Abstract
The compound 3-amino-1,2,4-benzotriazine 1,4-dioxide (tirapazamine, TPZ) is a clinically-promising anticancer agent that selectively kills the oxygen-poor (hypoxic) cells found in solid tumors. It has long been known that, under hypoxic conditions, TPZ causes DNA strand damage that is initiated by the abstraction of hydrogen atoms from the deoxyribose phosphate backbone of duplex DNA but exact chemical mechanisms underlying this process remain unclear. Here we describe detailed characterization of sugar-derived products arising from TPZ-mediated strand damage. We find that the action of TPZ on duplex DNA under hypoxic conditions generates 5-methylene-2-furanone (6), oligonucleotide 3′-phosphoglycolates (7), malondialdehyde equivalents (8 or 9), and furfural (10). These results provide evidence that TPZ-mediated strand damage arises via hydrogen atom abstraction from both the most hindered (C1′) and least hindered (C4′ and C5′) positions of the deoxyribose sugars in the double helix. The products observed are identical to those produced by hydroxyl radical. Additional experiments were conducted to better understand the chemical pathways by which TPZ generates the observed DNA-damage products. Consistent with previous work showing that TPZ can substitute for molecular oxygen in DNA damage reactions, it is found that, under anaerobic conditions, reaction of TPZ with a discrete, photogenerated C1′-radical in a DNA 2′-oligodeoxynucleotide cleanly generates the 2-deoxyribonolactone lesion (5) that serves as the precursor to 5-methylene-2-furanone (6). Overall, the results provide insight regarding the chemical structure of the DNA lesions that confront cellular repair, transcription, and replication machinery following exposure to TPZ and offer new information relevant to the chemical mechanisms underlying TPZ-mediated strand cleavage.
Solid tumors differ from most normal human tissue, in that they contain significant populations of oxygen-poor (hypoxic) cells.1-4 Accordingly, in the pursuit of improved anticancer drugs, medicinal chemists have long sought compounds that selectively generate cell-killing reactive intermediates under hypoxic conditions.5-10 The compound 3-amino-1,2,4-benzotriazine 1,4-dioxide (tirapazamine, TPZ, 1, Scheme 1) may be the most promising hypoxia-selective antitumor agent identified to date.5,7,11 TPZ is between 50-300 times more toxic to hypoxic cells versus normally-oxygenated cells.5,7,12 The anticancer properties of TPZ stem from its ability to cause oxidative DNA damage selectively in hypoxic tumor cells.7,11,13-19 This drug is currently undergoing a variety of phase II and III clinical trials for the treatment of human cancer.20
Scheme 1.
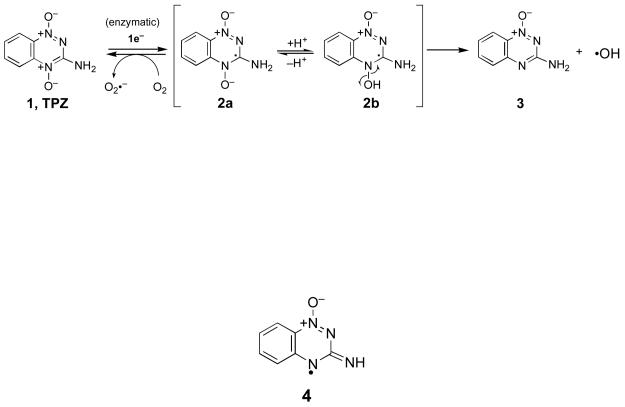
Upon entering cells, TPZ is metabolized via one-electron enzymatic reduction to the radical anion (2a, Scheme 1) that exists in equilibrium with its protonated neutral form, (2b, pKa = 6).13,21,22 In normally oxygenated cells, TPZ is relatively non-toxic because 2a is rapidly reoxidized to 1 (Scheme 1).13,21 This futile cycle of enzymatic reduction and O2-mediated back-oxidation generates superoxide radical (O2•−), which can cause cytotoxicity.23-26 However, enzyme systems including superoxide dismutase, catalase, glutathione peroxidase, and peroxiredoxins provide the cell with protection against superoxide radical and the reactive oxygen species derived from it.27-29 Under hypoxic conditions, the radical intermediate (2) causes oxidative DNA damage7,11,18 and the identity of the reactive intermediate(s) responsible for this medicinally-relevant process remains a subject of active investigation. Data from our previous investigations support a mechanism in which the neutral drug radical 2b undergoes homolytic fragmentation to release the well known DNA-damaging agent, hydroxyl radical (HO•).15 Consistent with this view, TPZ causes extensive oxidative damage to the nucleobases of DNA, including the generation of hydroxylated base analogs.16,17 Also in accord with the involvement of hydroxyl radical, TPZ causes sequence-independent DNA strand cleavage via abstraction of hydrogen atoms from the deoxyribose sugars of the DNA backbone.15 Neutralization-reionization mass spectrometry experiments30 and computational studies31 indicate that homolytic fragmentation of the N-O bond in 2b is chemically feasible. Furthermore, homolytic fragmentation of analogous N-O bonds is well precedented.32-38 Others have proposed39 that TPZ-mediated DNA strand cleavage arises through direct hydrogen atom abstraction by 2b and, more recently, it has been suggested40 that 2b undergoes dehydration to yield the benzotriazinyl radical 4 as the ultimate DNA-damaging intermediate. Finally, in addition to its ability to initiate the generation of DNA radicals, TPZ and its mono-N-oxide metabolites react with DNA radicals in a manner that mimics molecular oxygen, facilitating conversion of the radical intermediates into direct strand breaks and base-labile lesions under low-oxygen conditions.41-44
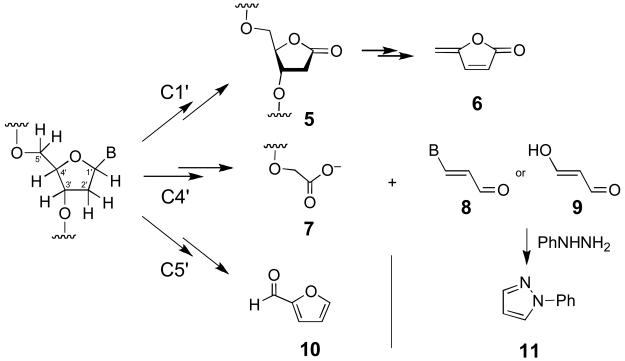
While it is clear that enzymatic activation of TPZ under hypoxic conditions leads to DNA strand damage via abstraction of hydrogen atoms from the sugar-phosphate backbone,15,19 the exact chemical mechanisms underlying this process remain uncertain. For example, the identity of the specific deoxyribose hydrogens involved in these strand damage reactions has not been determined. In general, hydrogen atom abstraction from the deoxyribose backbone of DNA leads to strand cleavage via complex and chemically interesting reaction cascades.45-49 Importantly, abstraction of each deoxyribose hydrogen atom yields a diagnostic set of products.47 Therefore, careful product analysis provides insight into the specific deoxyribose hydrogens involved in radical-mediated DNA strand damage by a given agent. For example, hydrogen atom abstraction from the C1′-position of DNA under aerobic conditions yields 5-methylene-2-furanone (6) upon heating, while abstraction of the C4′-hydrogen generates oligonucleotide 3′-phosphoglycolates (7), and an oxidized, three-carbon sugar fragment (either base propenal, 8, or malondialdehyde, 9) as characteristic products (Scheme 2).47,50-53 Abstraction of the C5′-hydrogen yields furfural (10) as a characteristic aerobic degradation product (Scheme 2).47 Small, highly reactive radicals such as hydroxyl radical (HO•) cause direct strand cleavage via reactions involving hydrogen atom abstraction at all positions on the deoxyribose backbone, with the extent of reaction at each site dictated largely by steric accessibility of the various hydrogens.47,54-56 In duplex DNA, the C4′ and C5′ hydrogens of deoxyribose are the most accessible, while the C1′ hydrogens are the least accessible, residing deep in the minor groove. Interestingly, recent results suggest that abstraction of C1′-hydrogens by hydroxyl radical also occurs via an indirect pathway involving secondary reactions of initially-formed pyrimidine base radical adducts.56,57
Scheme 2.
