Abstract
The simulation of cardiac electrical function is an example of a successful integrative multiscale modelling approach that is directly relevant to human disease. Today we stand at the threshold of a new era, in which anatomically detailed, tomographically reconstructed models are being developed that integrate from the ion channel to the electromechanical interactions in the intact heart. Such models hold high promise for interpretation of clinical and physiological measurements, for improving the basic understanding of the mechanisms of dysfunction in disease, such as arrhythmias, myocardial ischaemia and heart failure, and for the development and performance optimization of medical devices. The goal of this article is to present an overview of current state-of-art advances towards predictive computational modelling of the heart as developed recently by the authors of this article. We first outline the methodology for constructing electrophysiological models of the heart. We then provide three examples that demonstrate the use of these models, focusing specifically on the mechanisms for arrhythmogenesis and defibrillation in the heart. These include: (1) uncovering the role of ventricular structure in defibrillation; (2) examining the contribution of Purkinje fibres to the failure of the shock; and (3) using magnetic resonance imaging reconstructed heart models to investigate the re-entrant circuits formed in the presence of an infarct scar.
The heart is characterized by a complex electrometrical activity essential for the sustenance of body function. Cardiac disease is the leading cause of morbidity and mortality in the industrialized world (Rodgers & Vaughan, 2002). A comprehensive understanding of cardiac structure and function in health and disease is the central focus of basic and clinical heart research. Experimental investigations study animal and human cardiac behaviour in health and disease, from the molecular to the whole body levels. A major challenge remains the integration of insight, obtained at various levels of structural hierarchy, such as from ion channel activation and calcium cycling to cardiac propagation and regional strain formation. A key strategy in integrating from the individual building blocks to the whole heart is to characterize the emergent properties that arise at each successive scale in the hierarchy of biological complexity (Rudy et al. 2008). With the increase in complexity at each successive scale, new and often unexpected properties arise from the co-operative interactions between individual components. Since these properties are often not intuitive, detailed knowledge of the individual components themselves may provide little mechanistic insight into the organ function. Integrative approaches hold the highest promise for elucidation of the complexity of the structure–function relationships in the heart. In recent years, computational modelling has become an essential approach in the quest for integration of knowledge regarding the behaviour of the heart (Xie et al. 2004; Panfilov et al. 2007; Hunter et al. 2008; Kerckhoffs et al. 2008). Advanced computational approaches to cardiac electromechanical function have made significant contributions to data integration, extraction of information, interpretation and mechanistic insights. In addition, the importance of computational approaches is underscored by the fact that a host of relevant processes is not accessible by direct experimental manipulation or observation, and methods for three-dimensional assessment of transmembrane potential distribution are not yet available (Efimov et al. 2004). In contrast, experimentally validated computational models are capable of predicting three-dimensional behaviour in cases where the experiment fails (Rodriguez et al. 2005; Ten Tusscher et al. 2007; Ashihara et al. 2008). Finally, the invasive nature of many observational techniques prevents their application to human subjects, highlighting the need for new quantitative tools to enable the integration of knowledge and the prediction of behaviour of the human heart. Computational modelling is at the threshold of being able to address this need, ushering in a new era of computational medicine.
The goal of this article is to present an overview of current state-of-art advances towards predictive computational modelling of the heart as developed recently by the authors of this article. We first outline the methodology for constructing electrophysiological models of the heart and then provide three examples that demonstrate the use of these models, focusing specifically on the mechanisms for arrhythmogenesis in the heart.
Methodology
Construction of geometrical models
Two types of geometrical models are typically used in organ studies of electrophysiology and electromechanics: models of coarse representation of cardiac geometry; and high-resolution image-based models of cardiac structure and geometry. The specifics of these geometrical models are described below.
Models of coarse representation of cardiac geometry
The development of the first type of geometrical models was pioneered by Hunter and co-workers at the University of Auckland (Nielsen et al. 1991; dog ventricular model) and by McCulloch and co-workers at the University of California, San Diego (UCSD; Vetter & McCulloch, 1998; rabbit ventricular model). The data for the canine ventricular model were obtained using a specially designed mechanical rig, while for the rabbitmodel, digital imaging of histological sections was used. The models were generated by fitting the nodal parameters of a hexahedral finite element mesh to the measured data using least squares. Smooth estimates of the geometry and the fibrous structure of the ventricles were obtained using Hermite interpolation. For use in electrophysiological studies, meshes at an appropriate resolution needed to be generated from these models. The methodology by which this was done in our laboratory is described below; details can be found in the study of Trayanova (2006).
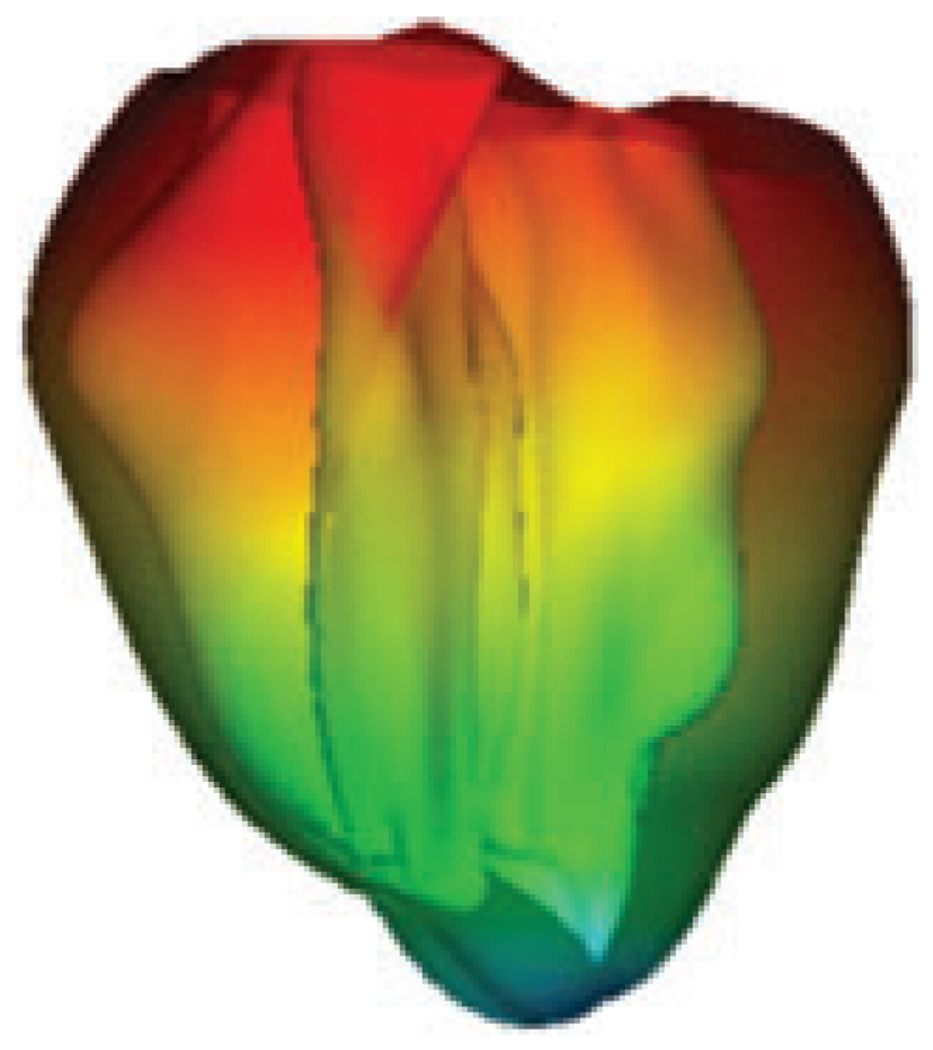
Specifically, this approach served to generate our three-dimensional (3-D) model of rabbit ventricular geometry and fibre orientation (Fig. 1), which used the UCSD geometry of the rabbit ventricles (Vetter & McCulloch, 1998). The original geometry, defined in prolate spheroidal co-ordinates, was translated into Cartesian co-ordinates. We used MSC Patran (MSC Software Corp., Santa Ana, CA, USA) to generate the computational grids. The surfaces and the ‘solids’ of the mesh were created with Patran functions. Additional ‘solids’ were generated to represent the ventricular cavities and the perfusing bath. Using Patran, surfaces and solids were meshed to create unstructured triangular surfaces and tetrahedral meshes, respectively, with an average element edge length of 300–500 µm in the tissue and 1 mm in cavities and perfusing bath. Once the entire mesh was complete, files were generated containing node co-ordinates, elemental connectivity and the original finite element to which each tetrahedral element in the new mesh belongs. This information was then used to determine, with the combination of two multidimensional root-finding algorithms, the fibre orientation at the centroid of each tetrahedron. Local material properties were assigned to each element using the fibre orientations. A detailed description of this procedure can be found in the study of Trayanova et al. (2002).
Figure 1. Coarse geometry 3-D model of rabbit ventricular anatomy.
Colours represent a propagating wavefront.
