Abstract
S100B and S100A10 are dimeric, EF-hand proteins. S100B undergoes a calcium-dependant conformational change allowing it to interact with a short contiguous sequence from the actin-capping protein CapZ (TRTK12). S100A10 does not bind calcium but is able to recruit the N-terminus of annexin A2 important for membrane fusion events, and to form larger multiprotein complexes such as that with the cation channel proteins TRPV5/6. In this work, we have designed, expressed, purified, and characterized two S100-target peptide hybrid proteins comprised of S100A10 and S100B linked in tandem to annexin A2 (residues 1–15) and CapZ (TRTK12), respectively. Different protease cleavage sites (tobacco etch virus, PreScission) were incorporated into the linkers of the hybrid proteins. In situ proteolytic cleavage monitored by 1H-15N HSQC spectra showed the linker did not perturb the structures of the S100A10-annexin A2 or S100B-TRTK12 complexes. Furthermore, the analysis of the chemical shift assignments (1H, 15N, and 13C) showed that residues T102-S108 of annexin A2 formed a well-defined α-helix in the S100A10 hybrid while the TRTK12 region was unstructured at the N-terminus with a single turn of α-helix from D108-K111 in the S100B hybrid protein. The two S100 hybrid proteins provide a simple yet extremely efficient method for obtaining high yields of intact S100 target peptides. Since cleavage of the S100 hybrid protein is not necessary for structural characterization, this approach may be useful as a scaffold for larger S100 complexes.
Keywords: S100, hybrid protein, EF-hand, protein interactions, NMR spectroscopy
Introduction
S100 proteins are small (10–12 kDa), acidic proteins that are expressed in a cell- and tissue-specific fashion, indicating a well-conserved biological role. With at least 25 identified members in humans, the S100 proteins comprise the largest subgroup of the EF-hand calcium-binding superfamily. Structurally, the S100s contain two helix-loop-helix calcium-binding sites connected by a flexible linker region and can exist as homo- or heterodimers under physiological conditions.1–3 Overall, S100 proteins function as calcium-signaling molecules involved in cell growth, differentiation and cytoskeletal development.4,5
The N-terminal “pseudo” EF-hand loop (helix I, loop, helix II) has a weak affinity for calcium and undergoes only a minor conformational change upon binding.6–8 Conversely, the canonical C-terminal EF-hand loop (helix III, loop, helix IV) ligates calcium with higher affinity that leads to a significant conformational change primarily through a reorientation of helix III and lengthening of helix IV. The movement of helix III results in the exposure of previously buried hydrophobic residues and formation of a recognition surface for protein-target interactions.5,9 One distinct member of the S100 family is S100A10, which has lost its ability to coordinate calcium ions due to alterations in both of its calcium-binding sites.10 Even though a calcium-induced conformational change does not occur for S100A10, it adopts a structure in its calcium-free state that is very similar to the calcium-bound form of other S100 proteins.11,12 S100A10 is able to interact and function with a variety of target proteins such as annexin A2,12 the cation channel proteins TRVP5/613,14 and TASK-1,15,16 and the blue-tongue viral protein NS317 in a calcium-insensitive manner. Interestingly S100A10 has been shown to form ternary complexes with at least two other proteins (AHNAK, TRVP5/6) along with annexin A2 although little structural information about these interactions is available.13,14,18,19
A notable trend is that many S100 proteins interact with members of the annexin family.5,20 For example, S100B, S100A1, S100A6, and S100A11 have been shown to form complexes with annexin A6.21–25 Each annexin protein contains a core domain comprised of four (annexins A1–A5, A7–A11, and A13) or eight (annexin A6) homologous tandem repeats (∼70-residues), each composed of five α-helices. An N-terminal “tail” region of variable length is also present in most annexins. Calcium binding to one face of the annexin core domains allows the proteins to interact with membrane phospholipids. For some annexin proteins, such as annexins A1 and A2, calcium binding leads to the release of the buried N-terminal tail to an exposed position available for protein interactions. S100A10 has been shown to form a tight heterotetrameric complex through the first 14 residues of the N-terminal tail region of annexin A2.26–28 Calcium-bound S100A11 (Ca2+-S100A11) is able to interact with a similar region in either annexins A1 and A2.29,30 It has been proposed that each S100A10 and Ca2+-S100A11 dimer bridges a pair of annexin proteins bringing two phospholipid membranes into close proximity during membrane fusion or reorganization processes.26
Although some structures of different S100 proteins bound to target peptides have been determined, only two crystal structures are available (S100A10-annexin A2, Ca2+-S100A11-annexin A1).11,12 This is likely due to the difficulties associated with crystal formation of the S100-target complexes. Better progress has been made for S100-target complexes by NMR spectroscopy where structures such as calcium-bound S100B (Ca2+-S100B) with peptide binding regions from p53,31 Ndr kinase,32 and CapZ,33,34 have been determined. However, some of these structures suffer from imprecision of the side chains, especially at the protein-peptide interface making details of the interaction difficult to identify. This could be improved by 15N, 13C-labeling of the target peptide although this is very costly for a synthetic peptide and can suffer from low yields and proteolysis when expressed as a fusion protein. Only one example of a biosynthetic, isotopically labeled peptide exists for the 30-residue Siah-1 interacting region for Ca2+-S100A6.35 Here, we describe a method where we use S100B and S100A10 as the carrier proteins for target peptides from CapZ and annexin A2, respectively. The premise is that the S100 protein will protect the target peptide from degradation during biosynthesis and purification. Human S100A10 and S100B proteins were linked to the N-terminal region of annexin A2 (residues 1–15) and the central region of CapZ (TRTK12; residues 1–12) using linkers containing different protease sites for cleavage and isolation of the peptides. Using NMR spectroscopy we show that the spectra for the S100A10-annexin A2 (A10A2) and S100B-TRTK12 (BT12) hybrid proteins are nearly identical to those obtained for the complexes formed between the individual S100 proteins and synthetic peptides. Further, we were able to cleave the hybrid proteins in situ and examine the protein and peptide structures without further purification. This approach should prove useful for examining other S100 protein complexes by crystallography or NMR methods. Further, the A10A2 may provide a foundation for the assembly and examination of ternary or larger S100 protein complexes.
