Abstract
During development, multipotent neural precursors give rise to oligodendrocyte progenitor cells (OPCs), which migrate and divide to produce additional OPCs. Near the end of embryogenesis and during postnatal stages, many OPCs stop dividing and differentiate as myelinating oligodendrocytes, whereas others persist as nonmyelinating cells. Investigations of oligodendrocyte development in mice indicated that the Nkx2.2 transcription factor both limits the number of OPCs that are formed and subsequently promotes their differentiation, raising the possibility that Nkx2.2 plays a key role in determining myelinating versus nonmyelinating fate. We used in vivo time-lapse imaging and loss-of-function experiments in zebrafish to further explore formation and differentiation of oligodendrocyte lineage cells. Our data show that newly specified OPCs are heterogeneous with respect to gene expression and fate. Whereas some OPCs express the nkx2.2a gene and differentiate as oligodendrocytes, others that do not express nkx2.2a mostly remain as nonmyelinating OPCs. Similarly to mouse, loss of nkx2.2a function results in excess OPCs and delayed oligodendrocyte differentiation. Notably, excess OPCs are formed as a consequence of prolonged OPC production from neural precursor cells. We conclude that Nkx2.2 promotes timely specification and differentiation of myelinating oligodendrocyte lineage cells from species representing different vertebrate taxa.
Keywords: Olig2, zebrafish, neural precursors, glia
INTRODUCTION
One of the key problems in oligodendrocyte biology concerns the nature of the genetic programs that direct formation and differentiation of oligodendrocytes to match the timing of myelination to the mode of embryonic and postembryonic development in different species. For example, animals such as amphibians and fish that develop outside the mother are subject to predation during embryonic and larval stages. These species have evolved simple nerve networks that rapidly transmit action potentials to enable escape from potential predators. Two general strategies have evolved to increase the speed of electrical impulse conduction and thereby facilitate escape (Hartline and Colman, 2007). First, organisms have evolved giant axons because the resistance of the axon to electrical current drops as the square of the interior diameter of the axon, which increases transmission speed. Second, organisms have evolved mechanisms to encase axons in myelin sheaths produced by glial cells, which promotes rapid, saltatory conduction of action potentials. Understanding the mechanisms that direct myelination in species that form early-developing escape responses may provide additional insights to mechanisms that regulate timing of myelination in all species.
Zebrafish have emerged over recent years as an investigative system with strengths in imaging and genetics that are complementary to those of other standard laboratory models. Zebrafish larvae, like larvae of other fish species, have a rapid escape response network formed by a small set of early-born primary neurons including the Mauthner neuron, which resides in the hindbrain and projects a large diameter axon contrallaterally down the spinal cord (Eaton et al., 2001; Korn and Faber, 2005). Numerous studies have addressed the genetic mechanisms that direct development of zebrafish primary neurons (Lewis and Eisen, 2003). More recently, zebrafish have begun to be used as a model for investigation of myelination (Brosamle and Halpern, 2002; Kirby et al., 2006; Pogoda et al., 2006).
One gene that might influence the timing of oligodendrocyte formation, differentiation and myelination in different species is Nkx2.2, which encodes a transcription factor. In spinal cords of different species, many oligodendrocytes are produced by ventral precursors called pMN cells. pMN cells express the bHLH transcription factor Olig2. Following a period of motor neuron production, ventral precursors give rise to oligodendrocyte progenitor cells (OPCs), which divide and migrate to become dispersed throughout the gray and white matter of the spinal cord and, hence, available to myelinate widely dispersed axons (Miller, 2002; Rowitch, 2004). Near the end of embryogenesis and during early postnatal life, many OPCs stop dividing, wrap multiple axons with fine membrane extensions and express myelin genes whereas other OPCs remain in a slowly dividing, immature, nonmyelinating state (Wolswijk and Noble, 1989; Nishiyama et al., 1999). During early stages of neural development in rodents, birds and fish, p3 precursors, which lie just ventral to pMN precursors, express Nkx2.2 (Ericson et al., 1997; Briscoe et al., 1999, 2000; Soula et al., 2001; Schafer et al., 2007). In chick embryos, Nkx2.2+ cells disperse from their ventral neuroepithelial origin and express markers of the oligodendrocyte lineage (Xu et al., 2000; Soula et al., 2001). Additionally, Nkx2.2 expression appears to expand dorsally from the p3 domain concomitant with completion of motor neuron production so that some Olig2+ cells begin to express Nkx2.2 (Zhou et al., 2001; Fu et al., 2002). These cells also appear to migrate from the ventricular zone and develop as oligodendrocytes. In mouse embryos Olig2+ cells seem to express Nkx2.2 only after they are specified for oligodendrocyte development and migrate from the ventricular zone (Fu et al., 2002). Therefore, whereas the chick data raise the possibility that Nkx2.2 promotes oligodendrocyte specification, the mouse data point to a later and possibly more limited role for Nkx2.2 in oligodendrocyte differentiation.
Overexpression of Nkx2.2 with Olig2 in chick embryos promotes ectopic expression of Sox10, an OPC marker, and ectopic and precocious expression of Myelin Basic Protein (MBP), suggesting that these factors together promote both specification and differentiation of oligodendrocytes (Zhou et al., 2001). By contrast, mouse embryos that lack Nkx2.2 function express Pdgfrα, a marker of newly specified OPCs, on schedule but delay expression of PLP-DM20 and MBP (Qi et al., 2001) consistent with the idea that, in mice, Nkx2.2 promotes oligodendrocyte differentiation but not specification. In fact, more cells express Pdgfrα in mutant embryos than in wild type indicating that Nkx2.2 limits formation of OPCs, perhaps by limiting the ventral boundary of the Olig2 domain, which gives rise to Pdgfrα+ OPCs. These observations point to apparent differences in expression and function of Nkx2.2 in chick and mice leaving its role in oligodendrocyte development unresolved.
In this study we used zebrafish as a model to investigate the role of nkx2.2a, which is an ortholog (83% identical) of rodent and chick Nkx2.2, in oligodendrocyte specification and differentiation. Our expression data and in vivo time-lapse imaging indicate that nkx2.2a expression distinguishes myelinating from nonmyelinating oligodendrocyte lineage cells and some presumptive OPCs express nkx2.2a as they are produced by neuroepithelial precursors. We also found evidence that the nkx2.2a+ subset of OPCs fails to form in the absence of nkx2.2a function but that these cells are then replenished during prolonged periods of neural precursor division. Similar to mice, loss of nkx2.2a function delays but does not prevent oligodendrocyte differentiation. We propose that nkx2.2a specifically promotes development of oligodendrocyte lineage cells that have myelinating fate.
OBJECTIVE
Investigations of chick and mouse embryos revealed apparent differences in the time at which oligodendrocyte lineage cells express the transcription factor Nkx2.2, leaving the role of Nkx2.2 in oligodendrocyte specification and differentiation unclear. We used in vivo time-lapse imaging of transgenic zebrafish embryos to watch the formation of OPCs from spinal cord precursors and antisense morpholino oligonucleotides (MOs) to test how loss of nkx2.2a function affects oligodendrocyte development.
METHODS
Fish husbandry
All animal studies were approved by the Vanderbilt University Institutional Animal Care and Use Committee. Zebrafish strains used in this study included AB, Tg(nkx2.2a:megfp)vu17 (Kirby et al., 2006; Kucenas et al., 2008), Tg(olig2:dsred2)vu19 (Kucenas et al., 2008) and Tg(sox10(7.2):mrfp)vu234 (Kucenas et al., 2008). Embryos were produced by pair-wise matings, raised at 28.5°C in egg water or embryo medium and staged according to hours post-fertilization (hpf). Embryos used for in situ hybridization, immunocytochemistry and microscopy were treated with 0.003% phenylthiourea in egg water to reduce pigmentation.
In vivo time-lapse imaging
At 24 hpf, all embryos used for live imaging were manually dechorionated and transferred to egg water containing phenylthiourea to block formation of pigment. At specified stages, embryos were anesthetized using 3-aminobenzoic acid ester (Tricaine), immersed in 0.8% low-melting point agarose and mounted on their sides in glass-bottomed 35-mm Petri dishes (World Precision Instruments). All images were captured using a 40× oil-immersion objective (NA = 1.3) mounted on a motorized Zeiss Axiovert 200 microscope equipped with a PerkinElmer ERS spinning-disk confocal system. A heated stage chamber was used to maintain embryos at 28.5°C. Z image stacks were collected every 10–15 min, and three-dimensional datasets were compiled using Sorenson 3 video compression (Sorenson Media) and exported to QuickTime (Apple) to create movies.