The use of this coarse geometrical model of the rabbit ventricles in electrophysiological studies is presented in the section on ‘Applications of the Modelling Methodology’. Simulations with such models have enabled us (Li et al. 2004; Rodriguez et al. 2005; Arevalo et al. 2007; Ashihara et al. 2008) and other researchers (Panfilov, 1999; Fenton et al. 2005) to address issues of arrhythmogenesis and anti-arrhythmia therapy and thus to showcase their predictive power. While being compact and thus enabling efficient numerical solutions, these models provide, however, only a coarse representation of the overall cardiac structure, and are lacking in fine detail, such as endocardial trabeculations and papillary muscles, as well as structural detail regarding cardiac disease, which might play an important role in a host of problems in cardiac electrophysiology and mechanics. Below we describe the generation of such high-resolution image-based models of the heart from structural and diffusion tensor (DT) magnetic resonance (MR) images.
Image-based model generation
In this section, we outline the new pipeline we have developed recently to generate models of the heart from structural and DTMR images. This pipeline allows one to generate heart models with high throughput, combining known elements of image-based model generation (segmentation, level set thresholding and fractional anisotropy) with new procedures (image-based mesh generation, a procedure for delineation of ventricles fromatria and automatic fibre mapping) developed by our team.
To generate models of the heart from structural and DTMR images, of both normal and structurally altered (i.e. infarcted) hearts, it is necessary to classify (or segment) the voxels in the structural image into different groups, such as normal tissue, infarct and background. From the segmented image, a finite element mesh is generated, and fibre orientations are assigned to the elements. Figure 2 shows the processing of a generic pipeline, which we use to generate image-based models of the heart. The figure illustrates the pipeline by showing the results of processing of an example image slice using the six steps in the pipeline. The steps are briefly explained below. The slice used for illustration is from the heart employed in the study presented in the section on ‘Modelling ventricular tachycardia associated with an infarct’. It is important to note that the models that our methodology generates retain fine structural details, such as endocardial trabeculations and papillary muscles, provided that such structural detail is resolved by the magnetic resonance imaging (MRI) acquisition.
Figure 2. The application of the high-resolution image-based model generation pipeline to an example image slice.
The original image slice is shown at the top. The sequence of rectangular blocks illustrates the results as the slice is processed through steps 1 to 6 in the pipeline.
Step 1: suspension medium removal
In this step, the structural MR image is processed to label and ‘remove’ the voxels corresponding to the cavity content and the medium in which the heart was suspended during the image acquisition (Burton et al. 2006). First, the edges in the image are detected (Law&Chung, 2007) by computing the intensity variance of the local neighbourhood of each voxel and selecting voxels with a variance above a certain threshold. From the detected edges, themyocardial boundary of the whole heart is extracted using a region-growing algorithm (Adams & Bischof, 1994). This algorithm, when initialized with a user-defined voxel, identifies a voxel set such that the set forms a connected structure which includes the user-defined voxel and the members of the set that have the same image intensity. Next, from the image that represents the myocardial boundary, voxels that correspond to the suspension and cavity medium are extracted using the region-growing algorithm. Finally, the suspension medium is removed from the original structural MR image by assigning the background intensity to all voxels that correspond to the medium. The result is an image of the myocardium. Step 1 in Fig. 2 shows the myocardial boundary, suspension medium, and myocardium for the example slice.
Step 2: level set segmentation
In the next step, a level set method is applied to the image of the myocardium to separate the larger coronary arteries and interlaminar clefts. In the level set method, the segmentation is achieved through the evolution of a surface, Γ, which is implicitly represented as the zero level set of a time-dependent 3-D function, Φ(x, y, z, t), i.e. Γ(t) = {x, y, z |Φ(x, y, z, t) = 0}, where x, y, = and z are the Cartesian co-ordinates, and t is the time. The evolution of Φ is formulated as a partial differential equation, which contains terms for advancing the surface based on image intensities, smoothing the surface and attracting the surface to the edges in the image. For detailed information on the level set segmentation, the reader is referred to Sethian (1996) and Ibanez et al. (2003). Step 2 in Fig. 2 shows the level set segmentation for the example slice.
Step 3: segmentation of the ventricles
In the third step of our model generation pipeline, segmentation of the ventricular myocardium is performed in order to account for the presence of an insulating layer of connective tissue between the atria and the ventricles (annulus fibrosus), as well as to assign different electrophysiological properties (atrial versus ventricular) to the tissue on either side of this boundary. To perform this step developed by our team, in each slice, the ventricular portion of the tissue is labelled by fitting a closed spline curve through spline points placed around the ventricles and along the atrioventricular border. All voxels that belong to tissue inside the curve are marked as ventricular. Step 3 in Fig. 2 shows the spline points, spline and ventricular myocardium for the example slice. The identification of spline points is performed manually for a number of slices that are evenly distributed in the image. The spline points for the remaining slices are obtained by linearly interpolating the manually identified points. To illustrate, consider an example image with 20 slices, where the spline points are manually identified on slices 1, 10 and 20. If x1 and x10 denote the x-co-ordinates of two corresponding spline points on slices 1 and 10, respectively, then the x-co-ordinate of the matching spline point on slice 4 is x1 + (x10 − x1)/3.
Step 4: infarct segmentation
As part of the pipeline, we developed a procedure to segment structural remodelling of the tissue, such as an infarct. After the delineation of the ventricles, any infarct tissue present is labelled. In this procedure, the DTMR image is interpolated (Chen et al. 2005) to the resolution of the structural MR image, if the resolution of the latter is higher. From the interpolated DTMR image, a 3-D fractional anisotropy image is generated by computing the fractional anisotropy of the diffusion tensor at each voxel (Basser & Pierpaoli, 1996). The fractional anisotropy is a measure of the directional diffusivity of water, and its values range from 0 to 1. Avalue of 0 indicates perfectly isotropic diffusion, and 1 indicates perfectly anisotropic diffusion. Based on the difference in fractional anisotropy values, the infarct region is separated from the normal myocardium by applying a level set segmentation to the fractional anisotropy image (Wu et al. 2006, 2007). Step 4 in Fig. 2 shows the segmentation of the fractional anisotropy image slice, which corresponds to the example slice. Next, the infarct region is subdivided into two areas, namely, a core, which is assumed to contain inexcitable scar tissue, and a border zone, which is assumed to contain excitable but pathologically remodelled tissue, by thresholding the structural MR image based on the intensity values of the voxels (Yan et al. 2006; Schmidt et al. 2007). Regions of high (<75%) and low grey-level intensities (<25%) are segmented as core scar, while tissue of medium intensity is segmented as border zone. Once any infarct areas present are identified, segmentation of the structural MR image is complete. Step 4 in Fig. 2 illustrates the final segmentation of the example slice.
Step 5: mesh generation
Next, a finite element mesh is generated from the segmented structural MR image. A novel automatic grid-based mesh generation approach was developed recently by our team (Prassl et al. 2009) to overcome limitations of traditional mesh generation approaches that are derived from engineering applications where descriptions of the geometry based on computer-aided-design are required. Guidelines followed in the mesh generation software development included the following: (1) a fully automatic mesh generation technique was desired, with no user interaction at all, to avoid the time-consuming training needed for familiarization with interactive tools and to enable batch-controlled high-throughput mesh generation; (2) finding discrete description of the cardiac surfaces from a segmented image stack, a non-trivial first step typically required in traditional mesh generation (Young et al. 2008), had to be avoided so that segmented image stacks could serve directly as the input for the mesh generation; and (3) the mesh generator had to meet certain technical requirements to be able to deal with the memory demands and computational load associated with the generation of large-scale meshes of up to 50 million nodes and 250 million elements.
Step 5 in Fig. 2 shows the mesh for the example slice. The software is very robust, produces boundary-fitted, locally refined, smooth conformal meshes, and allows the user to select the desired resolution in various regions. Hexahedra-dominant meshes are first generated, which are then split into purely tetrahedral meshes in a postprocessing step. Generated meshes are accurate volume-preserving representations of arbitrarily complex geometries with smooth surfaces. The software (Tarantula; http://www.meshing.at) automatically assigns region identifiers to the elements based on the segmentation, thereby differentiating the elements that belong to the infarct regions, the ventricles or the rest of the tissue. As Fig. 2 illustrates, the interior tissue volume is meshed at a lower resolution, while at the interface between tissue and non-tissue the mesh is refined by a factor of about two. This local adaptation of the resolution significantly reduces the number of elements in the mesh without compromising the geometric detail.
Step 6: fibre mapping
In the final step of the model generation pipeline, our team developed methodology to map fibre orientations onto our anatomical meshes by interpolating the primary diffusion vectors on the centroids of the elements. In this procedure, a reference vector field is constructed by computing the primary eigenvector of each water diffusion tensor in the previously interpolated DTMR image. This vector field is in the same co-ordinate system as the finite element mesh. The fibre orientation assigned to an element in the mesh is the direction of that vector in the reference field nearest to the centroid of the element. Step 6 in Fig. 2 displays the fibre orientations for the example slice.
Simulation of electrical activity
This section describes the methodology associated with simulating the electrophysiological behaviour of the heart developed by our team (detail on some of the aspects of this methodology can be found in the study of Plank et al. 2008). It can be used with both coarse geometrical models of ventricular geometry and image-based models of the heart.