Results
Design, expression, and purification of hybrid S100-target proteins
To generate hybrid S100-target protein complexes, two systems that have been well characterized by structural methods were chosen. The approach taken was to analyze the relative positions of the S100 proteins and their target peptides in the structures and then incorporate a linker between the two components to create a hybrid protein. The first structure employed S100A10 in complex with the N-terminus of annexin A2 (1BT6),12 and a second structure utilized Ca2+-S100B complexed to a portion of the alpha-1 subunit of F-actin capping protein (TRTK12), CapZ (1MWN, 1MQ1).33,34 In both these structures (Fig. 1), the C-termini of helix IV in the S100 protein are proximal to the N-terminus of the binding peptide. Although S100A10 and Ca2+-S100B are structurally similar, the orientations and distances of the annexin A2 and TRTK12 peptides with respect to the S100 protein are quite different. Further, in the S100A10-annexin A2 complex the distance from the C-terminal carboxyl carbon of S100A10 (K91) to the N-terminal nitrogen of annexin A2 is about 16 Å. For the Ca2+-S100B complex with TRTK12 the distance ranges from about 14–26 Å in the families of NMR structures. The choice of these two proteins allowed us to test our approach with one protein (S100B) that undergoes a calcium-induced conformational change to interact with a target, and one protein (S100A10) that is calcium insensitive. Further, the structures of the target peptides in both structures are quite different. Annexin A2 adopts an α-helix peptide for most of its sequence while the TRTK12 peptide is unstructured at its N-terminus followed by a single turn of helix.
Figure 1.
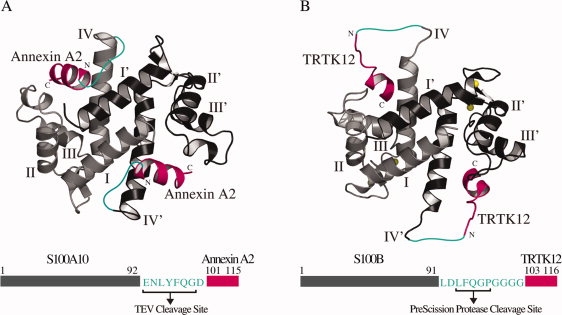
Models of the S100A10-annexin A2 (A10A2) and Ca2+-S100B-TRTK12 (BT12) hybrid constructs. Ribbon representations of S100A10 in complex with (A) annexin A2 (1BT6)12 and (B) Ca2+-S100B bound to TRTK12 (1MWN)33 are shown in similar orientations to demonstrate how the annexin A2 and TRTK12 peptides (magenta) are joined to the S100 proteins through a linker (cyan). In each model, one of the protomers is presented in light gray (helices labeled as I–IV) and the other protomer shaded in black (helices labeled as I′–IV′). Four bound calcium ions are shown in yellow spheres for Ca2+-S100B. The corresponding schematic diagrams of the constructs used for A10A2 and BT12 hybrid proteins are also illustrated with S100 protein located at the N-terminus (gray), annexin A2 and TRTK12 peptides positioned at the C-terminus (magenta), and the linker (cyan) connecting the S100 proteins to the target peptides. The amino acid composition of the linker region and the corresponding protease cleavage sites are indicated. All numbering is consecutive based on the S100 protein sequences.
The A10A2 comprised a linker spanning the measured protein-peptide distance (16 Å) in the S100A10-annexin A2 complex connecting the two proteins. Although this distance is sufficient to accommodate about five amino acid residues linked in a linear fashion, we found through modeling that a further four residues were required for the A10A2 hybrid to allow favorable ϕ, ψ angles in the linker. Within the nine-residue linker, a tobacco etch virus (TEV) protease cleavage site (ENLYFQ/G) was introduced. Finally, an aspartic acid at the N-terminus of the annexin A2 sequence was added to stabilize its α-helical structure [Fig. 1(A)] in lieu of the acetyl group found in the native protein.28 For the BT12 the maximum linker length observed in the NMR structures (∼26 Å) allowed about 11 residues to bridge the protein and target. Within this a PreScission protease cut site (LFQ/GP) and four glycine residues were included [Fig. 1(B)]. The PreScission protease site allowed us to test a different protease for the BT12 hybrid compared to the TEV site in the A10A2 hybrid protein. As the Ca2+-S100B complexes with TRTK12 are ensembles of NMR structures, the N-terminus of TRTK12 exhibits a wide range of positions with respect to Ca2+-S100B. This may be due to the flexibility of this region in TRTK12 not observed for annexin A2 in the S100A10 complex. The glycine residues were added in BT12 to maintain backbone flexibility of the linker. During the design process, the linkers for both proteins were incorporated into the existing structures by modeling. Energy minimization was used to determine that the hybrid proteins were not constrained or distorted, or that the linker did not make any significant contacts with the S100 protein that might mask the protease cleavage sites.
Both the A10A2 and BT12 hybrid proteins were expressed using a BL21 (DE3) codon-plus Escherichia coli strain. Following purification, we typically obtained 15–25 mg of 15N, 13C-labeled A10A2 or BT12 protein from 1L M9 minimal media cultures for heteronuclear NMR spectroscopy experiments. SDS–PAGE and MALDI–TOF mass spectroscopy analyses showed that the proteins were homogeneous and free of degradation products (Fig. 2), similar to the parent S100B and S100A10 proteins. In both A10A2 and BT12, the accessibility of the protease cleavage sites was tested by proteolysis using TEV and PreScission proteases, respectively. In both the cases, the cleavage proceeded to completion allowing a separation of S100 protein and peptide partner (Fig. 2).
Figure 2.
