Abstract
A lingering issue in the area of protein engineering is the optimal design of β motifs. In this regard, the framework provided by intestinal fatty acid binding protein (IFABP) was successfully chosen to explore the consequences on structure and function of the redesign of natural motifs. A truncated form of IFABP (Δ98Δ) served to illustrate the nonintuitive notion that the integrity of the β-barrel can indeed be compromised with no effect on the ability to attain a native-like fold. This is most likely the outcome of the key role played by the preservation of essential core residues. In the search for the minimal structural determinants of this fold, Δ98Δ offered room for further intervention. A dissection of this protein leads to a new abridged variant, Δ78Δ, containing 60% of the amino acids of IFABP. Spectroscopic analyses indicate that Δ78Δ retains substantial β-sheet content and preserves tertiary interactions, displaying cooperative unfolding and binding activity. Most strikingly, this construct adopts a remarkably stable dimeric structure in solution. This phenomenon takes advantage of the inherent structural plasticity of this motif, likely profitting from edge-to-edge interactions between β-sheets, whereas avoiding the most commonly occurring outcome represented by aggregation.
Keywords: abridged variant, β-barrel, IFABP, dimerization, protein truncation
Introduction
The β-sheet proteins are produced and folded up successfully in the cell, with only a few isolated cases of aggregation or insolubility. The understanding of the stability and folding mechanism of this class of proteins is becoming increasingly relevant because many “conformational diseases” with severe consequences on animal and human health are based on the generation of β-sheet structures.1
Although fatty acid binding proteins (FABP) display variable sequence identity, they share a common 3D structure consisting of a β-barrel composed of two five-stranded β-sheets (βA–βE and βF–βJ) arranged in a nearly orthogonal orientation (Fig. 1). Therefore, FABPs arise as attractive models for the study of β-sheet proteins.2–7 This structure differs from most globular proteins in that its interior is occupied by a large solvent-filled cavity that binds nonpolar ligands, while the hydrophobic core is small and somewhat displaced from the protein geometrical centre. All β-strands are connected by β-turns, with the exception of strands βA and βB, where an intervening helix-turn-helix motif appears.8
Figure 1.
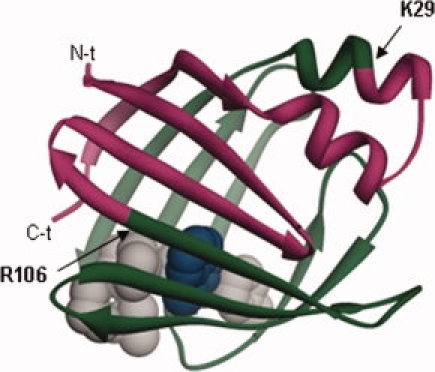
Ribbon structure of IFABP (PDB 2IFB) where the Δ78Δ construct is indicated. The excised N- and C-termini stretches are shown in pink, Δ78Δ is painted in green, and residues belonging to the hydrophobic core are depicted in grey with their side chains in CPK representation, except for W82 that is indicated in blue.
Intestinal FABP (IFABP) is an intracellular protein of 15 kDa that binds exclusively long-chain fatty acids.9 It is abundantly expressed in small intestine enterocytes, and together with liver FABP, represents a high proportion of the total cellular protein. Extensive work has been done to elucidate the ligand-binding mechanism of this protein. The structures of apo- and holo-forms of IFABP solved by NMR10,11 and X-ray diffraction12,13 reveal a disordered region in the absence of the fatty acid, which comprises the distal half of αII, the αII/βB linker, and the βC-βD and βE-βF turns.11 These findings agree with previous evidence derived from partial proteolysis experiments. This technique uncovers the differential exposure of proteolytic sites, thus providing information about the location of ligand-binding sites or conformational changes in proteins.14,15 Interestingly, the proteolytic pattern of IFABP with clostripain (Arg C) displays a high level of protection when bound to oleic acid, suggesting that smaller fragments preserve the ability to bind fatty acids.16 In this regard, a main proteolytic fragment named Δ98Δ was recently cloned, expressed, and structurally characterized in our laboratory.6,7 Although lacking one quarter of the sequence of the parent protein (a stretch of 98 amino acids corresponding to fragment 29–126 of IFABP), Δ98Δ is a stable and functional form of IFABP. Interestingly, this truncation leads to a loss of residues involved in the closure of the β-barrel. Despite this fact, cumulative evidence, including circular dichroism (CD), ultraviolet (UV) absorption, and intrinsic fluorescence, indicates that this fragment retains substantial β-sheet content and tertiary interactions. The reasons for this remarkable behavior lie in the ancillary role played by the segments deleted and in the conservation of the critical residues of the hydrophobic core.7 These amino acids have been implicated in the initial nucleation event leading to the folded state.17
Further investigations on the stabilizing role played by the binding event led us to challenge the Δ98Δ variant in a second round of proteolysis. Here again, the holo-form gives rise to a main fragment highly resistant to further degradation, which was named Δ78Δ. This outcome led us to investigate this new abridged variant of IFABP. The Δ78Δ was cloned, purified, and biophysically characterized, which revealed a well-folded and functional protein. Remarkably, this protein adopts a very stable dimeric structure in solution.
Results
Production and purification of Δ78Δ
Partial digestion of holo-Δ98Δ with clostripain generates a main fragment of about 8 kDa, which is resistant to further proteolysis. The mass of this fragment named Δ78Δ was indeed 8807.4 Da by ESI-MS, corresponding to fragment 29–106 of IFABP and an additional M residue at the N-terminus. When compared with IFABP, this new abridged variant contains only 60% of the amino acids of the full-length protein (Fig. 1). Specifically, it lacks the whole α-helical subdomain plus βA, βI, and βJ. The Δ78Δ was cloned, overexpressed in Escherichia coli, and purified from inclusion bodies (Supporting Information Fig. S1).
Aggregation state
The hydrodynamic properties of Δ78Δ were studied by size exclusion chromatography (SEC) and compared with those of intact IFABP and Δ98Δ (Fig. 2). As the predicted Stokes radius (Rs) for a monomer of Δ78Δ is 16 Å, it should exhibit a higher elution volume than IFABP and Δ98Δ. Nevertheless, Δ78Δ displays the lowest elution volume of the three proteins assayed. Unlike Δ98Δ that represents an expanded monomeric species,6 the “anomalous” behavior shown by Δ78Δ suggests that it adopts an oligomeric form. The Rs of Δ78Δ (23.6 ± 0.3 Å) is about 15% larger than that predicted for a globular protein of a molecular weight corresponding to that of a hypothetical dimer: 20.5 Å (see Materials and Methods). To get further insight into the nature of this oligomeric state, chemical cross-linking experiments were performed using disuccinimidyl suberate (DSS), an amine-reactive homo-bifunctional reagent. Remarkably, unlike IFABP, the fragment shows a clear propensity to exist as a dimer under identical experimental conditions and over a wide range of cross-linker concentrations (Fig. 3). This behavior does not change in the presence of oleic acid. Moreover, the progressive dilution of Δ78Δ (from 0.25 to 0.02 mg/mL) exerts no effect on the elution profile, as assessed by SEC (Superdex-75, data not shown). These results reveal the existence of tight interactions between protomers.
