five-year incidence of caries, salivary and microbial conditions in 60-, 70- and 80-year-old Swedish individuals. Caries Res. 1998;32:166–174. doi: 10.1159/000016449. [DOI] [PubMed] [Google Scholar]
- 6.Percival RS, Challacombe SJ, Marsh PD. Age-related microbiological changes in the salivary and plaque microflora of healthy adults. J Med Microbiol. 1991;35:5–11. doi: 10.1099/00222615-35-1-5. [DOI] [PubMed] [Google Scholar]
- 7.Marsh PD, Percival RS. The oral microflora--friend or foe? Can we decide? Int Dent J. 2006;56:233–239. doi: 10.1111/j.1875-595x.2006.tb00107.x. [DOI] [PubMed] [Google Scholar]
- 8.Ohazama J, Motegi E, Nomura M, Miyazaki H, Takane Y, Harazaki M, Yamaguchi H, Ishihara K, Okuda K, Matsuda I. Oral flora in independent over 80-year-olds with more than 20 teeth. Bull Tokyo Dent Coll. 2006;47:1–4. doi: 10.2209/tdcpublication.47.1. [DOI] [PubMed] [Google Scholar]
- 9.Paster BJ, Boches SK, Galvin JL, Ericson RE, Lau CN, Levanos VA, Sahasrabudhe A, Dewhirst FE. Bacterial diversity in human subgingival plaque. J Bacteriol. 2001;183:3770–3783. doi: 10.1128/JB.183.12.3770-3783.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zambon JJ, Kasprzak SA. The microbiology and histopathology of human root caries. Am J Dent. 1995;8:323–328. [PubMed] [Google Scholar]
- 11.Aas JA, Paster BJ, Stokes LN, Olsen I, Dewhirst FE. Defining the normal bacterial flora of the oral cavity. J Clin Microbiol. 2005;43:5721–5732. doi: 10.1128/JCM.43.11.5721-5732.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kaklamanos EG, Charalampidou M, Menexes G, Topitsoglou V, Kalfas S. Transient oral microflora in Greeks attending day centres for the elderly and residents in homes for the elderly. Gerodontology. 2005;22:158–167. doi: 10.1111/j.1741-2358.2005.00069.x. [DOI] [PubMed] [Google Scholar]
- 13.Miura H, Kariyasu M, Yamasaki K, Arai Y, Sumi Y. Relationship between general health status and the change in chewing ability: a longitudinal study of the frail elderly in Japan over a 3-year period. Gerodontology. 2005;22:200–205. doi: 10.1111/j.1741-2358.2005.00074.x. [DOI] [PubMed] [Google Scholar]
- 14.Sumi Y, Miura H, Nagaya M, Michiwaki Y, Uematsu H. Colonisation on the tongue surface by respiratory pathogens in residents of a nursing home - a pilot study. Gerodontology. 2006;23:55–59. doi: 10.1111/j.1741-2358.2006.00093.x. [DOI] [PubMed] [Google Scholar]
- 15.HOMIM. posting date . 2008 http://mim.forsyth.org/. [Online]
- 16.Preza D, Olsen I, Aas JA, Willumsen T, Grinde B, Paster BJ. Bacterial profiles of root caries in elderly patients. J Clin Microbiol. 2008;46:2015–2021. doi: 10.1128/JCM.02411-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Preza D, Olsen I, Willumsen T, Boches SK, Cotton SL, Grinde B, Paster BJ. Microarray analysis of the microflora of root caries in elderly. Eur J Clin Microbiol Infect Dis. 2008 doi: 10.1007/s10096-008-0662-8. (Ms. No. EJCMID-D-08–00421R1) In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Berbari EF, Cockerill FR, 3rd, Steckelberg JM. Infective endocarditis due to unusual or fastidious microorganisms. Mayo Clin Proc. 1997;72:532–542. doi: 10.4065/72.6.532. [DOI] [PubMed] [Google Scholar]
- 19.Li X, Kolltveit KM, Tronstad L, Olsen I. Systemic diseases caused by oral infection. Clin Microbiol Rev. 2000;13:547–558. doi: 10.1128/cmr.13.4.547-558.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shenep JL. Viridans-group streptococcal infections in immunocompromised hosts. Int J Antimicrob Agents. 2000;14:129–135. doi: 10.1016/s0924-8579(99)00172-7. [DOI] [PubMed] [Google Scholar]
- 21.Kononen E, Asikainen S, Jousimies-Somer H. The early colonization of gram-negative anaerobic bacteria in edentulous infants. Oral Microbiol Immunol. 1992;7:28–31. doi: 10.1111/j.1399-302x.1992.tb00016.x. [DOI] [PubMed] [Google Scholar]
- 22.Simons D, Kidd EA, Beighton D. Oral health of elderly occupants in residential homes. Lancet. 1999;353:1761. doi: 10.1016/s0140-6736(99)01343-4. [DOI] [PubMed] [Google Scholar]
- 23.Ten Cate JM, Larsen MJ, Pearce EIF, Fejerskov O. Chemical interactions between the tooth and oral fluids. In: Fejerskov O, Kidd EA, Nyvad B, Baelum V, editors. Dental Caries. 2. Blackwell Munksgaard; Oxford: 2008. p. 224. [Google Scholar]
- 24.van Houte J, Lopman J, Kent R. The final pH of bacteria comprising the predominant flora on sound and carious human root and enamel surfaces. J Dent Res. 1996;75:1008–1014. doi: 10.1177/00220345960750040201. [DOI] [PubMed] [Google Scholar]
- 25.Akhondi H, Rahimi AR. Haemophilus aphrophilus endocarditis after tongue piercing. Emerg Infect Dis. 2002;8:850–851. doi: 10.3201/eid0808.010458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Redmond AM, Meiklejohn C, Kidd TJ, Horvath R, Coulter C. Endocarditis after use of tongue scraper. Emerg Infect Dis. 2007;13:1440–1441. doi: 10.3201/eid1309.070544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Niederman MS, Mantovani R, Schoch P, Papas J, Fein AM. Patterns and routes of tracheobronchial colonization in mechanically ventilated patients. The role of nutritional status in colonization of the lower airway by Pseudomonas species. Chest. 1989;95:155–161. doi: 10.1378/chest.95.1.155. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.