In situ RNA hybridization
In situ RNA hybridization was performed as described previously (Hauptmann and Gerster, 2000). Probes included olig2 (Park et al., 2002), nkx2.2a (Barth and Wilson, 1995), plp/dm20 and mbp (Brosamle and Halpern, 2002). Plasmids were linearized with appropriate restriction enzymes and cRNA preparation was carried out using Roche DIG-labeling reagents and T3, T7 or SP6 RNA polymerases (New England Biolabs). After staining, embryos were embedded in 1.5% agar/30% sucrose and frozen in 2-methyl butane chilled by immersion in liquid nitrogen. Ten micrometer transverse sections were cut using a cryostat microtome, collected on microscope slides and covered with 75% glycerol. Images were obtained using a Retiga Exi-cooled CCD camera (Qimaging) mounted on an Olympus AX70 microscope equipped with Openlab software (Improvision). All images were imported into Adobe Photoshop. Image adjustments were limited to levels, contrast, color matching settings and cropping.
Immunocytochemistry
Embryos and larvae were fixed in AB Fix (4% paraformaldehyde, 8% sucrose, PBS) for 3 h at 23°C or overnight at 4°C and embedded as described above. We collected 10-μm transverse sections using a cryostat microtome. Sections were re-hydrated in 1 × PBS for 60 min at 23°C and preblocked in 2% goat serum/BSA/1 × PBS for 30 min. Sections were incubated in primary antibody overnight at 4°C. The primary antibodies used included rabbit anti-Sox10 at 1:500 dilution (Park et al., 2005), mouse anti-BrdU (1:1000, Developmental Studies Hybridoma Bank) and rabbit anti-Phospho-Histone H3 (1:1000, Upstate Biotechnology). Sections were washed extensively with 1 × PBS, incubated for 3 h at 23°C with either Alexa Fluor 647 goat anti-rabbit or Alexa Fluor 568 goat anti-mouse secondary antibodies (Molecular Probes) and washed with 1 × PBS for 30 min. Sections were mounted in Vectashield (Vector Laboratories) and imaged using the confocal microscope described above. Image adjustments were limited to contrast enhancement and levels settings using Volocity software and Adobe Photoshop.
Morpholino injections
Antisense MOs were purchased from Gene Tools, LLC (Philomath, OR). nkx2.2aMO1 (5′-CCG TCT TTG TGT TGG TCA ACG ACA T-3′) was complementary to a sequence spanning just before and including the translation start codon (Schafer et al., 2007; Kucenas et al., 2008). nkx2.2aMO2 (5′-AAG TTG CTG CAC CAG TTT GAC AAT C-3′) was complementary to a sequence in the 5′ UTR. MOs were dissolved in water to create a stock solution of 3 mM and diluted in 2 × injection buffer (5 mg ml−1 Phenol red, 40 mM HEPES, 240 mM KCl) to create a working injection concentration of 0.5 mM for nkx2.2aMO1 and 0.75 mM for nkx2.2aMO2. We injected 2–4 nl into the yolk just below the blastodisc of fertilized eggs. All MO-injected embryos were raised in embryo medium at 28.5°C.
Data quantification and statistical analyses
To count cells in MO-injected and wild-type embryos and larvae, composite Z image stacks were compiled using Volocity software. Cell counts were taken from consecutive, transverse trunk sections between somites 8 and 13. Individual Z images were sequentially observed and cells counted within the entire stack. All graphically presented data represent the mean of the analyzed data. Statistical analyses were performed with Prism software. The level of significance was determined by either paired or unpaired t-tests using a confidence interval of 95%.
BrdU labeling
Embryos and larvae were dechorionated, incubated in a 0.5% BrdU/10% DMSO solution for 20 min on ice and subsequently washed 4 × 5 min with embryo medium. Embryos and larvae were allowed to develop until desired stages in embryo medium. Samples were fixed, embedded, sectioned and prepared for immunocytochemistry as described above.
RESULTS
OPCs arise from both nkx2.2a+ and nkx2.2a− ventral spinal cord precursors
Previous gene expression studies demonstrated that chick and mouse embryos express Nkx2.2 differently in OPCs. Whereas in chick OPCs appear to arise from Nkx2.2+ spinal cord precursors (Soula et al., 2001), mouse OPCs express Nkx2.2 only after they initiate migration from their ventral spinal cord origin (Qi et al., 2001; Zhou et al., 2001; Fu et al., 2002). Zebrafish express two nkx2.2 paralogs, nkx2.2a and nkx2.2b (Barth and Wilson, 1995; Schafer et al., 2005) and our previous work showed that at least some OPCs express an nkx2.2a transgene (Kirby et al., 2006). Furthermore, zebrafish Nkx2.2a has greater amino acid identity with mammalian Nkx2.2 than Nkx2.2b. Therefore, to more fully investigate nkx2.2 gene expression in zebrafish OPCs, we focused on nkx2.2a, beginning with in situ RNA hybridization. At 24 and 36 hpf, nkx2.2a expression was restricted to cells of the lateral floor plate or p3 domain (Fig. 1A,B), ventral to olig2+ cells of the pMN domain (Fig. 1E,F). By 48 hpf and continuing through 72 hpf, nkx2.2a expression expanded dorsally (Fig. 1C,D) to overlap with some olig2+ cells of the pMN domain (Fig. 1G,H). A similar merger of Nkx2.2 and Olig2 expression domains occurs in chick embryos (Sun et al., 2001; Zhou et al., 2001; Fu et al., 2002, 2003; Liu and Rao, 2004). nkx2.2a+ cells also appeared in dorsal spinal cord in a pattern similar to the distribution of differentiating and myelinating oligodendrocytes in the white matter (Fig. 1C,D).
Fig. 1. nkx2.2a and olig2 RNA expression patterns in spinal cord.
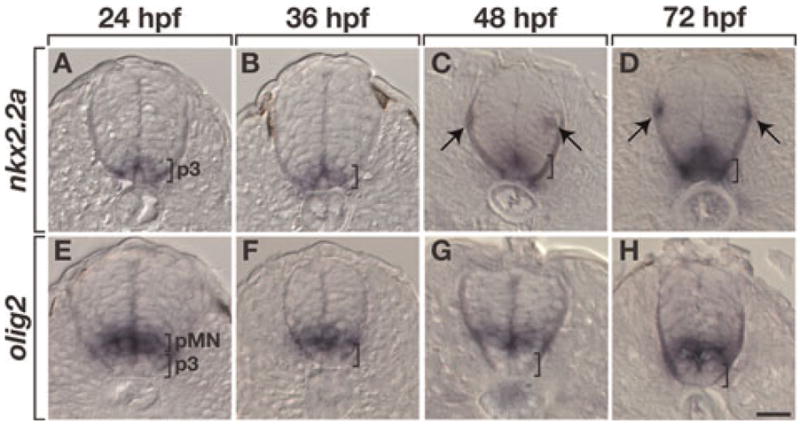
All panels show transverse sections with dorsal to the top. (A–D) At 24 and 36 hpf (A,B) nkx2.2a expression is restricted to the p3 precursor domain (brackets). At 48 and 72 hpf (C,D) the nkx2.2a ventral expression domain (brackets) expands dorsally and nkx2.2a+ cells appear in dorsal spinal cord near the pial surface (arrows). (E–H) olig2 expression marks the pMN precursor domain. Scale bar: (in H), 20 μm.