Figure 2.
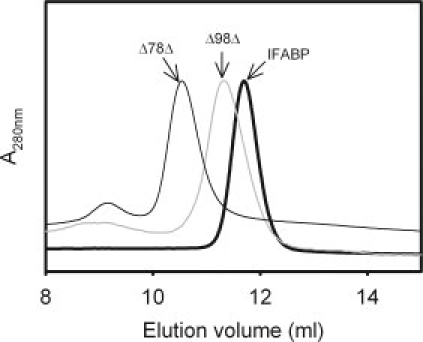
SEC of Δ78Δ compared with IFABP and Δ98Δ. The Δ78Δ, Δ98Δ, and IFABP were sampled separately onto a Superdex-75 column equilibrated and eluted with buffer A added with 100 mM NaCl.
Figure 3.
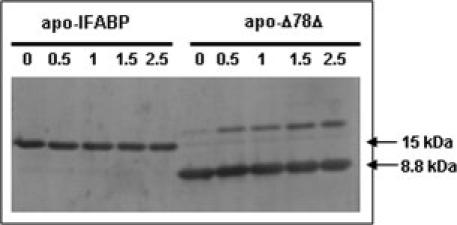
SDS-PAGE separation of covalently linked products after reaction of Δ78Δ or IFABP with DSS. Proteins were assayed in their apo- (ligand free) forms. No difference was observed in the presence of oleic acid (holo-forms, results not shown). The number on each lane indicates the DSS concentration used (in μg/mL).
Confirmatory evidence for these results came from light scattering measurements. This technique uncovers a monodisperse population corresponding to a species of molecular weight 19,160 ± 1916 Da. Taken together, these data are consistent with Δ78Δ adopting a dimeric, but slightly expanded/asymmetric conformational state. The fact that Δ78Δ behaves as a stable dimer in solution emerges as the most distinctive feature of this variant.
Fourth-derivative UV spectra
The fourth-derivative UV absorption spectrum provides insights into the protein conformation, as has been proven for IFABP.4,16 The spectra recorded for both IFABP and Δ78Δ show a similar fine structure, suggesting a preserved environment for the aromatic residues (Fig. 4). One should be reminded that each protomer of Δ78Δ conserves 1 of 4 Y, 5 of 8 F, and 1 of 2 W residues. Characteristically, a decrease in the intensity of the signal λ4 is observed for Δ78Δ when compared with the parent protein,6,18 a fact consistent with the absence of W6 in the construct. In addition, apo- and holo-IFABP spectra are almost superimposable, in agreement with previously published data.6,19 In this regard, there are minor spectral differences between the apo- and holo-forms of Δ78Δ. Specifically, the λ3 and λ4 peaks are slightly blue shifted in the holo-form, suggesting a more hydrophobic environment for the single remaining W residue (W82 in IFABP), which is the main chromophore contributing to the spectrum in that region.
Figure 4.
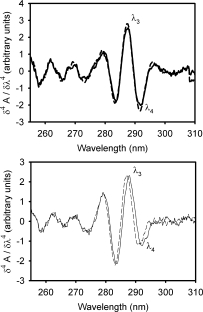
Fourth-derivative absorption spectra of (A) apo-  and holo-
and holo-  IFABP; and (B) apo- (—) and holo- (——) Δ78Δ.
IFABP; and (B) apo- (—) and holo- (——) Δ78Δ.
Intrinsic fluorescence measurements
The centre of mass of the spectrum20 of apo-Δ78Δ indicates a W residue located in a hydrophobic environment (336.3 nm). This signal is blue shifted with respect to apo-IFABP [338.1 nm, Fig. 5(A)]. In addition, binding of oleic acid to Δ78Δ causes an increase in the intensity and an extra 1.2 nm blue shift of the centre of mass with respect to the apo-form [inset in Fig. 5(A)].
Figure 5.
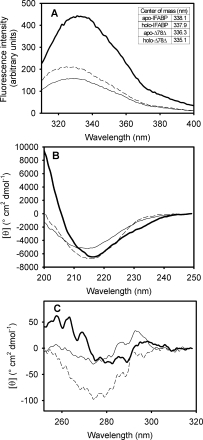
(A) Intrinsic fluorescence emission spectra of IFABP and Δ78Δ in their apo- and holo-forms. The wavelengths of the centre of mass are shown as an inset. The far-UV CD (B) and near-UV CD (C) spectra of IFABP and Δ78Δ are shown: apo-IFABP  , apo-Δ78Δ (—), and holo-Δ78Δ (——). The spectra of holo-IFABP are not displayed because they superimpose exactly over those of the apo-form.
, apo-Δ78Δ (—), and holo-Δ78Δ (——). The spectra of holo-IFABP are not displayed because they superimpose exactly over those of the apo-form.
Considering the fact that Δ78Δ is a stable dimer, we should take into account that, in principle, each protomer of the complex is contributing to the fluorescence spectrum. For this reason, the comparison of the emission spectra of IFABP and Δ78Δ is presented on a protomer basis. The maximum fluorescence intensity of apo-Δ78Δ is ∼36 % of that observed for IFABP. On the addition of oleic acid, the intensity increases ∼10% with respect to the apo-form. The reduced fluorescence emission of Δ78Δ can to some extent be justified by the absence of W6, which accounts for 35% of the fluorescence intensity of the wild-type protein, as previously described for the W6Y-IFABP mutant by Klimtchuk et al.21 In addition, the loss of W6 by itself causes only a 1-nm red shift in the maximum of the spectrum, as demonstrated for this same mutant by Dalessio et al.22 Nevertheless, to fully explain this observation, small perturbations in the closely packed hydrophobic environment around W82 appears to be a likely possibility. In summary, these results suggest a transition to a slightly more hydrophobic environment for W82 in Δ78Δ.
Circular dichroism
The far-UV CD spectra of apo- and holo-Δ78Δ show minima at ∼216 nm, a characteristic feature of β-sheet proteins. As is well known, IFABP shows similar spectra, regardless of the presence of the ligand oleic acid [Figs. 5(B); Ref. 6]. Nevertheless, the absolute ellipticity value of apo-Δ78Δ is about 80% that of apo-IFABP, but—at variance with the full-length protein—when oleic acid is bound to Δ78Δ, a 30% increase in the absolute value of the signal at 216 nm is observed. Remarkably, this enhanced signal reaches a value very close to that observed for IFABP. This fact is accompanied by a narrowing of the spectrum, suggesting a ligand-induced ordering effect in the abridged variant.
