Abstract
Studies on survivin over the past 2–3 years have shown that survivin possesses multiple subcellular localizations and is a multi-functional molecule involved in many aspects of cellular processes and/or behaviors. The subcellular localization and function of the survivin splice variants, however, have not yet been well elucidated. We have, therefore, provided additional observations on several survivin splice variants for further exploration. This review article will update the role of survivin, and its splice variants in the mitosis/cell cycle, apoptosis, tumorigenesis, chemoprevention, drug/radiation resistance, and cancer therapeutics.
Recent studies have indicated that the survivin molecule is not only localized on the mitotic apparatus and the centromere/kinetochore of chromosomes, but it is also localized on mitochondria. The multi-subcellular localization of survivin is consistent with its multiple functions in cellular metabolisms and responses. Alternative splicing of the survivin gene transcript produces a number of different survivin splice variant mRNAs (Fig. 1A), which encode different proteins (Fig. 1B). These survivin splice variants/isoforms appear to have unique subcellular localizations and functions as well. Moreover, new and yet to be characterized survivin splice variants have been found from EST databases (Fig. 2). The presence of these survivin splice variants may make the survivin gene even more functionally diverse and more strictly regulated. In the present review, we will update the role of survivin and its splice variants in the mitosis/cell cycle, apoptosis, tumorigenesis, chemoprevention, drug/radiation resistance, and cancer therapeutics.
Fig. 1.
Structural diagram of the five known survivin splice variants. A: Diagram of the survivin gene/pre-mRNA and its mature mRNA structures. There are four dominant (E1, E2, E3, and E4) and three hidden (2a, 2b, and 3b) exons for the survivin gene. Differential splicing of survivin pre-mRNA produces five known survivin splice variant mRNA as shown. The orange “→” represents the translation start codon (ATG) and the red “▽” represents the translation stop codon for the corresponding protein. B: Diagram of the protein structures for the survivin splice variants. As shown, while survivin and survivin-3B possess the intact baculovirus IAP repeat (BIR) domain, the BIR domains are truncated in survivin-ΔEx3 and survivin-2α, and disrupted by the 2b exon (coding 23 aa) in the survivin-2B protein. aa, amino acid(s).
Fig. 2.
Structural diagram of the new/bizarre survivin-related splice variants. A: Comparison of the survivin mRNA structure with the mRNA structures of the new/bizarre survivin-related splice variants. A complete search of EST databases revealed several meaning new survivin-related splice variants. (1) SurvivinΔptEx2/3 (BP338529, BP338711): The last 14 bp from exon 2 and the first 107 bp from exon 3 were spliced into intron 2. (2) Survivin-ΔptEx1/2 (DA000819): The last 39 bp from exon 1 and the first 49 bp from exon 2 were spliced into intron 1. (3) Survivin-ΔptEx1/2G/T (CB149872): This variant is derived from survivin-ΔptEx1/2 with two G to T/U mutations at the position 120 and 135, and one A to T/U mutation at 209 as shown. (4) Survivin-ΔptEx2 (AU099904, BP273122): The RNA base sequence from the position 30 to 55 in exon 2 is spliced out as a small intron. (5) Survivin-image (AY830084): This is a hybrid splice variant of survivin with a part of the mRNA sequence from a clone IMAGE EST (349–503 bp, BC013858). The survivin-image contains multiple mutations in survivin exons 2 and 3 as well as a seven base sequence (GACGTTG, the “blue part”) from an unknown origin between the survivin exon 3 and the image exon as shown (Zheng et al., 2005). Translation start/stop sites, relative exon sizes/positions, and mutations in some of these survivin-related splice variants are indicated. B: Comparison of the survivin protein structure with the protein structures derived from the new/bizarre survivin-related splice variant mRNAs. Mutations and/or novel protein sequences/domains derived from the mRNAs of survivin-ΔptEx2/3, survivin-ΔptEx1/2, survivin-ΔptEx1/2G/T, survivin-ΔptEx2, and survivin-image are indicated, respectively.
ROLE OF SURVIVIN IN THE CONTROL OF MITOSIS AND CELL CYCLE
Although the precise mechanism remains to be elucidated, survivin exerts a role in cell division control by its association with Aurora B and INCENP on the centromere/kinetochore to form a chromosome passage protein complex (Li, 2003). A significant progression has been made in this area since then. Honda et al. (2003) reported that RNAi depletion of either Aurora B, INCENP, or survivin impaired the localization of the entire Aurora B/INCENP/survivin complex to centromeres and the central spindle, and severely disturbed mitotic progression. Using recombinant proteins, however, it has been shown that INCENP, but not survivin, stimulated Aurora B kinase activity in vitro (Honda et al., 2003). This finding disagrees with an earlier finding that the presence of survivin enhances Aurora B kinase activity in vitro [see review (Li, 2003)]. Nevertheless, Wheatley et al. (2004) found that survivin is specifically phosphorylated in vitro by Aurora-B kinase at the Thr117 site, and that a survivin-T117A mutant prevents the in vitro survivin phosphorylation by Aurora B. Interestingly, while the survivin-T117A mutant did not alter its centromere’s localization in HeLa cells, the survivin-T117E phospho-mimic mutant was unable correctly to localize at any stage during mitosis (Wheatley et al., 2004), indicating a possibly negative role of Aurora B-mediated survivin phosphorylation at the T117 site for the centromere localization of survivin during mitosis. Importantly, Vong et al. recently reported that the ubiquitin-selective chaperone complex, p97-NP14-Ufd1, stimulates K63-linked ubiquitination on survivin. This process is not required for survivin protein degradation, but is required for survivin and Aurora B to attach to the centromere/kinetochore structure of chromosomes, while deubiquitination mediated by the deubiquitinating enzyme, hFAM is required for the dissociation of survivin from centromeres (Vong et al., 2005). Their results indicate that a timely balanced K63-linked ubiquitination status on survivin controls the dynamic association/dissociation of survivin and Aurora B to/from centromeres, which is important for proper chromosome alignment and segregation (Vong et al., 2005). Thus, this raises an interesting question whether the dynamic K63-linked ubiquitination of survivin has a functional relationship with its phosphorylation at the T117 site. For example, the K63-linked deubiquitination from survivin may represent an early signal for triggering the survivin T117 site phosphorylation. Alternatively, Aurora B-mediated survivin phosphorylation at the T117 site may be required for the deubiquitination of survivin. Regardless of their relationship, it is possible that both K63-linked deubiquitination and T117 phosphorylation of survivin are required for survivin dissociation from centromeres.
Consistent with the model that dynamic K63-linked ubiquitination of survivin controls survivin-centromere association, chromosome alignment, and segregation (Vong et al., 2005), cells with RNAi-mediated survivin depletion were shown to be delayed in prometaphase with misaligned chromosomes, and INCENP and Aurora-B were absent from the centromeres (Carvalho et al., 2003; Lens and Medema, 2003). A portion of survivin-depleted cells exited mitosis without completing cytokinesis resulting in multi-nucleated cells (Carvalho et al., 2003; Lens and Medema, 2003). Furthermore, in survivin-depleted cells, the kinetochore-attachment checkpoint protein BubR1 was prematurely displaced from kinetochores without generation of tension, suggesting that the stable association of BubR1 to kinetochores in response to the lack of tension depends upon survivin (Carvalho et al., 2003; Lens and Medema, 2003). Analysis of live cell imaging of Aurora B-GFP and survivin-GFP by FRAP has revealed that survivin-GFP, but not Aurora B-GFP, is highly mobile at prometaphase and metaphase, and ablation of Aurora B by siRNA decreased survivin-GFP mobility (Delacour-Larose et al., 2004), suggesting that Aurora B is able to modulate the association of survivin with centromeric chromatin. Similarly, photobleaching experiments have also revealed a rapid survivin turnover at centromeres during mitosis (Beardmore et al., 2004). These studies indicate a dynamic association of survivin with centromeres controlled by K63-linked ubiquitination and deubiquitination. Thus, survivin appears to play an important role in the chromosomal passenger complex localization during mitosis.
Recent studies have also revealed that at least in cancer cells there is an additional component (Dasra B/borealin) that appears to play a role in survivin stabilization and effective binding to INCENP. SiRNA-mediated depletion of Dasra B/borealin has been shown to induce a reduction of survivin level by 60%–70% (Sampath et al., 2004) and has resulted in the failure of survivin–INCENP interactions and mitotic defects (Vader et al., 2005). A survivin-INCENP hybrid molecule, however, was able to recruit Aurora B and localized normally during mitosis in the absence of borealin and the centromere-targeting domain (amino acid 1–46) of INCENP (Vader et al., 2005). Thus, a critical leading role for survivin in the proper localization of the chromosomal passenger complex during mitosis has been strikingly strengthened. A model to reflect the regulation and function of survivin in the chromosomal passenger complex during mitosis has been recently summarized (Earnshaw, 2005).