To investigate the relationship between gene expression and OPC formation more directly, we examined transverse sections of Tg(nkx2.2a:megfp);Tg(olig2:dsred2) embryos, which express membrane-tethered EGFP under control of nkx2.2a regulatory DNA (Kirby et al., 2006; Kucenas et al., 2008) and DsRed2 under control of olig2 (Kucenas et al., 2008), respectively. At 24 hpf, EGFP+ and DsRed2+ spinal cord cells occupied distinct, but contiguous domains (Fig. 2A) similar to the corresponding nkx2.2a and olig2 RNA expression patterns. By 48 hpf, the expression domains began to overlap so that some cells expressed both EGFP and DsRed2 (Fig. 2B), again consistent with nkx2.2a and olig2 RNAs. We next labeled sections of double transgenic embryos and larvae with an antibody specific to zebrafish Sox10 (Park et al., 2005), which is the earliest known marker for OPCs. From 48 hpf, at the onset of OPC migration, through 96 hpf, when OPCs wrap axons and differentiate into mature oligodendrocytes (Kirby et al., 2006), we found two classes of Sox10+ cells: nkx2.2a+ olig2+ Sox10+ cells and nkx2.2a− olig2+ Sox10+ cells (Fig. 2C–E). Consistent with the nkx2.2a RNA expression pattern, nkx2.2a+ olig2+ Sox10+ cells were usually within the axon-rich pial region of the spinal cord (Fig. 2C–E). Because myelinating oligodendrocytes occupy only the peripheral white matter and not the medial gray matter of the spinal cord, these data raise the possibility that a myelinating subset of OPCs selectively express nkx2.2a.
Fig. 2. nkx2.2a and olig2 expression define two distinct classes of spinal cord OPCs.
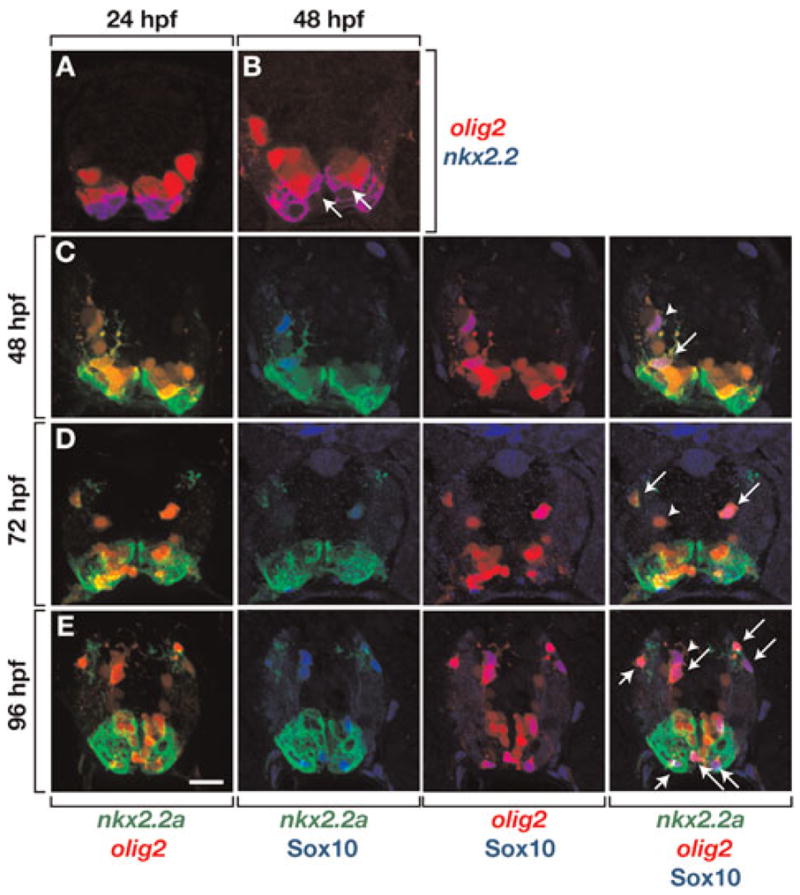
All panels show images of 10 μm transverse sections, dorsal to the top, of Tg(nkx2.2a:megfp);Tg(olig2:dsred2) embryos and larvae. (A) At 24 hpf, cells that express nkx2.2a, revealed by EGFP fluorescence (blue) and olig2, revealed by DsRed2 fluorescence (red) occupy distinct domains within the ventral spinal cord. (B) By 48 hpf, EGFP expression expands dorsally and some cells express both EGFP and DsRed2 (arrows). (C–E) Sections labeled with anti-Sox10 antibody (blue) to reveal OPCs. Sox10+ OPCs are both nkx2.2a+ olig2+ (arrows) and nkx2.2a− olig2+ (arrowheads). Scale bar: (in E), 20 μm.
We further investigated the correlation of nkx2.2a expression with OPC behavior and fate using in vivo time-lapse imaging. We first imaged larvae carrying both a Tg(sox10(7.2):mrfp) transgene, which expresses a membrane tethered Red Fluorescent Protein (mRFP) under the control of sox10 regulatory DNA (Kirby et al., 2006; Kucenas et al., 2008) and the Tg(nkx.2.a:megfp) transgene. Consistent with the above expression data, some OPCs were nkx2.2a+ sox10+ while other OPCs were nkx2.2a− sox10+ (Fig. 3A; see Supplementary Movie 1 online). Usually, nkx2.2a+ sox10+ OPCs wrapped axons whereas nkx2.2a− sox10+ OPCs did not, although we have occasionally observed axon wrapping by nkx2.2a− OPCs (data not shown). Time-lapse imaging of Tg(nkx2.2a:megfp);Tg(olig2:dsred2) larvae similarly revealed two distinct subsets of OPCs consisting of nkx2.2a+ olig2+ and nkx2.2a− olig2+ cells (Fig. 3B and see Supplementary Movie 2 online).
Fig. 3. In vivo, time-lapse imaging reveals two classes of spinal cord OPCs.
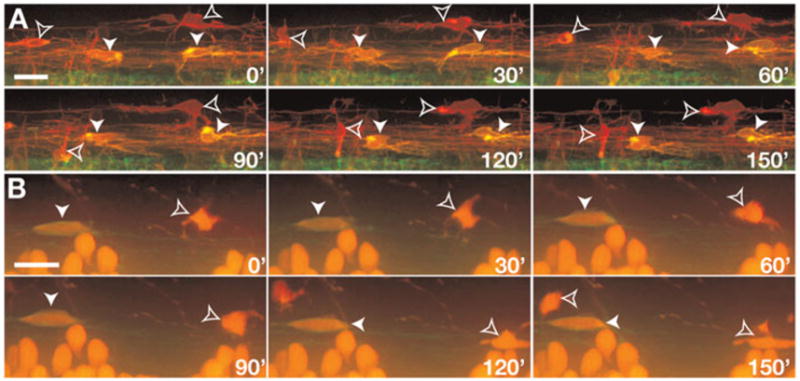
All panels show images from the side of the spinal cord with dorsal up. Numbers in lower right corners denote time elapsed from the first frame. (A) Frames captured from a 24 h time-lapse sequence (Supplemental Video 1) of a Tg(nkx2.2a:megfp);Tg(sox10(7.2):mrfp) embryo beginning at 52 hpf. Cells with OPC morphologies and behaviors are nkx2.2a+ sox10+ (yellow, arrowheads) and nkx2.2a− sox10+ (red, outlined arrowheads). (B) Frames captured from a 56 h time-lapse sequence (Supplemental Video 2) of a Tg(nkx2.2a:megfp);Tg(olig2:dsred2) embryo beginning at 60 hpf. OPCs are nkx2.2a+ olig2+ (red outlined by green membrane, arrowheads) and nkx2.2a− olig2+ (red cytoplasm, outlined arrowheads). Scale bars: 10 μm.
There are at least two possible explanations for differential expression of nkx2.2a among OPCs. First, OPCs might be specified as a homogenous nkx2.2a− population of cells, some of which later begin to express nkx2.2a. Alternatively, OPCs might arise from distinct nkx2.2a+ and nkx2.2a− precursors. We attempted to distinguish between these possibilities by observing OPC formation directly by time-lapse imaging Tg(nkx2.2a:megfp);Tg(sox10(7.2):mrfp) embryos. Altogether, we have observed three different patterns of OPC gene expression. First, at approximately 40 hpf, some presumptive OPCs appeared as nkx2.2a+ neuroepithelial cells in the ventral spinal cord (Fig. 4A and see Supplementary Movie 3 online). These cells then began to express sox10 and turned yellow from co-expression of these transgenes, and subsequently extended membrane processes and migrated dorsally. Second, some OPCs appeared to initially express sox10 and only later expressed nkx2.2a (Fig. 4A and see Supplementary Movie 3 online). Therefore, nkx2.2a+ sox10+ OPCs actually begin as distinct nkx2.2a+ and nkx2.2a− populations. Third, some OPCs expressed only sox10 (Fig. 4B and see Supplementary Movie 3 online) consistent with our data obtained from analysis of fixed tissues above. We conclude that OPCs that arise from the ventral spinal cord are heterogeneous, consisting of both nkx2.2a+ sox10+ and nkx2.2a− sox10+ cells.
