Abstract
True catalases are tyrosine-liganded, usually tetrameric, hemoproteins with subunit sizes of ∼55–84 kDa. Recently characterized hemoproteins with a catalase-related structure, yet lacking in catalatic activity, include the 40–43 kDa allene oxide synthases of marine invertebrates and cyanobacteria. Herein, we describe the 1.8 Å X-ray crystal structure of a 33 kDa subunit hemoprotein from Mycobacterium avium ssp. paratuberculosis (annotated as MAP-2744c), that retains the core elements of the catalase fold and exhibits an organic peroxide-dependent peroxidase activity. MAP-2744c exhibits negligible catalatic activity, weak peroxidatic activity using hydrogen peroxide (20/s) and strong peroxidase activity (∼300/s) using organic hydroperoxides as co-substrate. Key amino acid differences significantly impact prosthetic group conformation and placement and confer a distinct activity to this prototypical member of a group of conserved bacterial “minicatalases”. Its structural features and the result of the enzyme assays support a role for MAP-2744c and its close homologues in mitigating challenge by a variety of reactive oxygen species.
Keywords: X-ray crystallography, heme enzyme, catalase, peroxidase
Introduction
The catalases are among the earliest studied of all enzymes, exhibiting one of the prototypical activities among hemoproteins: the rapid dismutation of hydrogen peroxide to oxygen and water. By the 1980s, the first crystal structures were available1–3 and the current submissions in the Protein Data Bank now include multiple mammalian, yeast, and bacterial catalase structures.4–11 Analysis of the proteins with known catalatic activity combined with searches of available DNA databases allowed the classification of several hundred available sequences into three major groups: mono-functional catalases, heme-containing catalase-peroxidases, and nonheme catalases.12–14 The most abundant group is that of the mono-functional catalases, and the best characterized of these are homotetramers with one heme per monomer. This group is further subdivided into small (55–69 kDa) and large (74–84 kDa) subunit enzymes.14 Human catalase, a 240 kDa enzyme comprised of four 60 kDa monomers, is typical of the small subunit mono-functional catalases. The active site environment is characterized by a tyrosine as the proximal heme ligand, and by a conserved histidine and asparagine on the distal side of the heme. A long (∼20 Å) narrow channel leads from the surface of the protein down to the distal face of the heme, and this restricted access is considered to contribute to the enzyme selectivity for H2O2 as substrate.
Over a decade ago the heme domain of a peroxidase-lipoxygenase fusion protein from the prostaglandin-containing coral Plexaura homomalla was shown to exhibit weak sequence identity to mono-functional catalases. While the overall identity between catalases and the domain is <20%, sequence alignments indicated that the proximal Tyr, distal His, and vicinal Asn that flank the catalase heme were also conserved in the P. homomalla heme domain.15 This domain was shown to possess allene oxide synthase activity as it selectively metabolizes a specific fatty acid hydroperoxide to an unstable allene epoxide. The lipoxygenase domain of the fusion protein converted arachidonic acid to an 8(R)-hydroperoxide and this was shown to be the substrate for the hemoprotein domain now referred to as catalase-related allene oxide synthase (cAOS).16 The recent determination of the crystal structure of cAOS revealed a catalase-like architecture with a highly conserved heme environment.17 Despite the structural homology, which is particularly striking at the active site heme, and a large access channel that cannot exclude heme access by H2O2, cAOS shows no reaction with hydrogen peroxide.18 The cAOS domain is only 43 kDa in size, ∼120 amino acids shorter than the small mono-functional catalases, and is homodimeric when expressed without the C-terminal LOX domain.
The structural and functional information on cAOS established the possibility that other hemoproteins with catalase-related structures might exist and exhibit distinctive catalytic activities. Upon searching the DNA databases using the cAOS protein sequence as “bait”, we identified a number of small catalase-related sequences. One of these, that also existed in a fusion protein with a lipoxygenase, was found to specifically convert hydroperoxy-C18 fatty acids (the products of the LOX domain) to allylic epoxides.19,20 Another of the “hits” in these database searches was a genomic sequence from Mycobacterium avium ssp. paratuberculosis. The open reading frame encodes a 33 kDa protein, particularly small by the standards of catalase-related proteins, and the structure and function of this protein is the subject of the present report.
Mycobacterium avium ssp. paratuberculosis is a pathogen in cattle and other ruminants and is the causative agent of the gastrointestinal infections that are the basis of Johne's disease.21 Accordingly, it was targeted for whole genome sequencing and the results of these efforts were released 4 years ago.22 Like other prokaryotes, the M. avium genome encodes multiple catalase-related sequences, including among them the typical large subunit mono-functional catalase, and the catalase-peroxidase KatG. Moreover, like nearly all other prokaryotes, the M. avium genome does not encode a lipoxygenase gene, and these bacteria do not contain the polyunsaturated lipids that are suitable lipoxygenase substrates. Thus, unless the bacteria encounter the lipids from a host organism, they lack the type of hydroperoxide substrates that are utilized by cAOS and its close counterparts.16,19 These observations suggest novel function(s) for this prokaryotic catalase-related enzyme. In this manuscript, we present the crystal structure and activity of the catalase-related enzyme from M. avium ssp. paratuberculosis MAP_2744c. This structure suggests that while the heme-environments of the catalase-related heme enzymes are highly conserved, both key amino acid and active site shape differences may significantly effect heme conformation and enzyme activity.