In the work reported here, we characterized products arising from TPZ-mediated damage of DNA. We find that the action of activated TPZ on duplex DNA under hypoxic conditions generates 5-methylene-2-furanone (6) upon heating, as well as oligonucleotide 3′-phosphoglycolates (7), malondialdehyde equivalents (8/9), and furfural (10). These results provide evidence that TPZ-mediated strand cleavage arises via hydrogen atom abstraction from both the most hindered (C1′) and least hindered (C4′ and C5′) positions of the deoxyribose sugars in the double helix. The products observed are identical to those produced by hydroxyl radical.47 Additional experiments were conducted in the context of the C1′ radical to gain insight regarding the chemical pathways by which TPZ generates its DNA strand damage products. Consistent with previous work showing that TPZ can substitute for molecular oxygen in DNA damage reactions,41-43 these experiments reveal that, under anaerobic conditions, reaction of TPZ with a discrete, photogenerated C1′-radical in a DNA oligonucleotide cleanly yields the 2-deoxyribonolactone lesion (5) that serves as the precursor to 5-methylene-2-furanone (6, Scheme 2).
The results provide insight regarding the chemical structure of the DNA lesions that confront cellular repair, transcription, and replication machinery following exposure to TPZ and provides new information relevant to the chemical mechanisms underlying TPZ-mediated strand cleavage.
Result and Discussion
DNA-Damage Reactions
Calf-thymus DNA was used as a source of mixed sequence, double-stranded DNA. Either NADPH:cytochrome P450 reductase or xanthine/xanthine oxidase enzyme systems were used to carry out the one-electron reduction of TPZ to its activated form (2). NADPH:cytochrome P450 reductase is thought to be responsible for in vivo activation of TPZ14,58,59 and the xanthine/xanthine oxidase system has been used successfully as a reagent for the one-electron activation of TPZ in a variety of in vitro studies.13,15,17,43 Molecular oxygen was removed from stock solutions via freeze-pump-thaw degassing and final assay mixtures prepared and incubated in an inert atmosphere glove bag.
Evidence for Hydrogen Atom Abstraction from the C1′- and C5′-Positions of Deoxyribose in Duplex DNA: Identification of 5-Methylene-2-furanone (6) and Furfural (10) As Products Arising From Tirapazamine-Mediated DNA Strand Damage
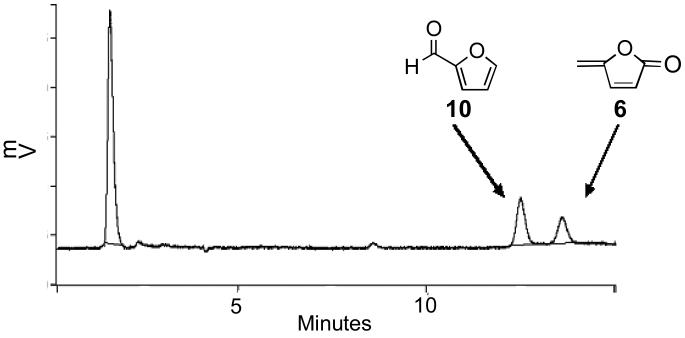
Compounds 6 and 10 are characteristic products stemming from the abstraction of hydrogen atoms from the C1′- and C5′-positions of deoxyribose, respectively.47 These products can be simultaneously detected using an HPLC assay.47,60-62 Accordingly, TPZ-damaged DNA was isolated by ethanol precipitation and heated at 90 °C for 15 min in a sealed tube to release 6 and 10. The resulting products were extracted into methylene chloride, concentrated, and analyzed by reverse-phase HPLC (Figure 1). Two peaks that co-migrate with authentic samples of furfural (10, eluting at ~12 min) and 5-methylenefuranone (6, eluting at ~13 min) were observed. Negligible amounts of these compounds were observed in control samples of untreated DNA, DNA treated with TPZ alone (no enzyme), or DNA treated with the NADPH:cytochrome P450 reductase enzyme system (no TPZ). The results provide evidence that TPZ-mediated DNA strand damage is initiated, at least in part, by abstraction of hydrogen atoms from both the C1′- and C5′-positions of the deoxyribose backbone.
Figure 1. HPLC detection of 5-methylenefuranone (6) and furfural (10) isolated from TPZ-damaged DNA.
TPZ-damaged DNA was heated to release 6 and 10, extracted with methylene chloride, the extract evaporated, and the products analyzed by reverse-phase HPLC as described in the Experimental Section.