Figure 6.
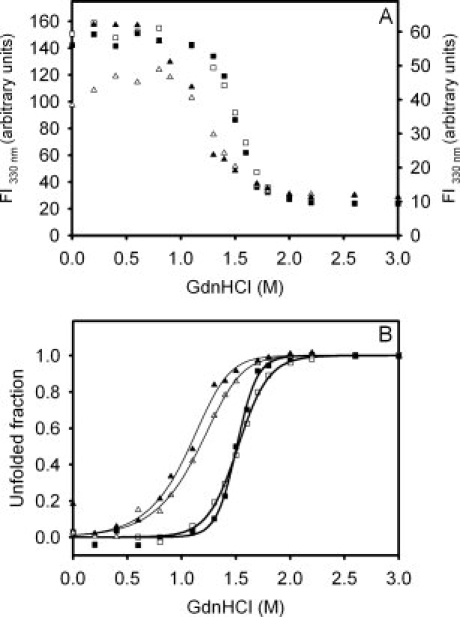
Equilibrium unfolding transitions of Δ78Δ and IFABP monitored by the change in fluorescence intensity: apo-IFABP (□), holo-IFABP (▪), apo-Δ78Δ (Δ), and holo-Δ78Δ (▴). (A) The fluorescence intensity at 330 nm, where the left and right ordinate axes correspond to IFABP and Δ78Δ, respectively. (B) Molar fraction of unfolded form derived from the GdnHCl-induced equilibrium unfolding transitions. The protein concentrations used were 39 and 24 μM for apo- and holo-Δ78Δ, respectively, and in the latter, a 4:1 oleic acid:protein molar ratio was maintained.
Two features of the spectrum of IFABP are conspicuously absent in that of Δ78Δ: (i) the strong positive band occurring below 200 nm, and (ii) the negative shoulder located at around 230 nm. Most likely, the former trait (similarly to the reduced intensity at 216 nm) can be attributed to the absence of the helical domain present in IFABP, whose contribution to ellipticity is almost twice that of a β-sheet on a molar basis.23 In fact, it is possible that the construct might exhibit a substantially high β-sheet content, despite the absence of strands βA, βI, and βJ. It is noteworthy that apo-Δ98Δ, lacking only βA and half of βJ, shows a 65% of the absolute ellipticity signal at 216 nm of apo-IFABP, whereas apo-Δ78Δ, putatively lacking a larger portion of the β-structure, presents a stronger signal than that of Δ98Δ.6 This apparent paradox arises from considering the structure of a protomer in isolation, that is, the dimeric nature of Δ78Δ where interchain interaction is possible could likely make its own contribution to the far-UV spectrum as well. On the other hand, although secondary structure elements are the main contributors to far-UV CD, in many instances, aromatic residues add signal to this region as well.24 For IFABP, it has been described that W6 contributes two negative bands at 202 and 225 nm,19 thus explaining the missing shoulder at around 230 nm in the abridged form.
Near-UV CD is a very sensitive tool to follow changes in the environment around aromatic residues. Remarkably, the presence of a structured spectrum in this region [Fig. 5(C)] shows that Δ78Δ adopts a folded state quite distinct from a classical molten globule.25 However, Δ78Δ presents a weaker signal in the range 250–270 nm when compared with IFABP. More precisely, loss of fine structure and reduced magnitude of the bands occur. This region could be assigned, in principle, to the dichroic absorption of F residues, although H residues could contribute as well.26 The changes observed could arise from the loss of F residues at 2, 17, and 128 in Δ78Δ and/or from reduced asymmetry around the remaining F residues. Interestingly, on oleic acid binding, the magnitude of the spectral signals of Δ78Δ is significantly enhanced, suggesting again a ligand-induced ordering phenomenon. However, in this case, additional signals arising from rearranged interactions between protomers in the dimer can add to this effect as well.
Equilibrium unfolding transition
The guanidinium chloride (GdnHCl)-induced equilibrium unfolding transition for Δ78Δ was monitored by the change in the intrinsic fluorescence emission. The corresponding experiment for IFABP was included for the sake of comparison. Significantly, the unfolding of the abridged variant Δ78Δ exhibits a cooperative transition akin to that of a well-folded protein [Fig. 6(A)].
As a first approximation, nonlinear least-squares fits describing a two-state model to the equilibrium data were tested [Fig. 6(B)]. In the case of Δ78Δ, this model considers that the only native species is a dimer that dissociates and unfolds concomitantly. The free energies of the overall process (expressed in protomer equivalents) for apo- and holo-Δ78Δ are 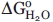 = 5.5 ± 0.3 kcal mol−1 (m = 2.2 ± 0.3 kcal mol−1 M−1, PT = 39 μM) and 5.6 ± 0.7 kcal mol−1 (m = 2.3 ± 0.5 kcal mol−1 M−1, PT = 24 μM), corresponding to the transition midpoints of 1.17 ± 0.03 and 1.10 ± 0.04M GdnHCl, respectively. These results do not support any significant stabilization effect exerted by ligand binding. By comparison, IFABP unfolds following cooperative transitions centered at 1.51 ± 0.02M GdnHCl (apo- and holo-forms), indicating that no significant stabilization occurs on oleic acid binding, although a small increase in cooperativity might be taking place (m = 4.2 ± 0.4 and 6.3 ± 0.7 kcal/mol/M, for the apo- and holo-forms, respectively).
= 5.5 ± 0.3 kcal mol−1 (m = 2.2 ± 0.3 kcal mol−1 M−1, PT = 39 μM) and 5.6 ± 0.7 kcal mol−1 (m = 2.3 ± 0.5 kcal mol−1 M−1, PT = 24 μM), corresponding to the transition midpoints of 1.17 ± 0.03 and 1.10 ± 0.04M GdnHCl, respectively. These results do not support any significant stabilization effect exerted by ligand binding. By comparison, IFABP unfolds following cooperative transitions centered at 1.51 ± 0.02M GdnHCl (apo- and holo-forms), indicating that no significant stabilization occurs on oleic acid binding, although a small increase in cooperativity might be taking place (m = 4.2 ± 0.4 and 6.3 ± 0.7 kcal/mol/M, for the apo- and holo-forms, respectively).
Binding activity
The binding activity of Δ78Δ was assayed with three ligands dissimilar in their chemical nature: the natural ligand oleic acid and two fluorescent probes, namely 1-anilino naphthalene-8-sulfonic acid (ANS) and trans-parinaric acid. Oleic acid binding was monitored by changes in the tryptophan fluorescence emission of the protein, whereas ANS binding was followed by changes in the fluorescence of this probe. Lastly, trans-parinaric acid binding was studied by the appearance of induced bands in the far-UV CD spectrum.
