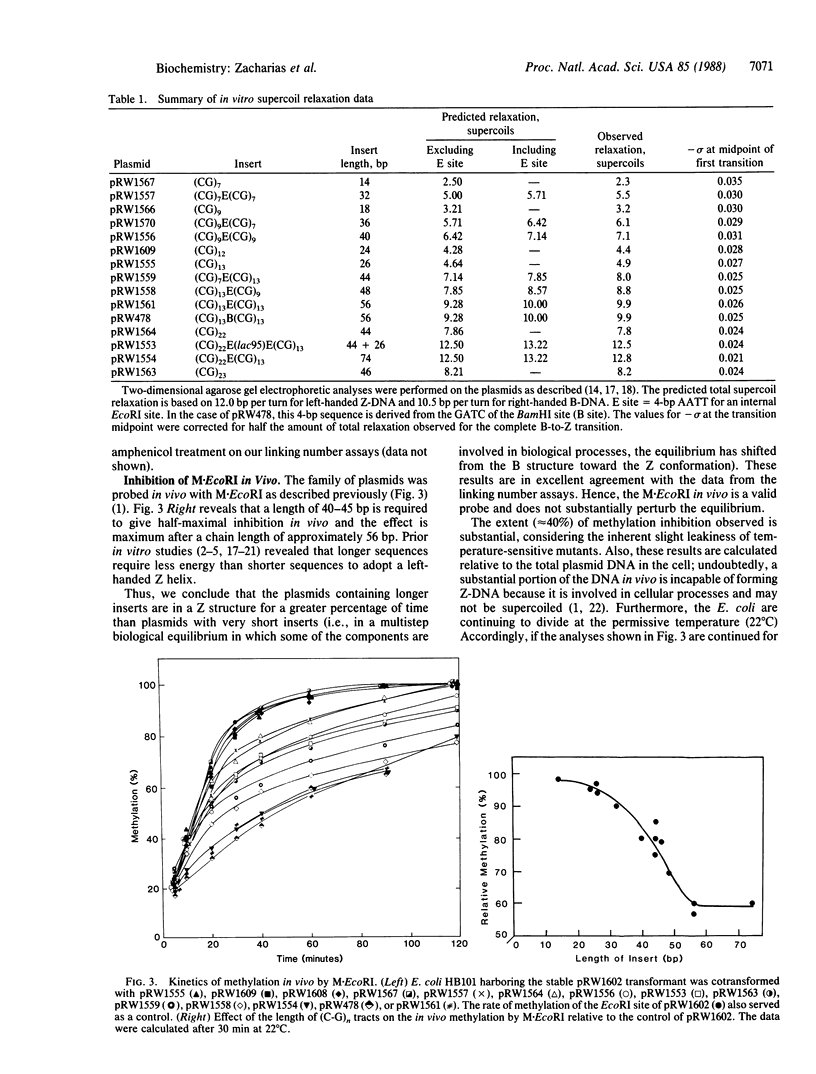
Abstract
Right-handed B and left-handed Z conformations coexist in equilibrium in portions of plasmids in Escherichia coli. The equilibria are influenced by the length of the sequences that undergo the structural transitions and are perturbed by biological processes. The composite results of three types of determinations indicate a supercoil density of -0.025 in vivo. The coexistence of alternative DNA conformations in living cells implies the potential of these structures or their transitions for important functions in genetic regulatory processes.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Balke V. L., Gralla J. D. Changes in the linking number of supercoiled DNA accompany growth transitions in Escherichia coli. J Bacteriol. 1987 Oct;169(10):4499–4506. doi: 10.1128/jb.169.10.4499-4506.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bliska J. B., Cozzarelli N. R. Use of site-specific recombination as a probe of DNA structure and metabolism in vivo. J Mol Biol. 1987 Mar 20;194(2):205–218. doi: 10.1016/0022-2836(87)90369-x. [DOI] [PubMed] [Google Scholar]
- Esposito F., Sinden R. R. Supercoiling in prokaryotic and eukaryotic DNA: changes in response to topological perturbation of plasmids in E. coli and SV40 in vitro, in nuclei and in CV-1 cells. Nucleic Acids Res. 1987 Jul 10;15(13):5105–5124. doi: 10.1093/nar/15.13.5105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstein E., Drlica K. Regulation of bacterial DNA supercoiling: plasmid linking numbers vary with growth temperature. Proc Natl Acad Sci U S A. 1984 Jul;81(13):4046–4050. doi: 10.1073/pnas.81.13.4046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haniford D. B., Pulleyblank D. E. Facile transition of poly[d(TG) x d(CA)] into a left-handed helix in physiological conditions. Nature. 1983 Apr 14;302(5909):632–634. doi: 10.1038/302632a0. [DOI] [PubMed] [Google Scholar]
- Haniford D. B., Pulleyblank D. E. The in-vivo occurrence of Z DNA. J Biomol Struct Dyn. 1983 Dec;1(3):593–609. doi: 10.1080/07391102.1983.10507467. [DOI] [PubMed] [Google Scholar]
- Jaworski A., Hsieh W. T., Blaho J. A., Larson J. E., Wells R. D. Left-handed DNA in vivo. Science. 1987 Nov 6;238(4828):773–777. doi: 10.1126/science.3313728. [DOI] [PubMed] [Google Scholar]
- Lilley D. Bacterial chromatin. A new twist to an old story. Nature. 1986 Mar 6;320(6057):14–15. doi: 10.1038/320014a0. [DOI] [PubMed] [Google Scholar]
- Lyons S. M., Schendel P. F. Kinetics of methylation in Escherichia coli K-12. J Bacteriol. 1984 Jul;159(1):421–423. doi: 10.1128/jb.159.1.421-423.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLean M. J., Blaho J. A., Kilpatrick M. W., Wells R. D. Consecutive A X T pairs can adopt a left-handed DNA structure. Proc Natl Acad Sci U S A. 1986 Aug;83(16):5884–5888. doi: 10.1073/pnas.83.16.5884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLean M. J., Wells R. D. The role of DNA sequence in the formation of Z-DNA versus cruciforms in plasmids. J Biol Chem. 1988 May 25;263(15):7370–7377. [PubMed] [Google Scholar]
- O'Connor T. R., Kang D. S., Wells R. D. Thermodynamic parameters are sequence-dependent for the supercoil-induced B to Z transition in recombinant plasmids. J Biol Chem. 1986 Oct 5;261(28):13302–13308. [PubMed] [Google Scholar]