In light of the hypothesis that activated tirapazamine cleaves DNA via the release of hydroxyl radical, we felt it would be useful to compare the relative yields of 6 and 10 produced by tirapazamine-mediated DNA damage with those produced hydroxyl radical generated by the well-studied Fe(II)/ETDA/H2O2/ascorbate system.47,54,63 We found that the relative yields of 10 and 6 produced by activated TPZ and authentic hydroxyl radical were identical within experimental error (10:6 = 1.6, based on HPLC peak area). The relatively large yield of 6 is unexpected based upon the notion that steric accessibility solely determines the extent of reaction at each 2′-deoxyribose hydrogen, but is consistent with recent studies indicating that hydroxyl radical adducts16,17 generated at pyrimidine nucleobases yield secondary oxygen-centered radicals that effectively abstract hydrogen atoms from the adjacent nucleotide on the 5′-side.56,57
Interestingly, when xanthine/xanthine oxidase was used to generate activated TPZ, only 5-methylenefuranone was detected. Likely this is due to the oxidation of furfural by xanthine oxidase. Aldehydes are known to be substrates of xanthine oxidase and both base propenals and malondialdehyde are oxidized by this enzyme.64-66
Products Arising from C4′-Hydrogen Atom Abstraction. Evidence for the Formation of Malondialdehyde Equivalents (8/9)
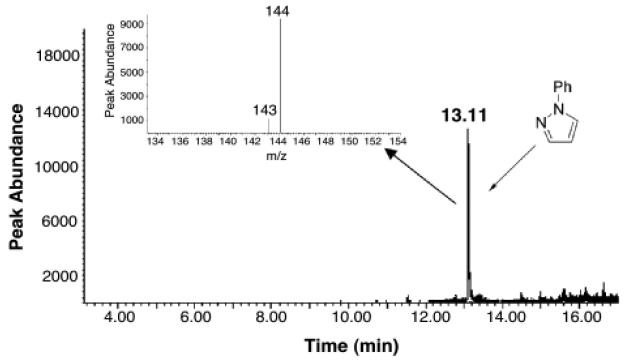
Hydrogen atom abstraction from the C4′-position of 2′-deoxyribose yields a three-carbon, oxidized 2′-deoxyribose fragment as a characteristic product. This oxidized sugar fragment can take the form of base propenals (8) or malondialdehyde (9), products collectively referred to as “malondialdehyde equivalents”.47,53,67,68 A sensitive method for the detection of these products involves derivatization with phenylhydrazine to yield phenylpyrazole (11, Scheme 2).53,69 An advantage of this particular assay is that it detects intact base propenals along with malondialdehyde and DNA base adducts generated by these species.69 Accordingly, TPZ-damaged duplex DNA was treated with phenylhydrazine (20 mM), incubated for 15 h, and then extracted with hexane. GC-MS-SIM revealed a significant peak with m/z of 144 eluting at 13 min, identical to an authentic sample of phenylpyrazole 11 (Figure 2). In contrast, the yield of phenylpyrazole was negligible in control samples of untreated DNA, DNA treated with TPZ alone (no enzyme), or DNA treated with the NADPH:cytochrome P450 reductase enzyme system (no TPZ). As expected,47,63 a positive control reaction involving treatment of DNA with the Fe(II)/ETDA/H2O2/ascorbate system, followed by derivatization as described above, generates phenylpyrazole 11. In general, our observation that TPZ generates malondialdehyde equivalents is consistent with early observations that the drug generates so-called thiobarbituric acid-reactive substances (TBARS) when it attacks cellular DNA.70 Base propenals and malondialdehyde are the primary TBARS generated in the oxidative degradation of DNA.47
Figure 2. Detection of phenylpyrazole (11) provides evidence for the formation of malondialdehyde equivalents in TPZ-damaged DNA.
TPZ-damaged DNA was treated with phenylhydrazine (20 mM) for 15 h to convert base propenals and other malondialdehyde equivalents into phenylpyrazole. The reactions were extracted with hexane, concentrated, and analyzed by GC-MS as described in the Experimental Section.
Further Evidence for C4′-Hydrogen Atom Abstraction: Observation of Oligonucleotide 3′-Phosphoglycolates (7)
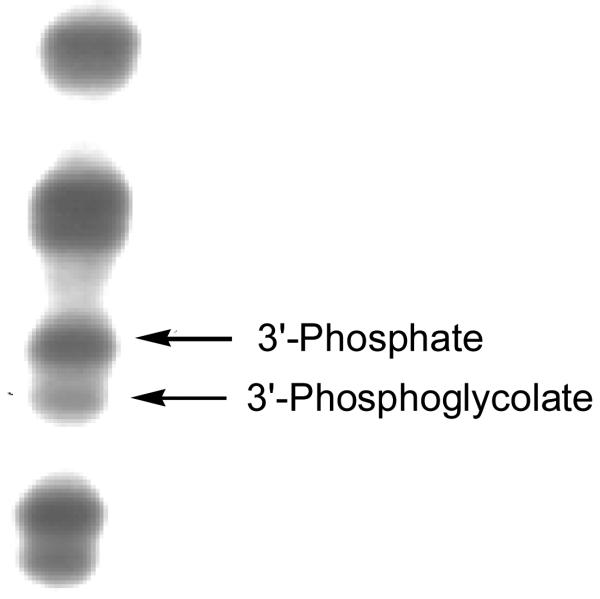
Abstraction of C4′-hydrogen atoms from DNA typically generates strand cleavage products bearing a 3′-phosphoglycolate end group (7, Scheme 2) at the cleavage sites.47,71 Gel electrophoresis can be used to detect these 3′-phosphoglycolate termini.47 Short 2′-oligodeoxynucleotide fragments containing a 3′-phosphoglycolate terminus migrate slightly faster in a 25% denaturing gel than do the same DNA fragments containing a 3′-phosphate terminus.47,63,72 Accordingly, we employed high-resolution denaturing 25% polyacrylamide gel electrophoretic analysis to investigate whether 3′-phosphoglycolate termini are produced by TPZ-mediated DNA cleavage.
A 5′-32P-labeled 377 bp restriction fragment was exposed to xanthine/xanthine oxidase-activated TPZ under anaerobic conditions. The DNA was isolated by ethanol precipitation, denatured, and the resulting cleavage products analyzed by denaturing 25% polyacrylamide gel electrophoresis. Visualization of the labeled fragments by phosphorimager analysis revealed that TPZ-derived DNA-cleavage sites consist of two closely-spaced bands characteristic of the 3′-phosphate/3′-phosphoglycolate pair of end products (Figure 3).47,73 This finding is in complete agreement with the earlier work of Jones and Weinfeld who used 32P-postlabeling of DNA digests to detect 3′-phosphoglycolate products in TPZ-damaged DNA.43
Figure 3. Gel electrophoretic detection of 3′-phosphoglycolate termini (7) in TPZ-damaged DNA.
A 32P-5′-labeled 377 bp restriction fragment was damaged by xanthine/xanthine oxidase-activated TPZ under anaerobic conditions, the DNA precipitated, washed, and the labeled DNA fragments resolved on a high resolution (25%), denaturing polyacrylamide gel as described in the Experimental Section. The labeled DNA fragments were visualized by phosophorimager analysis.
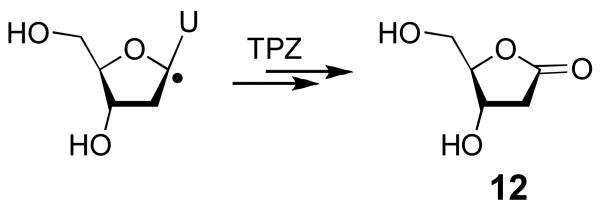
On the Origin of the 5-Methylene-2-furanone Product (6): TPZ Efficiently Oxidizes a Photogenerated C1′-Radical to the 2-Deoxyribonolactone Lesion in a 2′-Oligodeoxynucleotide
In radical-mediated DNA-damage processes that occur under aerobic conditions, the initially-formed 2′-deoxyribose radicals rapidly combine with molecular oxygen to yield peroxyl radical intermediates (ROO•) that subsequently decompose via complex reaction cascades to yield the final products of strand damage.46-49,74 Thus, O2 profoundly influences the spectrum of products generated during radical-mediated DNA degradation. The reactions described in this work involve TPZ-mediated DNA damage under anaerobic conditions, yet the products observed are those typically associated with the reaction of radicals with the deoxyribose backbone under aerobic conditions.46-49 How does the action of TPZ under anaerobic conditions lead to strand cleavage products typically associated with radical-mediated DNA damage under aerobic conditions? As noted in the introduction, the answer to this question was provided by the observation that TPZ and its mono-N-oxide metabolites can substitute for molecular oxygen in the conversion of deoxyribose radicals to DNA strand breaks.41-43 For example, previous work demonstrated that TPZ and its mono-N-oxide metabolites rapidly react with a C1′-radical in duplex DNA, thereby converting the deoxyribose radical to a base-labile strand cleavage site.41,42 The structural nature of the base-labile lesion generated in polymeric DNA was not determined in this early work, although model studies showed that reaction of a C1′-nucleoside radical with TPZ generates 2-deoxyribonolactone (12, Scheme 3).41,42 In order to gain a better general understanding of the chemical pathways involved in generation of the strand-cleavage products reported above, we set out to determine whether the reaction of TPZ with a photogenerated C1′-radical in a 2′-oligodeoxynucleotide gives rise to the 2-deoxyribonolactone lesion (5) that serves as a precursor to the 5-methylene-2-furanone product (6, Scheme 2).