Fig. 4. OPCs are born as nkx2.2a+ and nkx2.2a− cells.
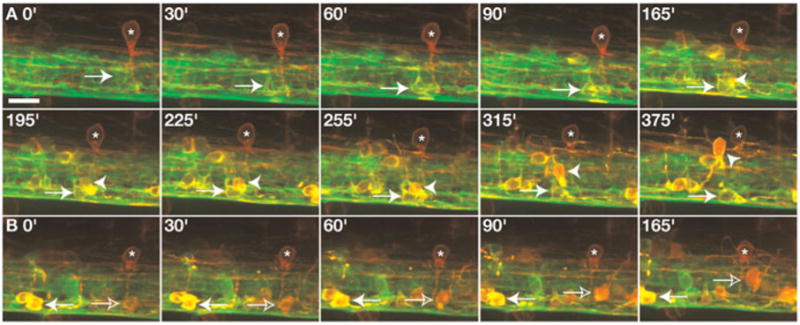
All panels show images from the side of the spinal cord with dorsal up. Frames were captured from a 24 h time-lapse sequence (Supplemental Video 3) of a Tg(nkx2.2a:megfp);Tg(sox10(7.2):mrfp) embryo beginning at 40 hpf. (A) Arrows indicate a green nkx2.2a+ cell that begins to turn yellow from co-expression of sox10. This OPC remained in ventral spinal cord whereas a neighboring nkx2.2a+ sox10+ OPC migrated dorsally (arrowheads). (B) Frames captured from the same time-lapse sequence showing a nkx2.2a+ sox10+ OPC (yellow, solid arrow) and a nkx2.2a− sox10+ OPC (red, outlined arrow). White asterisks mark ectopic expression of the sox10 reporter gene in interneurons. Scale bar: (in A), 10 μm.
nkx2.2a function is necessary for specification and differentiation of OPCs
Mouse embryos that are homozygous for a loss-of-function mutation of Nkx2.2 have elevated numbers of OPCs but reduced and delayed expression of myelin genes (Qi et al., 2001). These observations were interpreted to mean that Nkx2.2 expression in p3 cells limits the size of the overlying pMN precursor population, which gives rise to OPCs, and then later promotes oligodendrocyte differentiation (Qi et al., 2001; Lu et al., 2002). The formation of some OPCs from Nkx2.2+ precursors in chick (Soula et al., 2001) and zebra-fish raises the possibility that Nkx2.2 also promotes specification of a subset of OPCs. We tested this using antisense MOs designed to interfere with nkx2.2a translation. Previous work showed that nkx2.2a MOs reduce or eliminate expression of genes that mark cells born within the p3 domain (Schafer et al., 2007; Kucenas et al., 2008). Accordingly, in nkx2.2a MO-injected embryos, olig2 RNA expression expanded ventrally to include cells adjacent to medial floor plate (see Supplementary Fig. 1 online), similar to the ventral expansion of Olig2 expression in Nkx2.2 mutant mice (Qi et al., 2001).
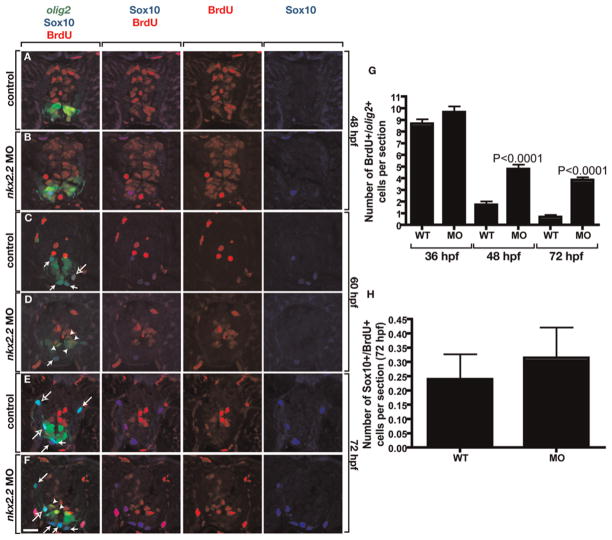
To investigate the role of Nkx2.2a in OPC specification, we injected nkx2.2a MOs into single-cell Tg(olig2:dsred2); Tg(nkx2.2a:megfp) embryos. Because one of our MOs (nkx2.2aMO1) was designed to block translation of endogenous nkx2.2a transcripts but not of egfp mRNA expressed by the transgene, we were able to use EGFP fluorescence as a marker of spinal cord patterning and OPC development. At 48 hpf, the total number of Sox10+ OPCs was substantially reduced in nkx2.2aMO1-injected embryos compared to control embryos (Fig. 5A,B,E and see Supplementary Table 1 online). Embryos injected with nkx2.2aMO2, designed to block translation by binding to 5′ UTR sequence, had a similar decrease in the number of Sox10+ OPCs (data not shown; data in Tables 1–4 present combined MO1 and MO2 data). This decrease in total Sox10+ OPC number was mostly attributable to a large deficit in the nkx2.2a+ olig2+ subset of OPCs (Fig. 5F and see Supplementary Table 1 online). Notably, these same embryos had a small but statistically significant increase in the number of nkx2.2a− olig2+ OPCs (Fig. 5F and see Supplementary Table 1 online). At 72 hpf, MO-injected larvae had excess Sox10+ OPCs compared to control, revealing a large expansion in OPC number between 48 and 72 hpf in MO-injected animals (Fig. 5C–E and see Supplementary Table 1 online). Similar to 48 hpf, at 72 hpf MO-injected larvae had significantly more nkx2.2a− olig2+ OPCs (Fig. 5G and see Supplementary Table 1 online). However, these larvae no longer had a deficit of nkx2.2a+ olig2+ OPCs, instead, the size of this subset was now equal to control (Fig. 5G and see Supplementary Table 1 online). In summary, these observations indicate that loss of nkx2.2a function causes a transient deficit of OPCs, which is followed by a large expansion in the nonmyelinating subpopulation of OPCs producing an excess of total OPCs.
Fig. 5. Loss of nkx2.2a function causes a transient deficit of OPCs.
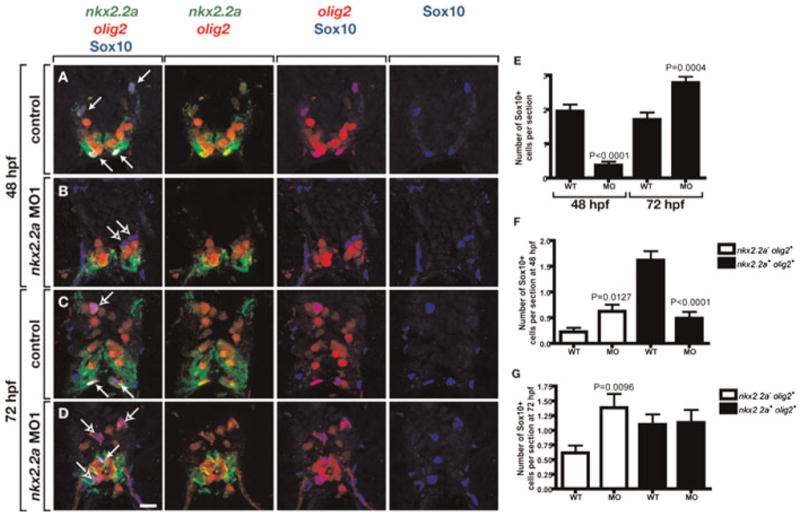
(A–D) Transverse sections, dorsal to the top, of Tg(nkx2.2a:megfp);Tg(olig2:dsred2) embryos and larvae labeled with a Sox10 antibody. Solid arrows mark nkx2.2a+ olig2+ OPCs and outlined arrows indicate nkx2.2a− olig2+ OPCs. At 48 nkx2.2a MO-injected embryos (B) have fewer nkx2.2a+ OPCs and fewer total OPCs than control embryos (A). By 72 hpf, nkx2.2a MO-injected larvae (D) have more nkx2.2a− OPCs and more total OPCs than control larvae (C). (E) Quantification of total OPC number revealing a deficit at 48 hpf and excess at 72 hpf. (F) Quantification of OPC subclasses at 48 hpf revealing a deficit of nkx2.2a− olig2+ OPCs and small excess of nkx2.2a+ olig2+ OPCs in nkx2.2a MO-injected embryos compared to wild-type (WT) controls. (G) Quantification of OPC subclasses at 72 hpf revealing equal numbers of nkx2.2a− olig2+ OPCs and excess nkx2.2a+ olig2+ OPCs in nkx2.2a MO-injected embryos compared to WT. Error bars represent standard error of the mean. P values represent comparisons between MO-injected animals and corresponding controls. Statistical significance was determined using the paired t-test. Scale bar: (in D), 20 μm.