Governing equations
The bidomain equations (Plonsey, 1988), a continuum approximation of cardiac electrical properties based on the representation of the tissue as a functional syncytium, are widely accepted as one of the most complete approaches to describe cardiac bioelectric activity. For a particular class of applications, such as extracellular stimulation and defibrillation, studies involving bath loading effects or calculation of the magnetic field, these equations are the only approach available today. Based on a charge conservation principle, the bidomain equations postulate that all currents entering the intracellular space, by crossing the cell membrane, must leave the extracellular space and vice versa. The transmembrane currents density, Im, represents the source for both the intracellular potential, ϕi, and the extracellular potential, ϕe. We use a recast version of the equations (Pollard et al. 1992) to work directly with the primary variable of interest, the transmembrane voltage, Vm. The entire system of equations is as follows:
| (1) |
| (2) |
| (3) |
where eqn (1) is an elliptic equation, eqn (2) is the parabolic portion, are the intracellular and extracellular conductivity tensors, respectively, β is the membrane surface-to-volume ratio, Itr is the current density of the transmembrane stimulus used to initiate an action potential, Ie is the current density of the extracellular stimulus, Cm is the membrane capacitance per unit area, g⃗ (Vm, η⃗) is a non-linear function describing the temporal evolution of the state variables, η⃗, and Iion is the total current density flowing through the membrane ionic channels, pumps and exchangers. At the tissue boundaries, electrical isolation is assumed by imposing no-flux boundary conditions on ϕe and ϕi.
In many cases of practical interest, cardiac tissue is surrounded by a conductive medium. For instance, when modelling an in vivo setting, the heart is embedded in a torso, and the cavities of the heart are filled with blood. In an in vitro context, a Langendorff-perfused heart is either covered by a thin liquid layer or it is immersed in a fluid bath (Tyrode solution). In these cases, the elliptic equation has to be extended as follows:
| (4) |
where σb is the isotropic conductivity of the conductive medium. In this case, no-flux boundary conditions are imposed at the boundaries of the conductive medium, whereas continuity of the normal component of the extracellular current and continuity of ϕe are enforced at the tissue–bath interface. The no-flux boundary conditions for ϕi remain.
Both monodomain and bidomain simulation were conducted, depending on the particular electrophysiological scenario and the study hypothesis and goals (see examples in the following sections). The numerical scheme in the monodomain case is essentially the same as in the bidomain case, with the difference that the step to solve the elliptic partial differential equation (PDE) and the term including ϕe in eqns (1) and (2) are omitted, and the conductivity tensor is replaced with (Roth, 1997).
Temporal discretization
Temporal discretization of eqns (1) and (2) involves an operator splitting technique (Keener & Bogar, 1998), which decouples the computing schemeinto three components: an elliptic PDE; a parabolic PDE; and a set of non-linear ordinary differential equations (ODEs). Solutions are then found by leapfrogging between the decoupled components, where either Vm in eqn (1) or ϕe in eqn (2) is considered constant. In the simplest case, the parabolic PDE can be solved with a forward Euler scheme (Vigmond et al. 2002), but for our studies, owing to the fine discretization of the mesh, it was advantageous to employ the slightly more expensive semi-implicit Crank–Nicolson scheme, since it is unconditionally stable and therefore allows larger time steps. Details can be found in the study of Vigmond et al. (2008).
Numerical solution
Owing to the problem sizes typical for our whole-heart studies, all simulations were executed in parallel to provide a sufficiently large memory space and to reduce execution times. All solver techniques described below showed very good parallel scaling within the range of four to 64 processors. The solver of the elliptic PDE employed an iterative conjugate gradient (CG) method in conjuction with an algebraic multigrid method preconditioner (Plank et al. 2007). The parabolic problem was solved then using a Block–Jacobi preconditioner for the CG solver together with an Incomplete Cholesky ICC(0) subblock preconditioner with a zero fill-in level to preserve the sparsity pattern. A subset of the ODEs were solved either by employing a non-standard finite difference technique (Maclachlan et al. 2007) based on an approach proposed by Rush & Larsen (1978), or by a simple forward Euler method. Details of the ODE solver technique are described in detail elsewhere (Plank et al. 2008).
Representation of ionic currents and membrane electroporation
For defibrillation studies, all membrane models, i.e. the specific representation of the ionic currents, need to be modified to ensure stability, since the shock produces a dramatic change in transmembrane potential. It is important to understand, however, that defibrillation shocks induce complex changes in transmembrane potential, some of which have been consistently observed in experiments, but never reproduced by membrane models. We therefore developed an augmented version of the Luo–Rudy dynamic model (aLRd;Ashihara&Trayanova, 2004) in which we were able to reproduce the experimentally observed rectangularly shaped positive ΔVm transient, negative-to-positive ΔVm ratio near two (Cheek et al. 2000; Fast et al. 2000), stronger electroporation at the anode (Cheek & Fast, 2004), and dependence of the ΔVm magnitude on field strength (Cheek et al. 2000; Fast et al. 2000). To conduct simulations with the rabbit ventricular model, similar changes were incorporated in the Puglisi–Bers ventricular myocyte ionic membrane model (Puglisi & Bers, 2001). Being equipped with a membrane model that can accurately reproduce the membrane responses to shocks is essential to our ability to simulate the tissue and organ behaviour observed experimentally.
Simulating the Purkinje network
Our team has developed a unique methodology to incorporate the Purkinje network into ventricular models which is detailed by Vigmond & Clements (2007). The Purkinje system (PS) was constructed as a set of cables laid upon the endocardium. Electrical conduction into the myocardium occurred only at the terminals of the network, through specialized Purkinje–myocyte junctions (PMJs). To describe the intracellular potential along each cable segment, one-dimensional cubic Hermite finite elements were used. These elements describe the potential as a cubic polynomial along the element, which is determined by the values of potential and longitudinal current, IL, at the element ends. Since IL is an explicit variable, its conservation is explicitly enforced at PMJs. The cubic Hermite formulation allows discrete gap junctions to be placed between cable segments, as is found physiologically. Transmembrane voltages are tracked at both ends of each element, with nodes shared by elements considered to be two nodes separated by an infinitely thin gap junction.
Originally, to construct the Purkinje network, the endocardial surface of each ventricle was flattened onto a plane, and a conduction system based on generalized descriptions of heart anatomy and excitation mappings (Durrer et al. 1970; Netter et al. 1997) was manually laid out. Since there was a high degree of variability among these sources, only important features were implemented, such as the major areas of activation. The PS was unflattened, and the left and right branches were joined by the bundle of His. The terminals of the Purkinje network were extended into the ventricular wall by appending elements, which made an angle of approximately 45 deg with the surface tangent to avoid any sharp discontinuities, and continued for several hundred micrometres into the tissue. Extraction of Purkinje systems from MRI images is a current topic of research in our laboratory.
In summary: (1) ϕe is computed from the ventricular model only; (2) Vm is updated through the DiFrancesco–Noble model of the Purkinje cell (DiFrancesco & Noble, 1985); (3) IL is computed; (4) the governing parabolic equation for ϕi (eqn (2)) is solved by the finite element method; and (5) transmembrane voltages of myocytes attached to PMJs are modified by the current flowing to/from the PS.
Applications of the modelling methodology
This section provides three examples of the use of the models described above to address various issues in cardiac electrophysiology. We present examples of the use of both coarse geometrical models and high-resolution image-based models of cardiac geometry and structure in cardiac electrophysiology, as well as of the Purkinje system model and the electromechanics model. Each example follows the same exposition structure: rationale; simulation protocol; and findings. The section ‘Mechanisms of cardiac vulnerability and defibrillation’ is based on research published elsewhere (Ashihara et al. 2008), while the rest of the examples present current research towards the development of these predictive models and their applications.
Mechanisms of cardiac vulnerability and defibrillation
Rationale
Defibrillation by high-energy electric shocks is the only reliable procedure for termination of ventricular fibrillation (VF); however, it can result in myocardial dysfunction and damage (Runsio et al. 1997). Furthermore, recent meta-analysis of industrial reports (Maisel, 2006) concluded that thousands of patients have been affected by malfunctions of high-voltage components in implantable cardioverter -defibrillators, causing psychological trauma. Comprehensive knowledge and appreciation of the mechanisms by which a shock interacts with the heart is the approach most likely to succeed in reducing shock energy.
The presence of an isoelectric window (IW), following unsuccessful defibrillation attempts (Chen et al. 1986; Shibata et al. 1988a; Wang et al. 2001) led to the understanding that an electric shock terminates ongoing VF, but then reinitiates it, hence the mechanisms of VF induction and its re-initiation (unsuccessful defibrillation) are the same. In this example of predictive modelling, we propose new mechanisms for postshock propagation and shock-induced arrhythmogenesis following monophasic and biphasic shocks and address the origin of the IW. Specifically, we propose that submerged ‘tunnel’ propagation of postshock activations through shock-induced intramural excitable areas underlies VF induction with monophasic and biphasic shock and the existence of IW. The goal of this study was to test this hypothesis and provide a comprehensive set of mechanisms for VF induction with electric shocks for all ranges of shock strengths and the waveforms used in clinical practice.