Scheme 3.
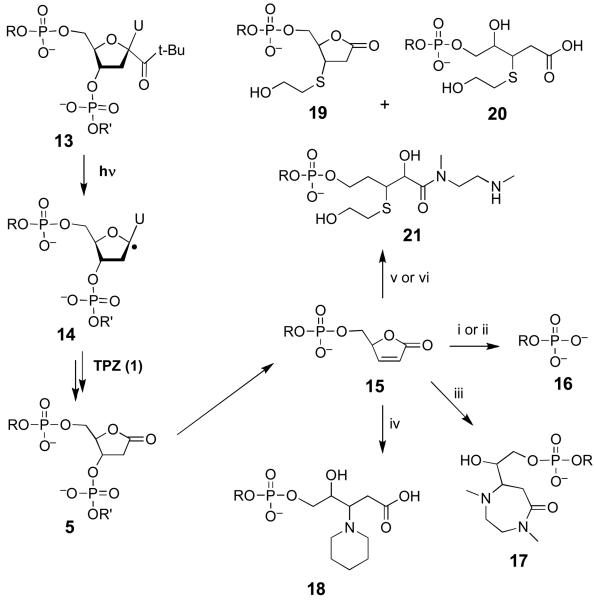
The 2-deoxyribonolactone lesion (5) in DNA is unstable and readily undergoes β-elimination of phosphate to yield the butenolide 15.75,76 Under physiological conditions, the butenolide does not accumulate because it rapidly undergoes γ-elimination to yield 5-methylene-2-furanone (6) and a 3′-oligonucleotide phosphate (16, Scheme 4). Nonetheless, the butenolide 15 can be trapped by a variety of agents (Scheme 4).75,76 The characteristic shifts in electrophoretic gel mobility induced by formation of the resulting products provide a diagnostic “fingerprint” that can be used to demonstrate the presence of a 3′-butenolide terminus at a DNA-cleavage site.75 We employed this approach to probe whether the reaction of TPZ with a photogenerated C1′-radical in 2′-oligodeoxynucleotide produces the 2-deoxyribonolactone lesion (5).
Scheme 4.
i. 100 mM piperidine, 90ÞC, 15 min; ii. 100 mM DMEDA, 90ÞC, 15 min; iii. 100 mM DMEDA, 37ÞC, 20 min; iv. 100 mM piperidine, 37ÞC, 20 min; v. 50 mM 2-mercaptoethanol, 100 DMEDA, 37ÞC, 20 min; vi. 50 mM 2-mercaptoethanol, 37ÞC, 20 min (yields 19).
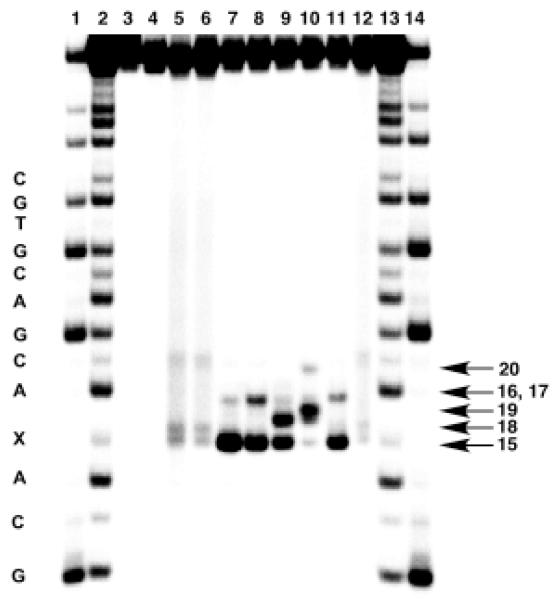
These studies employed a 5′-32P-labeled 2′-oligodeoxynucleotide containing a single t-butylketone-containing residue 13 that generates a C1′-radical (14) upon photolysis (Scheme 4).77,78 The 30-mer 2′-oligodeoxynucleotide, 5′-GTCACGTGCTGCA-13-ACGACGTGCTGAGCCT, was photolyzed under anaerobic conditions to generate the C1′-radical in the presence of TPZ.75 The resulting mixture was treated with one of the “fingerprinting” reagents piperidine, dimethylethylenediamine (DMEDA), and 2-mercaptoethanol and the products analyzed by gel electrophoresis. Visualization of the 32P-labeled DNA fragments in the gel by phosphorimager analysis reveals the characteristic set of gel-shifted products expected from the reaction of 15 with these various reagents (Figure 4, and Scheme 4).75,76 These results provide evidence that interaction of the C1′-radical with TPZ does, indeed, generate the 2-deoxyribonolactone lesion that serves as a precursor to 5-methylene-2-furanone (6). In the absence of TPZ, strand cleavage is very weak, thus confirming that efficient formation of the 2-deoxyribonolactone lesion under the anaerobic conditions employed here requires reaction of the C1′-radical with the drug. Importantly, there are no unidentified bands in the gel, suggesting that the reaction is “clean” – that is, the butenolide (15) is the only major base-labile lesion generated in these reactions.
Figure 4. Gel electrophoretic detection of 2-deoxyribonolactone lesion (5) generated by the reaction of TPZ with a C1′-radical in a 2′-oligodeoxynucleotide.
The C1′-radical was generated in the presence of TPZ by photolysis of the 2′-oligodeoxynucleotide radical precursor 32P-GTCACGTGCTGCA-12-ACGACGTGCTGAGCCT in sodium phosphate buffer (10 mM, pH 7) containing NaCl (100 mM). The product was then treated with N,N′-dimethylethylenediamine (100 mM), piperidine (100 mM), or piperidine (100 mM)/β-mercaptoethanol (50 mM) as described previously.75 Lanes 1 and 14, Maxam-Gilbert G specific cleavage reaction; lanes 2 and 13, Maxam-Gilbert A+G specific reaction; lanes 3-5 contain samples treated under aerobic conditions: lane 3, non-photolysed; lane 4, non photolysed + piperidine (100 mM), 30 min, 90 °C; lane 5, photolysed; lanes 6-12 contain samples that were photolysed with TPZ (1 mM) under anaerobic conditions: lane 6, no further treatment; lane 7, piperidine (100 mM), 20 min, 90 °C; lane 8, piperidine (100 mM), 20 min, 37 °C; lane 9, piperidine (100 mM), β-mercaptoethanol (50 mM), 20 min, 37 °C; lane 10, N,N′-dimethylethylenediamine (100 mM), β-mercaptoethanol (50 mM), 20 min, 37 °C; lane 11, N,N′-dimethylethylenediamine (100 mM) 20 min, 37 °C; lane 12, β-mercaptoethanol (50 mM), 20 min, 37 °C.