Our data raise the possibility that nkx2.2a function promotes formation of myelinating oligodendrocytes. Therefore, we next tested expression of two genes commonly used as markers of oligodendrocyte differentiation. In rodents, RNA for the DM20 splicing variant of PLP, which encodes proteolipid protein, is expressed in newly formed OPCs, well before myelination (Timsit et al., 1995; Dickinson et al., 1996; Peyron et al., 1997). Consequently, PLP/DM20 expression marks oligodendrocyte lineage cells as they progress from progenitor to myelinating stages (Baumann and Pham-Dinh, 2001). RNA expressed by the MBP gene, which encodes myelin basic protein, becomes evident after initiation of PLP/DM20 expression and marks myelinating oligodendrocytes (Trapp et al., 1987; Peyron et al., 1997). In zebrafish, although expression of plp/dm20 is never evident at the proliferative ventricular zone of the spinal cord, it slightly precedes expression of mbp (data not shown). Therefore, as in rodents, plp/dm20 expression is initiated at an earlier stage of oligodendrocyte maturation than mbp. Notably, we found a striking difference in the effect of nkx2.2a loss of function on plp/dm20 and mbp expression. At 3 and 4 dpf, control larvae similarly express plp/dm20 and mbp in cells positioned within the ventral and dorsal spinal cord white matter (Fig. 6A,C,E,G). By contrast, at 3 dpf MO-injected larvae had an excess of plp/dm20+ cells but a deficit of mbp+ cells (Fig. 6B,F,I,J and see Supplementary Table 2 online). By 4 dpf, the number of plp/dm20+ cells in MO-injected larvae was approximately equal to control embryos (Fig. 6D,I and see Supplementary Table 2 online) whereas the number of mbp+ cells, although more similar to wild-type number than at 3 dpf, remained at a deficit (Fig. 6H,J and see Supplementary Table 2 online). The increase in the number of plp/dm20+ cells at 3 dpf in MO-injected larvae correlates with the increase in total Sox10+ OPCs and the nkx2.2a− olig2+ OPC subpopulation. These data suggest that nkx2.2a is required to promote the progression of myelinating oligodendrocyte lineage cells from a plp/dm20+ state to a more mature mbp+ state.
Fig. 6. nkx2.2a promotes oligodendrocyte differentiation.
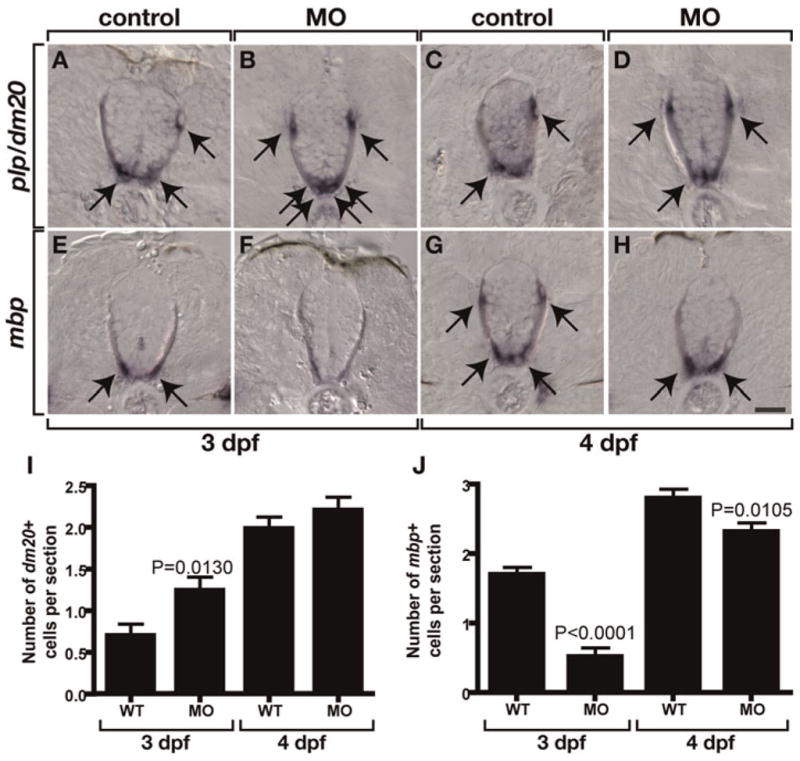
(A–E) Transverse sections of larvae processed for in situ RNA hybridization with dorsal to the top. Arrows mark differentiating oligodendrocytes. At 3 dpf, nkx2.2a MO-injected larvae have more plp/dm20+ cells (B) but fewer mbp+ cells (F) than corresponding controls (A,E). At 4 dpf, nkx2.2a MO-injected larvae have comparable number of plp/dm20+ cells (D) but fewer mbp+ cells (H) than corresponding controls (C,G). (I) Quantification of plp/dm20+ cells. The difference between WT and MO-injected larvae at 4 dpf is not statistically significant. (J) Quantification of mbp+ cells. Error bars represent standard error of the mean. P values represent comparisons between MO-injected animals and corresponding controls. Statistical significance was determined using the paired t-test. Scale bar: (in H), 20 μm.
olig2+ precursor cell division is increased in embryos lacking Nkx2.2a function
What accounts for the increase in the number of nkx2.2a− olig2+ OPCs in nkx2.2a MO-injected larvae? We previously showed that oligodendrocytes removed by laser microsurgery are rapidly replaced by neighboring OPCs that divide and migrate into the ablated region (Kirby et al., 2006). Therefore, one possibility is that nkx2.2a− olig2+ OPCs divide with increased frequency to compensate for the loss of the nkx2.2a+ olig2+ OPC subpopulation in MO-injected embryos. Alternatively, loss of nkx2.2a function might cause formation of excess nkx2.2a− olig2+ OPCs from ventricular zone precursors. To discriminate between these possibilities, we injected Tg(olig2:egfp) embryos with either nkx2.2aMO1 or nkx2.2aMO2 and treated them at different developmental stages with the thymidine analog BrdU to label cells in S-phase. Between 36 and 48 hpf, wild-type and nkx2.2a MO-injected embryos had equal numbers of replicating, olig2+ precursors (Fig. 7A,B,G and see Supplementary Table 3 online). Between 48 and 60 hpf, and continuing through 72 hpf, wild-type embryos had fewer olig2+ BrdU+ cells and fewer total BrdU+ cells than at earlier stages, indicating a reduction in the dividing cell population (Fig. 7C,E,G and see Supplementary Table 3 online). By contrast, nkx2.2a MO-injected animals had significantly more olig2+ BrdU+ cells than comparably staged wild types (Fig. 7D,F,G and see Supplementary Table 3 online). At all stages, the numbers of olig2+ BrdU+ Sox10+ cells in wild-type and nkx2.2a MO-injected embryos were similar (Fig. 7H and see Supplementary Table 3 online) indicating that OPCs did not change their frequency of division. Labeling wild-type and nkx2.2a MO-injected embryos with anti-phospho-Histone H3 antibody, to label M-phase cells, similarly revealed more dividing olig2+ precursors at 60 and 72 hpf (see Supplementary Table 4 online).
Fig. 7. Loss of nkx2.2a function extends the proliferative period of olig2+ precursors.