Simulation protocol
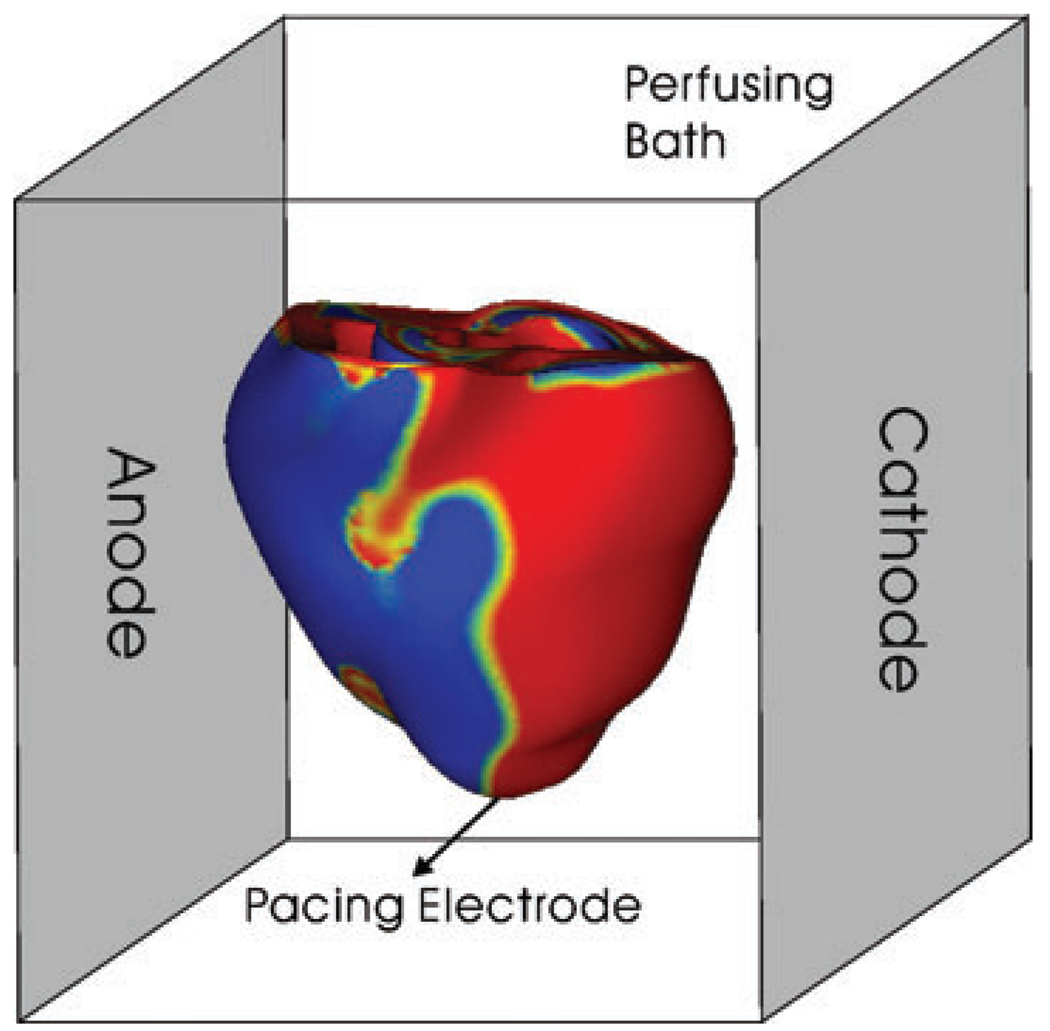
This study used the coarse-geometry rabbit ventricular model described in the section ‘Models of coarse representation of cardiac geometry’. The ventricles were paced at the apex, and shocks were delivered through plate electrodes occupying the vertical borders of the perfusing chamber (distance 3.92 cm), as depicted in Fig. 3. To model the extracellular field stimulation, a bidomain representation of the tissue was used. This set-up mimics experimental protocols against which model results have been validated (Rodriguez et al. 2005).
Figure 3. Anterior view of the rabbit ventricular model in the perfusing bath with plate electrodes that generate a uniformly applied electric field.
Pacing stimuli were applied at the LV apex. The shock induced regions of positive and negative virtual electrode polarization (red and blue regions).
Eight pacing stimuli of 2ms duration and strength twice diastolic threshold were applied at a basic cycle length (BCL) of 300 ms to achieve steady state. Shocks were delivered at various coupling intervals between onset of the last pacing stimulus (assumed as time zero) and shock onset. Monophasic shock duration was 10 ms and biphasic was 6/4 ms (results for biphasic shocks are not shown here, but can be found; Ashihara, 2008). Tilt for both waveforms was 62.0%. Applied field at shock onset (leading edge) represented shock strength. Polarity was anodal, i.e. electrode near left ventricle (LV) was the anode (Fig. 3).
Shock-induced arrhythmias were considered ‘sustained’ or ‘non-sustained’ if the shock induced ≥4 or <4 postshock beats, respectively (Chattipakorn et al. 2000). We defined the ‘initiating postshock activation’ as the first postshock activation to originate within the ventricles after shock end, and the ‘earliest-propagated postshock activation’ as the postshock activation that appeared on the epicardium at the earliest timing following shock end. Clearly, the initiating postshock activation may not become the earliest-propagated postshock activation, since it may die out before reaching the epicardium. The IW, observed on the epicardium, was defined as the time interval between shock end and appearance of earliest globally propagated postshock activation (Chen et al. 1986; Shibata et al. 1988a,b; Wang et al. 2001).
Findings
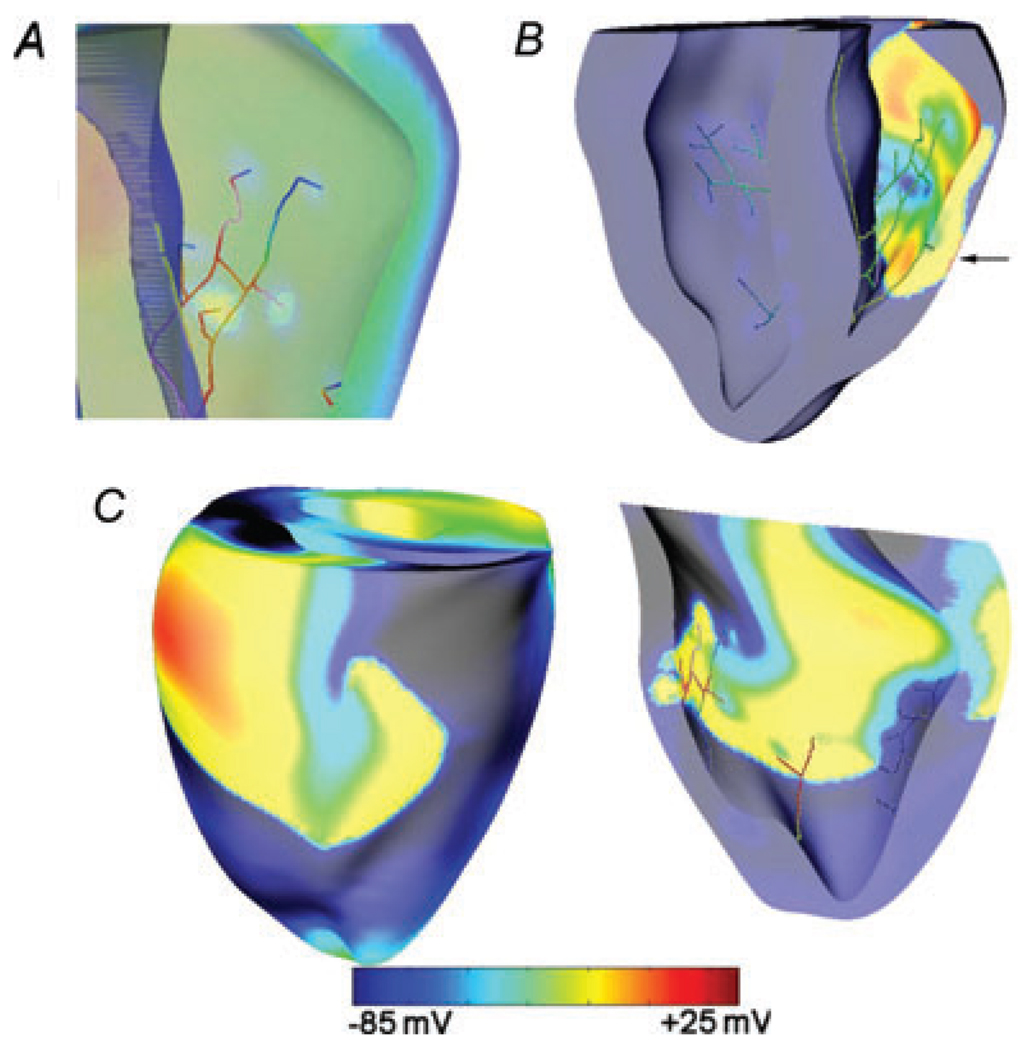
Figure 4A presents transmembrane potential distributions in an apex-to-base cross-section at the end of (230 ms) and 10 ms after (240 ms) 16 V cm−1 monophasic shock at a coupling interval of 220 ms. The shock episode was just below the upper limit of vulnerability and resulted in arrhythmia (Fig. 4C), with the earliest-propagated postshock activation appearing on the epicardial surface as a breakthrough after an IW of about 80ms and degrading into VF. At shock end, a distribution of virtual electrode polarization (VEP) was formed, consistent with previous studies of vulnerability and defibrillation (Rodriguez & Trayanova, 2003; Rodriguez et al. 2005; Ashihara et al. 2008). A large intramural excitable area was formed in the LV wall (230 ms).
Figure 4. Post-shock tunnel propagation.