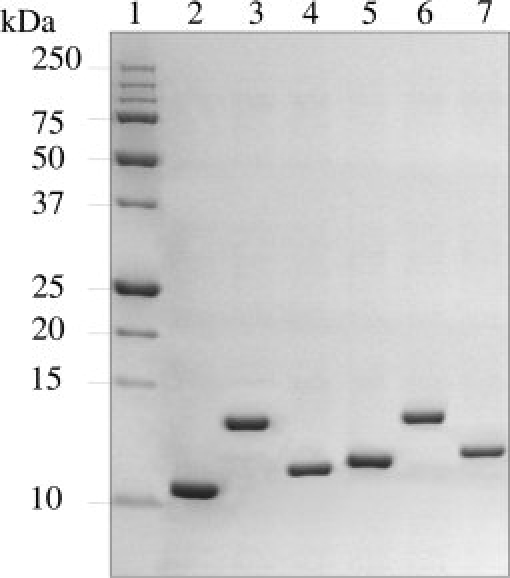
Coomassie-stained SDS–PAGE gel (16.5%) depicting the purification of S100A10-annexin A2 and S100B-TRTK12 constructs. Lane 1 contains the protein molecular weight markers, with molecular weights labeled on the left of the gel. Lanes 2 and 5 contain S100B and S100A10, while BT12 and A10A2 hybrid constructs are presented in lanes 3 and 6, respectively. Lanes 4 and 7 show S100B and S100A10 following cleavage of BT12 and A10A2 by PreScission and TEV proteases, respectively. Each of these proteins runs at a higher molecular weight than the parent due to the presence of the linker that remains appended to the C-terminus of the S100 protein. The TRTK12 and annexin A2 peptides are not observed.
The S100A10-annexin A2 hybrid protein adopts an active configuration
The A10A2 hybrid protein was examined by NMR spectroscopy to determine if the complex retained the similar structure observed in the crystal structure for S100A10 in combination with the N-terminal annexin A2 peptide. Using the standard heteronuclear multiple dimensional NMR spectroscopy, chemical shift assignments (1H, 15N, and 13C) for A10A2 and BT12 proteins were completed. Figure 3(A) shows a representative 1H-15N HSQC spectrum of A10A2 that is well dispersed and characteristic of a folded S100 protein. The spectrum of A10A2 is remarkably similar to that of 15N, 13C-labeled S100A10 in complex with the unlabeled annexin A2 [Fig. 3(B)] indicating that no major structural changes have occurred in the S100A10 protein by incorporating the C-terminal linker and annexin A2 sequences. The 1H-15N correlations for all residues in the annexin A2 segment (S101-D115) and the TEV cleavage site (E93-G99) were easily identified in A10A2 [Fig. 3(A)]. These residues are not visible in Figure 3(B) where the annexin A2 peptide is unlabeled. The resonances for residues S101-D115 are well separated ranging between 7.1 (S108) and 8.9 ppm (T102) indicating the annexin A2 portion of the hybrid is well folded. In contrast, the linker residues form a tight cluster between 7.9 and 8.3 ppm indicating this region of the A10A2 hybrid adopts little regular secondary structure. For BT12 a specific interaction between the S100B and TRTK12 segments was not observed in the absence of calcium (data not shown). However, upon calcium addition to BT12, a spectrum was obtained that was nearly identical to that for Ca2+-S100B in complex with a synthetic TRTK12 peptide. For BT12 all resonances in the linker (L92-G102) and TRTK (T103-E116) sequences were identified with the exception of three glycine residues (G99–G101) in the linker that were absent from the spectra due to the fast exchange with the solvent.
Figure 3.
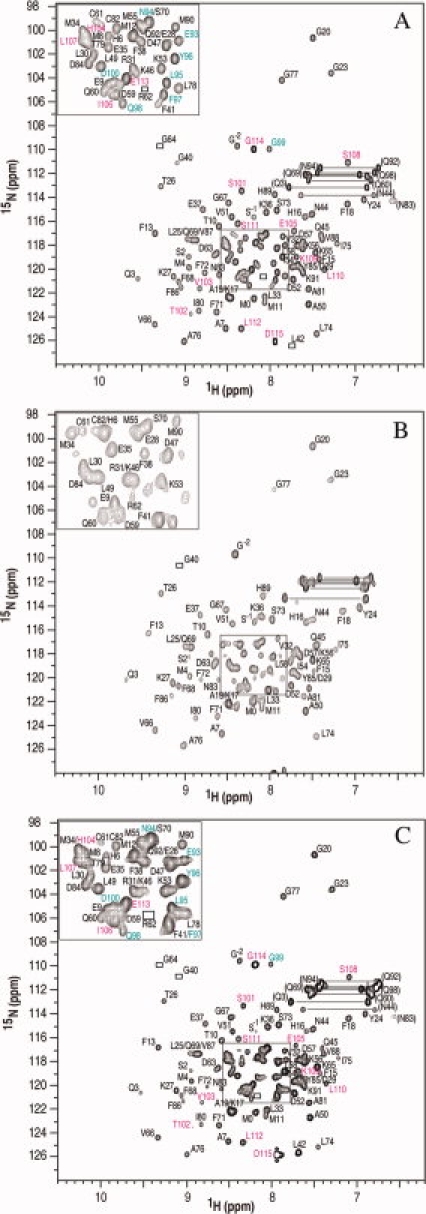
1H-15N HSQC spectra of 0.5 mM (monomer) uniformly (A) 15N, 13C-labeled A10A2 hybrid protein, (B) 15N, 13C-labeled S100A10 in complex with unlabeled annexin A2 peptide, and (C) the 15N, 13C-labeled A10A2 hybrid protein following TEV cleavage. All spectra were collected in 20 mM MOPS, 1 mM EDTA, 1 mM DTT, 50 mM arginine, 50 mM glutamic acid, 100 mM NaCl, pH 7.0 and 35°C. Assigned backbone amide cross peaks are indicated with their one letter amino acid code and number. In (A) and (C) the residues from 15N, 13C-labeled annexin A2 are presented in pink and those from the linker region in cyan. Pairs of resonances for side chain amide cross peaks are connected by horizontal lines. Peaks visible at lower contour levels are indicated by boxes.
One advantage of the A10A2 and BT12 hybrid proteins is that they allow in situ cleavage of the linker using TEV or PreScission enzymes, respectively. Figure 3(C) shows the 1H-15N HSQC spectrum of the A10A2 hybrid protein following TEV cleavage. The positions of the amide resonances for the S100A10 and annexin A2 components in this spectrum are nearly identical to those found in the presence of the linker [Fig. 3(A)]. Importantly, the NMR spectrum shows a tight complex is maintained between S100A10 and annexin A2 despite the absence of N-terminal acetylation of the annexin protein. Parallel experiments for the BT12 hybrid protein following cleavage with PreScission protease yielded similar results (data not shown).