On the other hand, in certain specialized physiological situations, survivin may not be involved in mitosis. For example, it was reported that survivin was not detected at any stage of the endomitotic cell cycle in polyploid forming bone marrow megakaryocytes, even though a low-survivin mRNA appeared to be there (Zhang et al., 2004b; Gurbuxani et al., 2005). Proteasome inhibition did not rescue this phenotype in megakaryocytes (Zhang et al., 2004b), suggesting that loss of survivin protein was not resulted from protein degradation. In the case of endomitosis of vascular smooth muscle cells, Nagata et al. (2005) reported that survivin does not colocalize with either Aurora B or INCENP, although a low level of diffuse survivin expression was detected. Interestingly, ectopic expression of survivin inhibits polyploidization in these cells (Nagata et al., 2005). These specialized physiological phenomena are in contrast to the upregulation of endogenous survivin during mitosis in cancer cells and warrant further investigation of the potential different role of survivin in cancer cells versus normal cells. Yang et al. (2004) recently reported that while the role of survivin in cell division is conserved in normal human primary lung fibroblasts and retinal pigment epithelial cells, normal human cells do not require survivin for survival. This observation provides additional evidence supporting the notion that the role and regulation or context for signaling interactions by survivin may differ in normal versus cancer cells, which would make survivin an advantageous target for the treatment of cancer and/or other diseases.
Finally, initial observation indicated that survivin is regulated by the cell cycle with a robust expression in G2/M phase (Li et al., 1998). An important question is whether survivin plays a role in cell-cycle progression outside of mitosis in cancer cells. Evidence suggests that survivin plays a role in G1/S transition and cell-cycle progression in cancer cells. The ectopic expression of survivin blocked vitamin D3-midiated G1 arrest and resulted in increased S and G2/M cell populations in MCF-7E breast cancer cells (Li et al., 2005), although the underlying mechanism remains to be explored. Here, we should point out that induction of survivin by growth factors/cytokines appears to be important during hematopoietic progenitor cell (HPC) maturation and G0/G1 to S transition as well (Fukuda and Pelus, 2001; Fukuda et al., 2002). However, the expression of survivin in cancer cells appears to be constitutive. Its signaling interactions may be different in cancer cells versus normal cells as a function of various stimuli (Li and Brattain, 2006).
ROLE AND LOCALIZATION OF SURVIVIN SPLICE VARIANTS IN MITOSIS AND APOPTOSIS CONTROLS
It is known that survivin homodimerizes in solution. We previously proposed a model that survivin-2B or survivin-ΔEx3 may heterodimerize with survivin to modulate the role of survivin in control of mitosis and/or apoptosis (Li, 2003). Noton et al. (2005) recently showed that in GST-pulldown experiments, survivin or Aurora B interacts with survivin-2B and survivin-ΔEx3, respectively, while borealin/Dasra B is unable to interact with survivin-ΔEx3 but does interact with survivin-2B. In the cotransfection experiments, survivin, Aurora B, or borealin immunoprecipitated survivin-2B but not survivin-ΔEx3 (Noton et al., 2005). However, the authors found that in single transfection experiments, survivin-2B or survivin-ΔEx3 was unable to immunoprecipitate the endogenous survivin or Aurora B (Noton et al., 2005). The fluorescence microscopic data also did not find the colocalization of either survivin-2B or survivin-ΔEx3 on the metaphase chromosome DNA (Noton et al., 2005). Using stable cell lines (U2OS or HeLa) over-expressed survivin, survivin-2B, or survivin-ΔEx3, these authors did not find differences in cell growth rates among these cell lines. They pointed out, however, that experiments encountered difficulties in establishing the survivin-2B-GFP expressing cell line (Noton et al., 2005), suggesting that survivin-2B expression is toxic to cells. Their observations are consistent with the recent report that transient expression of survivin-2B inhibited lung cancer cell growth and induced apoptosis (Ling et al., 2005), although the underlying mechanism remains to be elucidated. In any cases, based on the finding reported by Noton et al. (2005), inhibition of cell growth and induction of apoptosis by survivin-2B (Ling et al., 2005) is unlikely resulted from interferences of survivin function on centromeres.
Survivin-2B and survivin-ΔEx3 appear to play different roles in cancer development (Li, 2005a). Recent studies also showed that the expression ratio of survivin-2B/survivin was significantly higher in the colorectal ademocarcinoma samples than in the normal ones. Nevertheless, a higher ratio of survivin-2B/survivin correlated with a better prognosis, and the expression ratio of survivin-2B/survivin for Stage III and IV was lower than that for Stage I and II (Suga et al., 2005), suggesting a negative role of survivin-2B in cancer progression. The multi-variate Cox’s proportional hazards regression model revealed a 7.3-fold increased risk of tumor-related death for soft tissue sarcoma patients with survivin-ΔEx3 overexpressing tumors, while the effect of survivin and survivin-2B showed less pronounced increases in risk (2.2- and 1.9-fold) (Taubert et al., 2005). The studies from our laboratory indicated that high levels of survivin-2B are associated with patients showing of “no relapse and alive” while high levels of survivin-ΔEx3 is associated with patients who have died in association with disease relapse in non-small cell lung cancer (Ling et al., 2005). Consistent with this observation, forced expression of survivin-2B in A549 lung cancer cells inhibited cell growth, disrupted mitochondria potential, and induced cell death (Ling et al., 2005). However, the mechanisms by which survivin-2B and survivin-ΔEx3 differentially affect cancer development are unclear. This is in part owing to the uncertainty of their subcellular localization in cancer cells. Mahotka et al. (2002) initially showed that survivin-2B localized to cytoplasm in the HepG2 heptoma cell line. Using a computer-based PSORT II program, however, they predicted a 13% of survivin-2B localizes on mitochondria (Mahotka et al., 2002). On the other hand, Caldas et al. recently showed that survivin-2B does not co-fractionated with mitochondrial fraction but with cytosol fraction in HeLa cells, although these authors pointed out that survivin-2B colocalizes to the perinuclear membrane and to mitochondria(Caldas et al., 2005b). These observations and statements triggered our great interests to scrutinize the subcellular localization of survivin-2B. Our initial observation indicated that transiently expressed survivin-2B in the early time (before serious apoptosis occurs) localized to punctate structures in the cytoplasm, a pattern which contrasted with the even distribution of EGFP throughout the cell (Fig. 3A). Previous studies have shown that survivin localizes on mitotic spindles, midbodies, and chromosomal DNA (centromere) during mitosis [see review (Li, 2003)]. In contrast to the localization of survivin during mitosis, confocal microscopy experiments revealed that survivin-2B did not co-localize with the mitotic spindle, midbody, and chromosomal DNA (arrows) in mitotic cells, but punctately expressed in the cytoplasm (Fig. 3B). We therefore checked whether survivin-2B localizes to mitochondria as predicted by the computer-based PSORT II program (Mahotka et al., 2002). Confocal fluorescence microscopy analysis showed that the green dots reflecting the subcellular distribution of EGFP-survivin-2B well coincided with the signal derived from Mito-Tracker staining (Fig. 3C). We further generated a survivin-2B-specific antibody (Fig. 3D). Immunofluorescence microscopy experiments with this antibody obtained a similar mitochondrial localization pattern (Fig. 3E). We have further confirmed the localization of survivin-2B in mitochondria by subcellular fractionation experiments (Fig. 3F). Interestingly, we performed a search for sequence similarity and did not find a peptide motif in survivin-2B that is similar to the C-terminal sequence of survivin-ΔEx3 that was reported containing a mitochondrial-targeting sequence (Wang et al., 2002). Here, we should point out that the localization of survivin-2B on mitochondria might vary in different cell types. We observed that there are less obvious signals of survivin-2B on mitochondria in human embryonic kidney 293 (HEK293) cells in comparison with that in HeLa cells. In addition, we found that after cells went into serious apoptosis up on survivin-2B overexpression, it was more difficult to find the mitochondrial localization of survivin-2B. Instead, due to the condensation of cells and cell nuclei during apoptosis, survivin-2B mainly localized in the nuclei in the apoptotic cells. As to the mechanism by which survivin-2B induces apoptosis, Dohi et al. (2004a) recently demonstrated that there is a mitochondrial pool of survivin in cancer cells (but not in the normal survivin-positive tissues such as testis), which is essential for the inhibition of mitochondrial-mediated apoptosis. These authors further showed that in response to cell death stimuli, mitochondrial survivin is rapidly released into the cytosol, where it prevents caspase activation and inhibits caspase 9-mediated apoptosis (Dohi et al., 2004a). Thus, one mechanism for survivin-2B induction of apoptosis is to block mitochondrial-localized survivin release into the cytosol by their physical interaction (Fig. 6).
Fig. 3.