(A–F) Ten micrometer transverse sections, dorsal to the top, of Tg(olig2:egfp) embryos and larvae treated with BrdU and labeled with anti-Sox10 (blue) and anti-BrdU (red) antibodies. olig2 expression is revealed by EGFP fluorescence (green). Wild-type control and nkx2.2a MO-injected embryos have similar numbers of total BrdU+ cells and olig2+ BrdU+ cells at 48 hpf (A,B). By 60 hpf, control larvae (C) have many fewer BrdU+ cells than at 48 hpf but nkx2.2a MO-injected larvae (D) have substantial numbers of olig2+ BrdU+ cells (arrowheads). By 72 hpf, control larvae (E) have few olig2+ cells that incorporate BrdU whereas olig2+ cells in nkx2.2a MO-injected larvae (F) continue to incorporate BrdU. Solid and outlined arrows mark BrdU− and BrdU+ OPCs, respectively. (G) Quantification of olig2+ BrdU+ precursor cells. The difference between 48 hpf wild-type (WT) control and MO-injected embryos is not significant. (H) Quantification of Sox10+ BrdU+ OPCs in 72 hpf larvae. The difference is not statistically significant. Error bars represent standard error of the mean. P values represent comparisons between MO-injected animals and corresponding controls. Statistical significance was determined using the paired t-test. Scale bar: (in F), 20 μm.
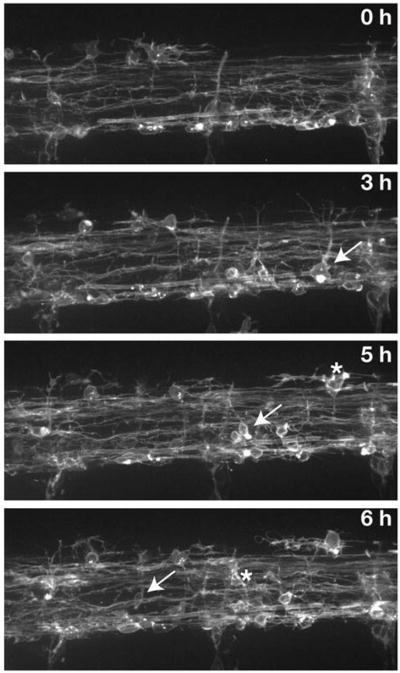
These data favor the idea that, in the absence of nkx2.2a function, a transient deficit of OPCs is overcome by prolonged production of OPCs from ventral spinal cord precursors. To test this more directly, we performed time-lapse imaging of nkx2.2a MO-injected Tg(olig2:egfp) and Tg(sox10(7.2):mrfp) embryos. In wild-type embryos, OPCs migrate dorsally between about 40 and 60 hpf (Kirby et al., 2006). In nkx2.2a MO-injected embryos most OPC migration was delayed, beginning instead about 60 hpf (data not shown), consistent with our observation that nkx2.2a MO-injected embryos have a deficit of Sox10+ OPCs at 48 hpf (Fig. 5). OPC migration into dorsal spinal cord of nkx2.2a MO-injected embryos continued until about 80 hpf (Fig. 8; see Supplementary Movie 4 online), matching the prolonged period of olig2+ precursor division revealed by BrdU incorporation (Fig. 7). OPC membrane process extension and retraction appeared normal in MO-injected embryos, but little axon wrapping was evident suggesting that nkx2.2a function promotes axon ensheathment. OPCs did not divide more frequently than in control embryos, again consistent with our BrdU labeling data, and indicating that the excess OPCs that form in MO-injected embryos do not result from elevated levels of OPC division. We conclude that the loss of nkx2.2a function results in an initial deficit of myelinating oligodendrocytes, which are then replaced by an extended period of production of nonmyelinating OPCs from olig2+ neural precursors.
Fig. 8. Loss of nkx2.2a function extends the period of OPC production and dorsal migration.
Panels show frames captured from a time-lapse sequence, beginning at 72 hpf, of a Tg(sox10(7.2):mrfp) larvae injected with nkx2.2a MO. Images are shown from the side with dorsal up. Numbers in upper right corners indicate time elapsed from beginning of sequence. Arrows mark OPCs initiating dorsal migration. Asterisks mark migrating OPCs in subsequent frame.
CONCLUSIONS
In the zebrafish spinal cord, OPCs arise from both nkx2.2a+ and nkx2.2a− ventral precursors.
The majority of myelinating oligodendrocytes develop from nkx2.2a+ OPCs.
Specification and differentiation of myelinating oligodendrocytes is delayed in the absence of nkx2.2a function.
After a transient deficit of OPCs in nkx2.2a deficient embryos, olig2+ precursors produce new OPCs at elevated rates.
DISCUSSION
OPCs arise from nkx2.2a+ and nkx2.2a− spinal cord precursors
Previous studies revealed apparent differences between chick and mouse embryos with respect to oligodendrocyte cell lineage and Nkx2.2 expression. In both chicks and mice, the p3 and pMN populations of ventral spinal cord precursor cells express Nkx2.2 and Olig2, respectively, through the period of motor neuron development (Mizuguchi et al., 2001; Novitch et al., 2001; Soula et al., 2001; Sun et al., 2001; Zhou et al., 2001; Fu et al., 2002). In chick embryos, Nkx2.2+ cells then begin to disperse throughout the spinal cord in a pattern characteristic of OPCs (Xu et al., 2000). At about the same time, Nkx2.2 expression expands dorsally into the Olig2 expression domain (Mizuguchi et al., 2001; Novitch et al., 2001; Sun et al., 2001; Zhou et al., 2001; Fu et al., 2002). Double labeling experiments showed that Nkx2.2+ dispersing cells initially are both Olig2+ and Olig2− but that later, most cells in the spinal cord white matter are Nkx2.2+ Olig2+ (Zhou et al., 2001; Fu et al., 2002). Therefore, in chick, Nkx2.2 expression in OPCs precedes their emigration from the ventricular zone. By contrast, in mouse embryos, Olig2+ OPCs express Nkx2.2 only after they migrate away from the ventricular zone (Fu et al., 2002).
Our analysis reveals that, like chick, zebrafish nkx2.2a expression expands dorsally concomitant with the onset of OPC migration so that some precursors express both nkx2.2a and olig2, raising the possibility that nkx2.2a is required for OPC specification. We also found that some OPCs do not express nkx2.2a. Following migration, nkx2.2a+ cells occupy white matter and not gray matter and our transgenic reporters show clearly that nkx2.2a+ cells wrap axons. Although most dispersed Nkx2.2+ cells in chick and mouse express oligodendrocyte lineage markers (Zhou et al., 2001; Fu et al., 2002), a careful examination of whether all oligodendrocyte lineage cells express Nkx2.2 has not been reported. However, only one-third of A2B5 immunoreactive OPCs isolated from E14 rat spinal cord were Nkx2.2+ in culture (Qi et al., 2001), suggesting rodent OPCs are heterogeneous with respect to Nkx2.2 expression. Our data indicates that, in zebrafish, nkx2.2a expression defines the myelinating subpopulation of oligodendrocyte lineage cells. Consistent with this interpretation, mouse Nkx2.2 can drive expression from the promoter of the myelin gene PLP/DM20 in transient transfection experiments (Qi et al., 2001).
nkx2.2a promotes oligodendrocyte differentiation
Targeted mutation of mouse Nkx2.2 causes a delay in onset of expression of the myelin genes MBP and PLP/DM20 and reduction in the number of spinal cord cells that express them at P8 (Qi et al., 2001). Our loss-of-function experiments are generally consistent with these data. At 3 dpf, nkx2.2a MO-injected larvae have significantly fewer mbp+ cells than controls. By 4 dpf, a significant deficit remains, but the difference to control is less than at 3 dpf, suggesting some recovery of mbp expression. Because MOs become depleted as the animal grows, we cannot be certain whether recovery results from gradual loss of MO blocking effect or because nkx2.2a is not absolutely required for mbp expression. Surprisingly, at 3 dpf we found the opposite effect on plp/dm20 expression, with a statistically significant increase in plp/dm20+ cells. This increase in plp/dm20+ cell number is coincident with the increase in the number of Sox10+ cells. This raises the possibility that, in the absence of nkx2.2a function, oligodendrocyte differentiation stalls at a stage marked by plp/dm20 expression and fails to progress to a more mature stage marked by mbp expression. Consistent with this, time-lapse imaging revealed that axon wrapping is substantially reduced in nkx2.2a MO-injected larvae. Oligodendrocytes can express MBP in the absence of axons (Mirsky et al., 1980; Bradel and Prince, 1983; Zeller et al., 1985; Dubois-Dalcq et al., 1986). Therefore, the reduction of axon wrapping and delay in mbp expression might reflect distinct roles for nkx2.2a function in myelination.