Results and Discussion
Protein structure
Crystals of MAP_2744c were obtained in two orthorhombic space groups (P212121 and I212121), and the structures were determined by multi-wavelength anomalous dispersion (MAD) phasing of the P212121 diffraction data using the Fe absorption edge. As expected from its sequence identity with catalase and cAOS, the 313 amino acid monomer of the enzyme has a catalase α + β “core” (Fig. 1) comprised of an eight-stranded antiparallel β-barrel and seven α-helices. In comparison to cAOS (373 amino acids; 1U5U), the only other catalase-related enzyme structure that has been reported, MAP_2744c has significantly less helical structure: there are 13 α-helices in cAOS, as opposed to the seven in MAP_2744c. The largest difference between the two proteins is at the amino terminus, which in MAP_2744c is 40 amino acids shorter than that of cAOS. The amino terminal extension in cAOS, relative to MAP_2744c, serves as a dimerization interface. Although two MAP_2744c monomers pack as a dimer in both crystal forms, the relatively small surface area buried (∼830 Å2) by this interaction, the paucity of hydrophobic interactions, and the presence of multiple solvent molecules in the interface, suggest that the dimer is not stable in solution. In fact, the protein elutes at a volume consistent with its monomeric molecular weight in size-exclusion chromatography (data not shown). Mono-functional catalases have N-terminal extensions of at least 100 amino acids relative to MAP_2744c and the extended termini pack onto neighboring monomers and provide contacts for tetramer formation.
Figure 1.
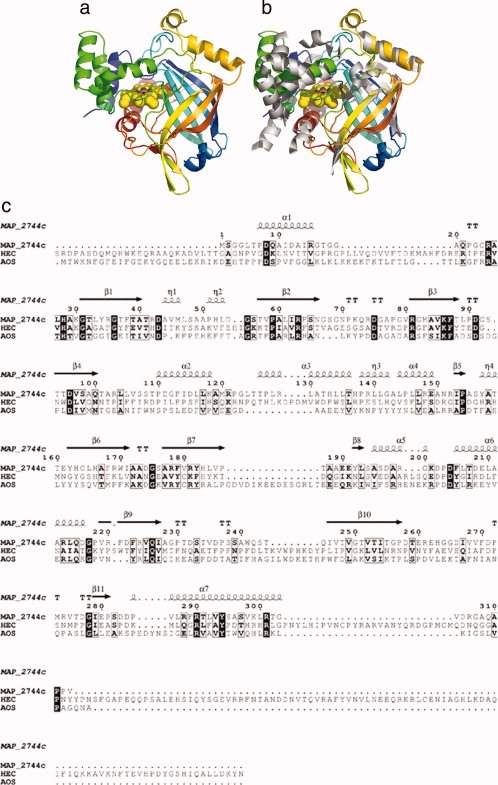
The structure of MAP_2744c, a catalase-related enzyme. (a) Cartoon rendering of MAP_2744c. Coloring is N→C blue→red. The heme is depicted in space-filled rendering. (b) cAOS (white) superimposed on MAP_2744c. For clarity only the secondary structural elements of cAOS are drawn. (c) A structure based sequence alignment for MAP_2744c, cAOS, and HEC sequences. Those amino acids which are identical in the sequences are indicated in black boxes, while empty boxes indicate conservative substitutions. The elements of secondary structure are labeled according to their location in MAP_2744c.
In addition to the fact that MAP_2744c has fewer helices than cAOS, those helices which are common to the proteins do not necessarily superimpose, while the core β-barrel structure is maintained. Note that in the structure-based sequence alignment of MAP_2744c, human erythrocyte catalase (HEC) and cAOS [Fig. 1(c)] there is more sequence identity among the enzymes in regions of β-sheet rather than α-helical segments. In fact, only the two carboxy terminal helices (α6 and α7) display significant sequence identity between MAP_2744c and cAOS, and only for α7 does the homology extend to include HEC. The C-terminal helix (α7) provides the proximal heme ligand as well as other amino acids that buttress the heme, while the penultimate helix (α6) packs at one end of the β-barrel and does not contribute directly to the binding site for the prosthetic group. The heme is positioned between the β-barrel and the amino terminal helix (α1) which runs perpendicular to the heme plane. The carboxy terminal helix is positioned “below” the prosthetic group, where it runs roughly parallel to the heme plane. The proximal Tyr (Y294) emanates from this helix. Substrate can gain access to the heme via a large cavity. At the surface, the oblong opening is ∼17 × 10 Å. The cavity, which is ∼13 Å deep, tapers to some extent so that at the heme it is ∼14 × 8 Å in dimensions. This cavity is substantially larger than the access channel described for cAOS, which is only 9 Å across at its longest dimension. Mono-functional catalases have even narrower access channels to restrict heme access to H2O2 and other small molecules such as ethanol. A surface rendering in which the cavity of MAP_2744c is depicted in presented in Figure 2. The contrast between this cavity and that in cAOS is striking: note the additional mass in cAOS in the region that superposes with the heme access opening in MAP_2744c [Fig. 2(b)].
Figure 2.
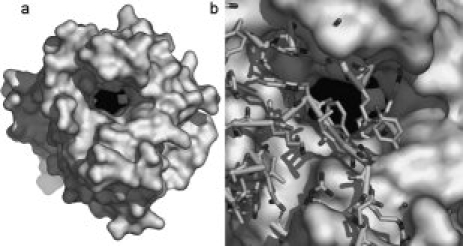
Access to heme. (a) A surface rendering of the MAP_2744c enzyme. The heme, in sphere rendering (Fe, white; C, N black) lies at the bottom of a large, deep cavity. (b) A stick rendering of cAOS is superposed on the MAP_2744c surface rendering. The presence of the additional side chain and main chain atoms in cAOS obstructs heme access.
The heme environment
When MAP_2744c and cAOS are superposed, the rmsd between 243 equivalent alpha carbons is 2.1 Å. The sequence identity between the structurally equivalent alpha carbons is 23%. MAP_2744c is as similar to cAOS as it is to HEC. The MAP_2744c and catalase (1DGB)23 structures superpose 264 equivalent CA with an rmsd of 2.1 Å and 25% sequence identity. However, roughly half the mass of this low molecular weight catalase monomer has no counterpart in the MAP_2744c structure.