Survivin-2B co-localizes to mitochondria and does not co-localize with the mitotic spindle, midbody, and chromosomal DNA. A: Survivin-2B localized to punctate structures in cytoplasm, which is in contrast to the evenly expressed EGFP proteins in the cells. HeLa cells were transfected with empty EGFP vectors (left part) or pEGFP-survivin-2B expression vectors (right part), and fixed with 4% paraformaldehyde in PBS 18 h after transfection. Images were captured under a Zeiss Axiovert 100M Fluorescence Microscopy System. B: Survivin-2B did not colocalize with the mitotic spindle, midbody, or chromosomal DNA. HeLa cells were transfected with pEGFP-survivin-2B (green), fixed as in A and stained with 4′, 6-diamidino-2-phenylindole (DAPI) for DNA (blue), or β-tubulin antibodies (red). Images were taken under a Leica confocal fluorescence microscopy. Arrows indicate the midbody, mitotic spindle, and chromosomal DNA, respectively. C: Survivin-2B co-localized with mitochondria in transfected cells. HeLa cells transfected with pEGFP-survivin-2B (green) were first incubated in 50 nM Mito Tracker Red CM-H2Xros for 1 h, then fixed as in A and stained with DAPI (DNA, blue) and Mito-Tracker (mitochondria, red). Images were taken as in B. D: Characterization of survivin-2B-specific antibodies. HeLa cells were transfected with or without various expression vectors as shown and analyzed by Western blotting using survivin-2B antibodies in the absence (left part) or presence (right part) of survivin-2B-specific peptides. The expression of various survivin splice variants was confirmed by Western blots shown at the bottom (left part). Actin is a loading control. E: Mitochondrial localization of endogenous survivin-2B. HeLa cells grown on circular coverslips were first incubated in 50 nM Mito Tracker Red CM-H2Xros for 1 h. Cells were then fixed with 4% paraformaldehyde in PBS, blocked/permeabilized with PBS containing 2% BSA and 0.2% Triton X-100, and then incubated in PBS containing 1% BSA, chicken anti-survivin-2B (1:500) antibodies for 60 min at 37°C. After washing with PBS, cells were incubated in PBS containing fluorescein-goat anti-chicken IgY (1:500) antibodies for 45 min at room temperature, followed by staining with DAPI at a final concentration of 0.5 μmg/ml in PBS for 10 min. Coverslips were then mounted on glass slides with Gel/Mount™ solution (Biomedia, Foster City, CA). Images were captured under a Zeiss Axiovert 100M Fluorescence Microscopy System. Note: DAPI (blue), Mito-Tracker (red), and survivin-2B (green). F. Survivin-2B existed in the mitochondrial fraction but not in the cytosol. HeLa cells were fractionated into cytosol and mitochondria 18 h after transfection. Survivin-2B and cytochrome c (internal control) were determined by immunoblotting. Note: Survivin-2B-specific IgY antibodies were purified by the corresponding peptide-specific affinity column from total IgY isolated from the KLH-conjugated peptides-immunized hen eggs (Aves Lab, Tigard, Oregon).
Fig. 6.
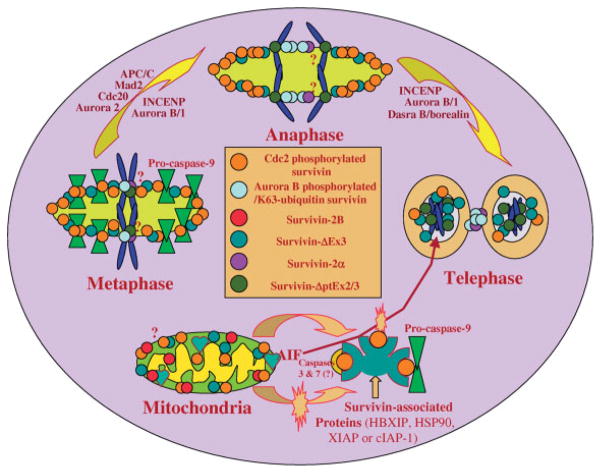
A diagrammatic model to summarize the role and subcellular localization of survivin and its splice variants in the regulation of apoptosis and control of mitosis in cancer cells. (1) The dynamic Cdc2-phosphorylated survivin localizes on the mitotic spindle and centrosome to inhibit a “default” apoptotic pathway during mitosis, and to facilitate microtubule polymerization and spindle formation. (2) The dynamic Aurora B-phosphorylated/K63-ubiquitin survivin complexes with Aurora B and INCENP, and plays a leading role in the binding of the chromosomal passenger protein complex to the centromere/kinetochore of chromosomes to control the precise completion of chromosome segregation and cytokinesis. (3) Survivin also localizes in mitochondria and is released into the cytoplasm upon apoptotic insult. The released survivin and/or other cytoplasmic survivin may interact with many other proteins to exert its role in apoptosis inhibition, G1/S transition/cell-cycle progression and possible other functions. Some of the survivin-associated proteins identified include HBXIP, HSP90, XIAP, and cIAP-1. Additionally, mitochondrial-associated survivin may block apoptosis-inducing factor (AIF) release and nuclear translocation as a mechanism for caspase-independent apoptosis control. (4) Survivin-2B localizes in mitochondria. One potential way for survivin-2B to induce cell death is that increased survivin-2B may be able to physically interact with and block the release of survivin from mitochondria for caspase inhibition. Alternatively, survivin-2B may also promote the AIF release and nuclear translocation. (5) It has been shown that survivin-ΔEx3 is localized in mitochondria and nuclei in interphase cells, and translocated to the mitotic spindle during mitosis. However, whether survivin-ΔEx3 plays a role in the spindle checkpoint and apoptosis control remains to be determined. It was shown that survivin-ΔEx3 is degraded in the nucleolus through the ubiquitination pathway although the significance of this phenomenon remains to be explored. (6) Survivin-ΔptEx2/3 and survivin-2 α were shown to be associated with the chromosomal DNA during mitosis in certain conditions. However, whether survivin-ΔptEx2/3 and survivin-2 α play a role during mitosis or whether the function of centromere-associated survivin can be modulated by survivin-ΔptEx2/3 and/or survivin-2 α remains to be determined in the future study. Note: The subcellular localization of survivin-3B is not clear currently and remains to be determined as well.
Several laboratories have showed that exogenously expressed survivin-ΔEx3 localized in nuclei (Mahotka et al., 2002; Song and Wu, 2005; Caldas et al., 2005b). PSORT II program predicted none of survivin-ΔEx3 on mitochondria (Mahotka et al., 2002). However, Caldas et al. (2005b) showed that while majority of survivin-ΔEx3 was in the nuclear fraction, there was some (one fifth or less by eye) of survivin-ΔEx3 in the mitochondrial but not cytosol fraction. Wang et al. (2002) reported that survivin-ΔEx3 contains a mitochondrial-targeting sequence, and these authors demonstrated that an K7 anti-apoptotic glycoprotein encoded by Kaposi’s sarcoma-associated herpesvirus structurally resembles survivin-ΔEx3 and localizes on mitochondria, endoplasmic reticulum, and the nuclear membrane. Recently, Song and Wu (2005) reported that survivin-ΔEx3 localizes in the nucleoli where it degrades rapidly through the ubiquitin-proteasome pathway. Interestingly, they showed that the protein motifs/sequences involved in the nucleolar localization and degradation of survivin-ΔEx3 is required for its anti-apoptotic function (Song and Wu, 2005), although the molecular mechanism is unclear. Thus, the subcellular localization and function of survivin-ΔEx3 remain to be elucidated. We have generated a survivin-ΔEx3-specific antibody using KLH-conjugated survivin-ΔEx3-specific peptides as immunogens (Fig. 4A). Immunofluorescence microscopy experiments with this survivin-ΔEx3-specific antibody showed that in the interphase HeLa cells, survivin-ΔEx3 colocalizes in both mitochondria and nuclei (Fig. 4B). However, in mitotic cells, survivin-ΔEx3 appeared to be translocated to and colocalized with the mitotic spindle (Fig. 4C). Consistent with the previous findings discussed above, we also observed that exogenously expressed survivin-ΔEx3 mainly localized in the peri-nuclei and nuclei (Fig. 4D). Together, these results would be important for helping to further delineate the mechanisms by which survivin-2B and survivin-ΔEx3 play different roles in the regulation of cell cycle and apoptosis (Fig. 6).
Fig. 4.
The subcellular localization of survivin-ΔEx3 in cancer cells. A: Characterization of the specificity of survivin-ΔEx3 IgY antibodies. HeLa cells were transfected with expression vectors for survivin-2B, survivin, survivin-ΔEx3, or empty vectors as shown, cells were lysed 36 h after transfection for Western blots using the survivin antibody (Santa Cruz, upper part, the weaker band at about 16 kD is the endogenous survivin), survivin-ΔEx3-specific antibody (middle part, the weak band with a similar size to the HA-survivin-ΔEx3 represents the endogenous survivin-ΔEx3), or actin antibody (lower part), respectively. Actin is the internal control. B: Subcellular localization of endogenous survivin-ΔEx3. HeLa cells were processed as described in Figure 3E with survivin-ΔEx3 antibodies. Note: Survivin-ΔEx3 (green), mitochondria (red), and DNA (blue). C: Subcellular localization of endogenous survivin-ΔEx3 in mitotic cells. Cells were processed as in (B). Note: Survivin-ΔEx3-specific IgY antibodies were purified through the peptide-specific affinity column from total IgY isolated from the KLH-conjugated peptides-immunized hen eggs (Aves Lab., Tigard, Oregon). D: Localization of exogenously expressed survivin-ΔEx3 in interphase cells. HeLa cells were transfected with pEGFP-survivin-ΔEx3. Cells were fixed with 4% paraformaldehyde in PBS and stained with DAPI 18 h after transfection. Images were taken under a Zeiss Axiovert 100M Fluorescence Microscopy System.