Spinal cord precursors produce more nkx2.2a− OPCs in the absence of nkx2.2a+ OPCs
Notably, Nkx2.2 mutant mice have excess Pdgrα+ OPCs, which was attributed to ventral expansion of the Olig2+ pMN domain to take the place of the p3 domain (Qi et al., 2001). Our data similarly reveal formation of excess Sox10+ OPCs in nkx2.2a-deficient zebrafish. However, our time-lapse imaging provides direct evidence that some OPCs arise from nkx2.2a+ precursors in zebrafish, which complicates interpretation of the role of Nkx2.2 genes in oligodendrocyte specification. There are at least two other possibilities that could account for an increase in OPC number in the absence of nkx2.2a function. First, if nkx2.2a is required for specification of only a subset of OPCs, compensatory divisions by nkx2.2a− OPCs could replace them. This possibility is supported by laser ablation of oligodendrocyte lineage cells that revealed effective replacement by nearby OPCs (Kirby et al., 2006). However, in these experiments, oligodendrocytes were restored to their normal number and not produced in excess. A second possibility is that ventricular zone precursors divide more frequently or for a longer period of time to replace OPCs. Our data are consistent with the latter hypothesis. During late embryogenesis, wild-type zebrafish have a sharp decline in the number of dividing spinal cord cells, including olig2+ precursors (Park et al., 2007). In nkx2.2a MO-injected embryos this decline was less substantial, so that at 72 hpf, an early larval stage, significant numbers of olig2+ cells remained in the cell cycle. This suggested that OPCs are replaced by an extended period of OPC production by olig2+ precursors. Indeed, time-lapse imaging revealed first a delay and then extension in the period of OPC production and dorsal migration in nkx2.2a deficient embryos and larvae.
What accounts for the extended proliferative response of olig2+ precursors to loss of nkx2.2a function? One possibility is that nkx2.2a promotes the timely transition of neural precursors to OPCs. We showed previously that a subset of dividing olig2+ spinal cord precursors persist through embryogenesis into adulthood (Park et al., 2007). We assume that at least some of these cells undergo asymmetric divisions to produce an OPC and a self-renewing precursor. In this study we learned that loss of nkx2.2a function causes a delay in OPC formation, which is followed by production of excess OPCs. This might occur if the absence of nkx2.2a function shifts the balance of precursor divisions from asymmetric to symmetric proliferative, whereby cells divide to make two new precursors. This could result in formation of excess precursors that, after a delay, produce excess OPCs. Time-lapse imaging of olig2+ precursors in nkx2.2a-deficient embryos might provide a good test of this possibility.
Supplementary Material
Acknowledgments
We thank M. Langworthy for comments on the manuscript. The anti-BrdU antibody, developed by S.J. Kaufman, was obtained from the Developmental Studies Hybridoma Bank developed under the auspices of the NICHD and maintained by The University of Iowa, Department of Biological Sciences, Iowa City, IA 52242. Confocal microscopy was performed using equipment obtained with funds provided by the Vanderbilt Academic Venture Capital Fund. This work was supported in part by NIH grant NS046668 and a grant from the National Multiple Sclerosis Society (B.A.), the Post Doctoral Training Program in Neurogenomics-MH65215 (S.K.) and a NIH Ruth L. Kirschstein NRSA HD056760 (S.K.)
Footnotes
Statement of interest
None
The supplementary material referred in this article can be found online at journals.cambridge.org/ngb.
References
- Barth KA, Wilson SW. Expression of zebrafish nk2.2 is influenced by sonic hedgehog/vertebrate hedgehog-1 and demarcates a zone of neuronal differentiation in the embryonic forebrain. Development. 1995;121:1755–1768. doi: 10.1242/dev.121.6.1755. [DOI] [PubMed] [Google Scholar]
- Baumann N, Pham-Dinh D. Biology of oligodendrocyte and myelin in the mammalian central nervous system. Physiological Review. 2001;81:871–927. doi: 10.1152/physrev.2001.81.2.871. [DOI] [PubMed] [Google Scholar]
- Bradel EJ, Prince FP. Cultured neonatal rat oligodendrocytes elaborate myelin membrane in the absence of neurons. Journal of Neuroscience Research. 1983;9:381–392. doi: 10.1002/jnr.490090404. [DOI] [PubMed] [Google Scholar]
- Briscoe J, Pierani A, Jessell TM, Ericson J. A homeodomain protein code specifies progenitor cell identity and neuronal fate in the ventral neural tube. Cell. 2000;101:435–445. doi: 10.1016/s0092-8674(00)80853-3. [DOI] [PubMed] [Google Scholar]
- Briscoe J, Sussel L, Serup P, Hartigan-O’Connor D, Jessell TM, Rubenstein JL, et al. Homeobox gene Nkx2.2 and specification of neuronal identity by graded Sonic hedgehog signalling. Nature. 1999;398:622–627. doi: 10.1038/19315. [DOI] [PubMed] [Google Scholar]
- Brosamle C, Halpern ME. Characterization of myelination in the developing zebrafish. Glia. 2002;39:47–57. doi: 10.1002/glia.10088. [DOI] [PubMed] [Google Scholar]
- Dickinson PJ, Fanarraga ML, Griffiths IR, Barrie JM, Kyriakides E, Montague P. Oligodendrocyte progenitors in the embryonic spinal cord express DM-20. Neuropathology and Applied Neurobiology. 1996;22:188–198. [PubMed] [Google Scholar]
- Dubois-Dalcq M, Behar T, Hudson L, Lazzarini RA. Emergence of three myelin proteins in oligodendrocytes cultured without neurons. Journal of Cell Biology. 1986;102:384–392. doi: 10.1083/jcb.102.2.384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eaton RC, Lee RK, Foreman MB. The Mauthner cell and other identified neurons of the brainstem escape network of fish. Progress in Neurobiology. 2001;63:467–485. doi: 10.1016/s0301-0082(00)00047-2. [DOI] [PubMed] [Google Scholar]
- Ericson J, Rashbass P, Schedl A, Brenner-Morton S, Kawakami A, van Heyningen V, et al. Pax6 controls progenitor cell identity and neuronal fate in response to graded Shh signaling. Cell. 1997;90:169–180. doi: 10.1016/s0092-8674(00)80323-2. [DOI] [PubMed] [Google Scholar]
- Fu H, Cai J, Rutledge M, Hu X, Qiu M. Oligodendrocytes can be generated from the local ventricular and subventricular zones of embryonic chicken midbrain. Brain Research. Developmental Brain Research. 2003;143:161–165. doi: 10.1016/s0165-3806(03)00108-1. [DOI] [PubMed] [Google Scholar]
- Fu H, Qi Y, Tan M, Cai J, Takebayashi H, Nakafuku M, et al. Dual origin of spinal oligodendrocyte progenitors and evidence for the cooperative role of Olig2 and Nkx2.2 in the control of oligodendrocyte differentiation. Development. 2002;129:681–693. doi: 10.1242/dev.129.3.681. [DOI] [PubMed] [Google Scholar]
- Hartline DK, Colman DR. Rapid conduction and the evolution of giant axons and myelinated fibers. Current Biology. 2007;17:R29–R35. doi: 10.1016/j.cub.2006.11.042. [DOI] [PubMed] [Google Scholar]
- Hauptmann G, Gerster T. Multicolor whole-mount in situ hybridization. Methods of Molecular Biology. 2000;137:139–148. doi: 10.1385/1-59259-066-7:139. [DOI] [PubMed] [Google Scholar]