A, distributions of transmembrane potential in a ventricular cross-section at shock end (230 ms) and 10 ms after (240 ms) for 16 V cm−1 monophasic shock at a 220 ms coupling interval. B, propagation, in the region indicated in by the box in A, of initiating postshock activation following the shock. Black open triangles indicate immediate postshock excitation. Asterisk denotes initiating postshock activation arising from intramural virtual electrodes. C, epicardial breakthrough of tunnel propagation (310 ms) and fibrillation initiation (350 ms). In B and C, white arrows mark propagation direction.
Following this 16 V cm−1 shock, since subepicardial layers were strongly hyperpolarized, they became quickly excited by fast propagation (indicated by open triangles at 245–250 ms; Fig. 4B). Meanwhile, an initiating postshock activation (* at 240 ms in Fig. 4B), which originated at the high transmembrane potential gradient boundary between oppositely polarized virtual electrodes around the apex, propagated at a slower velocity towards the base through the LV intramural excitable area, i.e. within the excitable ‘tunnel’ (white arrows; 240–260 ms in Fig. 4B) until it broke onto the LV epicardium as the earliest-propagated postshock activation. This breakthrough was possible because at that time the epicardial transmembrane potentials had decreased and transmembrane potential gradients were low, thus the wave was able to find excitable tissue to propagate through. The period of propagation of the postshock activation, from its origination in the intramural layers of the LV wall to its breakthrough on the epicardial surface, constitutes the mechanism for the existence of the IW.
Initiation of the postshock activation origin, tunnel propagation and epicardial breakthrough mechanisms presented here are fundamental, since their existence is underpinned only by ventricular structure and virtual electrode formation mechanisms. Figure 5 provides evidence of the robustness of these mechanisms and their validity for defibrillation failure following near-defibrillation threshold shocks. This is due to the fundamental nature of the mechanisms involved and because the state of refractoriness in the ventricles determines only the minimal shock strength needed for tunnel propagation to occur (Ashihara et al. 2008). At the time of shock delivery, the ventricles were in VF, characterized by the presence of numerous scroll waves (see filaments shown in pink in Fig. 5). The shock induced VEP similar to that in the previous figure. Postshock propagation appeared as a breakthrough on the epicardium, and the shock failed by identical mechanisms, with an IW of ~55 ms.
Figure 5. Distributions of transmembrane potential on the epicardial surface for (from left to right) preshock VF, shock end (0 ms), and 37 and 55 ms postshock for 16 V cm−1 monophasic shock.
Preshock VF scroll-wave filaments are shown in pink in the semi-transparent apical view. Epicardial breakthrough of intramural propagation occurs after an isoelectric window of 55 ms.
This study focused on the poorly understood three-dimensional aspects of postshock propagation in the heart, providing novel analysis of the mechanisms underlying postshock activation origin and its subsurface propagation in the process of VF induction by shocks. The study used the capabilities of state-of-the-art 3-D realistic heart modelling, which, already experimentally validated (Rodriguez et al. 2005), provides a unique opportunity to explore behaviour in the depth of the ventricular walls not thus far achievable by any recording technique.
The role of the Purkinje network in cardiac vulnerability and defibrillation
Rationale
The PS is the fast conduction system, responsible for rapid activation of the ventricles. However, it has largely been excluded from modelling studies of ventricular defibrillation. There are several reasons why it will behave differently from the myocardium in response to defibrillation shocks, as follows: (1) the PS possesses different electrophysiological properties from the working myocardium; (2) the shock response of the tissue is dependent on the angle of the muscle fibres with respect to the applied field, and the PS lies on top of the myocardium and has completely different fibre orientation; and (3) the PS may provide alternative propagation pathways and excitations during the immediate postshock response, which may affect re-entry reinitiation. Optical imaging of the Purkinje system at the organ level to measure these effects is difficult because the size of the Purkinje fibres lies below the camera resolution and the signal is swamped by the myocardial signal. Modelling offers a way to gain insight into the shock response of the PS.
Simulation protocol
The Purkinje system model was adjusted so that propagation times across the PMJs matched those observed experimentally, of the order of 5 ms in the anterograde direction and 1.5 ms in the retrograde direction (Wiedmann et al. 1996). These times varied depending on the exact PMJ, owing to geometrically induced myocardial load differences. Furthermore, for His activation, whole ventricular activation times and sites of epicarial breakthrough were compared with experimental observations (Ramanathan et al. 2006).
A uniform applied electric field was applied, with two planar electrodes placed in an anterior–posterior configuration, similar to the model configuration used in the section ‘Mechanisms of cardiac vulnerability and defibrillation’. A train of rapid shocks was applied to a quiescent heart (burst pacing) through the planar electrodes. Shock strength was varied between 2 and 7 V cm−1, and the BCL was varied from 100 to 200 ms. In all runs, tachyarrhythmias were considered sustained if a shock induced a re-entry that lasted for 400 ms, as in the section ‘Mechanisms of cardiac vulnerability and defibrillation’, and IW was also defined as in that section.
Findings
Figure 6 shows the vulnerability window to re-entry during burst pacing. Re-entry was induced with shock strengths ranging from 3.6 to 5.8 V cm−1 and BCLs from 125 to 135 ms. The dark-shaded regions in the figure exhibited non-sustained re-entry. In the presence of the PS, the ventricles are prone to re-entry induction with weaker shocks (3.6 V cm−1). In the absence of the PS, sustained re-entry was obtained with shock strengths greater than 6 V cm−1 (not shown in Fig. 6). The IWs ranged from 20 to 50 ms over the vulnerability window region, with the maximal IW corresponding to a BCL of 125 ms. The IW decreased with increasing BCL. No IWs were observed above the upper limit of vulnerability.
Figure 6. Region of vulnerability determined from the burst pacing protocol.
The shaded region represents successful re-entry induction. The darker shaded region corresponds to a non-sustained re-entry.
Activations were always observed in the PS immediately after the shock. Figure 7A shows the first endocardial postshock activations emanating from the PS strands in the right ventricle (RV) 10ms after the shock (strength, 3.6 V cm−1; and BCL, 130 ms). These endocardial activations propagated transmurally and appeared on the epicardium as the earliest-propagated focal postshock activations (shown in Fig. 7B) 40ms after the shock delivery. This breakthrough marked the end of IW. Note that the breakthrough site was situated exactly opposite a PS insertion point in all the simulations. Often, these activations degraded into re-entrant activity as shown in Fig. 7C. Rotors on the endocardium and epicardium are visible in Fig. 7, which might constitute a transmural re-entry. Initial stages of the re-entry were composed of frequent focal breakthroughs provided by the PS, which helped in stabilizing the rotors. The PS was active throughout the re-entry duration and showed both anterograde and retrograde conductions through PMJs.
Figure 7. Post-shock Purkinje system activation determines isoelectric window.
A, the earliest endocardial activations following the 3.6 V cm−1 shock during the burst pacing protocol (BCL 130 ms) are induced by Purkinje strands. B, these activations propagate transmurally to appear as the first propagated postshock breakthrough on the epicardium that marked the end of IW. Black arrow shows the breakthrough location. C, postshock activations degrade into re-entry; rotors are seen on RV epicardium (left) and endocardium (right).
These results indicate that the PS might have to be taken into account when modelling ventricular defibrillation. The presence of the PS leads to re-entry initiation at lower shock strengths, and could be the source of the first post-shock activations, providing an alternative mechanism which may coexist with the one presented in the section ‘Mechanisms of cardiac vulnerability and defibrillation’.
Modelling ventricular tachycardia associated with an infarct
Rationale
The role of the complex infarct geometry and altered electrophysiology of the infarct zone in initiation and maintenance of ventricular tachycardia (VT) is not fully understood. Current clinical practice relies on catheter recordings of endocardial electrical activity to map the location of infarct-related re-entrant circuits (Stevenson et al. 1998). Unfortunately, this technique is capable of providing insight into a subset of post-infarction re-entry only. Ventricular tachycardia can arise from re-entry circuits that may traverse transmural and epicardial regions of the myocardium (Kaltenbrunner et al. 1991). This suggests that the complex architecture and composition of the infarct within the ventricular wall results in the induction of multiple morphologies of VT.
The goal of this study was to elucidate how specific infarct geometry and composition result in the formation of non-endocardially located re-entrant circuits. The study used a high-resolution image-based canine heart model (Arevalo et al. 2008a,b) that incorporates accurate infarct geometry and distribution generated from MR and DTMR scans (scans acquired by Dr P.A. Helm, University of Virginia) to explore and characterize post-infarction VT formation and maintenance. The model is currently being validated with sock epicardial data recordings at Johns Hopkins University School of Medicine.