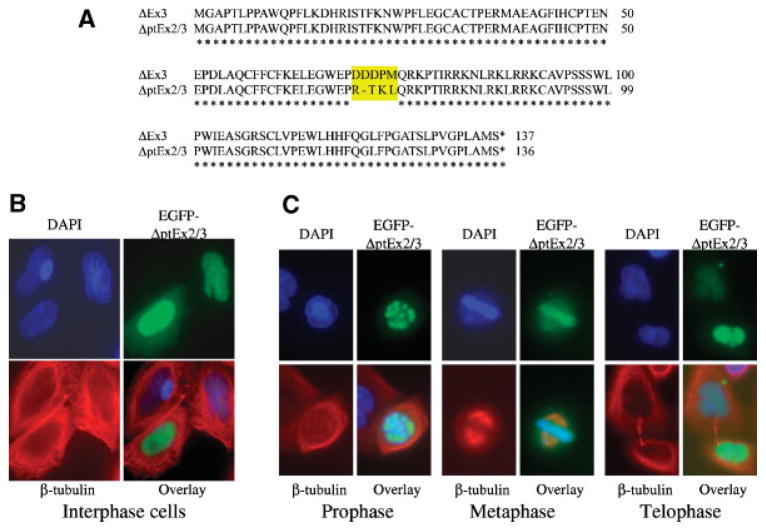
Bioinformatics search revealed several new survivin splice variants (Fig. 2), including the one that lacks a part of both survivin exons 2 and 3 (designated survivin-ΔptEx2/3). Sequence alignment indicated that survivin-ΔptE2/3 has an identical protein sequence to survivin-ΔEx3 with the exception of 4 amino acids from 70 to 73 (Fig. 5A). However, in contrast to the mitochondrial/nuclear and mitotic spindle localization of survivin-ΔEx3 in the interphase and metaphase cells, respectively, exogenous expression of the EGFP-tagged survivin-ΔptE2/3 appears to be mainly colocalized in nuclei and chromosomal DNA throughout the cell cycle (Fig. 5B,C). These observations imply that the role and functional mechanism for survivin-ΔptE2/3 may actually be different from those for survivin-ΔEx3, even though their high protein sequence similarity. In addition, a recent report showed that a truncated survivin splice variant, survivin-2α (Fig. 1) interacts with survivin and is recruited onto the centromere and midbody after ectopic expression of both survivin and survivin-2α (Caldas et al., 2005a). It will be interesting to determine whether survivin-ΔptE2/3 and survivin-2α are potential survivin modulators during mitosis under certain conditions, as proposed in the model (Fig. 6). Finally, survivin-3B was initially identified in bone marrow mononuclear cells from patients with myelo-dysplastic syndrome or acute myelogenous leukemia, and survivin-3B was also detected in human colon and gastric adenocarcinoma cell lines (Badran et al., 2004). The subcellular localization and function of survivin-3B remains to be determined. Vegran et al. (2005) recently reported that survivin-3B was more frequent in high-grade breast carcinomas and correlated with the p53 gene mutation, suggesting a positive role of survivivn-3B in apoptosis inhibition.
Fig. 5.
Subcellular localization of survivin-ΔptEx2/3. A: Protein sequence alignments of survivin-ΔptE2/3 with survivin-ΔEx3. B: Localization of survivin-ΔptEx2/3 in the interphase cells. HeLa cells were transfected with pEGFP-survivin-ΔptEx2/3. Cells were then fixed with 4% paraformaldehyde in PBS and stained with DAPI and β-tubulin antibodies 18 h after transfection. C: Localization of survivin-ΔptEx2/3 during mitosis (prophase, metaphase, and telophase). HeLa cells were transfected and stained as in (B). Images were taken under a Zeiss Axiovert 100M Fluorescence Microscopy System for both B and C. Note: Tubulin was stabilized by a brief taxol treatment in these experiments.
ROLE OF SURVIVIN IN APOPTOSIS CONTROL
Growing evidence indicated that survivin controls both caspase-dependent and caspase-independent apoptosis (Fig. 6) (Li, 2003). Recent progress in this area includes: (1) elucidation of the role of mitochondria-localized survivin in caspase inhibition upon apoptotic insult (Dohi et al., 2004a); (2) involvement of survivin in caspase-independent apoptosis control (Chakravarti et al., 2004; Liu et al., 2004); (3) survivin interaction and cooperation with HBXIP (Marusawa et al., 2003), HSP90 (Fortugno et al., 2003), XIAP (Dohi et al., 2004b), and c-IAP-1 (Samuel et al., 2005) for the modulation of anti-apoptotic function; and (4) survivin mediated increase of human telomerase reverse transcriptase (hTERT) expression and activity (Endoh et al., 2005).
Dohi et al. (2004a) showed that while survivin did not localize on mitochondria in survivin-positive cells from normal adult tissues, survivin localized on mitochondria in various cancer cell lines, indicating a potential unique role for survivin in cancer etiology. In response to inducers of cell death, mitochondrial survivin was rapidly discharged and released into the cytosol, where it prevents caspase activation and inhibits apoptosis (Dohi et al., 2004a). Adenovirus mediated, mitochondria-specific expression of survivin increased colony formation in soft agar, accelerated xenograft tumor growth in SCID mice, and abolishes tumor cell apoptosis in vivo (Dohi et al., 2004a). These studies provide new perspectives for survivin and has extended the model previously proposed for survivin function in apoptosis control (Li, 2003) (Fig. 6).
Consistent with the previous observation that survivin was possibly involved in both caspase-dependent and -independent apoptosis controls [see review (Li, 2003)], using a melanoma cell model harboring a tetracycline-controlled survivin dominant-negative mutant (T34A), Liu et al. (2004) demonstrated that apoptosis induced by interference of survivin function was only partially reduced by pan-caspase inhibitors. Survivin blockade resulted in mitochondrial events including nuclear translocation of mitochondrial apoptosis-inducing factor (AIF), and depolarization and release of cytochrome c and Smac prior to caspase activation (Liu et al., 2004). These observations argue for a protective role by survivin in both caspase-dependent and -independent apoptosis.
A very interesting study recently reported by Endoh et al. showed that ectopic expression of survivin enhanced telomerase activity by upregulation of hTERT expression in LS180 human colon cancer cells. Survivin enhanced DNA-binding activities of Sp1 and c-Myc to the hTERT core promoter as well as the phosphorylation of Sp1 and c-Myc at serine and threonine residues without changing total amounts of these proteins, In contrast, siRNA knockdown of survivin decreased Sp1 and c-Myc phosphorylation (Endoh et al., 2005). This observation suggests that survivin may have an additional role in gene transcription controls acting as a coactivator. Based on their observations above, the authors concluded that survivin participates not only in apoptosis inhibition, but also in prolonging cellular lifespan (Endoh et al., 2005). Whether this is an exclusive role for survivin in cancer remains to be explored. Normal cells, however, show low or no telomerase activity and survivin expression, suggesting a specific role for survivin in cancer cells. This characteristic would provide additional advantages for survivin as a cancer therapeutic target.
SURVIVIN IN TUMORIGENESIS AND CHEMOPREVENTION
Many studies indicated that survivin plays an important role in tumorigenesis, especially in colorectal tumorigenesis and virus infection-induced carcino-genesis (Li, 2005a). It is possible that some cancer prevention agents may function by inhibiting survivin expression (Li, 2005b). We demonstrated that survivin inhibition by vitamin D3 compounds is essential for vitamin D3-mediated cell growth arrest in G1 and apoptosis induction (Li et al., 2005). Aziz et al. (2005a,b) reported that resveratrol, a naturally occurring dietary compound with cancer prevention activity, imparts chemopreventive effects against UVB exposure-mediated damage in SKH-1 hairless mouse skin by inhibiting survivin and Zhang et al. (2004a) showed that sulindac, a colorectal cancer chemopreventive agent, induced a sustained decrease in survivin expression. These observations indicate an important role for survivin as a target for cancer prevention.