Despite the low level of sequence identity among MAP_2744c, cAOS and HEC, the heme environments are highly conserved. The heme environments of MAP_2744c and cAOS are superposed in Figure 3. Like cAOS and HEC, MAP_2744c has the proximal Tyr (Y294), distal histidine (H28) and a trio of Arginines (R25, –63, and –301) positioned to neutralize the heme carboxylates. All enzymes have as well a C-terminal Arg (R290) that has been proposed to “tune” the reactivity of the heme iron.24 However, those amino acids that flank the distal His in cAOS are not found in MAP_2744c. MAP_2744c has a Gln (Q100) in place of the Asn found in cAOS and catalase. While the substitution of a Gln for Asn is conservative, the longer side chain of Gln seems to have an important structural consequence. In both cAOS and HEC the corresponding Asn is positioned to H-bond to the distal His. But in MAP_2744c the Gln bends back away from the distal His and heme, leaving more of the heme exposed. The side chain of Q100 is H-bonded to the main chain NH and CO of A100. Although sequence line-ups indicate that Asn is generally found at this position, Gln at this position is not unique to MAP_2744c. The sequences of homologous proteins from Deinococcus radiodurans and Pseudomonas aeruginosa both have a Gln at this position. On the opposite side of the distal His, the Thr found in cAOS has been replaced by a Leu. In the classic mono-functional catalases this amino acid is an invariant Val. The first 100 sequences retrieved from the protein sequence database with MAP2744_c as “bait” suggest a preference for small amino acids at this position (Ala or Thr). Mutation of Thr to Val in cAOS confers cAOS with reactivity towards H2O2, albeit with an activity significantly lower than that of wild-type catalase.18 Thus, the variability at this position may contribute to the diversity of catalytic activities associated with catalase-related enzymes. As was described for the cAOS structure, the heme group of MAP_2744c does not show the distortion typical of catalase.25,26 The deviation from planarity of the MAP_2744c heme is ∼0.2, in comparison to ∼0.7 Å observed in HEC.25
Figure 3.
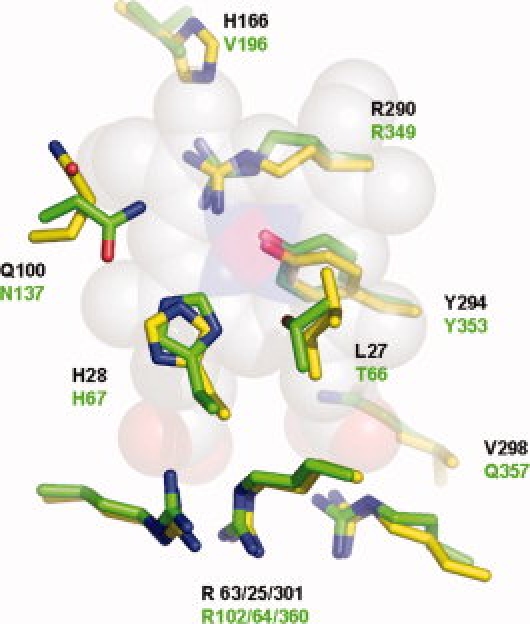
The heme environment. The side chains of MAP_2744c (C, yellow) and cAOS (C, green) that surround the heme (space-filled rendering, C, white, N, blue, O, red, and Fe, magenta).
The most distinct feature of MAP_2744c, when compared with cAOS and HEC, is not in the heme environment per se, but heme placement within this environment. As one can see from Figure 4, the heme in MAP_2744c is displaced “upwards” such that the Tyr-Fe distance is longer than those observed in HEC and cAOS. The Tyr-O-Fe distance is 2.35 Å in the 1.8 Å resolution MAP_2744c structure. However, this distance is 1.8 Å in HEC (1DGB) and 2.16 Å in cAOS. Heme displacement may be a consequence of a repositioning of the C-terminal helix that contains the proximal Tyr. Alternatively, or perhaps additionally, the disposition of Phe rings in the different structures in the enzymes could determine how the heme is anchored in the fold. Phe residues within ∼4 Å of the MAP_2744c, cAOS and HEC hemes are rendered in Figure 4(b). (There are no Trp or Tyr side chains, besides the proximal Tyr, within van der Waals distance of any of the hemes.). Note that both cAOS and HEC have multiple phenylalanines positioned above the heme plane, while in MAP_2744c a single Phe (F113) is positioned above the plane of the prosthetic group, with its ring roughly parallel to the heme. Neither cAOS nor HEC has a Phe below the heme plane, while MAP_2744c has Phe (F289) with it ring perpendicular to the heme. In a multi-sequence line-up of 100 sequences obtained from available sequence databases when Map_2744c is used as the “bait,” only 12 unique protein sequences have a Phe or Tyr at this position. The presence of a Phe here could prevent positioning the heme closer to the proximal Tyr.
Figure 4.
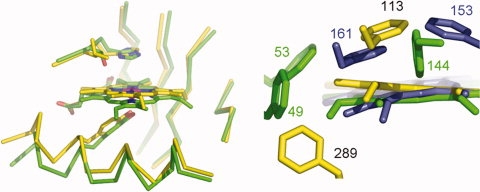
Heme displacement. (a) The heme in MAP_2744c (C, yellow) is displaced from its position in the cAOS structure (C, green). (b) Phe residues that impact heme placement. MAP_2744c, yellow, cAOS, green, catalase (HEC), blue.
Enzyme activity
Given the structural similarity to catalase, the catalase activity of MAP_2744c was measured. The enzyme catalyzes the dismutation of H2O2, but at a rate (23 ± 6 s–1) at least three orders of magnitude lower than that of mono-functional catalases.27 By contrast, the enzyme has significant peroxidase activity; an 2,2'-azino-bis(3-ethylbenzthiazoline-6-sulphonic acid) (ABTS) activation assay in the presence of t-butyl hydroperoxide gives a kcat/Km value ∼15,000M–1 s–1 (Table I). For comparison, the peroxidase activity of the catalase-peroxidase KatG from Mycobacterium partuberculosis in an equivalent assay (ABTS, t-butyl hydroperoxide) was reported to have a kcat/Km value of 7.4M–1s–1 and catalase activity of ∼6000 s–1.28 Thus, the catalase and peroxidase activities are “reversed” from those of KatG: MAP_2744c is a poor catalase, but a robust peroxidase.
Table I.