Simulation protocol
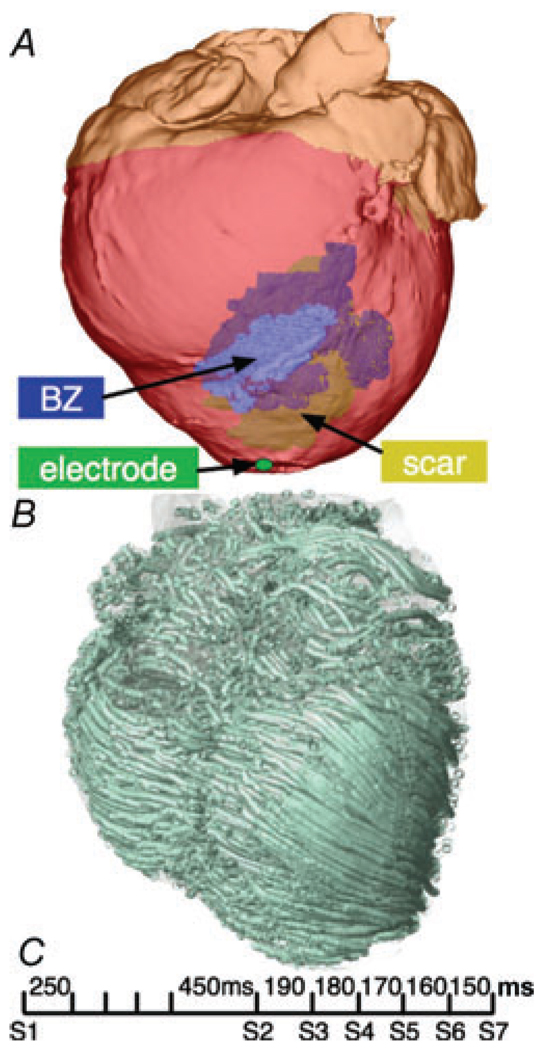
Figure 8A and B shows the model generated from high-resolution MR and DTMR scans of a canine heart ~4 weeks post-infarction scanned at a resolution of 300 µm × 300 µm × 800 µm, using the new image-based model generation pipeline described in the section ‘Construction of geometrical models’. The model incorporates accurate cardiac geometry, including representations of the different regions of the infarct, namely, a non-excitable scar and the partly viable border zone (Fig. 8A), and realistic fibre orientation obtained from the DTMR data (Fig. 8B). Border zone conductivity values were assigned based on previous studies reporting a 90% decrease in transverse coupling due to gap junctional remodelling (Yao et al. 2003). The infarct scar and atria were assumed non-excitable.
Figure 8. MRI-derived infarcted canine heart model.
A, anterior view of the infarcted canine heart with inexcitable scar (yellow) and partly viable border zone (BZ; blue). B, the corresponding fibre orientation calculated from the DTMR, visualized as streamlines. C, aggressive pacing protocol delivered at the LV apex to induce VT.
The ionic kinetics in the normal and border zone myocardium were represented by the Luo–Rudy dynamic model (Luo & Rudy, 1994). Membrane kinetics in the border zone was modified based on data from the literature. Previous studies of the border zone in infarcted canine hearts have reported a reduction in peak sodium current to 38% of the normal value (Pu & Boyden, 1997), in peak L-type calcium current to 31% of normal (Dun et al. 2004), and in peak potassium currents IKr and IKs to 30 and 20% of the maximum (Jiang et al. 2000), respectively. These modifications result in longer action potential duration and decreased excitability compared with the normal myocardium. The mathematical description of current flow in cardiac tissue was based on the monodomain representation.
To examine the arrhythmogenic propensity of the infarct substrate, an aggressive pacing protocol was delivered from the apex (Fig. 8C), similar to protocols used for clinical evaluation of patients with myocardial infarction. Pacing commenced at a BCL of 250 ms for five beats (S1); 450 ms after the last S1, six stimuli were delivered at progressively shorter coupling intervals, starting at 190 ms and decreasing in steps of 10 ms. The induced activity was monitored for an additional 2.5 s. The model findings, as described below, are in good agreement with sock recordings on the epicardial surface (Ashikaga et al. 2007).
Findings
Figure 9 illustrates the events that lead to VT induction. It depicts isochrones of epicardial activation times for the time period from the onset of a given stimulus (S4, S5 and S6) to the time of delivery of the subsequent stimulus. Images on the right present the intramural activation pattern on a slice through the heart, the location of which is indicated by the white line on the S4 epicardium.
Figure 9. VT induction in anatomically detailed canine heart model.
Anterior views (left) of the epicardial activation maps of figure-of-eight re-entry anchored on the border zone 200 ms following aggressive apical pacing (S4, S5 and S6) and the corresponding long axis transmural views of the activation maps (right; view indicated by dashed line in S4).
Judging from the fibre orientations shown in Fig. 8, the wavefront on the epicardium propagates in a direction transverse to the fibres. Upon reaching the border zone, the decreased transverse conductivity there results in a significantly slower conduction compared with the surrounding normal tissue. This can be seen from the shorter distance between isochrones in Fig. 9. Faster wavefronts from the normal myocardium converge into the border zone laterally (white arrows), activating the entire border zone. In the S5 panel of Fig. 9, the transmural view also shows late activation of the border zone due to the wavefront propagating from the normal myocardium. Since the border zone has a longer action potential duration, it remains refractory while the surrounding myocardium is fully recovered.
At slow pacing rates, the refractory border zone does not result in re-entry formation, since it recovers before the arrival of the subsequent beat. As the pacing rate is increased, the wavefront encounters refractory tissue, resulting in conduction block. In the S6 panel of Fig. 9, a sizeable portion of border zone does not activate because of conduction block of the preceding beat (black region), providing a pathway for re-entry. This region of block later becomes the conduit for wavefront propagation from the intramural border zone towards the surface. When pacing is completed, the activation from within the border zone tissue develops into an epicardial quatrefoil re-entry. The re-entry core remains within the border zone and is sustained throughout the simulation with a rotation frequency of 5Hz.
Previous experimental studies of infarcted canine hearts have reported the induction of VT with epicardial re-entry morphology (Ashikaga et al. 2007; Ciaccio et al. 2007). The novel image-based modelling technique developed for this study revealed that decreased excitability, longer action potential duration and reduced conduction velocity throughout the border zone promoted conduction block and wave break that develops into epicardial re-entry. Furthermore, the simulation showed that the site of wave break and re-entry formation occurred in both the epicardial and the intramural portions of the border zone. Owing to the relative position of the scar, the arrhythmogenic substrate would be undetectable by electrical mapping of the endocardium. Thus, this study showcased the utility of computational methods in predicting sites of re-entry formation and maintenance.
Concluding remarks
This article presented the state-of-art in computational modelling of the heart as developed by our team, both in terms of methodology and in the use of these models to address issues in cardiac electrophysiology pertinent to arrhythmogenesis and the response of the heart to electric shocks. It examined both the geometrical assembly of the models, presenting the new pipeline for developing image-based models of the heart, and the numerical techniques employed to solve the governing equations. The studies presented here demonstrate that simulating cardiac electrical function is a successful integrative multiscale modelling approach pertinent to human disease. While general principles of cardiac propagation were seen in these studies, similar to previous modelling work in the field (Berenfeld & Jalife, 1998; Ten Tusscher et al. 2007; Clayton, 2008), specific outcomes were only manifest here because of the detailed anatomical representation of the organ. With a large number of accurate heart models, as available through our pipeline, we can uncover important phenomena and identify structures in which pathologies are likely to arise.
Although not detailed in this paper, the heart is ultimately a mechanical pump with important electromechanical feedback mechanisms. Integration of mechanical and electrical function in computer models is needed to fully address cardiac performance (Hunter et al. 2003; Vigmond et al. 2007; Kerckhoffs et al. 2008; Niederer & Smith, 2008; Gurev et al. 2008). Today we stand at the threshold of a new era in computational modelling of the heart. Anatomically detailed, tomographically reconstructed models of hearts from various species are being developed, as demonstrated here, that integrate functions from the ion channel to the electromechanical interactions in the intact heart (Hunter et al. 2008). Such models hold great promise for enhanced interpretation of clinical and physiological measurements in terms of cellular mechanisms and for improving the basic understanding of the mechanisms of dysfunction in disease conditions, such as re-entrant arrhythmias, myocardial ischaemia and heart failure. Although this progress has been encouraging, efforts to develop comprehensive human heart models have lagged well behind those aimed at simulating animal models (Rudy et al. 2008), and a concerted effort is needed to understand the differences and similarities between human and animal cardiac electrophysiological behaviour. Furthermore, we need to make better use of available human data to develop integrative, physiologically realistic multiscale models of the human heart. We also need to combine these models with patient-specific clinical information towards the goal of developing new tools and technologies that can inform clinical decisions and improve healthcare delivery.
Acknowledgements
This work was supported by American Heart Association postdoctoral fellowship no. 0725392U to V.G.; Marie Curie Fellowship MC-OIF 040190 funded by the European Commission and SFB grant F3210-N18 from the Austrian Science Fund (FWF) to G.P.; NIH grants R01-HL063195, R01- HL082729 and R01-HL067322 and NSF grant CBET-0601935 to N.T.; and Alberta Ingenuity Fund & Natural Sciences and Engineering Research Council of Canada grants to E.V.