The assignments of Hα, Cα, and C′ resonances of the annexin A2 and TRTK12 regions in the A10A2 and BT12 hybrid proteins allowed their secondary structures to be determined using the chemical shift index.36 This analysis (Fig. 4) shows that residues T102-S108 of annexin A2 form a well-defined α-helix. This compares very well with the crystal structure of S100A10 with the N-terminal peptide from annexin A2 (1BT6) that shows residues T2-K9 (T102-K109 in A10A2) adopt an α-helical structure.12 The secondary structure of TRTK12 in the BT12 hybrid protein shows an unstructured N-terminus and a single turn of α-helix from D108-K111. The position of the α-helix in BT12 agrees well with a previously reported structure of Ca2+-S100B in complex with TRTK12 (1MWN).33
Figure 4.

Secondary structure determinations for annexin A2 and TRTK12 peptides in the A10A2 and BT12 hybrid constructs using the chemical shift index36 for Cα, C′, and Hα atoms. The secondary structure identified for each atom is shown as α-helix (•), β-strand (▪) or coil conformation (○).
Discussion
Bacterial expression of peptides is a challenging task as unstructured peptides are prone to degradation by proteases during purification. This results in very low yields of purified peptides and can lead to inhomogeneity of the peptide due to non-specific proteolysis. Although successful methods have been developed using GB1-tagged37 or His-tagged35 peptides for S100B and S100A6, respectively, in our hands previous attempts to biosynthetically prepare annexin A1 and A2 peptides using GST-tagged constructs lead to <1 mg peptide/L culture and the isolated peptide was prone to proteolysis (Rintala-Dempsey, unpublished results). In the current work, we have developed a successful method for the high-yield bacterial expression of annexin A2 and TRTK12 peptides by linking them to S100A10 and S100B, respectively. In essence the target protein has been used as a carrier for the target peptide. Similar approaches have been previously used to produce hybrid molecules of calmodulin bound to the 26-residue binding region of myosin light-chain kinase (M13),38,39 and peptides from the calmodulin-binding domains of calcineurin,40 the olfactory nucleotide-gated ion-channel,41 tobacco MAPK phosphatase-1,42 and chromogranin A.43 In all of these studies, the linker was composed of glycine residues to maintain flexibility in the linker connecting the carrier protein to the target peptides. Here, we have demonstrated that this is not a necessary requirement and that alternate amino acids can be used. This is particularly important for the incorporation of proteolytic cleavage sites such as TEV or PreScission protease used in this work to obtain complete digestion of the target sites. In principle, this would allow purification of the target peptides (annexin A2 or TRTK12). For NMR studies, this would provide a rapid and facile method to obtain reasonable quantities of 15N, 13C-labeled peptides. Alternatively, we have chosen not to purify the cleaved peptides and examine the intact hybrid protein or the complex after in situ cleavage. This further simplifies the procedure and in the case of protein-peptide complexes offers the advantage of perfect 1:1 stoichiometry.
One requirement for the interaction between some S100 proteins and target peptides is the N-terminal acetylation of the peptides.28,34,44 For example, both annexins A1 and A2 are naturally acetylated. Although, crystal structures of S100A10 in complex with annexin A2 and Ca2+-S100A11 bound to annexin A1 do not illustrate direct contact between the S100 protein and the peptide acetyl groups,11,12 unacetylated peptides show ∼1000-fold decreased binding affinity for the respective S100 proteins.28 It has been suggested the acetyl group acts to stabilize the α-helical conformation of the peptide. In the A10A2 hybrid protein acetylation of the annexin A2 moiety is not possible. However, we have incorporated an acidic residue at N-terminus of the annexin A2 sequence to neutralize the helical dipole and stabilize the helix structure. From our results, it is clear the A10A2 hybrid maintains a tight association between the S100A10 and the annexin peptide indicating the acidic residue is sufficient to stabilize the interaction. In support of this approach, GST-annexin A2 fusion proteins also show tight binding to S100A10 presumably because the annexin N-terminus is extended (by GST) in place of acetylation.45 Similarly, an interaction between TRTK12 and S100B is enhanced upon acetylation of the TRTK12 peptide,46 even though the α-helix forms further down the peptide sequence.33 The hybrid BT12 protein would support this enhanced interaction by mimicking the neutralized TRTK12 sequence.
One of the limitations that could be associated with this method is that the hybrid proteins might suffer from reduced solubility in the unbound form. This was not a problem for A10A2 because binding of annexin A2 to S100A10 is not calcium sensitive. However, for BT12 the hybrid showed poorer solubility in the absence of calcium and exhibited NMR spectra suggestive of protein oligomerization. This is perhaps not surprising given that the TRTK12 peptide motif or accompanying linker could result in nonspecific interactions in the absence of calcium. However, upon calcium binding the BT12 showed excellent solubility and provided 1H-15N HSQC spectra that were better compared to the individual protein peptide complexes.
In summary, we have demonstrated a successful method of high-yield bacterial expression of S100 hybrid proteins. Two different protease cleavage sites were found to be successfully incorporated into the linker regions and could be used in situ to form the S100-peptide complex. This method could be applied for the synthesis of S100-binding peptides for biophysical or biological characterization. Further, the method may prove useful for the assembly of larger S100 complexes such as those involving ternary complexes with S100A10.
Materials and Methods
15NH4Cl and 13C6-glucose were obtained from Cambridge Isotope Laboratories (Andover, MA). The expression vector for rabbit S100A10 (pGEX-6P-1-derived vector; GE Healthcare, Buckinghamshire, UK) was generously provided by Dr. Michael Walsh (University of Calgary). Puratronic grade CaCl2 (99.9995% purity) was purchased from Johnson-Matthey (Ward Hill, MA). PreScission protease was obtained from GE Healthcare. Annexin A2 (STVHEILSKLSLEG) and TRTK12 (TRTKIDWNKILSLE) peptides were synthesized as previously described.29,34 All other reagents used in the following experiments were of the highest purity commercially available.
Molecular modeling of hybrid S100 proteins
The lengths of the linker connecting S100A10 to annexin A2 or S100B to TRTK12 were determined using Swiss-PDB Viewer.47 The coordinates for S100A10 bound to annexin A2 (PDB code: 1BT6)12 and Ca2+-S100B complexed with TRTK12 (PDB codes: 1MQ1, 1MWN)33,34 were obtained from the Protein Data Bank.48 The last four residues at the C-terminus of S100A10 were disordered and not visible in the structure. Initial linkers were built using glycine spacers with moderate rotations of ϕ, ψ angles. A total of nine glycine residues were required to connect the observed C-terminus of S100A10 (K91) to the N-terminus of the annexin A2 peptide (STVHEILSKLSLEG). Eleven glycine residues were added in the BT12 hybrid from the C-terminus of the S100B (E91) to the N-terminus of the TRTK12 peptide (TRTKIDWNKILS). Each structure was energy minimized using steepest descents minimization. Upon completion, glycine residues were swapped to those corresponding to the TEV (S100A10) or PreScission (S100B) cleavage sites. Subsequent energy minimization and visualization was used to insure there were no bad contacts between the linker residues and the corresponding S100 protein.