- Kirby BB, Takada N, Latimer AJ, Shin J, Carney TJ, Kelsh RN, et al. In vivo time-lapse imaging shows dynamic oligodendrocyte progenitor behavior during zebrafish development. Nature Neuroscience. 2006;9:1506–1511. doi: 10.1038/nn1803. [DOI] [PubMed] [Google Scholar]
- Korn H, Faber DS. The Mauthner cell half a century later: a neurobiological model for decision-making? Neuron. 2005;47:13–28. doi: 10.1016/j.neuron.2005.05.019. [DOI] [PubMed] [Google Scholar]
- Kucenas S, Takada N, Park HC, Woodruff E, Broadie K, Appel B. CNS-derived glia ensheath peripheral nerves and mediate motor root development. Nature Neuroscience. 2008;11:143–151. doi: 10.1038/nn2025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis KE, Eisen JS. From cells to circuits: development of the zebrafish spinal cord. Progress in Neurobiology. 2003;69:419–449. doi: 10.1016/s0301-0082(03)00052-2. [DOI] [PubMed] [Google Scholar]
- Liu Y, Rao MS. Olig genes are expressed in a heterogeneous population of precursor cells in the developing spinal cord. Glia. 2004;45:67–74. doi: 10.1002/glia.10303. [DOI] [PubMed] [Google Scholar]
- Lu QR, Sun T, Zhu Z, Ma N, Garcia M, Stiles CD, et al. Common developmental requirement for Olig function indicates a motor neuron/oligodendrocyte connection. Cell. 2002;109:75–86. doi: 10.1016/s0092-8674(02)00678-5. [DOI] [PubMed] [Google Scholar]
- Miller RH. Regulation of oligodendrocyte development in the vertebrate CNS. Progress in Neurobiology. 2002;67:451–467. doi: 10.1016/s0301-0082(02)00058-8. [DOI] [PubMed] [Google Scholar]
- Mirsky R, Winter J, Abney ER, Pruss RM, Gavrilovic J, Raff MC. Myelin-specific proteins and glycolipids in rat Schwann cells and oligodendrocytes in culture. Journal of Cell Biology. 1980;84:483–494. doi: 10.1083/jcb.84.3.483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuguchi R, Sugimori M, Takebayashi H, Kosako H, Nagao M, Yoshida S, et al. Combinatorial roles of olig2 and neurogenin2 in the coordinated induction of pan-neuronal and subtype-specific properties of motoneurons. Neuron. 2001;31:757–771. doi: 10.1016/s0896-6273(01)00413-5. [DOI] [PubMed] [Google Scholar]
- Nishiyama A, Chang A, Trapp BD. NG2+ glial cells: a novel glial cell population in the adult brain. Journal of Neuropathology and Experimental Neurology. 1999;58:1113–1124. doi: 10.1097/00005072-199911000-00001. [DOI] [PubMed] [Google Scholar]
- Novitch BG, Chen AI, Jessell TM. Coordinate regulation of motor neuron subtype identity and pan-neuronal properties by the bHLH repressor Olig2. Neuron. 2001;31:773–789. doi: 10.1016/s0896-6273(01)00407-x. [DOI] [PubMed] [Google Scholar]
- Park H, Mehta A, Richardson JS, Appel B. olig2 is required for zebrafish primary motor neuron and oligodendrocyte development. Developmental Biology. 2002;248:356–368. doi: 10.1006/dbio.2002.0738. [DOI] [PubMed] [Google Scholar]
- Park HC, Boyce J, Shin J, Appel B. Oligodendrocyte specification in zebrafish requires notch-regulated cyclin-dependent kinase inhibitor function. Journal of Neuroscience. 2005;25:6836–6844. doi: 10.1523/JNEUROSCI.0981-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park HC, Shin J, Roberts RK, Appel B. An olig2 reporter gene marks oligodendrocyte precursors in the postembryonic spinal cord of zebrafish. Developmental Dynamics. 2007;236:3402–3407. doi: 10.1002/dvdy.21365. [DOI] [PubMed] [Google Scholar]
- Peyron F, Timsit S, Thomas JL, Kagawa T, Ikenaka K, Zalc B. In situ expression of PLP/DM-20, MBP, and CNP during embryonic and postnatal development of the jimpy mutant and of transgenic mice overexpressing PLP. Journal of Neuroscience Research. 1997;50:190–201. doi: 10.1002/(SICI)1097-4547(19971015)50:2<190::AID-JNR8>3.0.CO;2-A. [DOI] [PubMed] [Google Scholar]
- Pogoda HM, Sternheim N, Lyons DA, Diamond B, Hawkins TA, Woods IG, et al. A genetic screen identifies genes essential for development of myelinated axons in zebrafish. Developmental Biology. 2006;298:118–131. doi: 10.1016/j.ydbio.2006.06.021. [DOI] [PubMed] [Google Scholar]
- Qi Y, Cai J, Wu Y, Wu R, Lee J, Fu H, et al. Control of oligodendrocyte differentiation by the Nkx2.2 homeodomain transcription factor. Development. 2001;128:2723–2733. doi: 10.1242/dev.128.14.2723. [DOI] [PubMed] [Google Scholar]
- Rowitch DH. Glial specification in the vertebrate neural tube. Nature Reviews in Neuroscience. 2004;5:409–419. doi: 10.1038/nrn1389. [DOI] [PubMed] [Google Scholar]
- Schafer M, Kinzel D, Neuner C, Schartl M, Volff JN, Winkler C. Hedgehog and retinoid signalling confines nkx2.2b expression to the lateral floor plate of the zebrafish trunk. Mechanics Development. 2005;122:43–56. doi: 10.1016/j.mod.2004.09.002. [DOI] [PubMed] [Google Scholar]
- Schafer M, Kinzel D, Winkler C. Discontinuous organization and specification of the lateral floor plate in zebrafish. Developmental Biology. 2007;301:117–129. doi: 10.1016/j.ydbio.2006.09.018. [DOI] [PubMed] [Google Scholar]
- Soula C, Danesin C, Kan P, Grob M, Poncet C, Cochard P. Distinct sites of origin of oligodendrocytes and somatic motoneurons in the chick spinal cord: oligodendrocytes arise from Nkx2.2-expressing progenitors by a Shh-dependent mechanism. Development. 2001;128:1369–1379. doi: 10.1242/dev.128.8.1369. [DOI] [PubMed] [Google Scholar]
- Sun T, Echelard Y, Lu R, Yuk DI, Kaing S, Stiles CD, et al. Olig bHLH proteins interact with homeodomain proteins to regulate cell fate acquisition in progenitors of the ventral neural tube. Current Biology. 2001;11:1413–1420. doi: 10.1016/s0960-9822(01)00441-9. [DOI] [PubMed] [Google Scholar]
- Timsit S, Martinez S, Allinquant B, Peyron F, Puelles L, Zalc B. Oligodendrocytes originate in a restricted zone of the embryonic ventral neural tube defined by DM-20 mRNA expression. Journal of Neuroscience. 1995;15:1012–1024. doi: 10.1523/JNEUROSCI.15-02-01012.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapp BD, Moench T, Pulley M, Barbosa E, Tennekoon G, Griffin J. Spatial segregation of mRNA encoding myelin-specific proteins. Proceedings of the National Academy of Sciences USA. 1987;84:7773–7777. doi: 10.1073/pnas.84.21.7773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolswijk G, Noble M. Identification of an adult-specific glial progenitor cell. Development. 1989;105:387–400. doi: 10.1242/dev.105.2.387. [DOI] [PubMed] [Google Scholar]
- Xu X, Cai J, Fu H, Wu R, Qi Y, Modderman G, et al. Selective expression of Nkx-2.2 transcription factor in chicken oligodendrocyte progenitors and implications for the embryonic origin of oligodendrocytes. Molecular Cell and Neuroscience. 2000;16:740–753. doi: 10.1006/mcne.2000.0916. [DOI] [PubMed] [Google Scholar]
- Zeller NK, Behar TN, Dubois-Dalcq ME, Lazzarini RA. The timely expression of myelin basic protein gene in cultured rat brain oligodendrocytes is independent of continuous neuronal influences. Journal of Neuroscience. 1985;5:2955–2962. doi: 10.1523/JNEUROSCI.05-11-02955.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Q, Choi G, Anderson DJ. The bHLH transcription factor Olig2 promotes oligodendrocyte differentiation in collaboration with Nkx2.2. Neuron. 2001;31:791–807. doi: 10.1016/s0896-6273(01)00414-7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.