References
- Adams R, Bischof L. Seeded region growing. IEEE Trans Pattern Anal Mach Intell. 1994;16:641–647. [Google Scholar]
- Arevalo H, Helm PA, Trayanova N. Development of a model of the infarcted canine heart that predicts arrhythmia generation from specific cardiac geometry and scar distribution. Computers in Cardiology. 2008a;35:497–500. [Google Scholar]
- Arevalo H, Prassl A, Plank G, Helm P, Halperin HR, Trayanova N. Heart Rhythm. vol. 5. San Francisco: 2008b. Model of the infarcted canine heart predicts arrhythmia generation from specific cardiac geometry and scar distribution; p. S109. [Google Scholar]
- Arevalo H, Rodriguez B, Trayanova NA. Arrhythmogenesis in the heart: multiscale modeling of the effects of defibrillation shocks and the role of electrophysiological heterogeneity. Chaos. 2007;17 doi: 10.1063/1.2430637. 015103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashihara T, Constantino J, Trayanova NA. Tunnel propagation of postshock activations as a hypothesis for fibrillation induction and isoelectric window. Circ Res. 2008;102:737–745. doi: 10.1161/CIRCRESAHA.107.168112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashihara T, Trayanova NA. Asymmetry in membrane responses to electric shocks: insights from bidomain simulations. Biophys J. 2004;87:2271–2282. doi: 10.1529/biophysj.104.043091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashikaga H, Sasano T, Dong J, Zviman MM, Evers R, Hopenfeld B, Castro V, Helm RH, Dickfeld T, Nazarian S, Donahue JK, Berger RD, Calkins H, Abraham MR, Marban E, Lardo AC, McVeigh ER, Halperin HR. Magnetic resonance-based anatomical analysis of scar-related ventricular tachycardia: implications for catheter ablation. Circ Res. 2007;101:939–947. doi: 10.1161/CIRCRESAHA.107.158980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basser PJ, Pierpaoli C. Microstructural and physiological features of tissues elucidated by quantitative-diffusion-tensor MRI. J Magn Reson. 1996;111:209–219. doi: 10.1006/jmrb.1996.0086. [DOI] [PubMed] [Google Scholar]
- Berenfeld O, Jalife J. Purkinje-muscle reentry as a mechanism of polymorphic ventricular arrhythmias in a 3-dimensional model of the ventricles. Circ Res. 1998;82:1063–1077. doi: 10.1161/01.res.82.10.1063. [DOI] [PubMed] [Google Scholar]
- Burton RAB, Plank G, Schneider JE, Grau V, Ahammer H, Keeling SL, Lee J, Smith NP, Gavaghan D, Trayanova NA, Kohl P. Three-dimensional models of individual cardiac histoanatomy: tools and challenges. Ann N Y Acad of Sci. 2006;1080:301–319. doi: 10.1196/annals.1380.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chattipakorn N, Rogers JM, Ideker RE. Influence of postshock epicardial activation patterns on initiation of ventricular fibrillation by upper limit of vulnerability shocks. Circulation. 2000;101:1329–1336. doi: 10.1161/01.cir.101.11.1329. [DOI] [PubMed] [Google Scholar]
- Cheek ER, Fast VG. Nonlinear changes of transmembrane potential during electrical shocks: role of membrane electroporation. Circ Res. 2004;94:208–214. doi: 10.1161/01.RES.0000111526.69133.DE. [DOI] [PubMed] [Google Scholar]
- Cheek ER, Ideker RE, Fast VG. Nonlinear changes of transmembrane potential during defibrillation shocks: role of Ca2+ current. Circ Res. 2000;87:453–459. doi: 10.1161/01.res.87.6.453. [DOI] [PubMed] [Google Scholar]
- Chen J, Liu W, Zhang H, Lacy L, Yang X, Song S-K, Wickline SA, Yu X. Regional ventricular wall thickening reflects changes in cardiac fiber and sheet structure during contraction: quantification with diffusion tensor MRI. Am J Physiol Heart Circ Physiol. 2005;289:H1898–H1907. doi: 10.1152/ajpheart.00041.2005. [DOI] [PubMed] [Google Scholar]
- Chen P, Shibata N, Dixon E, Wolf P, Danieley N, Sweeney M, Smith W, Ideker R. Activation during ventricular defibrillation in open-chest dogs. Evidence of complete cessation and regeneration of ventricular fibrillation after unsuccessful shocks. J Clin Invest. 1986;77:810–823. doi: 10.1172/JCI112378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciaccio EJ, Ashikaga H, Kaba RA, Cervantes D, Hopenfeld B, Wit AL, Peters NS, McVeigh ER, Garan H, Coromilas J. Model of reentrant ventricular tachycardia based on infarct border zone geometry predicts reentrant circuit features as determined by activation mapping. Heart Rhythm. 2007;4:1034–1045. doi: 10.1016/j.hrthm.2007.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clayton RH. Vortex filament dynamics in computational models of ventricular fibrillation in the heart. Chaos. 2008;18 doi: 10.1063/1.3043805. 043127. [DOI] [PubMed] [Google Scholar]
- DiFrancesco D, Noble D. A model of cardiac electrical activity incorporating ionic pumps and concentration changes. Philos Trans R Soc Lond B Biol Sci. 1985;307:353–398. doi: 10.1098/rstb.1985.0001. [DOI] [PubMed] [Google Scholar]
- Dun W, Baba S, Yagi T, Boyden PA. Dynamic remodeling of K+ and Ca2+ currents in cells that survived in the epicardial border zone of canine healed infarcted heart. Am J Physiol Heart Circ Physiol. 2004;287:H1046–H1054. doi: 10.1152/ajpheart.00082.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durrer D, Van Freud GE, Janse MJ, Meijler FL, Arzbaecher RC. Total excitation of the isolated human heart. Circulation. 1970;41:899–912. doi: 10.1161/01.cir.41.6.899. [DOI] [PubMed] [Google Scholar]
- Efimov IR, Nikolski VP, Salama G. Optical imaging of the heart. Circ Res. 2004;95:21–33. doi: 10.1161/01.RES.0000130529.18016.35. [DOI] [PubMed] [Google Scholar]
- Fast VG, Rohr S, Ideker RE. Nonlinear changes of transmembrane potential caused by defibrillation shocks in strands of cultured myocytes. Am J Physiol Heart Circ Physiol. 2000;278:H688–H607. doi: 10.1152/ajpheart.2000.278.3.H688. [DOI] [PubMed] [Google Scholar]
- Fenton FH, Cherry EM, Karma A, Rappel WJ. Modeling wave propagation in realistic heart geometries using the phase-field method. Chaos. 2005;15:13502. doi: 10.1063/1.1840311. [DOI] [PubMed] [Google Scholar]
- Gurev V, Constantino J, Trayanova N. Transmural dyssynchrony of myofiber shortening is determined by depolarization sequence within myocardial layers. AHA Scientific Sessions. 2008;vol. 118:S349–S350. [Google Scholar]
- Hunter PJ, Crampin EJ, Nielsen PM. Bioinformatics, multiscale modeling and the IUPS Physiome Project. Brief Bioinform. 2008;9:333–343. doi: 10.1093/bib/bbn024. [DOI] [PubMed] [Google Scholar]
- Hunter PJ, Pullan AJ, Smaill BH. Modelling total heart function. Annu Rev Biomed Eng. 2003;5:147–177. doi: 10.1146/annurev.bioeng.5.040202.121537. [DOI] [PubMed] [Google Scholar]
- Ibanez L, Schroeder W, Ng L, Cates J. The ITK Software Guide: The Insight Segmentation and Registration Toolkit. New York: Kitware Inc.; 2003. [Google Scholar]
- Jiang M, Cabo C, Yao J, Boyden PA, Tseng G. Delayed rectifier K currents have reduced amplitudes and altered kinetics in myocytes from infarcted canine ventricle. Cardiovasc Res. 2000;48:34–43. doi: 10.1016/s0008-6363(00)00159-0. [DOI] [PubMed] [Google Scholar]
- Kaltenbrunner W, Cardinal R, Dubuc M, Shenasa M, Nadeau R, Tremblay G, Vermeulen M, Savard P, Page PL. Epicardial and endocardial mapping of ventricular tachycardia in patients with myocardial infarction. Is the origin of the tachycardia always subendocardially localized? Circulation. 1991;84:1058–1071. doi: 10.1161/01.cir.84.3.1058. [DOI] [PubMed] [Google Scholar]
- Keener JP, Bogar K. A numerical method for the solution of the bidomain equations in cardiac tissue. Chaos. 1998;8:234–241. doi: 10.1063/1.166300. [DOI] [PubMed] [Google Scholar]
- Kerckhoffs RC, McCulloch AD, Omens JH, Mulligan LJ. Effects of biventricular pacing and scar size in a computational model of the failing heart with left bundle branch block. Med Image Anal. 2008 doi: 10.1016/j.media.2008.06.013. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Law MWK, Chung ACS. Weighted local variance-based edge detection and its application to vascular segmentation in magnetic resonance angiography. IEEE Trans Med Imaging. 2007;26:1224–1241. doi: 10.1109/TMI.2007.903231. [DOI] [PubMed] [Google Scholar]
- Li W, Kohl P, Trayanova N. Induction of ventricular arrhythmias following mechanical impact: a simulation study in 3D. J Mol Histol. 2004;35:679–686. doi: 10.1007/s10735-004-2666-8. [DOI] [PubMed] [Google Scholar]