The dissociation constant (Kd) for oleic acid was estimated by fitting the binding data to an equation corresponding to a model considering a single class of sites (Fig. 7). Interestingly, Δ78Δ retains the ability to bind this ligand (Kd = 0.4 μM), displaying a lower affinity than IFABP (Kd = 0.07 μM) but higher than Δ98Δ.6
Figure 7.
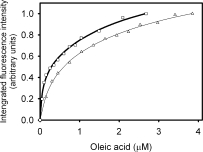
Fatty acid binding activity of Δ78Δ and IFABP. Binding isotherms for oleic acid evidenced by the change in the intrinsic fluorescence emission of the proteins: IFABP (□) and Δ78Δ (Δ).
The fluorescent probe ANS can shed light on the structure and polarity of the binding site environment. IFABP binds ANS with a high affinity and a 1:1 stoichiometry.16,18,27,28 The emission maximum of the Δ78Δ-ANS complex is 471 nm [Fig. 8(A)], indicating a significantly more hydrophobic environment than that observed for the IFABP complex (479 nm), but less than that corresponding to ANS bound to Δ98Δ (466 nm; Ref. 7). In addition, the binding isotherm for ANS to Δ78Δ yields a value for Kd [64 μM, Fig. 8(B)] considerably higher than that measured for IFABP (Kd ∼ 1–10 μM, see also Refs. 16, 28) and somewhat smaller than that reported for Δ98Δ Kd ∼ 133 μM; Ref. 7). To get further insight into the nature of the ANS binding site, a competition assay was run where increasing amounts of oleic acid are added to a preformed protein-ANS complex [Fig. 8(C)]. Previous evidence pointed to a dissimilar behavior between IFABP and the construct Δ98Δ: although the former presents a single binding site for ANS that is fully displaceable by the natural ligand oleic acid, the latter exhibits an additional binding site where oleic acid cannot be bound.7 As a result, Δ78Δ proved to behave in a similar fashion to Δ98Δ. A hyperbolic curve was fitted to the data, yielding a value for Kdapp of 4.8 μM and a ΔFres of 0.34. No significant difference exists in the wavelength corresponding to the maximum of fluorescence emission between samples assayed either in the absence or in the presence of an excess amount of oleic acid (result not shown).
Figure 8.
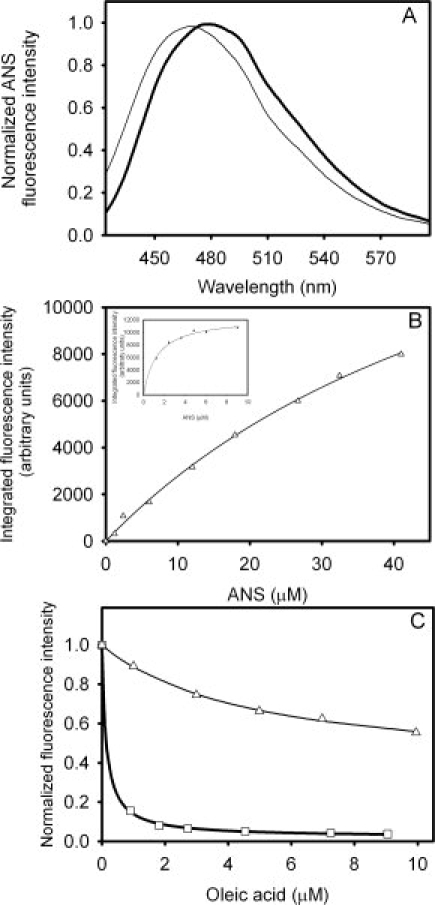
Binding of ANS to Δ78Δ and IFABP. (A) Intensity of fluorescence emission of ANS when bound to Δ78Δ (—) or IFABP  . (B) Titration of Δ78Δ (Δ) or IFABP (□, inset) with this fluorescent probe. (C) Competition of ANS bound to Δ78Δ (Δ) or IFABP (□) by oleic acid. The continuous lines correspond to the fitting of Eq. (2) (see Materials and Methods) to the data. The values of the parameters thus derived are the following: ΔF0 = 0.66, Kdapp = 4.8 μM, and ΔFres = 0.34 for Δ78Δ; and ΔF0 = 0.98, Kdapp = 0.14 μM, and ΔFres = 0.02 for IFABP.
. (B) Titration of Δ78Δ (Δ) or IFABP (□, inset) with this fluorescent probe. (C) Competition of ANS bound to Δ78Δ (Δ) or IFABP (□) by oleic acid. The continuous lines correspond to the fitting of Eq. (2) (see Materials and Methods) to the data. The values of the parameters thus derived are the following: ΔF0 = 0.66, Kdapp = 4.8 μM, and ΔFres = 0.34 for Δ78Δ; and ΔF0 = 0.98, Kdapp = 0.14 μM, and ΔFres = 0.02 for IFABP.
trans-Parinaric acid is a 18-carbon fluorescent polyunsaturated fatty acid (18:4)—derived from the naturally occurring (Z,E,E,Z) cis-parinaric acid—with advantageous spectroscopic properties for biophysical studies.29,30 trans-Parinaric acid exhibits a strong π→π* transition above 300 nm, a region where most proteins hardly absorb light, and because of its symmetrical nature, this molecule shows no optical activity either in organic or in aqueous solutions. These characteristics enabled its use as a probe to study protein binding sites by CD.7,31 The CD spectrum of the complex between Δ78Δ and trans-parinaric acid was compared with that of its ligand free form (Fig. 9). In the far-UV CD region, both protein and ligand absorb light. Therefore, the dichroic bands observed can most likely be attributed to induced bands of the bound chromophore. These are negative band centered at 247 nm and positive band centered at 235 nm, with a crossover point at 242 nm. Strikingly, Δ98Δ shows a remarkable similarity in this regard: a negative band centered at 247 nm and a positive band centered at 238 nm—albeit of smaller magnitude—with a crossover point at 243 nm.7 For comparison, two very weak bands of opposite sign with a crossover point at 240 nm also appear in the case of IFABP (result not shown). Relative differences in the conformational flexibility of the constructs Δ78Δ and Δ98Δ with respect to IFABP might explain this dissimilar behavior.
Figure 9.
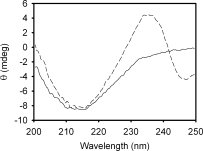
Far-UV CD spectra of Δ78Δ in the presence (dashed line) or in the absence (continuous line) of trans-parinaric acid. The protein:probe molar ratio is 1:0.6.