SURVIVIN AND CHEMOTHERAPY RESISTANCE
Growing evidence has indicated that survivin expression plays an essential role in drug resistance, and that genetic or pharmacological modulation of survivin expression affects drug effectiveness in apoptosis induction. Wang et al. (2003) reported that expression of survivin antisense RNA significantly reversed multiple drug resistance (MDR) in adriamycin-resistant HL-60 cells with a 5.36-fold increase in chemosensitivity as compared with adriamycin alone. Fulda et al. showed that depletion of survivin by resveratrol or by survivin antisense sensitized various tumor cells to TRAIL (Fulda and Debatin, 2004b) or chemotherapeutic drug (Fulda and Debatin, 2004a) induced apoptosis, but had no effect on normal human fibroblasts (Fulda and Debatin, 2004b). Similarly, quercetin, a ubiquitous bioactive plant flavonoid, increased survivin expression in nuclei of lung carcinoma cells, and transfection of a survivin antisense oligonucleotide enhanced quercetin-induced cell growth inhibition, and cytotoxicity (Kuo et al., 2004). Interestingly, the histone deacetylase inhibitors, SAHA and VPA differentially modulated survivin in melanoma cells. VPA enhanced survivin expression only in VPA-resistant cell lines, whereas VPA and SAHA downregulated survivin in VPA-sensitive cells (Facchetti et al., 2004), indicating a role for survivin in modulating VPA effectiveness. In non-small cell lung cancer cells, ectopic expression of Cox-2 upregulated survivin expression and increased resistance to radiation and drug-induced apoptosis, whereas ectopic expression of Cox-2 antisense downregulated survivin and sensitized cells to apoptosis (Krysan et al., 2004b). This observation was recapitulated by direct siRNA-mediated suppression of survivin (Krysan et al., 2004a). Mechanistically, Cox-2-dependent inhibition of the K48-linked ubiquitination of survivin led to survivin protein stabilization in Cox-2 overexpressing cells. Inhibition of Cox-2 expression by administration of Cox-2 inhibitors in SCID mouse models decreased tumorigenicity and inhibited survivin expression in vivo (Krysan et al., 2004b). The authors concluded that Cox-2-dependent expression of survivin is critical for apoptosis resistance in non-small cell lung cancer (Krysan et al., 2004a). In addition, Belyanskaya et al. (2005) recently reported that cisplatin activates Akt in small cell lung cancer cells and attenuates apoptosis by upregulation of survivin. Zhang et al. (2005a) observed that androgen stimulation with 5α-dihydrotestosterone increased survivin expression in LNCaP cells. Conversely, treatment with the anti-androgen, Flutamide decreased survivin expression and adenovirus-mediated overexpression of survivin increased resistance to death induced by Flutamide. Exogenous insulin-like growth factor-1 activated Akt and increased survivin expression in all cell-cycle phases as well as imparting resistance to Flutamide treatment. The authors concluded that targeted inhibition of survivin enhances the therapeutic effects of Flutamide (Zhang et al., 2005a), which may represent a novel strategy to enhance sensitivity to androgen ablation therapy. Using the same adenovirus-mediated infection system, these authors also showed that overexpression of survivin increased resistance to paclitaxel in both androgen receptor-positive (LNCaP) and negative (PC-3 and DU-145) prostate cancer cells, both in vitro and in vivo. In contrast, inhibition of survivin by the survivin-T34A mutant significantly increases the spontaneous and paclitaxel-induced apoptosis in all cell lines, both in vitro and in vivo. Similarly, Hayashi et al. (2005) recently reported that a combination of etoposide with adenovirus-mediated expression of survivin antisense dramatically inhibits DU145 xenograft growth without tumor recurrence (28 day). Sensitization of paclitaxel-induced apoptosis by interference of survivin was abolished by co-treatment with the pan-caspase inhibitor VAD-CHO, indicating that survivin mediates resistance to paclitaxel at least in part through suppression of caspase-mediated apoptosis (Zhang et al., 2005b). Based on these observations, the authors concluded that survivin mediates paclitaxel-resistance in prostate cancer cells, and that survivin inhibition sensitizes prostate cancer cells to paclitaxel-induced apoptosis through a caspase-dependent mechanism in vitro and in vivo (Zhang et al., 2005b). Here, it should be pointed out that paclitaxel per se induced survivin expression. This appears to be an innate paclitaxel-resistant factor.
Interestingly, using a paclitaxel-resistant ovarian cancer cell clone PTX10 with a β-tubulin mutation at the paclitaxel-binding site, Zhou et al. (2004) found that paclitaxel treatment was not able to induce mitotic arrest, and failed to induce survivin as well. However, ectopic expression of survivin restored paclitaxel-induced mitotic arrest in PTX10 cells. These observations suggest that upregulation of survivin is required for paclitaxel-mediated mitotic arrest in PTX10 cells. Thus, a critical question is whether mitotic arrest is essential for paclitaxel-induced PTX10 cell apoptosis. In other words, does the requirement of survivin in paclitaxel-mediated mitotic arrest mean that increases but not decreases of survivin help paclitaxel-mediated apoptosis induction? The data in Figure 6A from the report (Zhou et al., 2004) showed that ectopic survivin decreases apoptosis induced by paclitaxel from ~27% and ~64% in control cells (left part) to ~10% and ~50% in survivin-overexpressed cells (middle part) at 24 and 36 h points, respectively (Zhou et al., 2004). The data in the left part of Figure 6A indicated that although there is no mitotic arrest due to the absence of survivin, paclitaxel effectively induces apoptosis (Zhou et al., 2004). This suggests that mitotic arrest is not really required for paclitaxel-induced apoptosis. These observations are consistent with the recent findings that upregulation of survivin before (Ling et al., 2004) and/or during (O’Connor et al., 2002) mitosis is a paclitaxel-resistance factor. Unexpectedly, upregulation of survivin by paclitaxel appears to be an early event and independent of paclitaxel-mediated mitotic arrest in MCF-7 breast cancer cells (Ling et al., 2004). Determination of the mechanism for paclitaxel-mediated rapid upregulation of survivin before mitotic arrest would be important for understanding paclitaxel resistance.
SURVIVIN AND RADIATION THERAPY RESISTANCE
Evidence has also revealed an essential role for survivin in cancer cell resistance to radiation therapy. Rodel et al. (2003) showed that in colorectal cell lines of high (SW480) and low (SW48) radioresistance, higher constitutive expression and a more pronounced induction of survivin in SW480 cells was observed 48 h after irradiation treatment, whereas survivin expression was low and not induced in radiosensitive SW48 cells after irradiation. Downregulation of survivin by siRNA increased apoptosis and caspase 3/7 activity in SW480 and HCT-15 (intermediate radioresistance) colorectal cancer cells, which is in parallel with decreased cell viability and colonogenic survival (Rodel et al., 2005). Consistently, data from 59 rectal cancer patients treated with a combination of radiotherapy and 5-fluorouracil indicated that survivin level was inversely associated with the levels of apoptosis, and high survivin level showed a significantly higher risk of a local tumor recurrence (Rodel et al., 2005). Similarly, Chakravarti et al. (2004) showed that survivin expression is much higher in two radiation-resistant glioblastoma multiform cell lines (GM20 and GM21) as compared with two radiation-sensitive cell lines (GM22 and GM23), and that radiation treatment significantly induced survivin expression in GM20 and GM21 cells, but only slightly increased it in GM22 and GM23 cells. Survivin expressed in all phases of cell cycle in GM20 and GM21 cells but only in the G2/M phase of GM22 and GM23 cells (Chakravarti et al., 2004). Expression of the survivin-T34A mutant tended to enhance radiation sensitivity as compared to the corresponding controls. In contrast, adenovirus-mediated overexpression of survivin increased levels of radioresistance (Chakravarti et al., 2004), suggesting that survivin plays a critical role in mediating radiation resistance. Interestingly, pan-caspase inhibitor VAD-CHO treatment did not increase cell survival (Chakravarti et al., 2004), suggesting that survivin suppressed apoptotic cell death largely via a caspase-independent manner. Radiation treatment resulted in the translocation of survivin from the cytoplasm to nuclei, and survivin-T34A overexpression enhanced radiation-induced dsDNA damage (Chakravarti et al., 2004). This suggests that survivin may regulate dsDNA break repair. In addition, inhibition of survivin by an antisense approach improved radiotherapy in a mouse model of lung cancer (Cao et al., 2004), and radiation significantly increased survivin promoter activity and expression in pancreatic cancer cell lines, while silencing of survivin by siRNA in AsPC-1, the most radioresistant cell line, augmented caspase-3 activity (Kami et al., 2005). It was noted that at least in sarcoma cells, radiosensitization by survivin inhibition is p53 status dependent. SiRNA silencing of survivin caused radiosensitization and increased caspases 3/7 in A-204 with wild-type p53, but not in US 8–93 with mutant p53 (Kappler et al., 2005).
Finally, Sui and Fan (2005) showed that combination of vincristine or vinblastine with radiation did not provide any synergistic or additive effects. Instead, the colonogenic assays showed that the combination treatment increased cell survival rates up to 2.17-fold and 2.7-fold, respectively, in comparison with the vincristine or vinblastine treatment alone. One potential mechanism to explain the antagonistic effects of the drug/radiation combination treatment could be the double induction of survivin expression and/or survivin T34 phosphorylation by means of drug and radiotherapy. This possibility is consistent with the finding that vincristine or vinblastine increases survivin expression in cancer cells (unpublished observations). Thus, positive or negative modulation of survivin expression should be considered as a novel parameter to predict the potential outcomes in the drug/drug or drug/radiation combination therapy as well as a criterion for consideration in the combination of multiple drug or drug/radiation therapies.