Peroxidase Activity of MAP_2744c ABTS Oxidation Assay
| Substrate | Km (M) | kcat (s–1) | kcat/Km (M–1s–1) |
|---|---|---|---|
| H2O2 | 30 (4) | 13 (1) | 433 |
| t-Butyl hydroperoxide | 22 (2) | 320 (10) | 14.5 × 103 |
| Cumene hydroperoxide | 3 (0.5) | 330 (14) | 110 × 103 |
The active sites of MAP_2744c, cAOS and catalases appear distinctly different is terms of both substrate access and heme chemistry/reactivity. cAOS catalyzes the transformation of 8R-hydroperoxy-eicosatetraenoic acid (HPETE) to an allene oxide, a reaction proposed to involve an epoxy allylic carbocation intermediate.17 Thus, the mechanism is analogous to that for members of the CYP74 P450 subfamily29 and by extension other catalase-related enzymes might exhibit allene oxide synthase, hydroperoxide lyase, or divinyl ether synthase activities, as is true for P450s.30 The prediction for catalytic biosynthetic diversity of catalase-related enzymes has been confirmed by the identification of the product of an AOS ortholog from Anabaena. The catalase-related domain of this bi-functional enzyme catalyzes the synthesis of a highly unusual bicyclobutane containing fatty acid from a hydroperoxy intermediate.19 As mentioned above, in general prokaryotes lack lipoxygenase genes and polyunsaturated fatty acids and thus are unable to generate fatty acid hydroperoxides. Nonetheless, a possible role for MAP-2744c as part of a broader function in the elimination of reactive oxygen species is deactivation of hydroperoxides generated by its hosts. Accordingly, we tested the ability of MAP_2744c to metabolize four selected fatty acid hydroperoxides. When the initial rate of ABTS oxidation was measured at 60 μM fatty acid hydroperoxide concentration using a conventional spectrophotometer, the 9S- and 13S-hydroperoxy derivatives of linoleic acid [hydroperoxy-octadecadienoic acid (HPODE)] gave turnover numbers of 71 ± 5 and 52 ± 3 s–1, respectively, and the 15S and 8R derivatives of arachidonic acid (HPETE) gave 35 ± 3 s–1, and 20 ± 4 s–1, respectively. However, it was apparent from the curvature of the 417 nm recordings that the initially observed rates using fatty acid hydroperoxides were not maintained due to rapid enzyme inactivation. Indeed, when the reaction was repeated using a stopped flow spectrophotometer with a range of concentrations of 9S-HPODE (4–96 μM), kcat computed within the first second of reaction as 529 ± 27 s–1, KM as 49 ± 5 μM, and the catalytic efficiency kcat/KM as 11 × 106M–1 s–1. We conclude that MAP-2744c exhibits strong peroxidase activity using fatty acid hydroperoxides although reaction is rapidly blunted by enzyme inactivation, at least under these in vitro conditions. Neutralizing amino acids (R74, R148) to tether the substrate carboxylate are positioned at the top of the large substrate cavity, and the large size of the heme access cavity is consistent with a bulky substrate. These structural features and the result of the enzyme assays support a role for MAP-2744c in mitigating challenge by a variety of reactive oxygen species.
A function as a scavenger of reactive oxygen species is consistent with the large and accessible active site cavity. In contrast, the product of the catalase-related heme domain of the Anabaena ortholog is a fatty acid derivative with a highly constrained bicyclobutane moiety.19 The Anabaena enzyme is unique among the catalase-related enzymes as sequence alignments suggest that the proximal heme ligand is a histidine rather that a Tyr. Although the heme ligand will impact the chemistry of the reaction catalyzed, the highly unfavorable bond geometry of the product is more likely induced by an active site that forces the substrate to adopt a constrained, but productive, intermediate conformer. Thus this prediction, along with the structures of cAOS and MAP_2744c enzymes, suggest that the substrate binding cavities of the catalase related enzymes vary substantially in shape and size. This diversity of active sites size and shape is consistent with what has been described for the P450 family of heme enzymes, for which significantly more structures are available.31
Other Tyr-coordinated heme proteins
Recently, the structure of a novel tyrosine-coordinated heme enzyme has been described.32 The protein is a hexamer in which each 76 amino acid monomer binds heme. On the same side of the heme as the proximal tyrosine is an Arg positioned similarly to the “tuning” Arg conserved in catalases24 and catalase-related enzymes. However, beyond this highly localized structural similarity in heme coordination, there is no significant structural homology between catalase and the hexameric tyrosine-coordinated heme protein. Furthermore, the much smaller polypeptide of the hexameric protein leaves a significant portion of the heme exposed and the propionic acid groups are fully solvent accessible. The protein has measurable peroxidase and catalase activities, but the turnover numbers are significantly lower than those for MAP_2744c.
Concluding remarks
Catalase-related sequences that code for enzymes ∼350 amino acids in length are found in a wide range of prokaryotes and in selected eukaryotes (Fig. 5). Those sequences from eukaryotes that have thus far been functionally characterized transform fatty acid hydroperoxides in biosynthetic pathways. These pathways are not endogenous to prokaryotes, which for the most part lack the lipoxygenase activities. The structure of the catalase-related peroxidase described herein establishes a prototype for prokaryotic catalase-related heme enzymes, as it represents a novel branch of this protein superfamily, equally distant from the eukaryotic cAOS and catalase itself.
Figure 5.
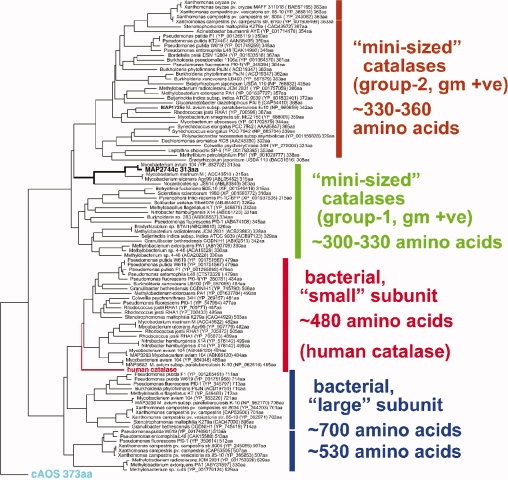
Phylogenetic tree of MAP-2744c and related proteins. A TBLASTN database search was conducted using the peptide sequence of MAP-2744c as bait. The amino acid sequences of the proteins identified were entered in the MegAlign program of DNAStar which compiled the phylogenetic tree. Human catalase and cAOS were not found in the TBLASTN search and were added prior to the alignment. The two lower groups correspond to the catalases with “small” and “large” subunits as deduced by Klotz and Loewen.12 On top are two new groups comprising mini-sized catalases with polypeptides of 330–360 and 300–330 amino acids. MAP-2744c segregates with the 300–330 amino acid subgroup from gram positive (+ve) bacteria.