Discussion
One main goal of our line of research is to learn about those structural determinants giving rise to folded β-barrel motifs. For this purpose, IFABP was chosen as a model system. Previous work has demonstrated that IFABP can withstand the deletion of about 25% of its primary structure while still displaying a well-preserved protein scaffold. This abridged variant, named Δ98Δ, has been described as the shortest form of IFABP preserving binding activity.6,7 In this work, we introduce Δ78Δ—a yet shorter version of IFABP—which was cloned, expressed in bacteria, and purified to yield a soluble and stable form. This fragment lacks both the 1–28 and the 107–131 stretches (according to the sequence numbering scheme accepted for the full-length protein).
Extensive work has been performed on the IFABP structure2–7,32 pointing toward establishing the importance of the α-helical subdomain and the region involving the closure of the β-barrel. The helix-turn-helix motif is neither required for the integrity of the β-clam nor for the nucleation event initiating the folding process. On the other hand, key residues belonging to the hydrophobic core—which is somewhat displaced from the centre of the protein—play a critical role for folding.7,17,33 These interactions are proposed to be present even under highly denaturing conditions, illustrating their importance as structural determinants.34 It is noteworthy that all the residues belonging to the hydrophobic core of IFABP are preserved in Δ78Δ, a fact reflected on its biophysical behavior. Diverse spectroscopic techniques support the fact that Δ78Δ bears a stable well-folded state.
The more distinctive feature of Δ78Δ is that it exists in solution as a stable dimer, either in the presence or in the absence of fatty acid ligands. This is based on the results obtained by SEC, static light scattering, and chemical cross-linking. The fine structure of the fourth-derivative spectrum reveals that Δ78Δ has its single tryptophan residue placed in a microenvironment similar to that of IFABP. However, the Stokes shift to a lower wavelength of the fluorescence emission might point to a slight rearrangement of the hydrophobic milieu around that tryptophan as a consequence of dimer formation. The far-UV CD spectrum of Δ78Δ is typical of a β-sheet protein, but shows 20% less intensity at 216 nm than that of IFABP. Considering that the Δ78Δ lacks the whole α-helical subdomain plus strands βA, βI, and βJ, one would expect a larger loss in the signal. Nevertheless, it is tempting to speculate that, if dimer formation involves a rearrangement of the β-barrel, an increased, β-sheet content would occur. Remarkably, this new species appears also to consolidate a stable tertiary fold, as revealed by the weak but distinct signals present in the near-UV CD spectrum.
Indeed, Δ78Δ unfolds through a cooperative transition where, in principle, dissociation and unfolding represent coupled phenomena. We believe that this construct illustrates a case of gain in thermodynamic stability, where the interaction energy between protomers partially compensates for the loss of intramolecular contacts. This effect might boost Δ78Δ to a value (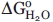 = 5.5 ± 0.3 kcal/mol) comparable with that of the full-length protein (
= 5.5 ± 0.3 kcal/mol) comparable with that of the full-length protein (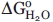 = 6.4 ± 0.6 kcal/mol). Proof of this statement is the fact that the monomeric form Δ98Δ exhibits a much lower stability (
= 6.4 ± 0.6 kcal/mol). Proof of this statement is the fact that the monomeric form Δ98Δ exhibits a much lower stability (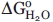 = 2.3 ± 0.3 kcal/mol; Ref. 7).
= 2.3 ± 0.3 kcal/mol; Ref. 7).
From a functional standpoint, Δ78Δ preserves the ability to bind fatty acids. In fact, the interaction with the ligand brings about a gain in structure, as attested by the observed increase in ellipticity in both near- and far-UV regions and the occurrence of a significant blue shift of the centre of mass of the fluorescence emission spectra on oleic acid addition. Here, one should recall that Δ78Δ was originated by digestion of holo-Δ98Δ with Arg-C, whereas the apo-form is completely degraded. In addition, the apo-form of Δ78Δ appears to be larger than expected from its molecular weight (of a dimer; Fig. 2). This cumulative evidence emphasizes the role played by the binding of the fatty acid on the consolidation of the structure, either by reducing the overall mobility or the geometrical asymmetry.
By its ability to bind to this protein, the fluorescent probe ANS sheds light on its structure. The affinity measured for this ligand is well above the range usually observed for typical molten globules.35 In addition, it is revealing to analyze the displacement of ANS from the complex with the natural ligand oleic acid. Indeed, the fatty acid effectively competes with ANS to cause the removal of a major part of the bound probe. However, the fact that about one third remains bound, even in the presence of a large excess of fatty acid, uncovers the existence of a binding site in the construct that is absent in the full-length protein. This behavior is quite similar to that observed for Δ98Δ.7 However, Δ78Δ consistently exhibits a higher binding affinity toward each ligand (oleic acid or ANS) than that measured for Δ98Δ.
On the other hand, binding of trans-parinaric acid uncovers an enhanced flexibility of Δ78Δ when compared with IFABP. This fact is underscored by the lack of defined induced bands giving rise to a fine-structured CD spectrum in the near-UV region and the occurrence, in turn, of a large spectral change in the far-UV region (Fig. 10). This behavior is not unlike that observed for Δ98Δ,7 albeit different in sign and of much higher magnitude than that seen for IFABP.
Natural β-sheets in proteins present different mechanisms to avoid edge-to-edge mediated aggregation. Particularly, β-barrel motifs escape this situation, because they tend not to expose free edges. To accomplish this, a continuous β hydrogen bonding network, all around the cylindrical barrel, is displayed.36 In principle, the extensive stretches deleted in Δ78Δ could determine the appearance of free edges prompting dimerization. The association of each protomer with the other might implicate an interface involving primarily a new set of backbone-backbone hydrogen bonds stabilizing the dimeric structure. The findings presented in this article illustrate the concept that removal of key segments in a monomeric protein triggers a switch mechanism that resorts to quaternary interactions to achieve stabilization. Efforts to further investigate this phenomenon of protein interaction elicited by truncation are currently underway. Here, one main goal points to the elucidation of those structural features conducive to the appearance of a protein-protein interface out of a monomeric parent molecule. In this fashion, lessons could be learned on key issues of protein remodeling leading to attainment of oligomeric character.
Materials and Methods
Materials
Rat IFABP cDNA, coded in the plasmid pET-11, was expressed in E. coli strain BL21(DE3), and the protein was purified as described previously.16 Recombinant Δ98Δ was expressed and purified as described elsewhere.7 DSS, GdnHCl, and buffers were purchased from Sigma-Aldrich (St. Louis, MO).