Conclusions
The product analysis described here shows that TPZ-mediated DNA strand damage proceeds, at least in part, via hydrogen atom abstraction from both the most hindered (C1′) and the least hindered (C4′ and C5′) positions of the deoxyribose sugars in the double helix. This type of nonselective hydrogen atom abstraction mirrors that by the highly reactive species, hydroxyl radical (HO•).54 Accordingly, the results provide new evidence consistent with the proposal that TPZ delivers hydroxyl radical, the active agent of radiation therapy, selectively to oxygen-poor tumor cells.15-17
Although DNA damage by TPZ occurs under low-oxygen conditions, we provide evidence here that the structural nature of the strand damage generated by this drug is typical of that observed for radical-mediated DNA damage by hydroxyl radical under aerobic conditions. The results presented here extend previous results41-43 by providing evidence that TPZ (and its N-oxide metabolites) are able to mediate the oxygenation of the C1′, C4′, and C5′ radicals in duplex DNA. In addition, to gain a better chemical understanding of the pathways by which these products arise, we investigated the reaction of TPZ with a photogenerated C1′-radical in a 2′-oligodeoxyribonucleotide. We find that the reaction cleanly generates the 2-deoxyribonolactone lesion (5) that serves as the precursor to the methylene lactone (6) detected as a product of TPZ-mediated DNA damage in these studies.
The chemical mechanism underlying the oxygenation of DNA radicals by TPZ remains a matter of active research.40-42,44 We previously observed that oxygenation of a C1′ nucleoside radical by 16O-containing TPZ in H218O leads to a mixture of 16O-containing (70%) and 18O-containing (30%) lactone 12.41,42 We proposed that the 16O-containing product arises via formation of a TPZ-deoxyribose radical adduct, followed by fragmentation to generate the C1′-alkoxyl radical. On the other hand, the 18O-containing product must arise through net oxidation of the radical intermediate, followed by hydrolysis to yield hydroxylation at the original site of the radical.41,42,44 With regard to these possible mechanisms, the observation of the 3′-phosphoglycolate termini (7) in the present work is of particular interest because this product is consistent with the involvement of alkoxyl radical intermediates. Specifically, a host of previous studies indicate that the C4′-alkoxyl radical is a precursor to 3′-phosphoglycolate end products.49,79-82 In contrast, the furfural product observed here (10) arises via a C5′-hydroxyl intermediate rather than an alkoxyl radical intermediate.49,83,84 Overall, the results support the view that the oxygenation of DNA radicals by TPZ is a complex process that proceeds via multiple mechanistic pathways.
The biological activities of any DNA-damaging agent are ultimately determined by the chemical structure of the DNA lesions that it generates. The characterization of TPZ-mediated DNA strand damage products reported here helps build a foundation for understanding the chemical basis of this compound's promising medicinal activity.
Experimental Section
Materials
TPZ was prepared as described previously.85,86 Materials were obtained from the following suppliers and were of the highest purity available: furfural, phenylhydrazine, phenylpyrazole, 5-hydroxymethyl-2-furanone, N,N′-dimethylethylenediamine, piperidine, β-mercaptoethanol, Aldrich Chemical Co. (Milwaukee, WI); xanthine, xanthine oxidase, NADPH, cytochrome P450 reductase, sodium acetate, N,N′-methylenebisacrylamide, Sigma Chemical Company (St. Louis, MO); alkaline phosphatase, catalase, SOD, acrylamide, Roche Diagnostics Corporation (Indianapolis, IN); acetonitrile (HPLC grade) Burdick and Jackson (Muskegon, MI); HPLC grade solvents acetonitrile, ethyl acetate, water Fisher (Pittsburgh, PA); T4 polynucleotide kinase, New England Biolabs (Beverly, MA); 5′-[γ-32P]dATP, Perkin Elmer Life Sciences (Boston, MA), and highly polymerized calf thymus DNA was from Sigma Chemical Company (St. Louis, MO). The compound 5-methylenefuranone was prepared for use as an authentic standard by the method of Alibas et al.87
DNA-damage reactions
Typical assays were conducted under hypoxic conditions and contained DNA (125 μg/mL of highly polymerized calf thymus DNA), TPZ (250 or 500 μM), desferal (1 mM), NADPH (500 μM), cytochrome P450 reductase (50 mU), catalase (100 μg/mL), SOD (10 μg/mL) and sodium phosphate buffer (50 mM, pH 7.0). The solutions of TPZ, desferal, and buffer were degassed by using three cycles of freeze-pump-thaw in Pyrex tubes. The tubes were torched sealed under high vacuum, scored and transferred to an argon-filled glove bag. The tubes were then opened and DNA, NADPH and enzymes were added. Desferal, SOD, and catalase were added to abrogate reactive oxygen species (O2•−, H2O2, and HO•) that could potentially be generated by the reaction of 2 with traces of molecular oxygen in the assay mixtures. The reactions were incubated inside the glove bag protected from light for 45 or 240 min for the detection of 5-methylene furanone and furfural or malondialdehyde equivalents, respectively.
Positive controls involving Fe-EDTA/H2O2/ascorbate damaged DNA were prepared by incubation of calf thymus DNA (125 μg/mL) FeCl3 (10 μM), EDTA (20 μM), ascorbate (1 mM), and H2O2 (0.03%) in sodium phosphate buffer (pH 7.0, 50 mM) at room temperature under aerobic conditions for 45 min. The DNA was then isolated by ethanol precipitation, washed with 70% cold ethanol and air dried before further analysis.
Detection of 5-methylenefuranone (6) and furfural (10)
TPZ (250 μM) or Fe-EDTA/ascorbate/H2O2 damaged DNA (100 μg) was dissolved in 100 μL of 10 mM sodium phosphate buffer. The samples were then heated for 15 min at 90 °C, cooled for 5 min at 4 °C or 30 min at 20 °C and then extracted with methylene chloride (3 × 100 μL). The organic layer was then evaporated by sitting at 24 °C in an open vial. Then, 5% acetonitrile-water (20 μL) was added and the mixture analyzed by reverse phase HPLC using a C-18 Rainin Microsorb-MV column (100 Ǻ sphere size, 5 mm pore size, 25 cm length, 4.6 mm i.d.) eluted with an isocratic mobile phase of 5% acetonitrile and 95% water. The compounds were detected by their absorbance at 260 nm.
Detection of malondialdehyde equivalents released from TPZ-damaged DNA: derivatization with phenylhydrazine to form phenylpyrazole (11)
DNA-damage reactions were carried out in silylated vials as described above. Following DNA damage, 50 μL of acetonitrile and 10 μL of 10% phenylhydrazine (in 10% aqueous acetonitrile) were added to 400 μL of reaction mixture, followed by the addition of 40 μL of water. The final concentration of phenylhydrazine was 20 mM. The reaction mixture was incubated for 15 h at 24 °C and then extracted with hexane (2 × 300 μL). Two such samples were pooled and the hexane layers dried under a gentle stream of nitrogen gas. The residue was then dissolved in 50 μL of hexane and a 4 μL aliquot analyzed by GC-MS using the following instrument settings: positive ion electron ionization (EI 70 eV); 250 °C inlet in the splitless mode; capillary column DB 624 (28.4m, 0.25mm, 1.4 μM); helium carrier flow 1 mL/min; oven temperature 70 °C, linear gradient to 220 °C at 10 °C/min, hold at 220 °C for 2 min; data was acquired in the SIM mode with a mass range of m/z 143–145 after a 3 min solvent delay.