SURVIVIN AND ITS SPLICING VARIANTS IN CANCER THERAPEUTICS
Survivin appears to be an attractive target due to its highly expression in various cancers. Studies from the area of cancer therapeutics using survivin as a target fall into four categories (Li, 2003, 2005c): (1) inhibition of survivin expression. This includes the use of survivin antisense oligonucleotides or survivin antisense expression vectors, RNA interference (RNAi: siRNA or shRNA), ribozymes, triplex DNA formation, and anti-cancer agents, individually or in combination; (2) suppression of survivin function using surviving-dominant negative mutants, pharmacological inhibitors, or survivin peptidomimetic, individually or in combination; (3) survivin immunotherapy using survivin cDNA, RNA, protein, or peptides; and (4) applications of the survivin promoter for cancer-specific expression of cytotoxic genes.
Growing evidence indicates that survivin also plays a role in certain normal physiology such as during HPC maturation (Fukuda and Pelus, 2001; Fukuda et al., 2002). Thus, a critical question is whether survivin cancer therapy is potentially toxic to normal tissues. Survivin is considered as a tumor-associated antigen, for example, while melanocytic cells express relatively high levels of Bcl-2, Bcl-XL, Mcl-1, c-IAP-1, c-IAP-2, XIAP and Livin, survivin expression is found only in melanoma cells but not in melanocytes (Bowen et al., 2003). Second, although several normal human adult tissues/cells express survivin, anti-survivin antibodies are not detected in sera from normal people. In contrast, anti-survivin antibodies are readily detected in sera from cancer patients (Li, 2003). Consistently, although survivin is expressed in CD34+ HPCs, survivin peptide-activated cytotoxic T lymphocytes did not significantly decrease the colony-forming ability of HPCs (Pisarev et al., 2003), indicating a lack of toxicity to normal cells. Third, signal interaction contexts for survivin in normal versus cancer cells may be different as discussed above. Finally, Plescia et al. (2005) recently reported that a survivin peptidomimetic, shepherdin, which targets the binding interface between Hsp90 and survivin, strikingly induces tumor cell death, but does not decrease the viability of normal cells, and does not affect colony formation of purified hematopoietic progenitors. Administration of shepherdin inhibited human xenograft tumor growth in mice without obvious toxicity (Plescia et al., 2005). Thus, there is a reason to believe that targeting survivin for the treatment of cancer or other diseases may produce little toxicity to normal adult tissues although the molecular mechanism for non-toxic to normal cells remains to be explored in the future.
Additionally, it was found that survivin and its splice variants could be differentially regulated. Islam et al. showed that while the expression of survivin was downregulated during retinoic acid-induced apoptosis in CHP134 neuroblastoma cells, survivin-2B expression was slightly increased during apoptosis (Li, 2005a). Zhu et al. (2004) showed that doxorubicin-activated p53 upregulated survivin-2B but downregulated survivin and survivin-ΔEx3. We found that survivin-2B localizes on mitochondria (Fig. 3) and forced expression of survivin-2B induces cancer cell death (Ling et al., 2005). On the basis of these observations, control of survivin-2B expression may represent a novel approach for cancer treatment (Fig. 6). It is no doubt that elucidation of the differential regulation and/or function of survivin and its splice variants in normal versus abnormal tissues should be essential for using survivin as a target for the treatment of cancer and other human diseases.
CONCLUDING REMARKS
Studies on survivin in the past 3 years have further confirmed the notion that survivin is a multi-functional molecule. In addition to the critical role for survivin in the localization of the chromosomal passenger complex and the regulation of mitosis (Fig. 6), evidence indicates that survivin not only plays a role in caspase-dependent but also caspase-independent apoptosis controls in cancer cells, and certain survivin splice variants such as survivin-2B may modulate survivin function (Fig. 6). Expression of survivin in cancer cells is strongly associated with drug and radiation resistance, and many chemoprevention agents may exert their cancer prevention effects by inhibiting the expression of survivin. In the future studies it would be important to elucidate further the differential mechanisms for the regulation and signal transduction controls of survivin, and its splice variants in cancerous/abnormal cells versus normal ones. This will help to develop better applications of survivin as a therapeutic target for prevention and treatment of cancer as well as other diseases.
Acknowledgments
Contract grant sponsor: NIH R01; Contract grant number: CA109481; Contract grant sponsor: Concern Foundation; Contract grant sponsor: NIH Cancer Center Support Grant; Contract grant number: CA16056.
We would like to thank Dr. Yutaka Suzuki (The university of Tokyo) for the survivin-ΔptE2/3 EST clone (BP338529), Lei Song for help in the bioinformatics search of new survivin splice variants, Dr. Gary Ciment (Aves Labs) for providing much help during generation of IgY antibodies, and Dr. Michael G. Brattain and Dr. Paul Spengler (RPCI) for critical reading of the manuscript. We also apologize that some of relevant reports may not be cited due to the space limitation. This work was sponsored in part by a NIH R01 Grant (CA109481) and a grant from Concern Foundation (Beverly Hill, CA) to F.L., and by shared resources supported by NIH Cancer Center Support Grant CA16056 to Roswell Park Cancer Institute.
LITERATURE CITED
- Aziz M, Afaq F, Ahmad N. Prevention of ultraviolet B radiation damage by resveratrol in mouse skin is mediated via modulation in Survivin. Photochem Photobiol. 2005a;81(1):25–31. doi: 10.1562/2004-08-13-RA-274. [DOI] [PubMed] [Google Scholar]
- Aziz MH, Reagan-Shaw S, Wu J, Longley BJ, Ahmad N. Chemoprevention of skin cancer by grape constituent resveratrol: Relevance to human disease? FASEB J. 2005b;19(9):1193–1195. doi: 10.1096/fj.04-3582fje. [DOI] [PubMed] [Google Scholar]
- Badran A, Yoshida A, Ishikawa K, Goi T, Yamaguchi A, Ueda T, Inuzuka M. Identification of a novel splice variant of the human anti-apoptopsis gene survivin. Biochem Biophys Res Commun. 2004;314(3):902–907. doi: 10.1016/j.bbrc.2003.12.178. [DOI] [PubMed] [Google Scholar]
- Beardmore VA, Ahonen LJ, Gorbsky GJ, Kallio MJ. Survivin dynamics increases at centromeres during G2/M phase transition and is regulated by microtubule-attachment and Aurora B kinase activity. J Cell Sci. 2004;117(Pt 18):4033–4042. doi: 10.1242/jcs.01242. [DOI] [PubMed] [Google Scholar]
- Belyanskaya LL, Hopkins-Donaldson S, Kurtz S, Simoes-Wust AP, Yousefi S, Simon HU, Stahel R, Zangemeister-Wittke U. Cisplatin activates Akt in small cell lung cancer cells and attenuates apoptosis by survivin upregulation. Int J Cancer. 2005;117(5):755–763. doi: 10.1002/ijc.21242. [DOI] [PubMed] [Google Scholar]
- Bowen AR, Hanks AN, Allen SM, Alexander A, Diedrich MJ, Grossman D. Apoptosis regulators and responses in human melanocytic and keratinocytic cells. J Invest Dermatol. 2003;120(1):48–55. doi: 10.1046/j.1523-1747.2003.12010.x. [DOI] [PubMed] [Google Scholar]
- Caldas H, Honsey LE, Altura RA. Survivin 2alpha: A novel Survivin splice variant expressed in human malignancies. Mol Cancer. 2005a;4(1):11. doi: 10.1186/1476-4598-4-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caldas H, Jiang Y, Holloway MP, Fangusaro J, Mahotka C, Conway EM, Altura RA. Survivin splice variants regulate the balance between proliferation and cell death. Oncogene. 2005b;24(12):1994–2007. doi: 10.1038/sj.onc.1208350. [DOI] [PubMed] [Google Scholar]
- Cao C, Mu Y, Hallahan DE, Lu B. XIAP and survivin as therapeutic targets for radiation sensitization in preclinical models of lung cancer. Oncogene. 2004;23(42):7047–7052. doi: 10.1038/sj.onc.1207929. [DOI] [PubMed] [Google Scholar]