Ligands
In experiments involving holo-proteins, a 10 mM solution of oleic acid in ethanol was added under stirring to the protein dissolved in buffer A (20 mM Tris-HCl, pH 8.0) in a 4:1 fatty acid:protein molar ratio. The mixture was incubated for at least 40 min at 37°C. ANS concentration was estimated by ultraviolet absorption (ɛ372 = 7800/M/cm, in ethanol). trans-Parinaric acid concentration was estimated by ultraviolet absorption (ɛ306 = 77,000/M/cm, in ethanol). The final concentration of ethanol in the assays never exceeded 2% (v/v).
Expression and purification of recombinant Δ78Δ
Recombinant Δ78Δ was cloned using primers 5'-GGAATTCCATATGAAGCTTGGAGCTCATG-3'and 5'-CGCGGATCCTCATCGGACAGCAATCAGC-3'. The PCR product was digested with both NdeI and BamHI and cloned in the pET-22b(+) vector linearized with the same enzymes. The protein was purified as described previously for Δ98Δ (Ref. 7 and Supporting Information).
Mass spectrometry analyses
ESI-MS analyses were carried out in a LCQ-Duo mass spectrometer (Thermo-Finnigan, San José, CA) with ion trap detector.
Fourth-derivative ultraviolet (UV) spectra
Spectra were collected on a Jasco 7850 spectrophotometer, using a 1-cm path length cuvette sealed with a Teflon cap. IFABP or Δ78Δ (20 μM) dissolved in buffer A were used. Scans were recorded from 250 to 320 nm, with a bandwidth of 1 nm at a 40 nm/min speed. Data were taken every 0.2 nm. Fourth-derivative spectra were calculated by applying two successive cycles of second-order derivation: Δ2A/Δλ2 = (Ai+20 − 2Ai + Ai−20)/2Δλ2.37
Fluorescence measurements
Steady-state fluorescence measurements were performed in an Aminco Bowman Series 2 spectrofluorimeter operating in the ratio mode and equipped with a thermostated cell holder connected to a circulating water bath set at 25°C. Either a 1-cm or a 0.3-cm path cuvette sealed with a Teflon cap were used. For ANS, excitation wavelength was 400 nm and emission was collected in the range 420–600 nm. Spectral slit widths were set to 4 and 8 nm, respectively. When the intrinsic fluorescence of protein was measured, excitation wavelength was set to 295 nm and emission was collected in the range 310–400 nm. In this case, the spectral slit widths were set to 4 nm for both monochromators. Data were corrected for dilution and inner filters effects.38 For intrinsic fluorescence measurements, IFABP or Δ78Δ (15–20 μM) dissolved in buffer A were used. For each spectrum, the wavelength of the centre of mass,20 the maximum intensity, the intensity at a given wavelength, or the integrated intensity were considered for further analysis.
Circular dichroism
Spectra were recorded on a Jasco J-810 spectropolarimeter. Data in the near-UV (250–320 nm) or in the far-UV (200–250 nm) regions were collected using a 10-mm or a 1-mm path cuvettes, respectively. A scan speed of 20 nm/min with a time constant of 1 s was used. IFABP or Δ78Δ (15–20 μM) were dissolved in buffer A. Each spectrum was measured at least three times, and the data were averaged to reduce noise. Molar ellipticity was calculated as described elsewhere,39 using mean residue weight values of 114.5 and 111.5 for IFABP and Δ78Δ, respectively.
Size exclusion chromatography
SEC experiments were carried out in an FPLC system (Amersham Biosciences) with a Superdex-75 HR10/30 (GE Healthcare Life Sciences, Uppsala, Sweden) column equilibrated in buffer A plus 100 mM NaCl. A flow rate of 0.5 mL/min was used, and elution profiles were recorded following the UV absorption at 280 nm. Rs were calculated from a calibration curve of standard proteins according to Uversky.40
Cross-linking experiments
A fresh solution of DSS in dimethyl sulfoxide was added to protein samples (0.2 mg/mL in 20 mM sodium phosphates buffer, pH 8.0) up to a final concentration of 2.5 μg/mL. The mixture was incubated for 1 h at 27°C under continuous stirring. The reaction was stopped by adding an excess amount of Tris (1 μL of 200 mM Tris-HCl, pH 8.0, per 125 μL of mixture). After this step, samples were dried in a Speed-Vac system (Savant, Farmingdale, NY) and analyzed by Tris-Tricine SDS-PAGE, as described earlier. Similarly to IFABP and Δ98Δ, the new variant Δ78Δ shows a normal electrophoretic behavior under SDS denaturing condition. It presents mobility slightly higher than that of a 10-kDa molecular weight marker (result not shown).
Light scattering
A sample of Δ78Δ was subjected to SEC on a Superdex-200 HR10/30 column (GE Healthcare Life Sciences). Eluted peaks were monitored at 690 nm with a light scattering detector (miniDAWN; Wyatt Technology, Santa Barbara, CA) and at 280 nm with a UV detector (Rainin Dynamax, NY). The molar mass of the protein sample was determined with the Astra software (Wyatt Technology).
Equilibrium unfolding studies
Conformational transitions were monitored as a function of denaturant concentration by measuring the change in the intrinsic fluorescence intensity of proteins. GdnHCl stock solutions were prepared on the same day of the experiment. Individual samples ranging in denaturant concentration from 0 to 3 M GdnHCl were obtained by dilution of a fixed volume of protein stock solution in mixtures of buffer A and 8M GdnHCl. Spectra were measured after incubation for at least 2 h to ensure that the equilibrium had been reached. Fluorescence data were corrected for the background signal of buffer and denaturant and expressed in arbitrary units. Nonlinear least-squares fits to the equilibrium data were achieved using an equation representing a two-state model for protein denaturation adapted from Santoro and Bolen.41 For Δ78Δ, a minimal two-state model considering simultaneous dissociation and unfolding (D ⇆ 2U) was used for the fitting. The following equation relates the equilibrium constant for the overall process (K) with the unfolded fraction (fU) and the total protein concentration assayed, expressed in terms of moles of protomer units per liter (PT):
| (1) |
In addition, one assumes that the relationship ΔGo = 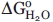 − m [GdnHCl] holds true for this process.
− m [GdnHCl] holds true for this process.
Binding experiments
Oleic acid binding
The binding of this ligand was monitored by measuring changes in the intensity of the intrinsic fluorescence of Δ78Δ or IFABP. For this assay, the concentration of each protein was 2 μM. The ligand was repeatedly added to each protein dissolved in buffer A or to buffer A alone (blank). Incubation for 2 min at 25°C ensured complete equilibration. Nonlinear regression fitting to the binding data was achieved with an equation corresponding to a model of two nonidentical, noninteracting sites, using the Microsoft Excel solver tool.