Detection of 3′-phosphoglycolate termini by gel electrophoretic analysis of TPZ-damaged DNA
Plasmid pBR322 DNA (20-30 μg) was digested with EcoRI (240 units) followed by 5′-end dephosphorylation with calf intestinal phosphatase (8-10 units). The DNA was then 5′-end labeled using T4 polynucleotide kinase (20 units) and 5′-[γ-32P]dATP (80 μCi). A second restriction enzyme digest was performed with BamHI (30 units) and the desired 377 base pair fragment was purified using gel electrophoresis on a 5% polyacrylamide gel. An autoradiograph of the gel was obtained and used to locate the position of the labeled DNA in the gel. The band corresponding to the 377 base pair DNA fragment was excised from the gel and crushed into an elution buffer88 containing NaCl and Na2-EDTA. The elution buffer mixture containing the crushed DNA gel slice was vortexed 1-2 hours then incubated at 37 °C for 8-10 hours. The gel slice/elution buffer was Centrex® filtered and the volume of the DNA solution was decreased via butanol extraction.88 When the volume of the DNA solution was decreased to approximately 200 μL, the 377 base pair DNA fragment was recovered via ethanol precipitation.88 The DNA pellet was air-dried and redissolved in 100 μL of deionized H2O and counted using liquid scintillation (expected yield is 1-10 million cpm).
5′-labeled 377 base pair DNA-cleavage reactions involving TPZ were carried out in Pyrex® glass tubes, freeze-pump-thawed degassed (3x) and sealed with rubber septa under a nitrogen atmosphere. A typical, degassed solution (50 μL final volume) contained the 5′-labeled 377 base pair restriction fragment (100,000-150,000 cpm), TPZ (2 mM), xanthine (2 mM), desferal (1 mM), superoxide dismutase (10 μg/mL), and catalase (100 μg/mL) in sodium phosphate (50 mM, pH 7.0). The xanthine oxidase (0.04 units) was added to the inside wall of the glass tube following the degassing procedure, sealed and vortexed to initiate the enzymatic activation of TPZ. The reactions were then phenol extracted,88 ethanol precipitated,88 briefly dried under vacuum in a Speed Vac concentrator, and redissolved in formamide loading buffer (10 mL). The samples were heated for 5 min at 90 °C, loaded onto a 25% high resolution, denaturing polyacrylamide gel (1:19 crosslinked, 0.4 mm thick, containing 7.5 M urea)63 and the gel electrophoresed at 1600 V for 5 h in 1 × TBE buffer. Following electrophoresis, radioactivity on the gels was visualized using Fuji RX X-ray film. Alternatively, radioactivity was visualized by exposing a PhosphorImager plate to the gel, followed by scanning of the plate with a Molecular Dynamics Model 400E PhosphorImager (Sunnyvale, CA).
Detection of the 2-deoxyribonolactone lesion (5) generated by the reaction of TPZ with a C1′-radical in a 2′-oligodeoxynucleotide
The oligonucleotide precursor 5′-GTCACGTGCTGCA-13-ACGACGTGCTGAGCCT, where 13 is C1′-radical precursor shown in Scheme 4, was 5′-end labeled using standard protocols.88 In a typical assay, this 5′-32P-labeled oligonucleotide (250,000 cpm) was dissolved in sodium phosphate buffer (10 mM, pH 7) containing NaCl (100 mM), and TPZ (1 mM). The mixture was placed in a Pyrex tube (6 mm i.d.) and degassed via three freeze-pump-thaw cycles. The tubes were sealed under vacuum and photolysed for 15 min using a high pressure quartz-mercury vapor lamp (Ace-Hanovia). After photolysis, the solution was transferred to an microcentrifuge tube and the photolysis tube washed with water (2 × 50 μL). Aliquots of the combined photolysate were then treated with N,N′-dimethylethylenediamine (100 mM), piperidine (100 mM), or piperidine (100 mM)/β-mercaptoethanol (50 mM) as described previously.75 The treated DNA was precipitated with 0.3 M sodium acetate (pH 5.2) in 70% ethanol,88 washed twice with cold 70% ethanol, air dried, and resuspended in formamide loading buffer by heating the sample at 50 °C for 5 min with intermittent vortex-mixing. Finally, the samples were loaded onto a 20% denaturing polyacrylamide gel and electrophoresed for 2 h at 1400 V. The radiolabeled oligonucleotide fragments in the gel were visualized by phosphorimager analysis (Molecular Dynamics). The Maxam-Gilbert G and A+G reactions were performed according to standard protocols.88
Acknowledgement
We thank the NIH for supporting this research (CA 100757 to KSG and GM 054996 to MMG).
References
- 1.Horsman MR. Int. J. Radiation Oncology Biol. Phys. 1998;42:701–704. doi: 10.1016/s0360-3016(98)00332-0. [DOI] [PubMed] [Google Scholar]
- 2.Siemann DW. Int. J. Radiation Oncology Biol. Phys. 1998;42:697–699. doi: 10.1016/s0360-3016(98)00336-8. [DOI] [PubMed] [Google Scholar]
- 3.Vaupel P, Kallinowski F, Okunieff P. Cancer Res. 1989;49:6449–6465. [PubMed] [Google Scholar]
- 4.Wilson WR, Waring MJ. Tumour hypoxia: challenges for cancer chemotherapy. In: Ponder BAJ, editor. The Search For New Anticancer Drugs. Kluwer Academic; Lancaster: 1992. [Google Scholar]
- 5.Brown JM, Wilson WR. Nature Rev. Cancer. 2004;4:437–447. doi: 10.1038/nrc1367. [DOI] [PubMed] [Google Scholar]
- 6.Denny WA. The Lancet Oncol. 2000;1:25–29. doi: 10.1016/S1470-2045(00)00006-1. [DOI] [PubMed] [Google Scholar]
- 7.Brown JM. Cancer Res. 1999;59:5863–5870. [PubMed] [Google Scholar]