- Carvalho A, Carmena M, Sambade C, Earnshaw WC, Wheatley SP. Survivin is required for stable checkpoint activation in taxol-treated HeLa cells. J Cell Sci. 2003;116(Pt 14):2987–2998. doi: 10.1242/jcs.00612. [DOI] [PubMed] [Google Scholar]
- Chakravarti A, Zhai GG, Zhang M, Malhotra R, Latham DE, Delaney MA, Robe P, Nestler U, Song Q, Loeffler J. Survivin enhances radiation resistance in primary human glioblastoma cells via caspase-independent mechanisms. Oncogene. 2004;23(45):7494–7506. doi: 10.1038/sj.onc.1208049. [DOI] [PubMed] [Google Scholar]
- Delacour-Larose M, Molla A, Skoufias DA, Margolis RL, Dimitrov S. Distinct dynamics of Aurora B and Survivin during Mitosis. Cell Cycle. 2004;3(11):1418–1426. doi: 10.4161/cc.3.11.1203. [DOI] [PubMed] [Google Scholar]
- Dohi T, Beltrami E, Wall NR, Plescia J, Altieri DC. Mitochondrial survivin inhibits apoptosis and promotes tumorigenesis. J Clin Invest. 2004a;114(8):1117–1127. doi: 10.1172/JCI22222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dohi T, Okada K, Xia F, Wilford CE, Samuel T, Welsh K, Marusawa H, Zou H, Armstrong R, Matsuzawa S, Salvesen GS, Reed JC, Altieri DC. An IAP-IAP complex inhibits apoptosis. J Biol Chem. 2004b;279(33):34087–34090. doi: 10.1074/jbc.C400236200. [DOI] [PubMed] [Google Scholar]
- Earnshaw WC. Cell biology. Keeping survivin nimble at centromeres in mitosis. Science. 2005;310(5753):1443–1444. doi: 10.1126/science.1121952. [DOI] [PubMed] [Google Scholar]
- Endoh T, Tsuji N, Asanuma K, Yagihashi A, Watanabe N. Survivin enhances telomerase activity via up-regulation of specificity protein 1- and c-Myc-mediated human telomerase reverse transcriptase gene transcription. Exp Cell Res. 2005;305(2):300–311. doi: 10.1016/j.yexcr.2004.12.014. [DOI] [PubMed] [Google Scholar]
- Facchetti F, Previdi S, Ballarini M, Minucci S, Perego P, Porta CA. Modulation of pro- and anti-apoptotic factors in human melanoma cells exposed to histone deacetylase inhibitors. Apoptosis. 2004;9(5):573–582. doi: 10.1023/B:APPT.0000038036.31271.50. [DOI] [PubMed] [Google Scholar]
- Fortugno P, Beltrami E, Plescia J, Fontana J, Pradhan D, Marchisio PC, Sessa WC, Altieri DC. Regulation of survivin function by Hsp90. Proc Natl Acad Sci USA. 2003;100(24):13791–13796. doi: 10.1073/pnas.2434345100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuda S, Pelus LM. Regulation of the inhibitor-of-apoptosis family member survivin in normal cord blood and bone marrow CD34(þ) cells by hematopoietic growth factors: Implication of survivin expression in normal hematopoiesis. Blood. 2001;98(7):2091–2100. doi: 10.1182/blood.v98.7.2091. [DOI] [PubMed] [Google Scholar]
- Fukuda S, Foster RG, Porter SB, Pelus LM. The antiapoptosis protein survivin is associated with cell cycle entry of normal cord blood CD34(þ) cells and modulates cell cycle and proliferation of mouse hematopoietic progenitor cells. Blood. 2002;100(7):2463–2471. doi: 10.1182/blood.V100.7.2463. [DOI] [PubMed] [Google Scholar]
- Fulda S, Debatin KM. Sensitization for anticancer drug-induced apoptosis by the chemopreventive agent resveratrol. Oncogene. 2004a;23(40):6702–6711. doi: 10.1038/sj.onc.1207630. [DOI] [PubMed] [Google Scholar]
- Fulda S, Debatin KM. Sensitization for tumor necrosis factor-related apoptosis-inducing ligand-induced apoptosis by the chemopreventive agent resveratrol. Cancer Res. 2004b;64(1):337–346. doi: 10.1158/0008-5472.can-03-1656. [DOI] [PubMed] [Google Scholar]
- Gurbuxani S, Xu Y, Keerthivasan G, Wickrema A, Crispino JD. Differential requirements for survivin in hematopoietic cell development. Proc Natl Acad Sci USA. 2005;102(32):11480–11485. doi: 10.1073/pnas.0500303102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi N, Asano K, Suzuki H, Yamamoto T, Tanigawa N, Egawa S, Manome Y. Adenoviral infection of survivin antisense sensitizes prostate cancer cells to etoposide in vivo. Prostate. 2005;65(1):10–19. doi: 10.1002/pros.20232. [DOI] [PubMed] [Google Scholar]
- Honda R, Korner R, Nigg EA. Exploring the functional interactions between Aurora B, INCENP, and Survivin in Mitosis. Mol Biol Cell. 2003;14(8):3325–3341. doi: 10.1091/mbc.E02-11-0769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kami K, Doi R, Koizumi M, Toyoda E, Mori T, Ito D, Kawaguchi Y, Fujimoto K, Wada M, Miyatake S, Imamura M. Downregulation of survivin by siRNA diminishes radioresistance of pancreatic cancer cells. Surgery. 2005;138(2):299–305. doi: 10.1016/j.surg.2005.05.009. [DOI] [PubMed] [Google Scholar]
- Kappler M, Taubert H, Bartel F, Blumke K, Panian M, Schmidt H, Dunst J, Bache M. Radiosensitization, after a combined treatment of survivin siRNA and irradiation, is correlated with the activation of caspases 3 and 7 in a wt-p53 sarcoma cell line, but not in a mt-p53 sarcoma cell line. Oncol Rep. 2005;13(1):167–172. [PubMed] [Google Scholar]
- Krysan K, Dalwadi H, Sharma S, Pold M, Dubinett S. Cyclooxygenase 2-dependent expression of survivin is critical for apoptosis resistance in non-small cell lung cancer. Cancer Res. 2004a;64(18):6359–6362. doi: 10.1158/0008-5472.CAN-04-1681. [DOI] [PubMed] [Google Scholar]
- Krysan K, Merchant FH, Zhu L, Dohadwala M, Luo J, Lin Y, Heuze-Vourc’h N, Pold M, Seligson D, Chia D, Goodglick L, Wang H, Strieter R, Sharma S, Dubinett S. COX-2-dependent stabilization of survivin in non-small cell lung cancer. FASEB J. 2004b;18(1):206–208. doi: 10.1096/fj.03-0369fje. [DOI] [PubMed] [Google Scholar]
- Kuo PC, Liu HF, Chao JL. Survivin and p53 modulate quercetin-induced cell growth inhibition and apoptosis in the human lung carcinoma cells. J Biol Chem. 2004;279(53):55875–55885. doi: 10.1074/jbc.M407985200. [DOI] [PubMed] [Google Scholar]
- Lens SM, Medema RH. The survivin/aurora B complex: Its role in coordinating tension and attachment. Cell Cycle. 2003;2(6):507–510. doi: 10.4161/cc.2.6.559. [DOI] [PubMed] [Google Scholar]
- Li F. Survivin Study: What is the next wave? J Cell Physiol. 2003;197(1):8–29. doi: 10.1002/jcp.10327. [DOI] [PubMed] [Google Scholar]
- Li F. Role of survivin and its splice variants in tumorigenesis. Br J Cancer. 2005a;92(2):212–216. doi: 10.1038/sj.bjc.6602340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F. Role of survivin in cancer chemoprevention. Assay Des-Simply Sci. 2005b;1(1):2–6. [Google Scholar]
- Li F. Survivin, other IAPs, Smac/DIABLO, and Omi/HtrA2—modulation of the advancing apoptotic process. In: Los M, Gibson SB, editors. Apoptotic pathways as target for novel therapies in cancer and other diseases. New York: Kluwer Press; 2005c. pp. 137–155. [Google Scholar]
- Li F, Brattain MG. Role of the survivin gene in pathophysiology. Am J Pathol. 2006 doi: 10.2353/ajpath.2006.060121. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F, Ambrosini G, Chu EY, Plescia J, Tognin S, Marchisio PC, Altieri DC. Control of apoptosis and mitotic spindle checkpoint by survivin. Nature. 1998;396(6711):580–584. doi: 10.1038/25141. [DOI] [PubMed] [Google Scholar]
- Li F, Ling X, Huang H, Brattain L, Apontes P, Wu J, Binderup L, Brattain MG. Differential regulation of survivin expression and apoptosis by vitamin D(3) compounds in two isogenic MCF-7 breast cancer cell sublines. Oncogene. 2005;24(8):1385–1395. doi: 10.1038/sj.onc.1208330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling X, Bernacki RJ, Brattain MG, Li F. Induction of survivin expression by taxol (paclitaxel) is an early event which is independent on taxol-mediated G2/M arrest. J Biol Chem. 2004;279(15):15196–15203. doi: 10.1074/jbc.M310947200. [DOI] [PubMed] [Google Scholar]