The MAP_2744c structure indicates that, while the heme-environments of the catalase-related enzymes are highly conserved, key amino acid differences may significantly effect prosthetic group conformation and reactivity. This is precisely the type of structural variability one would expect given that catalase-related enzymes are, as suggested, as diverse in the reactions they catalyze as members of the P450 superfamily.
Materials and Methods
Cloning, expression, and purification of MAP
MAP_2744c was cloned by PCR from genomic DNA. Forward and reverse primers in PCR reaction contained NdeI and EcoRI restriction sites, respectively, and an N-terminal (His)6-tag was engineered. The sequence of the forward primer was 5′-CATATGCATCACCATCACCATCACAGCGGTGGATTAACTCCCGACCAG-3′ and that of the reverse primer 5′-GAATTCTCACACCGGCGGGGCTTGCGCGCC-3′. The correct PCR product was subsequently cut with NdeI and EcoRI restriction enzymes and subcloned into the same sites of the expression vector pET17b. The DNA sequencing confirmed the identity to the published sequence in GenBank. Expression in Escherichia coli BL21 (DE3) cells (Novagen) was accomplished with methods described previously by Hoffman et al.33 and Boutaud and Brash16 The frozen pellet was resuspended in BugBuster® Protein Extraction Reagent (Novagen) in the presence of 1 mM phenylmethylsulfonyl fluoride, 1 μg/mL Benzonase and protease inhibitor mix. The 16,000g supernatant was loaded on a nickel-NTA column (0.5 mL bed volume, Qiagen) equilibrated with 50 mM potassium phosphate buffer, pH 7.2, 500 mM NaCl at 0.5 mL/min. The column was then washed with the equilibration buffer and the nonspecific bound proteins were eluted with 50 mM potassium phosphate buffer, pH 7.2, 1M NaCl, 70 mM glycine. A final wash was performed by 50 mM potassium phosphate buffer, pH 7.2, 500 mM NaCl, 20 mM imidazole. The His-tagged protein was then eluted with 50 mM potassium phosphate buffer, pH 7.2, 500 mM NaCl, 250 mM imidazole. Fractions containing protein, as judged by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS–PAGE), were pooled and dialyzed overnight against 50 mM Tris buffer, pH 7.5, 150 mM NaCl, 20% glycerol and aliquoted and frozen in –80°C for later use.
Protein purification for crystallization
Supernatant prepared as descried above was applied to a TALON Co2+ affinity (Clontech) pre-equilibrated with 50 mM potassium phosphate pH 7.2, 500 mM NaCl, 1% Triton and 20% glycerol. Protein was eluted with 20 mM imidazole. Further purification was accomplished with gel filtration chromatography on a Superdex 200 column (GE Healthcare, Uppsala) buffered with 10 mM Tris–HCl (8.0) to yield pure protein that displayed the characteristic ratio of A406/A280 of ∼3.
Crystallization
Crystals of the enzyme were obtained at 11°C using the hanging drop vapor-diffusion method by mixing equal volumes of protein at 10.5 mg/mL and the reservoir solution (1.5M 1,6-hexanediol, 100 mM sodium citrate pH 6.9, 13–15% polyethylene glycol (PEG) 20,000, 2.5–6.5% glycerol). Crystals of both crystal forms (P212121 and I212121) grew under the same conditions and often in the same drop.
Data collection, structure solution, and refinement
The structure of the MAP_2744c protein was solved with Fe-MAD data. Data at 100 K were collected at the protein crystallography beamline at the Center for Advanced Microstructures and Devices at Louisiana State University, with a Mar charge-coupled device detector. The images were processed and scaled using DENZO and SCALEPACK.34 Data collection and data processing statistics are summarized in Table II.
Table II.
Data Collection Statistics
| P212121 | I212121 | Peak (λ1) | Inflection (λ2) | Remote (λ3) | |
|---|---|---|---|---|---|
| Wavelength (Å) | 1.38079 | 1.5896 | 1.73838 | 1.74082 | 1.5896 |
| Resolution (Å) | 1.8 | 2.6 | 2.5 | 2.5 | 2.5 |
| Temperature (K) | 113 | 113 | 113 | 113 | |
| Space group | P212121 | I212121 | P212121 | ||
| Cell dimensions | |||||
| a (Å) | 68.72 | 69.60 | 69.18 | ||
| b (Å) | 81.62 | 92.68 | 81.95 | ||
| c (Å) | 163.11 | 165.50 | 164.09 | ||
| Molecules per asymmetric unit | 2 | 1 | 2 | ||
| No. of unique reflections | 75 154 | 15 361 | 61 237 | 61 304 | 61 970 |
| Rsyma,b (%) | 5.7 (47.5) | 8.8 (45.2) | 8.6 (39.0) | 9.3 (42.1) | 8.4 (34.6) |
| Completeness (%) | 87.6 (86.5) | 91.1 (67.4) | 98.8 (88.9) | 97.6 (78.2) | 99.8 (98.5) |
| Redundancies | 2.2 (2.1) | 4.3 (3.5) | 4.7 (2.8) | 5.6 (2.6) | 7.1 (5.1) |
| I/σ(I) | 19.5 (2.0) | 13.9 (2.0) | 15.9 (2.3) | 15.8 (2.0) | 23.0 (4.3) |
| Phasing statistics | |||||
| Resolution range (Å) | 30–2.5 | ||||
| Number of sites | 2 | ||||
| Phasing powerc acentric/centric | 1.30/1.45 | ||||
| Rcd centric | 0.64 | ||||
| Mean FOM after MAD phasing | 0.50 | ||||
| Refinement statistics | |||||
| Resolution range (Å) | 36.5–1.8 | 43.2–2.6 | |||
| No. of reflections used in refinement | 71 376 | 14 446 | |||
| σ cutoff used in refinement | None | None | |||
| R/Rfreee (%) | 19.74/21.01 | 20.15/25.70 | |||
| Number of refined atoms | |||||
| Protein | 4519 | 2273 | |||
| Heterogen atoms | 165 | 60 | |||
| Water | 367 | 122 | |||
| Average B-factors (Å2) | |||||
| Protein | 30.1 | 42.4 | |||
| Heme | 25.9 | 41.1 | |||
| Heterogen atoms | 55.0 | 44.2 | |||
| Water | 35.2 | 33.4 | |||
| R.M.S. deviations | |||||
| Bonds (Å) | 0.005 | 0.006 | |||
| Angles (°) | 1.2 | 1.2 | |||
| Ramachandran plot (%) | |||||
| Favored | 93.6 | 90.4 | |||
| Allowed | 5.8 | 8.8 | |||
| Generous | 0.2 | 0.4 | |||
| Disallowed | 0.4 | 0.4 | |||
Values in parentheses are for the highest-resolution shell.