- 8.Rauth AM, Melo T, Misra V. Int. J. Radiation Oncology Biol. Phys. 1998;42:755–762. doi: 10.1016/s0360-3016(98)00302-2. [DOI] [PubMed] [Google Scholar]
- 9.Sartorelli AC, Rockwell S. Oncol. Res. 1994;6:501–508. [PubMed] [Google Scholar]
- 10.Lin AJ, Cosby LA, Shansky CW, Sartorelli AC. J. Med. Chem. 1972;15:1247–1252. doi: 10.1021/jm00282a011. [DOI] [PubMed] [Google Scholar]
- 11.Zeman EM, Brown JM, Lemmon MJ, Hirst VK, Lee WW. Int. J. Radiat. Oncol. Biol. Phys. 1986;12:1239–1242. doi: 10.1016/0360-3016(86)90267-1. [DOI] [PubMed] [Google Scholar]
- 12.Brown JM, Wang L-H. Anti-Cancer Drug Design. 1998;13:529–539. [PubMed] [Google Scholar]
- 13.Laderoute KL, Wardman P, Rauth M. Biochem. Pharmacol. 1988;37:1487–1495. doi: 10.1016/0006-2952(88)90010-x. [DOI] [PubMed] [Google Scholar]
- 14.Fitzsimmons SA, Lewis AD, Riley RJ, Workman P. Carcinogenesis. 1994;15:1503–1510. doi: 10.1093/carcin/15.8.1503. [DOI] [PubMed] [Google Scholar]
- 15.Daniels JS, Gates KS. J. Am. Chem. Soc. 1996;118:3380–3385. [Google Scholar]
- 16.Kotandeniya D, Ganley B, Gates KS. Bioorg. Med. Chem. Lett. 2002;12:2325–2329. doi: 10.1016/s0960-894x(02)00468-7. [DOI] [PubMed] [Google Scholar]
- 17.Birincioglu M, Jaruga P, Chowdhury G, Rodriguez H, Dizdaroglu M, Gates KS. J. Am. Chem. Soc. 2003;125:11607–11615. doi: 10.1021/ja0352146. [DOI] [PubMed] [Google Scholar]
- 18.Biedermann KA, Wang J, Graham RP. Br. J. Cancer. 1991;63:358–362. doi: 10.1038/bjc.1991.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Siim BG, van Zijl PL, Brown JM. Br. J. Cancer. 1996;73:952–960. doi: 10.1038/bjc.1996.187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Marcu L, Olver I. Curr. Clin. Oncol. 2006;1:71–79. doi: 10.2174/157488406775268192. [DOI] [PubMed] [Google Scholar]
- 21.Wardman P, Priyadarsini KI, Dennis MF, Everett SA, Naylor MA, Patel KB, Stratford IJ, Stratford MRL, Tracy M. Br. J. Cancer. 1996;74:S70–S74. [PMC free article] [PubMed] [Google Scholar]
- 22.Priyadarsini KI, Tracy M, Wardman P. Free Rad. Res. 1996;25:393–399. doi: 10.3109/10715769609149061. [DOI] [PubMed] [Google Scholar]
- 23.Elwell JH, Siim BG, Evans JW, Brown JM. Biochem. Pharmacol. 1997;54:249–257. doi: 10.1016/s0006-2952(97)00171-8. [DOI] [PubMed] [Google Scholar]
- 24.Wouters BG. Cancer Res. 2001;61:145–152. [PubMed] [Google Scholar]
- 25.Lloyd RV, Duling DR, Rumyantseva GV, Mason RP, Bridson PK. Mol. Pharmacol. 1991;40:440–445. [PubMed] [Google Scholar]
- 26.Patterson LH, Taiwo FA. Biochem. Pharmacol. 2000;60:1933–1935. doi: 10.1016/s0006-2952(00)00487-1. [DOI] [PubMed] [Google Scholar]
- 27.Sies H. Angew. Chem. Int. Ed. Eng. 1986;25:1058–1071. [Google Scholar]
- 28.Finkel T, Holbrook NJ. Nature. 2000;408:239–247. doi: 10.1038/35041687. [DOI] [PubMed] [Google Scholar]
- 29.Wood ZA, Poole LB, Karplus PA. Science. 300:650–653. doi: 10.1126/science.1080405. [DOI] [PubMed] [Google Scholar]
- 30.Zagorevski D, Yuan Y, Fuchs T, Gates KS, Song M, Breneman C, Greenlief CM. J. Am. Soc. Mass Spec. 2003;14:881–892. doi: 10.1016/S1044-0305(03)00334-9. [DOI] [PubMed] [Google Scholar]
- 31.Li L-C, Zha D, Zhu Y-Q, Xu M-H, Wong N-B. Chem. Phys. Lett. 2005;408:329–334. [Google Scholar]
- 32.Barton DHR, Crich D, Motherwell WB. Tetrahedron. 1985;41:3901–3924. [Google Scholar]
- 33.Boivin J, Crepon E, Zard SZ. Tetrahedron Lett. 1990;31:6869–6872. [Google Scholar]
- 34.Adam W, Ballmaier D, Epe B, Grimm GN, Saha-Moller CR. Angew. Chem. Int. Ed. Eng. 1995;34:2156–2158. [Google Scholar]
- 35.Barton DHR, Jasberenyi JC, Morrell AI. Tetrahedron Lett. 1991;32:311–314. [Google Scholar]
- 36.Aveline BM, Kochevar IE, Redmond RW. J. Am. Chem. Soc. 1996;118:289–290. [Google Scholar]
- 37.Wölfle I, Lodays J, Sauerwein B, Schuster GB. J. Am. Chem. Soc. 1992;114:9304–9309. [Google Scholar]
- 38.Lorance ED, Kramer WH, Gould IR. J. Am. Chem. Soc. 2002;124:15225–15238. doi: 10.1021/ja020768e. [DOI] [PubMed] [Google Scholar]
- 39.Brown JM. Br. J. Cancer. 1993;67:1163–1170. doi: 10.1038/bjc.1993.220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Anderson RF, Shinde SS, Hay MP, Gamage SA, Denny WA. J. Am. Chem. Soc. 2003;125:748–756. doi: 10.1021/ja0209363. [DOI] [PubMed] [Google Scholar]
- 41.Daniels JS, Gates KS, Tronche C, Greenberg MM. Chem. Res. Toxicol. 1998;11:1254–1257. doi: 10.1021/tx980184j. [DOI] [PubMed] [Google Scholar]
- 42.Hwang J-T, Greenberg MM, Fuchs T, Gates KS. Biochemistry. 1999;38:14248–14255. doi: 10.1021/bi991488n. [DOI] [PubMed] [Google Scholar]
- 43.Jones GDD, Weinfeld M. Cancer Res. 1996;56:1584–1590. [PubMed] [Google Scholar]
- 44.Shi X, Mandel SM, Platz MS. J. Am. Chem. Soc. 2007;129:4542–4550. doi: 10.1021/ja0647405. [DOI] [PubMed] [Google Scholar]
- 45.Greenberg MM. Org. Biomol. Chem. 2007;5:18–30. doi: 10.1039/b612729k. [DOI] [PubMed] [Google Scholar]
- 46.Greenberg MM. Chem. Res. Toxicol. 1998;11:1235–1248. doi: 10.1021/tx980174i. [DOI] [PubMed] [Google Scholar]
- 47.Pogozelski WK, Tullius TD. Chem. Rev. 1998;98:1089–1107. doi: 10.1021/cr960437i. [DOI] [PubMed] [Google Scholar]
- 48.Breen AP, Murphy JA. Free Rad. Biol. Med. 1995;18:1033–1077. doi: 10.1016/0891-5849(94)00209-3. [DOI] [PubMed] [Google Scholar]
- 49.Pratviel G, Bernadou J, Meunier B. Angew. Chem. Int. Ed. Eng. 1995;34:746–769. [Google Scholar]