Construction of the S100A10-annexin A2 and S100B-TRTK12 hybrid genes
A derivative of the pGEX-6P-1 expression vector containing the rabbit S100A10 was used to construct the S100A10-annexin A2 hybrid gene. The QuikChange method (Stratagene, La Jolla, CA) was used in three consecutive steps to (a) substitute the last four amino acid residues of S100A10; K93E, G94N, K95L, and K96Y, (b) insert a TEV protease cleave site, and (c) generate the annexin A2 hybrid DNA fragment. The resulting vector was confirmed by DNA sequencing.
The pSS2 expression vector containing the human S100B was utilized to construct the S100B-TRTK12 hybrid gene.49 A unique XhoI reporter cut site was generated directly adjacent to the stop codon of the S100B coding sequence using the QuikChange kit (Stratagene). Forward and reverse primers (5′-phosphorylated; Sigma-Aldrich) corresponding to residues from the linker region, the entire TRTK12 peptide and appropriate XhoI site overhangs (69 bases in length) were annealed together and inserted into the unique XhoI site of the dephosphorylated vector containing the S100B gene. The correct orientation and a single insertion of the annealed primers in the resulting vector were confirmed by DNA sequencing.
Expression and purification of S100A10 and S100A10-annexin A2
Uniformly 15N, 13C-labeled GST-S100A10 and GST-S100A10-annexin A2 were overexpressed in the BL21 (DE3) codon-plus E. coli strain using M9 minimal medium supplemented with 1 g 15NH4Cl and 2 g 13C6-glucose as the sole nitrogen and carbon sources, respectively. The cultures were grown at 37°C in the presence of ampicillin (100 μg/mL) to a density of (A600) 0.8 AU, when they were induced by the addition of 1 mM isopropyl-beta-D-thiogalactopyranoside. Induction continued for another 8 h with constant shaking at 37°C. Cells were harvested by centrifugation at 6000 rpm for 15 min and lysed by French Pressure at 20,000 psi. Cell debris was removed by centrifugation at 38,000 rpm for 90 min. All fractionation steps were performed at 4°C. The supernatant was applied to a 5 mL glutathione-linked sepharose column equilibrated in buffer (140 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, and 1.8 mM KH2PO4, pH 7.3). The 15N, 13C-labeled GST-S100A10 or GST-S100A10-annexin A2 were eluted in elution buffer (50 mM Tris, 20 mM reduced glutathione, pH 8.0). Fractions containing the protein were pooled and extensively dialyzed against PreScission protease cleavage buffer (50 mM Tris, 150 mM NaCl, 1 mM EDTA, and 1 mM DTT, pH 7.0) overnight. S100A10 or S100A10-annexin A2 were cleaved from the glutathione-sepharose protein using 150 units of PreScission protease in 48 h. The cleaved protein was applied to a GST column in binding buffer and the flow-through fractions were collected. MALDI–TOF mass data for A10A2 (MWcalc = 14,517.9 Da; MWobs = 14,507.2 Da for 15N, 13C-labeled protein) confirmed the protein identities.
Expression and purification of S100B and S100B-TRTK12
Expression and purification of uniformly 15N, 13C-labeled S100B-TRTK12 (BT12) in E. coli strain N99 were conducted as described for human S100B by Smith et al.49 The protocol was modified by reducing the (NH4)2SO4 concentration to 50% saturation due to the decreased solubility of BT12 compared to S100B. Following dialysis (25 mM Tris, 100 mM NaCl, 1 mM DTT, and 5 mM CaCl2, pH 7.5), BT12 was applied to a phenyl-sepharose column equilibrated in the same buffer. The column was washed (25 mM Tris, 100 mM NaCl, 1 mM DTT, and 1 mM CaCl2, pH 7.5) and S100B-TRTK12 was eluted with EDTA buffer (25 mM Tris, 100 mM NaCl, 1 mM DTT, and 1 mM EGTA, pH 7.5). MALDI–TOF mass data for 15N-labeled BT12 (desformylmethionine MWcalc = 13,430.2 Da; MWobs = 13,427.3 Da, desformyl MWcalc = 13,563.2 Da; MWobs = 13,559.5 Da, and formylmethionine MWcalc = 13,591.2 Da; MWobs = 13,587.6 Da) confirmed the presence of the three chemically different forms of BT12 at its N-terminus similar to those previously characterized for S100B.50
NMR spectroscopy
All NMR experiments were acquired at 35°C on a Varian INOVA 600 MHz spectrometer equipped with a 13C-enhanced triple resonance cold probe. NMR samples of 0.5 mM (monomer) uniformly 15N, 13C-labeled S100A10 and A10A2 were prepared in 10% D2O, 20 mM MOPS, 1 mM EDTA, 1 mM DTT, 50 mM arginine, 50 mM glutamic acid, and 100 mM NaCl buffer at pH 7.0, and 100 μM DSS as an internal standard. Uniformly 15N, 13C-labeled NMR samples of 0.7 mM S100B and BT12 were prepared in a similar buffer using either 20 mM CaCl2 or 1 mM EDTA. Sequential 1H, 13C, and 15N resonance assignments of A10A2 and calcium saturated BT12 were completed using HNCACB,51 CBCA(CO)NH,52 HNCO,53 HNCA,54 and C(CO)NH experiments.55 Sensitivity-enhanced 1H-15N HSQC spectra56 for A10A2 were acquired using carrier frequencies of 4.705 (1H) and 114 ppm (15N) and spectral widths of 8000.0 and 1700.0 Hz, respectively. Similar spectra were generated for Ca2+-S100B-TRTK12 using carrier frequencies of 4.702 (1H) and 120 ppm (15N) and spectral widths of 8000.0 and 2000.0 Hz, respectively. All data were processed using NMRPipe and NMRDraw57 and spectra were analyzed by NMRView.58 The 1H-15N HSQC spectra of the 15N, 13C-labeled A10A2, and BT12 hybrid proteins cleaved with TEV and PreScission proteases were acquired 48 h following the addition of 16 units of each of the enzymes, respectively. The presence of the fully cleaved product in the NMR sample was verified on an SDS–PAGE.