Rsym = ∑|Ii – <Ii>|/∑Ii, where Ii is the intensity of the ith observation and <Ii> is the mean intensity of the reflection.
Phasing power = ∑<Fh>/E, where <Fh> is the root-mean-square structure factors and E is the residual lack of closure error.
Rc = ∑||FPH ± FP| – FHcalc|/∑FPH ± FP, where FPH and FP are the structure-factors amplitudes for the data collection on the iron absorption edge.
R = ∑||Fo| – |Fc||/∑|Fo|, where Fo and Fc are the observed and calculated structure factors amplitudes. Rfree is calculated using 6.9% and 5.9% of reflections omitted from the refinement for the P212121 and I212121 structures, respectively. The Rfree set from the P212121 crystal form was used for the refinement of the I212121 data set.
The 4 Å resolution anomalous difference Patterson revealed clear peaks attributable to the heme iron and these positions were refined with 2.5 Å data in CNS.35 The hand of the Fe positions was assigned by visual inspection of maps calculated with the refined and flipped Fe coordinates. Density modification was carried out in CNS. The structure was refined using the maximum likelihood refinement in CNS.35 No sigma cutoff was applied to the data. A bulk solvent correction was applied. The program O,36 was used to build models throughout the refinement. The final model consists of residues 4-306 and 4-305 for both protein monomers, two heme cofactors, six 1,6-hexanediol molecules, four phosphate ions, one glycerol molecule, and 367 water molecules. Alternate conformations were built for M83, V247, and D277 (monomer A) and D43, R169, and D277 (monomer B). The P212121 model served as a search model to solve the I212121 structure by the molecular replacement method. The refinement of the I212121 was performed similarly as in the case of the P212121 structure. The final model consists of protein residues 4-304, two glycerol molecules, one phosphate ion and 122 water molecules. Refinement statistics are listed in Table II.
The protein molecules display good stereochemistry in both crystal forms (Table II) with the exception of L165 which was found in the disallowed region. The corresponding residues in cAOS, as well as in all known catalase structures, are also in an energetically disallowed region.
Data deposition
The atomic coordinates have been deposited to the Protein Data Bank with the accession codes 3E4W (P212121) and 3E4Y (I212121).
Peroxidase activity assay
MAP-2744c peroxidase activity was assayed at room temperature by following the increase in absorbance at 417 nm due to oxidation of ABTS (1 mM) in 50 mM potassium phosphate buffer, pH 7, using a Lambda 35 UV/VIS spectrophotometer (PerkinElmer). The reaction was initiated by addition of MAP_2744c enzyme (50 nm) to 1 cm cuvette containing different concentrations of substrate [H2O2 (1–50 mM), t-butyl hydroperoxide (1–100 mM), and cumene hydroperoxide (0.5–20 mM)]. Due to the rapid inactivation of enzyme upon mixing enzyme with the fatty acid hydroperoxides, the activity measurement with fatty acid hydroperoxides was only at one concentration (20 μM). The initial rate was expressed as nanomoles of ABTS oxidized per nanomole of heme per second. Enzyme assays were performed in triplicate and the standard error was calculated from the fit to the Michaelis-Menten equation.
The peroxidase activity assay was also conducted using an SX-18MV stopped flow instrument (Applied Photophysics, Leatherhead, UK) equipped with a 20 μL observation cell (10-mm path length) or 5 μL (5-mm path length) cell. The ABTS concentration was 1 mM, MAP-2744c 0.043 μM, 9S-HPODE co-substrate 4–96 μM in 50 mM potassium phosphate buffer, pH 7.0. It was found that MAP-2744c tended to lose activity upon dilution into the mixing syringe and that this could be prevented by including 1 mM CHAPS detergent in the reaction buffer. Four individual stopped-flow experiments were recorded at each concentration of 9S-HPODE and averaged before subsequent data analyses and fitting was done with the manufacturer's software.
Catalase activity assay
MAP_2744c catalase activity was assayed at room temperature by measuring the decrease at 240 nm due to the consumption of H2O2 in 50 mM potassium phosphate buffer, pH 7.0, using a Lambda 35 UV/VIS spectrophotometer (PerkinElmer). Similarly, the initial rate was expressed as nanomoles of H2O2 decomposed per nanomole of heme per second.
Acknowledgments
This work was supported in part by the National Science Foundation (MCB-0818387) and the Louisiana Governor's Biotechnology Initiative (MEN) and the National Institutes of Health (GM 53638; ARB). Diffraction data used in this publication were collected at the Gulf Coast Protein Crystallography (GCPCC) Beamline at the Center for Advanced Microstructures and Devices (CAMD). This beamline is supported by the National Science Foundation grant DBI-9871464 with co-funding from the National Institute for General Medical Sciences (NIGMS). We thank Henry Bellamy and David Neau for assistance at the Center for Advanced Microstructures and Devices PX beamline, and Vivek Kapur (University of Minnesota) for the MAP_2744c DNA.