ANS binding
In this assay, the binding was monitored by changes in the probe emission. The ligand was repeatedly added from a stock solution to each protein dissolved in buffer A or to buffer A alone (blank). Incubation for 3 min at 25°C ensured complete equilibration. Initially, nonlinear regression fitting to the binding data was achieved to a model considering a single class of binding sites using the Microsoft Excel solver tool.
Competition experiments
Displacement of bound ANS (10 μM) to Δ78Δ or IFABP by oleic acid was measured by the decrease of fluorescence intensity with increasing oleic acid concentration. Protein concentration was 2 or 4 μM in 20 mM potassium phosphates buffer, pH 7.4, for IFABP or Δ78Δ, respectively. The spectra were recorded after equilibration for 3 min at 25°C. The apparent dissociation constant (Kdapp) was calculated by fitting the following equation to the data:
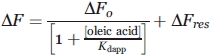 |
(2) |
where ΔF represents the value of the observed fluorescence intensity subtracted from the contribution of the probe at each concentration of oleic acid assayed; ΔF0 is the difference in fluorescence intensity measured in the absence or in the presence of an excess amount of oleic acid; and ΔFres is a term that accounts for the remnant fluorescence at high oleic acid concentration. All fluorescence data were normalized by dividing each difference by ΔFmax (=ΔF0 + ΔFres).
Binding to trans-parinaric acid followed by CD
The Δ78Δ or IFABP were incubated with trans-parinaric acid at a protein:fatty acid ratio of 1:0.6, and the CD spectra of the complexes were measured both in the far-UV and near-UV regions.
Acknowledgments
The authors thank Ms. Gabriela Gómez for her critical reading of the final version of this manuscript.
Glossary
Abbreviations:
- ANS
1-anilino naphthalene-8-sulfonic acid
- DSS
disuccinimidyl suberate
- GdnHCl
guanidinium chloride
- IFABP
intestinal fatty acid binding protein
- SEC
size exclusion chromatography
- Δ98Δ
a truncated variant of IFABP corresponding to the fragment 29–126 of the parent protein
- Δ78Δ
a truncated variant of IFABP corresponding to the fragment 29–106 of the parent protein
- apo- or holo-
prefixes that denote the absence or presence of fatty acid ligand, respectively.
References
- 1.Carrell RW, Lomas DA. Conformational diseases. Lancet. 1997;350:134–138. doi: 10.1016/S0140-6736(97)02073-4. [DOI] [PubMed] [Google Scholar]
- 2.Kim K, Cistola DP, Frieden C. Intestinal fatty acid-binding protethe structure and stability of a helix-less variant. Biochemistry. 1996;35:7553–7558. doi: 10.1021/bi9529115. [DOI] [PubMed] [Google Scholar]
- 3.Steel RA, Emmert DA, Kao J, Hodson ME, Frieden C, Cistola DP. The three-dimensional structure of a helix-less variant of intestinal fatty acid-binding protein. Protein Sci. 1998;7:1332–1339. doi: 10.1002/pro.5560070609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Clérico EM, Peisajovich SG, Ceolín M, Ghiringhelli PD, Ermácora MR. Engineering of a compact non-native state of intestinal fatty acid binding protein. Biochim Biophys Acta. 1999;1476:203–218. doi: 10.1016/s0167-4838(99)00247-2. [DOI] [PubMed] [Google Scholar]
- 5.Ogbay B, Dekoster GT, Cistola DP. The NMR structure of a stable compact all-β-sheet variant of intestinal fatty acid-binding protein. Protein Sci. 2003;13:1227–1237. doi: 10.1110/ps.03546204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Curto LM, Caramelo JJ, Delfino JM. Δ98Δ, a functional all-beta-sheet abridged form of intestinal fatty acid binding protein. Biochemistry. 2005;44:13847–13857. doi: 10.1021/bi051080s. [DOI] [PubMed] [Google Scholar]
- 7.Curto LM, Caramelo JJ, Franchini GR, Delfino JM. Δ98Δ, a minimalist model of antiparallel beta-sheet proteins based on intestinal fatty acid binding protein. Protein Sci. 2009;18:735–746. doi: 10.1002/pro.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Banaszak L, Winter N, Xu Z, Bernlohr DA, Cowan S, Jones A. Lipid-binding proteins: a family of fatty acids and retinoid transport proteins. Adv Protein Chem. 1994;45:89–151. doi: 10.1016/s0065-3233(08)60639-7. [DOI] [PubMed] [Google Scholar]
- 9.Bass NM. The cellular fatty acid binding proteins: aspects of structure, regulation, and function. Int Rev Cytol. 1988;3:143–184. doi: 10.1016/s0074-7696(08)61733-7. [DOI] [PubMed] [Google Scholar]
- 10.Hodsdon ME, Ponder JW, Cistola DP. The NMR solution structure of intestinal fatty acid-binding protein complexed with palmitate: application of a novel distance geometry algorithm. J Mol Biol. 1996;264:585–602. doi: 10.1006/jmbi.1996.0663. [DOI] [PubMed] [Google Scholar]
- 11.Hodsdon ME, Cistola DP. Discrete backbone disorder in the nuclear magnetic resonance structure of apo-intestinal fatty acid-binding proteimplications for the mechanism of ligand entry. Biochemistry. 1997;36:1450–1460. doi: 10.1021/bi961890r. [DOI] [PubMed] [Google Scholar]
- 12.Scapin G, Gordon JI, Sacchettini JC. Refinement of the structure of recombinant rat intestinal fatty acid-binding apoprotein at 1.2-Å resolution. J Biol Chem. 1992;267:4253–4269. doi: 10.2210/pdb1ifc/pdb. [DOI] [PubMed] [Google Scholar]
- 13.Sacchettini JC, Scapin G, Gopaul D, Gordon JI. Refinement of the structure of Escherichia coli-derived rat intestinal fatty acid binding protein with bound oleate to 1.75-Å resolution. Correlation with the structures of the apoprotein and the protein with bound palmitate. J Biol Chem. 1992;267:23534–23545. [PubMed] [Google Scholar]
- 14.Jamison RS, Newcomer ME, Ong DE. Cellular retinoid-binding proteins: limited proteolysis reveals a conformational change upon ligand binding. Biochemistry. 1994;33:2873–2879. doi: 10.1021/bi00176a017. [DOI] [PubMed] [Google Scholar]
- 15.Honma Y, Niimi M, Uchiumi T, Takahashi Y, Odani S. Evidence for conformational change of fatty acid-binding protein accompanying binding of hydrophobic ligands. J Biochem. 116:1025–1029. doi: 10.1093/oxfordjournals.jbchem.a124623. [DOI] [PubMed] [Google Scholar]