The 15N, 13C-labeled S100A10 complex with annexin A2 peptide was prepared by the titration of 0.5 mM (monomer) S100A10 with a 20 mM stock solution of annexin A2 peptide under constant buffer conditions. A final peptide concentration of 0.8 mM was used. The pH of the sample remained at 7.0 throughout the titration. The sample was mixed after each addition and equilibrated for 15 min. The total volume increase of the NMR sample was 24 μL (4%). 1H-15N HSQC spectra were recorded at each point to monitor the titration. A sample of 0.7 mM (monomer) apo-S100B prepared in 12 mM CaCl2 and titrated with a 20 mM stock solution of TRTK12 peptide in the same buffer (above) to a final peptide concentration of 0.9 mM was used. The pH of the sample remained at 7.0 throughout the titration.
Glossary
Abbreviations:
- TEV
tobacco etch virus
- Ca2+-S100B
calcium-bound S100B
- BT12
S100B-TRTK12 hybrid protein
- A10A2
S100A10-annexin A2 hybrid protein
- Ca2+-S100A11
calcium-bound S100A11
- MALDI-TOF
matrix-assisted laser desorption/ionization time-of-flight
- GST
glutathione S-transferase.
References
- 1.Deloulme JC, Assard N, Mbele GO, Mangin C, Kuwano R, Baudier J. S100A6 and S100A11 are specific targets of the calcium- and zinc-binding S100B protein in vivo. J Biol Chem. 2000;275:35302–35310. doi: 10.1074/jbc.M003943200. [DOI] [PubMed] [Google Scholar]
- 2.Propper C, Huang X, Roth J, Sorg C, Nacken W. Analysis of the MRP8-MRP14 protein-protein interaction by the two-hybrid system suggests a prominent role of the C-terminal domain of S100 proteins in dimer formation. J Biol Chem. 1999;274:183–188. doi: 10.1074/jbc.274.1.183. [DOI] [PubMed] [Google Scholar]
- 3.Wang G, Zhang S, Fernig DG, Martin-Fernandez M, Rudland PS, Barraclough R. Mutually antagonistic actions of S100A4 and S100A1 on normal and metastatic phenotypes. Oncogene. 2005;24:1445–1454. doi: 10.1038/sj.onc.1208291. [DOI] [PubMed] [Google Scholar]
- 4.Schafer BW, Heizmann CW. The S100 family of EF-hand calcium-binding proteins: functions and pathology. Trends Biochem Sci. 1996;21:134–140. doi: 10.1016/s0968-0004(96)80167-8. [DOI] [PubMed] [Google Scholar]
- 5.Santamaria-Kisiel L, Rintala-Dempsey AC, Shaw GS. Calcium-dependent and -independent interactions of the S100 protein family. Biochem J. 2006;396:201–214. doi: 10.1042/BJ20060195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Drohat AC, Baldisseri DM, Rustandi RR, Weber DJ. Solution structure of calcium-bound rat S100B(betabeta) as determined by nuclear magnetic resonance spectroscopy. Biochemistry. 1998;37:2729–2740. doi: 10.1021/bi972635p. [DOI] [PubMed] [Google Scholar]
- 7.Smith SP, Shaw GS. A novel calcium-sensitive switch revealed by the structure of human S100B in the calcium-bound form. Structure. 1998;6:211–222. doi: 10.1016/s0969-2126(98)00022-7. [DOI] [PubMed] [Google Scholar]
- 8.Sastry M, Ketchem RR, Crescenzi O, Weber C, Lubienski MJ, Hidaka H, Chazin WJ. The three-dimensional structure of Ca2+-bound calcyclimplications for Ca2+-signal transduction by S100 proteins. Structure. 1998;6:223–231. doi: 10.1016/s0969-2126(98)00023-9. [DOI] [PubMed] [Google Scholar]
- 9.Bhattacharya S, Bunick CG, Chazin WJ. Target selectivity in EF-hand calcium binding proteins. Biochim Biophys Acta. 2004;1742:69–79. doi: 10.1016/j.bbamcr.2004.09.002. [DOI] [PubMed] [Google Scholar]
- 10.Gerke V, Weber K. The regulatory chain in the p36-kd substrate complex of viral tyrosine-specific protein kinases is related in sequence to the S-100 protein of glial cells. EMBO J. 1985;4:2917–2920. doi: 10.1002/j.1460-2075.1985.tb04023.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rety S, Osterloh D, Arie JP, Tabaries S, Seeman J, Russo-Marie F, Gerke V, Lewit-Bentley A. Structural basis of the Ca(2+)-dependent association between S100C (S100A11) and its target, the N-terminal part of annexin I. Struct Fold Des. 2000;8:175–184. doi: 10.1016/s0969-2126(00)00093-9. [DOI] [PubMed] [Google Scholar]
- 12.Rety S, Sopkova J, Renouard M, Osterloh D, Gerke V, Tabaries S, Russo-Marie F, Lewit-Bentley A. The crystal structure of a complex of p11 with the annexin II N-terminal peptide. Nat Struct Biol. 1999;6:89–95. doi: 10.1038/4965. [DOI] [PubMed] [Google Scholar]
- 13.van de Graaf SF, Hoenderop JG, Gkika D, Lamers D, Prenen J, Rescher U, Gerke V, Staub O, Nilius B, Bindels RJ. Functional expression of the epithelial Ca(2+) channels (TRPV5 and TRPV6) requires association of the S100A10-annexin 2 complex. EMBO J. 2003;22:1478–1487. doi: 10.1093/emboj/cdg162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Borthwick LA, Neal A, Hobson L, Gerke V, Robson L, Muimo R. The annexin 2-S100A10 complex and its association with TRPV6 is regulated by cAMP/PKA/CnA in airway and gut epithelia. Cell Calcium. 2008;44:147–157. doi: 10.1016/j.ceca.2007.11.001. [DOI] [PubMed] [Google Scholar]
- 15.Renigunta V, Yuan H, Zuzarte M, Rinne S, Koch A, Wischmeyer E, Schlichthorl G, Gao Y, Karschin A, Jacob R, Schwappach B, Daut J, Preisig-Muller R. The retention factor p11 confers an endoplasmic reticulum-localization signal to the potassium channel TASK-1. Traffic. 2006;7:168–181. doi: 10.1111/j.1600-0854.2005.00375.x. [DOI] [PubMed] [Google Scholar]