Glossary
Abbreviations:
- ABTS
2,2'-azino-bis(3-ethylbenzthiazoline-6-sulphonic acid)
- cAOS
catalase-related allene oxide synthase
- HEC
human erythrocyte catalase
- HPETE
hydroperoxy-eicosatetraenoic acid
- HPODE
hydroperoxy-octadecadienoic acid.
References
- 1.Murthy MR, Reid TJ, 3rd, Sicignano A, Tanaka N, Rossmann MG. Structure of beef liver catalase. J Mol Biol. 1981;152:465–499. doi: 10.1016/0022-2836(81)90254-0. [DOI] [PubMed] [Google Scholar]
- 2.Reid TJd, Murthy MR, Sicignano A, Tanaka N, Musick WD, Rossmann MG. Structure and heme environment of beef liver catalase at 2.5 Å resolution. Proc Natl Acad Sci U S A. 1981;78:4767–4771. doi: 10.1073/pnas.78.8.4767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vainshtein BK, Melik-Adamyan WR, Barynin VV, Vagin AA, Grebenko AI, Borisov VV, Bartels KS, Fita I, Rossmann MG. Three-dimensional structure of catalase from Penicillium vitale at 2.0 Å resolution. J Mol Biol. 1986;188:49–61. doi: 10.1016/0022-2836(86)90479-1. [DOI] [PubMed] [Google Scholar]
- 4.Murshudov GN, Melik-Adamyan WR, Grebenko AI, Barynin VV, Vagin AA, Vainshtein BK, Dauter Z, Wilson KS. Three-dimensional structure of catalase from Micrococcus lysodeikticus at 1.5 Å resolution. FEBS Lett. 1992;312:127–131. doi: 10.1016/0014-5793(92)80919-8. [DOI] [PubMed] [Google Scholar]
- 5.Bravo J, Verdaguer N, Tormo J, Betzel C, Switala J, Loewen PC, Fita I. Crystal structure of catalase HPII from Escherichia coli. Structure. 1995;3:491–502. doi: 10.1016/s0969-2126(01)00182-4. [DOI] [PubMed] [Google Scholar]
- 6.Gouet P, Jouve HM, Dideberg O. Crystal structure of Proteus mirabilis PR catalase with and without bound NADPH. J Mol Biol. 1995;249:933–954. doi: 10.1006/jmbi.1995.0350. [DOI] [PubMed] [Google Scholar]
- 7.Bravo J, Mate MJ, Schneider T, Switala J, Wilson K, Loewen PC, Fita I. Structure of catalase HPII from Escherichia coli at 1.9 Å resolution. Proteins. 1999;34:155–166. doi: 10.1002/(sici)1097-0134(19990201)34:2<155::aid-prot1>3.0.co;2-p. [DOI] [PubMed] [Google Scholar]
- 8.Ko TP, Safo MK, Musayev FN, Di Salvo ML, Wang C, Wu SH, Abraham DJ. Structure of human erythrocyte catalase. Acta Crystallogr D Biol Crystallogr. 2000;56(Pt 2):241–245. doi: 10.1107/s0907444999015930. [DOI] [PubMed] [Google Scholar]
- 9.Carpena X, Soriano M, Klotz MG, Duckworth HW, Donald LJ, Melik-Adamyan W, Fita I, Loewen PC. Structure of the Clade 1 catalase, CatF of Pseudomonas syringae, at 1.8 Å resolution. Proteins. 2003;50:423–436. doi: 10.1002/prot.10284. [DOI] [PubMed] [Google Scholar]
- 10.Loewen PC, Carpena X, Rovira C, Ivancich A, Perez-Luque R, Haas R, Odenbreit S, Nicholls P, Fita I. Structure of Helicobacter pylori catalase, with and without formic acid bound, at 1.6 Å resolution. Biochemistry. 2004;43:3089–3103. doi: 10.1021/bi035663i. [DOI] [PubMed] [Google Scholar]
- 11.Putnam CD, Arvai AS, Bourne Y, Tainer JA. Active and inhibited human catalase structures: ligand and NADPH binding and catalytic mechanism. J Mol Biol. 2000;296:295–309. doi: 10.1006/jmbi.1999.3458. [DOI] [PubMed] [Google Scholar]
- 12.Klotz MG, Loewen PC. The molecular evolution of catalatic hydroperoxidases: evidence for multiple lateral transfer of genes between prokaryota and from bacteria into Eukaryota. Mol Biol Evol. 2003;20:1098–1112. doi: 10.1093/molbev/msg129. [DOI] [PubMed] [Google Scholar]
- 13.Zamocky M, Koller F. Understanding the structure and function of catalases: clues from molecular evolution and in vitro mutagenesis. Prog Biophys Mol Biol. 1999;72:19–66. doi: 10.1016/s0079-6107(98)00058-3. [DOI] [PubMed] [Google Scholar]
- 14.Klotz MG, Klassen GR, Loewen PC. Phylogenetic relationships among prokaryotic and eukaryotic catalases. Mol Biol Evol. 1997;14:951–958. doi: 10.1093/oxfordjournals.molbev.a025838. [DOI] [PubMed] [Google Scholar]
- 15.Koljak R, Boutaud O, Shieh BH, Samel N, Brash AR. Identification of a naturally occurring peroxidase-lipoxygenase fusion protein. Science. 1997;277:1994–1996. doi: 10.1126/science.277.5334.1994. [DOI] [PubMed] [Google Scholar]
- 16.Boutaud O, Brash AR. Purification and catalytic activities of the two domains of the allene oxide synthase-lipoxygenase fusion protein of the coral Plexaura homomalla [In Process Citation] J Biol Chem. 1999;274:33764–33770. doi: 10.1074/jbc.274.47.33764. [DOI] [PubMed] [Google Scholar]
- 17.Oldham ML, Brash AR, Newcomer ME. The structure of coral allene oxide synthase reveals a catalase adapted for metabolism of a fatty acid hydroperoxide. Proc Natl Acad Sci U S A. 2005;102:297–302. doi: 10.1073/pnas.0406352102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tosha T, Uchida T, Brash AR, Kitagawa T. On the relationship of coral allene oxide synthase to catalase. A single active site mutation that induces catalase activity in coral allene oxide synthase. J Biol Chem. 2006;281:12610–12617. doi: 10.1074/jbc.M600061200. [DOI] [PubMed] [Google Scholar]