- 50.Dizdaroglu M, Von Sonntag C, Schulte-Frohlinde D. J. Am. Chem. Soc. 1975;97:2277–2278. doi: 10.1021/ja00841a051. [DOI] [PubMed] [Google Scholar]
- 51.Rashid R, Langfinger D, Wagner R, Schuchmann H-P, Von Sonntag C. Int. J. Radiat. Biol. 1999;75:101–109. doi: 10.1080/095530099140852. [DOI] [PubMed] [Google Scholar]
- 52.Zhou X, Taghizadeh K, Dedon PC. J. Biol. Chem. 2005;280:25377–25382. doi: 10.1074/jbc.M503079200. [DOI] [PubMed] [Google Scholar]
- 53.Giloni L, Takashita M, Johnson F, Iden C, Grollman AP. J. Biol. Chem. 1981;256:8608–8615. [PubMed] [Google Scholar]
- 54.Balasubramanian B, Pogozelski WK, Tullius TD. Proc. Nat. Acad. Sci. USA. 1998;95:9738–9743. doi: 10.1073/pnas.95.17.9738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sy D, Savoye C, Begusova M, Michalik V, Charlier M, Spotheim-Maurizot M. Int. J. Radiat. Biol. 1997;72:147–155. doi: 10.1080/095530097143365. [DOI] [PubMed] [Google Scholar]
- 56.(a) Hong IS, Carter KN, Sato K, Greenberg MM. J. Am. Chem. Soc. 2007;129:4089–4098. doi: 10.1021/ja0692276. [DOI] [PubMed] [Google Scholar]; (b) Tallman KA, Greenberg MM. J. Am. Chem. Soc. 2001;123:5181–5187. doi: 10.1021/ja010180s. [DOI] [PubMed] [Google Scholar]
- 57.Carter KN, Greenberg MM. J. Am. Chem. Soc. 2003;125:13376–13378. doi: 10.1021/ja036629u. [DOI] [PubMed] [Google Scholar]
- 58.Patterson AV, Saunders MP, Chinje EC, Patterson LH, Stratford IJ. Anti-Cancer Drug Des. 1998;13:541–573. [PubMed] [Google Scholar]
- 59.Walton MI, Wolf CR, Workman P. Biochem. Pharmacol. 1992;44:251–259. doi: 10.1016/0006-2952(92)90007-6. [DOI] [PubMed] [Google Scholar]
- 60.Kuwabara M, Yoon C, Goyne T, Thedarahn T, Sigman DS. Biochemistry. 1986;25:7401–7408. doi: 10.1021/bi00371a023. [DOI] [PubMed] [Google Scholar]
- 61.Pitié M, Burrows CJ, Meunier B. Nucleic Acids Res. 2000;28:4856–4864. doi: 10.1093/nar/28.24.4856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Pratviel G, Petieé M, Bernadou J, Meunier B. Nucleic Acids Res. 1991;19:6283–6288. doi: 10.1093/nar/19.22.6283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Pogozelski WK, McNeese TJ, Tullius TD. J. Am. Chem. Soc. 1995;117:6428–6433. [Google Scholar]
- 64.Panoutsopoulos GI, Kouretas D, Beedham C. Chem. Res. Toxicol. 2004;17:1368–1376. doi: 10.1021/tx030059u. [DOI] [PubMed] [Google Scholar]
- 65.Morpeth FF. Biochim. Biophys. Acta. 1983;744:328–34. doi: 10.1016/0167-4838(83)90207-8. [DOI] [PubMed] [Google Scholar]
- 66.Gutteridge JMC, West ME,K, Floyd RA. Free Rad. Res. Commun. 1990;10:159–65. doi: 10.3109/10715769009149884. [DOI] [PubMed] [Google Scholar]
- 67.Murugesan N, Xu C, Ehrenfeld GM, Sugiyama H, Kilkuskie RE, Rodriguez LO, Chang LH, Hecht SM. Biochemistry. 1985;24:5735–44. doi: 10.1021/bi00342a008. [DOI] [PubMed] [Google Scholar]
- 68.Janicek MF, Haseltine WA, Henner WD. Nucleic Acids Res. 1985;13:9011–9029. doi: 10.1093/nar/13.24.9011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Otteneder M, Plastaras JP, Marnett LJ. Chem. Res. Toxicol. 2002;15:312–318. doi: 10.1021/tx010105v. [DOI] [PubMed] [Google Scholar]
- 70.Baker MA, Zeman EM, Hirst VK, Brown JM. Cancer Res. 1988;48:5947–5952. [PubMed] [Google Scholar]
- 71.Stubbe J, Kozarich JW. Chem. Rev. 1987;87:1107–1136. [Google Scholar]
- 72.Henner WD, Rodriguez LO, Hecht SM, Haseltine WA. J. Biol. Chem. 1983;258 [PubMed] [Google Scholar]
- 73.Williams RM, Flanagan ME, Tippie TN. Biochemistry. 1994;33:4086–4092. doi: 10.1021/bi00179a038. [DOI] [PubMed] [Google Scholar]
- 74.Emmanuel CJ, Newcomb M, Ferreri C, Chatgilialoglu C. J. Am. Chem. Soc. 1999;121:2927–2928. [Google Scholar]
- 75.Hwang J-T, Tallman KA, Greenberg MM. Nucleic Acids Res. 1999;27:3805–3810. doi: 10.1093/nar/27.19.3805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zheng Y, Sheppard TL. Chem. Res. Toxicol. 2004;17:197–207. doi: 10.1021/tx034197v. [DOI] [PubMed] [Google Scholar]
- 77.Goodman BK, Greenberg MM. J. Org. Chem. 1996;61:2–3. [Google Scholar]
- 78.Hwang J-T, Greenberg MM. J. Am. Chem. Soc. 1999;121:4311–4315. [Google Scholar]
- 79.Dedon PC, Jiang Z-W, Goldberg IH. Biochemistry. 1992;31:1917–1927. doi: 10.1021/bi00122a004. [DOI] [PubMed] [Google Scholar]
- 80.Kappen LS, Goldberg IH, Frank BL, Worth L, Christner DF, Kozarich JW, Stubbe J. Biochemistry. 1991;30:2034–2042. doi: 10.1021/bi00222a005. [DOI] [PubMed] [Google Scholar]
- 81.Kappen LS, Goldberg IH. Proc. Nat. Acad. Sci. USA. 1992;89:6706–6710. doi: 10.1073/pnas.89.15.6706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Kappen LS, Goldberg IH. Biochemistry. 1992;31:9081–9089. doi: 10.1021/bi00152a052. [DOI] [PubMed] [Google Scholar]
- 83.Chin D-H, Kappen LS, Goldberg IH. Proc. Nat. Acad. Sci. USA. 1987;84:7070–7074. doi: 10.1073/pnas.84.20.7070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Kappen LS, Lee TR, Yang C.-c., Goldberg IH. Biochemistry. 1989;28:4540–4542. doi: 10.1021/bi00437a004. [DOI] [PubMed] [Google Scholar]
- 85.Mason JC, Tennant G. J. Chem. Soc. B. 1970:911–916. [Google Scholar]
- 86.Fuchs T, Chowdhary G, Barnes CL, Gates KS. J. Org. Chem. 2001;66:107–114. doi: 10.1021/jo001232j. [DOI] [PubMed] [Google Scholar]
- 87.Alibes R, Font J, Mula A, Ortuno RM. Syn. Comm. 1990;20:2607–15. [Google Scholar]
- 88.Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Lab Manual. Cold Spring Harbor Press; Cold Spring Harbor, NY: 1989. [Google Scholar]