- 16.Arighi CN, Rossi JPF, Delfino JM. Temperature-induced conformational switch in intestinal fatty acid binding protein (IFABP) revealing an alternative mode for ligand binding. Biochemistry. 2003;42:7539–7551. doi: 10.1021/bi020680d. [DOI] [PubMed] [Google Scholar]
- 17.Yeh S, Ropson IJ, Rousseau DL. Hierarchical folding of intestinal fatty acid-binding protein. Biochemistry. 2001;40:4205–4210. doi: 10.1021/bi0155044. [DOI] [PubMed] [Google Scholar]
- 18.Arighi CN, Rossi JPF, Delfino JM. Temperature-induced conformational transition of intestinal fatty acid binding protein enhancing ligand binding: a functional, spectroscopic, and molecular modeling study. Biochemistry. 1998;37:16802–16814. doi: 10.1021/bi981827x. [DOI] [PubMed] [Google Scholar]
- 19.Clérico EM, Ermácora MR. Tryptophan mutants of intestinal fatty acid-binding proteultraviolet absorption and circular dichroism studies. Arch Biochem Biophys. 2001;395:215–224. doi: 10.1006/abbi.2001.2554. [DOI] [PubMed] [Google Scholar]
- 20.Weber G. Protein interactions. New York: Chapman and Hall Inc; 1992. [Google Scholar]
- 21.Klimtchuk E, Venyaminov S, Kurian E, Wessels W, Kirk W, Prendergast FG. Photophysics of ANS. I. Protein-ANS complexes: intestinal fatty acid binding protein and single-trp mutants. Biophys Chem. 2007;125:1–12. doi: 10.1016/j.bpc.2006.07.016. [DOI] [PubMed] [Google Scholar]
- 22.Dalessio PM, Fromholt SE, Ropson IJ. The role of Trp-82 in the folding of instestinal fatty acid binding protein. Proteins. 2005;61:176–183. doi: 10.1002/prot.20463. [DOI] [PubMed] [Google Scholar]
- 23.Creigthon TE. Proteins. Structural and molecular properties. 2nd ed. New York: WH Freeman and Company; 1993. [Google Scholar]
- 24.Clark PL, Liu ZP, Zhang J, Gierasch LM. Intrinsic tryptophan mutants as probes of structure and folding. Protein Sci. 1996;5:1108–1117. doi: 10.1002/pro.5560050613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Arai M, Kuwajima K. Role of the molten globule state in protein folding. Adv Protein Chem. 2000;53:209–282. doi: 10.1016/s0065-3233(00)53005-8. [DOI] [PubMed] [Google Scholar]
- 26.Goux WJ, Hooker TM., Jr The chiroptical properties of proteins. II. Near-ultraviolet circular dichroism of lysozyme. Biopolymers. 1980;19:2191–2208. doi: 10.1002/bip.1980.360191205. [DOI] [PubMed] [Google Scholar]
- 27.Kirk WR, Kurian E, Prendergast FG. Characterization of the sources of protein-ligand affinity: 1-sulfonato-8-(1') anilinonaphthalene binding to intestinal fatty acid binding protein. Biophys J. 1996;70:69–83. doi: 10.1016/S0006-3495(96)79592-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pastukhov AV, Ropson IJ. Fluorescent dyes as probes to study lipid-binding proteins. Proteins. 2003;53:607–615. doi: 10.1002/prot.10401. [DOI] [PubMed] [Google Scholar]
- 29.Sklar LA, Hudson BS, Simoni RD. Conjugated polyene fatty acids as membrane probes: preliminary characterization. Proc Natl Acad Sci U S A. 1975;72:1649–1653. doi: 10.1073/pnas.72.5.1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sklar LA, Hudson BS, Petersen M, Diamond J. Conjugated polyene fatty acids on fluorescent probes: spectroscopic characterization. Biochemistry. 1977;16:813–819. doi: 10.1021/bi00624a001. [DOI] [PubMed] [Google Scholar]
- 31.Zsila F, Bikádi Z. trans-Parinaric acid as a versatile spectroscopic label to study ligand binding properties of bovine β-lactoglobulin. Spectrochimica Acta. 2005;62:666–672. doi: 10.1016/j.saa.2005.02.037. [DOI] [PubMed] [Google Scholar]
- 32.Cistola DP, Kim K, Rogl H, Frieden C. Fatty acid interactions with a helix-less variant of intestinal fatty acid binding protein. Biochemistry. 1996;35:7559–7565. doi: 10.1021/bi952912x. [DOI] [PubMed] [Google Scholar]
- 33.Dalessio PM, Boyer JA, McGettigan JL, Ropson IJ. Swapping core residues in homologous proteins swaps folding mechanism. Biochemistry. 2005;44:3082–3090. doi: 10.1021/bi048125u. [DOI] [PubMed] [Google Scholar]
- 34.Ropson IJ, Boyer JA, Dalessio PM. A residual structure in unfolded intestinal fatty acid binding protein consists of amino acids that are neighbors in the native state. Biochemistry. 2006;45:2608–2617. doi: 10.1021/bi052091o. [DOI] [PubMed] [Google Scholar]
- 35.Semisotnov GV, Rodionova NA, Razgulyaev OI, Uversky VN, Gripas' AF, Gilmanshin RI. Study of the “molten globule” intermediate state in protein folding by a hydrophobic fluorescent probe. Biopolymers. 1991;31:119–128. doi: 10.1002/bip.360310111. [DOI] [PubMed] [Google Scholar]
- 36.Richardson JS, Richardson DC. Natural β-sheet proteins use negative design to avoid edge-to-edge aggregation. Proc Natl Acad Sci U S A. 2002;99:2754–2759. doi: 10.1073/pnas.052706099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nozaki Y. Determination of tryptophan, tyrosine and phenylalanine by second derivative spectrophotometry. Arch Biochem Biophys. 1990;277:324–333. doi: 10.1016/0003-9861(90)90587-o. [DOI] [PubMed] [Google Scholar]
- 38.Lakowicz JR. Principles of fluorescence spectroscopy. 3rd ed. New York: Springer Science and Business Media; 2006. [Google Scholar]
- 39.Schmid F. Optical spectroscopy to characterize protein conformation and conformational changes. In: Creighton TE, editor. Protein Structure: a practical approach. 2nd ed. Oxford, UK: IRL; 1989. pp. 251–296. [Google Scholar]
- 40.Uversky VN. Use of fast protein size-exclusion liquid chromatography to study the unfolding of proteins which denature through the molten globule. Biochemistry. 1993;32:13288–13298. doi: 10.1021/bi00211a042. [DOI] [PubMed] [Google Scholar]
- 41.Santoro MM, Bolen DW. Unfolding free energy changes determined by the linear extrapolation method. 1. Unfolding of α-chymotrypsin using different denaturants. Biochemistry. 1998;27:8063–8068. doi: 10.1021/bi00421a014. [DOI] [PubMed] [Google Scholar]