- 16.Girard C, Tinel N, Terrenoire C, Romey G, Lazdunski M, Borsotto M. p11, an annexin II subunit, an auxiliary protein associated with the background K+ channel, TASK-1. Embo J. 2002;21:4439–4448. doi: 10.1093/emboj/cdf469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Beaton AR, Rodriguez J, Reddy YK, Roy P. The membrane trafficking protein calpactin forms a complex with bluetongue virus protein NS3 and mediates virus release. Proc Natl Acad Sci USA. 2002;99:13154–13159. doi: 10.1073/pnas.192432299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Benaud C, Gentil BJ, Assard N, Court M, Garin J, Delphin C, Baudier J. AHNAK interaction with the annexin 2/S100A10 complex regulates cell membrane cytoarchitecture. J Cell Biol. 2004;164:133–144. doi: 10.1083/jcb.200307098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.De Seranno S, Benaud C, Assard N, Khediri S, Gerke V, Baudier J, Delphin C. Identification of an AHNAK binding motif specific for the Annexin2/S100A10 tetramer. J Biol Chem. 2006;281:35030–35038. doi: 10.1074/jbc.M606545200. [DOI] [PubMed] [Google Scholar]
- 20.Rintala-Dempsey AC, Rezvanpour A, Shaw GS. S100-annexin complexes—structural insights. FEBS J. 2008;275:4956–4966. doi: 10.1111/j.1742-4658.2008.06654.x. [DOI] [PubMed] [Google Scholar]
- 21.Chang N, Sutherland C, Hesse E, Winkfein R, Wiehler WB, Pho M, Veillette C, Li S, Wilson DP, Kiss E, Walsh MP. Identification of a novel interaction between the Ca(2+)-binding protein S100A11 and the Ca(2+)- and phospholipid-binding protein annexin A6. Am J Physiol Cell Physiol. 2007;292:C1417–C1430. doi: 10.1152/ajpcell.00439.2006. [DOI] [PubMed] [Google Scholar]
- 22.Arcuri C, Giambanco I, Bianchi R, Donato R. Annexin V, annexin VI, S100A1 and S100B in developing and adult avian skeletal muscles. Neuroscience. 2002;109:371–388. doi: 10.1016/s0306-4522(01)00330-x. [DOI] [PubMed] [Google Scholar]
- 23.Garbuglia M, Verzini M, Donato R. Annexin VI binds S100A1 and S100B and blocks the ability of S100A1 and S100B to inhibit desmin and GFAP assemblies into intermediate filaments. Cell Calcium. 1998;24:177–191. doi: 10.1016/s0143-4160(98)90127-0. [DOI] [PubMed] [Google Scholar]
- 24.Garbuglia M, Verzini M, Hofmann A, Huber R, Donato R. S100A1 and S100B interactions with annexins. Biochim Biophys Acta. 2000;1498:192–206. doi: 10.1016/s0167-4889(00)00096-3. [DOI] [PubMed] [Google Scholar]
- 25.Zeng FY, Gerke V, Gabius HJ. Identification of annexin II, annexin VI and glyceraldehyde-3-phosphate dehydrogenase as calcyclin-binding proteins in bovine heart. Int J Biochem. 1993;25:1019–1027. doi: 10.1016/0020-711x(93)90116-v. [DOI] [PubMed] [Google Scholar]
- 26.Gerke V, Moss SE. Annexins: from structure to function. Physiol Rev. 2002;82:331–371. doi: 10.1152/physrev.00030.2001. [DOI] [PubMed] [Google Scholar]
- 27.Johnsson N, Johnsson K, Weber K. A discontinuous epitope on p36, the major substrate of src tyrosine-protein-kinase, brings the phosphorylation site into the neighbourhood of a consensus sequence for Ca2+/lipid-binding. FEBS Lett. 1988;236:201–204. doi: 10.1016/0014-5793(88)80314-4. [DOI] [PubMed] [Google Scholar]
- 28.Johnsson N, Marriott G, Weber K. p36, the major cytoplasmic substrate of src tyrosine protein kinase, binds to its p11 regulatory subunit via a short amino-terminal amphiphatic helix. EMBO J. 1988;7:2435–2442. doi: 10.1002/j.1460-2075.1988.tb03089.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rintala-Dempsey AC, Santamaria-Kisiel L, Liao Y, Lajoie G, Shaw GS. Insights into S100 target specificity examined by a new interaction between S100A11 and annexin A2. Biochemistry. 2006;45:14695–14705. doi: 10.1021/bi061754e. [DOI] [PubMed] [Google Scholar]
- 30.Streicher WW, Lopez MM, Makhatadze GI. Annexin I and annexin II N-terminal peptides binding to S100 protein family members: specificity and thermodynamic characterization (dagger) Biochemistry. 2009;48:2788–2798. doi: 10.1021/bi8019959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rustandi RR, Baldisseri DM, Weber DJ. Structure of the negative regulatory domain of p53 bound to S100B (ββ) Nat Struct Biol. 2000;7:570–574. doi: 10.1038/76797. [DOI] [PubMed] [Google Scholar]
- 32.Bhattacharya S, Large E, Heizmann CW, Hemmings B, Chazin WJ. Structure of the Ca2+/S100B/NDR kinase peptide complex: insights into S100 target specificity and activation of the kinase. Biochemistry. 2003;42:14416–14426. doi: 10.1021/bi035089a. [DOI] [PubMed] [Google Scholar]
- 33.Inman KG, Yang R, Rustandi RR, Miller KE, Baldisseri DM, Weber DJ. Solution NMR structure of S100B bound to the high-affinity target peptide TRTK-12. J Mol Biol. 2002;324:1003–1014. doi: 10.1016/s0022-2836(02)01152-x. [DOI] [PubMed] [Google Scholar]
- 34.McClintock KA, Shaw GS. A novel S100 target conformation is revealed by the solution structure of the Ca2+-S100B-TRTK-12 complex. J Biol Chem. 2003;278:6251–6257. doi: 10.1074/jbc.M210622200. [DOI] [PubMed] [Google Scholar]