- Ling X, Yang J, Tan D, Ramnath N, Younis T, Bundy BN, Slocum HK, Yang L, Zhou M, Li F. Differential expression of survivin-2B and survivin-DeltaEx3 is inversely associated with disease relapse and patient survival in non-small-cell lung cancer (NSCLC) Lung Cancer. 2005;49:353–361. doi: 10.1016/j.lungcan.2005.03.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu T, Brouha B, Grossman D. Rapid induction of mitochondrial events and caspase-independent apoptosis in Survivin-targeted melanoma cells. Oncogene. 2004;23(1):39–48. doi: 10.1038/sj.onc.1206978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahotka C, Liebmann J, Wenzel M, Suschek CV, Schmitt M, Gabbert HE, Gerharz CD. Differential subcellular localization of functionally divergent survivin splice variants. Cell Death Differ. 2002;9(12):1334–1342. doi: 10.1038/sj.cdd.4401091. [DOI] [PubMed] [Google Scholar]
- Marusawa H, Matsuzawa S, Welsh K, Zou H, Armstrong R, Tamm I, Reed JC. HBXIP functions as a cofactor of survivin in apoptosis suppression. EMBO J. 2003;22(11):2729–2740. doi: 10.1093/emboj/cdg263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagata Y, Jones MR, Nguyen HG, McCrann DJ, St Hilaire C, Schreiber BM, Hashimoto A, Inagaki M, Earnshaw WC, Todokoro K, Ravid K. Vascular smooth muscle cell polyploidization involves changes in chromosome passenger proteins and an endomitotic cell cycle. Exp Cell Res. 2005;305(2):277–291. doi: 10.1016/j.yexcr.2004.12.028. [DOI] [PubMed] [Google Scholar]
- Noton EA, Colnaghi R, Tate S, Starck C, Carvalho A, Ko Ferrigno P, Wheatley SP. Molecular analysis of survivin isoforms: Evidence that alternatively spliced variants do not play a role in mitosis. J Biol Chem. 2006;281:1286–1295. doi: 10.1074/jbc.M508773200. [DOI] [PubMed] [Google Scholar]
- O’Connor DS, Wall NR, Porter AC, Altieri DC. A p34(cdc2) survival checkpoint in cancer. Cancer Cell. 2002;2(1):43–54. doi: 10.1016/s1535-6108(02)00084-3. [DOI] [PubMed] [Google Scholar]
- Pisarev V, Yu B, Salup R, Sherman S, Altieri DC, Gabrilovich DI. Full-length dominant-negative survivin for cancer immunotherapy. Clin Cancer Res. 2003;9(17):6523–6533. [PubMed] [Google Scholar]
- Plescia J, Salz W, Xia F, Pennati M, Zaffaroni N, Daidone MG, Meli M, Dohi T, Fortugno P, Nefedova Y, Gabrilovich DI, Colombo G, Altieri DC. Rational design of shepherdin, a novel anticancer agent. Cancer Cell. 2005;7(5):457–468. doi: 10.1016/j.ccr.2005.03.035. [DOI] [PubMed] [Google Scholar]
- Rodel C, Haas J, Groth A, Grabenbauer GG, Sauer R, Rodel F. Spontaneous and radiation-induced apoptosis in colorectal carcinoma cells with different intrinsic radiosensitivities: Survivin as a radioresistance factor. Int J Radiat Oncol Biol Phys. 2003;55(5):1341–1347. doi: 10.1016/s0360-3016(02)04618-7. [DOI] [PubMed] [Google Scholar]
- Rodel F, Hoffmann J, Distel L, Herrmann M, Noisternig T, Papadopoulos T, Sauer R, Rodel C. Survivin as a radioresistance factor, and prognostic and therapeutic target for radiotherapy in rectal cancer. Cancer Res. 2005;65(11):4881–4887. doi: 10.1158/0008-5472.CAN-04-3028. [DOI] [PubMed] [Google Scholar]
- Sampath SC, Ohi R, Leismann O, Salic A, Pozniakovski A, Funabiki H. The chromosomal passenger complex is required for chromatin-induced micro-tubule stabilization and spindle assembly. Cell. 2004;118(2):187–202. doi: 10.1016/j.cell.2004.06.026. [DOI] [PubMed] [Google Scholar]
- Samuel T, Okada K, Hyer M, Welsh K, Zapata JM, Reed JC. cIAP1 localizes to the nuclear compartment and modulates the cell cycle. Cancer Res. 2005;65(1):210–218. [PubMed] [Google Scholar]
- Song Z, Wu M. Identification of a novel nucleolar localization signal and a degradation signal in Survivin-deltaEx3: A potential link between nucleolus and protein degradation. Oncogene. 2005;24(16):2723–2734. doi: 10.1038/sj.onc.1208097. [DOI] [PubMed] [Google Scholar]
- Suga K, Yamamoto T, Yamada Y, Miyatake S, Nakagawa T, Tanigawa N. Correlation between transcriptional expression of survivin isoforms and clinicopathological findings in human colorectal carcinomas. Oncol Rep. 2005;13(5):891–897. [PubMed] [Google Scholar]
- Sui M, Fan W. Combination of gamma-radiation antagonizes the cytotoxic effects of vincristine and vinblastine on both mitotic arrest and apoptosis. Int J Radiat Oncol Biol Phys. 2005;61(4):1151–1158. doi: 10.1016/j.ijrobp.2004.12.028. [DOI] [PubMed] [Google Scholar]
- Taubert H, Kappler M, Bache M, Bartel F, Kohler T, Lautenschlager C, Blumke K, Wurl P, Schmidt H, Meye A, Hauptmann S. Elevated expression of survivin-splice variants predicts a poor outcome for soft-tissue sarcomas patients. Oncogene. 2005;24(33):5258–5261. doi: 10.1038/sj.onc.1208702. [DOI] [PubMed] [Google Scholar]
- Vader G, Kauw JJ, Medema RH, Lens SM. Survivin mediates targeting of the chromosomal passenger complex to the centromere and midbody. EMBO Rep. 2005;7:85–92. doi: 10.1038/sj.embor.7400562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vegran F, Boidot R, Oudin C, Riedinger JM, Lizard-Nacol S. Distinct expression of Survivin splice variants in breast carcinomas. Int J Oncol. 2005;27(4):1151–1157. [PubMed] [Google Scholar]
- Vong QP, Cao K, Li HY, Iglesias PA, Zheng Y. Chromosome alignment and segregation regulated by ubiquitination of survivin. Science. 2005;310(5753):1499–1504. doi: 10.1126/science.1120160. [DOI] [PubMed] [Google Scholar]
- Wang HW, Sharp TV, Koumi A, Koentges G, Boshoff C. Characterization of an anti-apoptotic glycoprotein encoded by Kaposi’s sarcoma-associated herpes-virus which resembles a spliced variant of human survivin. EMBO J. 2002;21(11):2602–2615. doi: 10.1093/emboj/21.11.2602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Zhang GM, Feng ZH. Down-regulation of survivin expression reversed multidrug resistance in adriamycin-resistant HL-60/ADR cell line. Acta Pharmacol Sin. 2003;24(12):1235–1240. [PubMed] [Google Scholar]
- Wheatley SP, Henzing AJ, Dodson H, Khaled W, Earnshaw WC. Aurora-B phosphorylation in vitro identifies a residue of Survivin that is essential for its localization and binding to INCENP in vivo. J Biol Chem. 2004;279(7):5655–5660. doi: 10.1074/jbc.M311299200. [DOI] [PubMed] [Google Scholar]
- Yang D, Welm A, Bishop JM. Cell division and cell survival in the absence of survivin. Proc Natl Acad Sci USA. 2004;101(42):15100–15105. doi: 10.1073/pnas.0406665101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang T, Fields JZ, Ehrlich SM, Boman BM. The chemopreventive agent sulindac attenuates expression of the antiapoptotic protein survivin in colorectal carcinoma cells. J Pharmacol Exp Ther. 2004a;308(2):434–437. doi: 10.1124/jpet.103.059378. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Nagata Y, Yu G, Nguyen HG, Jones MR, Toselli P, Jackson CW, Tatsuka M, Todokoro K, Ravid K. Aberrant quantity and localization of Aurora-B/AIM-1 and survivin during megakaryocyte polyploidization and the consequences of Aurora-B/AIM-1-deregulated expression. Blood. 2004b;103(10):3717–3726. doi: 10.1182/blood-2003-09-3365. [DOI] [PubMed] [Google Scholar]
- Zhang M, Latham DE, Delaney MA, Chakravarti A. Survivin mediates resistance to antiandrogen therapy in prostate cancer. Oncogene. 2005a;24(15):2474–2482. doi: 10.1038/sj.onc.1208490. [DOI] [PubMed] [Google Scholar]
- Zhang M, Mukherjee N, Bermudez RS, Latham DE, Delaney MA, Zietman AL, Shipley WU, Chakravarti A. Adenovirus-mediated inhibition of survivin expression sensitizes human prostate cancer cells to paclitaxel in vitro and in vivo. Prostate. 2005b;64(3):293–302. doi: 10.1002/pros.20263. [DOI] [PubMed] [Google Scholar]
- Zheng W, Ma X, Wei D, Wang T, Ma Y, Yang S. Molecular cloning and bioinformatics analysis of a novel spliced variant of survivin from human breast cancer cells. DNA Seq. 2005;16(5):321–328. doi: 10.1080/10425170500226490. [DOI] [PubMed] [Google Scholar]
- Zhou J, O’Brate A, Zelnak A, Giannakakou P. Survivin deregulation in beta-tubulin mutant ovarian cancer cells underlies their compromised mitotic response to taxol. Cancer Res. 2004;64(23):8708–8714. doi: 10.1158/0008-5472.CAN-04-2538. [DOI] [PubMed] [Google Scholar]
- Zhu N, Gu L, Findley HW, Li F, Zhou M. An alternatively spliced survivin variant is positively regulated by p53 and sensitizes leukemia cells to chemotherapy. Oncogene. 2004;23:7545–7551. doi: 10.1038/sj.onc.1208038. [DOI] [PubMed] [Google Scholar]