- Panayotatos N., Fontaine A. A native cruciform DNA structure probed in bacteria by recombinant T7 endonuclease. J Biol Chem. 1987 Aug 15;262(23):11364–11368. [PubMed] [Google Scholar]
- Peck L. J., Wang J. C. Energetics of B-to-Z transition in DNA. Proc Natl Acad Sci U S A. 1983 Oct;80(20):6206–6210. doi: 10.1073/pnas.80.20.6206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rich A., Nordheim A., Wang A. H. The chemistry and biology of left-handed Z-DNA. Annu Rev Biochem. 1984;53:791–846. doi: 10.1146/annurev.bi.53.070184.004043. [DOI] [PubMed] [Google Scholar]
- Rodriguez-Campos A., Ellison M. J., Pérez-Grau L., Azorin F. DNA conformation and chromatin organization of a d(CA/GT)30 sequence cloned in SV40 minichromosomes. EMBO J. 1986 Jul;5(7):1727–1734. doi: 10.1002/j.1460-2075.1986.tb04417.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singleton C. K., Kilpatrick M. W., Wells R. D. S1 nuclease recognizes DNA conformational junctions between left-handed helical (dT-dG n. dC-dA)n and contiguous right-handed sequences. J Biol Chem. 1984 Feb 10;259(3):1963–1967. [PubMed] [Google Scholar]
- Singleton C. K., Klysik J., Stirdivant S. M., Wells R. D. Left-handed Z-DNA is induced by supercoiling in physiological ionic conditions. Nature. 1982 Sep 23;299(5881):312–316. doi: 10.1038/299312a0. [DOI] [PubMed] [Google Scholar]
- Singleton C. K., Klysik J., Wells R. D. Conformational flexibility of junctions between contiguous B- and Z-DNAs in supercoiled plasmids. Proc Natl Acad Sci U S A. 1983 May;80(9):2447–2451. doi: 10.1073/pnas.80.9.2447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singleton C. K., Wells R. D. The facile generation of covalently closed, circular DNAs with defined negative superhelical densities. Anal Biochem. 1982 May 15;122(2):253–257. doi: 10.1016/0003-2697(82)90277-9. [DOI] [PubMed] [Google Scholar]
- Stirdivant S. M., Kłysik J., Wells R. D. Energetic and structural inter-relationship between DNA supercoiling and the right- to left-handed Z helix transitions in recombinant plasmids. J Biol Chem. 1982 Sep 10;257(17):10159–10165. [PubMed] [Google Scholar]
- Vardimon L., Rich A. In Z-DNA the sequence G-C-G-C is neither methylated by Hha I methyltransferase nor cleaved by Hha I restriction endonuclease. Proc Natl Acad Sci U S A. 1984 Jun;81(11):3268–3272. doi: 10.1073/pnas.81.11.3268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wells R. D., Goodman T. C., Hillen W., Horn G. T., Klein R. D., Larson J. E., Müller U. R., Neuendorf S. K., Panayotatos N., Stirdivant S. M. DNA structure and gene regulation. Prog Nucleic Acid Res Mol Biol. 1980;24:167–267. doi: 10.1016/s0079-6603(08)60674-1. [DOI] [PubMed] [Google Scholar]
- Wells R. D. Unusual DNA structures. J Biol Chem. 1988 Jan 25;263(3):1095–1098. [PubMed] [Google Scholar]
- Zacharias W., Larson J. E., Kilpatrick M. W., Wells R. D. HhaI methylase and restriction endonuclease as probes for B to Z DNA conformational changes in d(GCGC) sequences. Nucleic Acids Res. 1984 Oct 25;12(20):7677–7692. doi: 10.1093/nar/12.20.7677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zacharias W., O'Connor T. R., Larson J. E. Methylation of cytosine in the 5-position alters the structural and energetic properties of the supercoil-induced Z-helix and of B-Z junctions. Biochemistry. 1988 Apr 19;27(8):2970–2978. doi: 10.1021/bi00408a046. [DOI] [PubMed] [Google Scholar]