- 35.Lee YT, Dimitrova YN, Schneider G, Ridenour WB, Bhattacharya S, Soss SE, Caprioli RM, Filipek A, Chazin WJ. Structure of the S100A6 complex with a fragment from the C-terminal domain of Siah-1 interacting protea novel mode for S100 protein target recognition. Biochemistry. 2008;47:10921–10932. doi: 10.1021/bi801233z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wishart DS, Sykes BD, Richards FM. The Chemical Shift Index: a fast and simple method for the assignment of protein secondary structure through NMR spectroscopy. Biochemistry. 1992;31:1647–1651. doi: 10.1021/bi00121a010. [DOI] [PubMed] [Google Scholar]
- 37.Lindhout DA, Thiessen A, Schieve D, Sykes BD. High-yield expression of isotopically labeled peptides for use in NMR studies. Protein Sci. 2003;12:1786–1791. doi: 10.1110/ps.0376003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Porumb T, Yau P, Harvey TS, Ikura M. A calmodulin-target peptide hybrid molecule with unique calcium-binding properties. Prot Eng. 1994;7:109–115. doi: 10.1093/protein/7.1.109. [DOI] [PubMed] [Google Scholar]
- 39.Martin SR, Bayley PM, Brown SE, Porumb SE, Zhang M, Ikura M. Spectroscopic characterization of a high-affinity calmodulin-target peptide hybrid molecule. Biochemistry. 1996;35:3508–3517. doi: 10.1021/bi952522a. [DOI] [PubMed] [Google Scholar]
- 40.Ye Q, Li X, Wong A, Wei Q, Jia Z. Structure of calmodulin bound to a calcineurin peptide: a new way of making an old binding mode. Biochemistry. 2006;45:738–745. doi: 10.1021/bi0521801. [DOI] [PubMed] [Google Scholar]
- 41.Chyan CL, Huang PC, Lin TH, Huang JW, Lin SS, Huang HB, Chen YC. Purification, crystallization and preliminary crystallographic studies of a calmodulin-OLFp hybrid molecule. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2005;61:785–787. doi: 10.1107/S1744309105023006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ishida H, Rainaldi M, Vogel HJ. Structural studies of soybean calmodulin isoform 4 bound to the calmodulin-binding domain of tobacco MAPK phosphatase-1 provide insights into a sequential target binding mode. J Biol Chem. 2009;284:28292–28305. doi: 10.1074/jbc.M109.025080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kang S, Kang J, Yoo SH, Park S. Recombinant preparation and characterization of interactions for a calmodulin-binding chromogranin A peptide and calmodulin. J Pept Sci. 2007;13:237–244. doi: 10.1002/psc.837. [DOI] [PubMed] [Google Scholar]
- 44.Johnsson N, Vandekerckhove J, Van Damme J, Weber K. Binding sites for calcium, lipid and p11 on p36, the substrate of retroviral tyrosine-specific protein kinases. FEBS Lett. 1986;198:361–364. doi: 10.1016/0014-5793(86)80437-9. [DOI] [PubMed] [Google Scholar]
- 45.Mailliard WS, Haigler HT, Schlaepfer DD. Calcium-dependent binding of S100C to the N-terminal domain of annexin I. J Biol Chem. 1996;271:719–725. doi: 10.1074/jbc.271.2.719. [DOI] [PubMed] [Google Scholar]
- 46.McClintock KA, Van Eldik LJ, Shaw GS. The C-terminus and linker region of S100B exert dual control on protein-protein interactions with TRTK-12. Biochemistry. 2002;41:5421–5428. doi: 10.1021/bi011732m. [DOI] [PubMed] [Google Scholar]
- 47.Guex N, Peitsch MC. SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis. 1997;18:2714–2723. doi: 10.1002/elps.1150181505. [DOI] [PubMed] [Google Scholar]
- 48.Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE. The Protein Data Bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Smith SP, Barber KR, Dunn SD, Shaw GS. Structural influence of cation binding to recombinant human brain S100b: evidence for calcium-induced exposure of a hydrophobic surface. Biochemistry. 1996;35:8805–8814. doi: 10.1021/bi952698c. [DOI] [PubMed] [Google Scholar]
- 50.Smith SP, Barber KR, Shaw GS. Identification and structural influence of a differentially modified N-terminal methionine in human S100b. Protein Sci. 1997;6:1110–1113. doi: 10.1002/pro.5560060518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wittekind M, Mueller L. HNCACB, A high sensitivity 3D NMR experiment to correlate amide proton and nitrogen resonances with the α-carbon and β-carbon resonances in proteins. J Magn Reson Series B. 1993;101:171–180. [Google Scholar]
- 52.Grzesiek S, Bax A. Correlating backbone amide and side chain resonances in larger proteins by multiple relayed triple resonance NMR. J Am Chem Soc. 1992;114:6291–6293. [Google Scholar]
- 53.Kay LE, Ikura M, Tschudin R, Bax A. Three-dimensional triple-resonance NMR spectroscopy of isotopically enriched proteins. J Magn Reson. 1990;89:496–514. doi: 10.1016/j.jmr.2011.09.004. [DOI] [PubMed] [Google Scholar]
- 54.Grzesiek S, Bax A. An efficient experiment for sequential backbone assignment of medium-sized isotopically enriched proteins. J Magn Reson. 1992;99:201–207. [Google Scholar]
- 55.Kay LE. Pulsed-field gradient-enriched three-dimensional NMR experiment for correlating 13Cα/β, 13C', and 1Hα chemical shifts in uniformly 13C-labeled proteins dissolved in H2O. J Am Chem Soc. 1993;115:2055–2057. [Google Scholar]
- 56.Kay LE, Keifer P, Saarinen T. Pure absorption gradient enhanced heteronuclear single quantum correlation spectroscopy with improved sensitivity. J Am Chem Soc. 1992;114:10663–10665. [Google Scholar]
- 57.Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J Biomol NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 58.Johnson BA, Belvins RA. NMRView: a computer program for the visualization and analysis of NMR data. J Biomol NMR. 1994;4:603–614. doi: 10.1007/BF00404272. [DOI] [PubMed] [Google Scholar]