- 19.Schneider C, Niisuke K, Boeglin WE, Voehler M, Stec DF, Porter NA, Brash AR. Enzymatic synthesis of a bicyclobutane fatty acid by a hemoprotein lipoxygenase fusion protein from the cyanobacterium Anabaena PCC 7120. Proc Natl Acad Sci U S A. 2007;104:18941–18945. doi: 10.1073/pnas.0707148104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lang I, Gobel C, Porzel A, Heilmann I, Feussner I. A lipoxygenase with linoleate diol synthase activity from Nostoc sp. PCC 7120. Biochem J. 2008;410:347–357. doi: 10.1042/BJ20071277. [DOI] [PubMed] [Google Scholar]
- 21.Chacon O, Bermudez LE, Barletta RG. Johne's disease, inflammatory bowel disease, and Mycobacterium paratuberculosis. Annu Rev Microbiol. 2004;58:329–363. doi: 10.1146/annurev.micro.58.030603.123726. [DOI] [PubMed] [Google Scholar]
- 22.Li L, Bannantine JP, Zhang Q, Amonsin A, May BJ, Alt D, Banerji N, Kanjilal S, Kapur V. The complete genome sequence of Mycobacterium avium subspecies paratuberculosis. Proc Natl Acad Sci U S A. 2005;102:12344–12349. doi: 10.1073/pnas.0505662102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Putnam CD, Arvai AS, Bourne Y, Tainer JA. Active and inhibited human catalase structures: ligand and NADPH binding and catalytic mechanism. J Mol Biol. 2000;296:295–309. doi: 10.1006/jmbi.1999.3458. [DOI] [PubMed] [Google Scholar]
- 24.Green MT. The structure and spin coupling of catalase compound I: a study of noncovalent effects. J Am Chem Soc. 2001;123:9218–9219. doi: 10.1021/ja010105h. [DOI] [PubMed] [Google Scholar]
- 25.Jentzen W, Ma JG, Shelnutt JA. Conservation of the conformation of the porphyrin macrocycle in hemoproteins. Biophys J. 1998;74:753–763. doi: 10.1016/S0006-3495(98)74000-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shelnutt JA, Song XZ, Ma JG, Jia SL, Jentzen W, Medforth CJ. Nonplanar porphyrins and their significance in proteins. Chem Soc Rev. 1998;27:31–41. [Google Scholar]
- 27.Switala J, Loewen PC. Diversity of properties among catalases. Arch Biochem Biophys. 2002;401:145–154. doi: 10.1016/S0003-9861(02)00049-8. [DOI] [PubMed] [Google Scholar]
- 28.Ghiladi RA, Knudsen GM, Medzihradszky KF, Ortiz de Montellano PR. The Met-Tyr-Trp cross-link in Mycobacterium tuberculosis catalase-peroxidase (KatG): autocatalytic formation and effect on enzyme catalysis and spectroscopic properties. J Biol Chem. 2005;280:22651–22663. doi: 10.1074/jbc.M502486200. [DOI] [PubMed] [Google Scholar]
- 29.Tijet N, Schneider C, Muller BL, Brash AR. Biogenesis of volatile aldehydes from fatty acid hydroperoxides: molecular cloning of a hydroperoxide lyase (CYP74C) with specificity for both the 9-and 13-hydroperoxides of linoleic and linolenic acids. Arch Biochem Biophys. 2001;386:281–289. doi: 10.1006/abbi.2000.2218. [DOI] [PubMed] [Google Scholar]
- 30.Feussner I, Wasternack C. The lipoxygenase pathway. Annu Rev Plant Biol. 2002;53:275–297. doi: 10.1146/annurev.arplant.53.100301.135248. [DOI] [PubMed] [Google Scholar]
- 31.Otyepka M, Skopalik J, Anzenbacherova E, Anzenbacher P. What common structural features and variations of mammalian P450s are known to date? Biochim Biophys Acta. 2007;1770:376–389. doi: 10.1016/j.bbagen.2006.09.013. [DOI] [PubMed] [Google Scholar]
- 32.Jeoung JH, Pippig DA, Martins BM, Wagener N, Dobbek H. HTHP: a novel class of hexameric, tyrosine-coordinated heme proteins. J Mol Biol. 2007;368:1122–1131. doi: 10.1016/j.jmb.2007.02.079. [DOI] [PubMed] [Google Scholar]
- 33.Hoffman BJ, Broadwater JA, Johnson P, Harper J, Fox BG, Kenealy WR. Lactose fed-batch overexpression of recombinant metalloproteins in Escherichia coli BL21 (DE3): process control yielding high levels of metal-incorporated, soluble protein. Protein Expr Purif. 1995;6:646–654. doi: 10.1006/prep.1995.1085. [DOI] [PubMed] [Google Scholar]
- 34.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. In: Carter CW Jr, Sweet RM, editors. Vol. 276. New York: Academic Press; 1997. pp. 307–326. Methods in enzymology. [DOI] [PubMed] [Google Scholar]
- 35.Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, Read RJ, Rice LM, Simonson T, Warren GL. Crystallography & NMR System: a new software suite for macromolecular structure determination. Acta Crystallogr. Sect. D. 1998;54(Pt 5):905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 36.Jones TA, Zou JY, Cowan SW, Kjeldgaard M. Improved methods for binding protein models in electron density maps and the location of errors in these models. Acta Crystallogr A. 1991;47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]