- Luo C, Rudy Y. A dynamic model of the cardiac ventricular action potential. I. Simulations of ionic currents and concentration changes. Circ Res. 1994;74:1071–1096. doi: 10.1161/01.res.74.6.1071. [DOI] [PubMed] [Google Scholar]
- Maclachlan M, Sundnes J, Raymond S. A comparison of non-standard solvers for ODEs describing cellular reactions in the heart. Comput Methods Biomech Biomed Eng. 2007;10:317–326. doi: 10.1080/10255840701259301. [DOI] [PubMed] [Google Scholar]
- Maisel WH. Pacemaker and ICD generator reliability: meta-analysis of device registries. JAMA. 2006;295:1929–1934. doi: 10.1001/jama.295.16.1929. [DOI] [PubMed] [Google Scholar]
- Netter FH. Atlas of Human Anatomy. 2nd ed. New Jersey: Novartis; 1997. [Google Scholar]
- Niederer SA, Smith NP. An improved numerical method for strong coupling of excitation and contraction models in the heart. Prog Biophys Mol Biol. 2008;96:90–111. doi: 10.1016/j.pbiomolbio.2007.08.001. [DOI] [PubMed] [Google Scholar]
- Nielsen PM, Le Grice IJ, Smaill BH, Hunter PJ. Mathematical model of geometry and fibrous structure of the heart. Am J Physiol Heart Circ Physiol. 1991;260:H1365–H1378. doi: 10.1152/ajpheart.1991.260.4.H1365. [DOI] [PubMed] [Google Scholar]
- Panfilov AV. Three-dimensional organization of electrical turbulence in the heart. Phys Rev E Stat Phys Plasmas Fluids Relate Interdiscip Yopics. 1999;59:R6251–R6254. doi: 10.1103/physreve.59.r6251. [DOI] [PubMed] [Google Scholar]
- Panfilov AV, Keldermann RH, Nash MP. Drift and breakup of spiral waves in reaction–diffusion–mechanics systems. Proc Natl Acad Sci USA. 2007;104:7922–7926. doi: 10.1073/pnas.0701895104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plank G, Liebmann M, Weber dos Santos R, Vigmond EJ, Haase G. Algebraic multigrid preconditioner for the cardiac bidomain model. IEEE Trans Biomed Eng. 2007;54:585–596. doi: 10.1109/TBME.2006.889181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plank G, Zhou L, Greenstein JL, Cortassa S, Winslow RL, O’Rourke B, Trayanova NA. From mitochondrial ion channels to arrhythmias in the heart: computational techniques to bridge the spatio-temporal scales. Philos Transact A Math Phys Eng Sci. 2008;366:3381–3409. doi: 10.1098/rsta.2008.0112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plonsey R. Bioelectric sources arising in excitable fibers (ALZA lecture) Ann Biomed Eng. 1988;16:519–546. doi: 10.1007/BF02368014. [DOI] [PubMed] [Google Scholar]
- Pollard AE, Hooke N, Henriquez CS. Cardiac propagation simulation. Crit Rev Biomed Eng. 1992;20:171–210. [PubMed] [Google Scholar]
- Prassl AJ, Kickinger F, Ahammer H, Grau V, Schneider JE, Hofer E, Vigmond EJ, Trayanova NA, Plank G. Automatically generated, anatomically accurate meshes for cardiac electrophysiology problems. 2009 doi: 10.1109/TBME.2009.2014243. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pu J, Boyden PA. Alterations of Na+ currents in myocytes from epicardial border zone of the infarcted heart. A possible ionic mechanism for reduced excitability and postrepolarization refractoriness. Circ Res. 1997;81:110–119. doi: 10.1161/01.res.81.1.110. [DOI] [PubMed] [Google Scholar]
- Puglisi JL, Bers DM. Labheart: an interactive computer model of rabbit ventricular myocyte ion channels and ca transport. Am J Physiol Cell Physiol. 2001;281:C2049–C2060. doi: 10.1152/ajpcell.2001.281.6.C2049. [DOI] [PubMed] [Google Scholar]
- Ramanathan C, Jia P, Ghanem R, Ryu K, Rudy Y. Activation and repolarization of the normal human heart under complete physiological conditions. Proc Natl Acad Sci USA. 2006;103:6309–6314. doi: 10.1073/pnas.0601533103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodgers A, Vaughan P. The World Health Report: Reducing Risks, Promoting Healthy Life. Geneva, Switzerland: The World Health Organization; 2002
- Rodriguez B, Li L, Eason J, Efimov I, Trayanova NA. Differences between left and right ventricular chamber geometry affect cardiac vulnerability to electric shocks. Circ Res. 2005;97:168–175. doi: 10.1161/01.RES.0000174429.00987.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez B, Trayanova NA. Upper limit of vulnerability in a defibrillation model of the rabbit ventricles. J Electrocardiol. 2003;36 Suppl:51–56. doi: 10.1016/j.jelectrocard.2003.09.066. [DOI] [PubMed] [Google Scholar]
- Roth BJ. Electrical conducitivity values wth the bidomain model of cardiac tissue. IEEE Trans Biomed Eng. 1997;44:326–328. doi: 10.1109/10.563303. [DOI] [PubMed] [Google Scholar]
- Rudy Y, Ackerman MJ, Bers DM, Clancy CE, Houser SR, London B, McCulloch AD, Przywara DA, Rasmusson RL, Solaro RJ, Trayanova NA, Van Wagoner DR, Varro A, Weiss JN, Lathrop DA. Systems approach to understanding electromechanical activity in the human heart: a national heart, lung, and blood institute workshop summary. Circulation. 2008;118:1202–1211. doi: 10.1161/CIRCULATIONAHA.108.772715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Runsio M, Kallner A, Kallner G, Rosenqvist M, Bergfeldt L. Myocardial injury after electrical therapy for cardiac arrhythmias assessed by troponin-T release. Am J Cardiol. 1997;79:1241–1245. doi: 10.1016/s0002-9149(97)00090-8. [DOI] [PubMed] [Google Scholar]
- Rush S, Larsen H. A practical algorithm for solving dynamic membrane equations. IEEE Trans Biomed Eng. 1978;25:389–392. doi: 10.1109/TBME.1978.326270. [DOI] [PubMed] [Google Scholar]
- Schmidt A, Azevedo CF, Cheng A, Gupta SN, Bluemke DA, Foo TK, Gerstenblith G, Weiss RG, Marbán E, Tomaselli GF, Lima JAC, Wu KC. Infarct tissue heterogeneity by magnetic resonance imaging identifies enhanced cardiac arrhythmia susceptibility in patients with left ventricular dysfunction. Circulation. 2007;115:2006–2014. doi: 10.1161/CIRCULATIONAHA.106.653568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sethian JA. Level Set Methods: Evolving Interfaces in Computational Geometry, Fluid Mechanics, Computer Vision, and Materials Science. Cambridge: Cambridge University Press; 1996. [Google Scholar]
- Shibata N, Chen PS, Dixon EG, Wolf PD, Danieley ND, Smith WM, Ideker RE. Influence of shock strength and timing on induction of ventricular arrhythmias in dogs. Am J Physiol Heart Circ Physiol. 1988a;255:H891–H901. doi: 10.1152/ajpheart.1988.255.4.H891. [DOI] [PubMed] [Google Scholar]
- Shibata N, Chen PS, Dixon EG, Wolf PD, Danieley ND, Smith WM, Ideker RE. Epicardial activation after unsuccessful defibrillation shocks in dogs. Am J Physiol Heart Circ Physiol. 1988b;255:H902–H909. doi: 10.1152/ajpheart.1988.255.4.H902. [DOI] [PubMed] [Google Scholar]
- Stevenson WG, Friedman PL, Kocovic D, Sager PT, Saxon LA, Pavri B. Radiofrequency catheter ablation of ventricular tachycardia after myocardial infarction. Circulation. 1998;98:308–314. doi: 10.1161/01.cir.98.4.308. [DOI] [PubMed] [Google Scholar]
- Ten Tusscher KH, Hren R, Panfilov AV. Organization of ventricular fibrillation in the human heart. Circ Res. 2007;100:e87–e101. doi: 10.1161/CIRCRESAHA.107.150730. [DOI] [PubMed] [Google Scholar]
- Trayanova N. Defibrillation of the heart: insights into mechanisms from modelling studies. Exp Physiol. 2006;91:323–337. doi: 10.1113/expphysiol.2005.030973. [DOI] [PubMed] [Google Scholar]
- Trayanova N, Eason J, Aguel F. Computer simulations of cardiac defibrillation: a look inside the heart. Comput Visual Sci. 2002;V4:259–270. [Google Scholar]
- Vetter FJ, McCulloch AD. Three-dimensional analysis of regional cardiac function: a model of rabbit ventricular anatomy. Prog Biophys Mol Biol. 1998;69:157–183. doi: 10.1016/s0079-6107(98)00006-6. [DOI] [PubMed] [Google Scholar]
- Vigmond EJ, Aguel F, Trayanova NA. Computational techniques for solving the bidomain equations in three dimensions. IEEE Trans Biomed Eng. 2002;49:1260–1269. doi: 10.1109/TBME.2002.804597. [DOI] [PubMed] [Google Scholar]
- Vigmond EJ, Clements C. Construction of a computer model to investigate the sawtooth effect in the Purkije system. IEEE Trans Biomed Eng. 2007;54:389–399. doi: 10.1109/TBME.2006.888817. [DOI] [PubMed] [Google Scholar]
- Vigmond EJ, Clements C, McQueen DM, Peskin C. Effect of bundle branch block on cardiac output: a whole heart simulation study. Prog Biophys Mol Biol. 2007;97:520–542. doi: 10.1016/j.pbiomolbio.2008.02.022. [DOI] [PubMed] [Google Scholar]
- Vigmond EJ, Weber Dos Santos R, Prassl AJ, Deo M, Plank G. Solvers for the cardiac bidomain equations. Prog Biophys Mol Biol. 2008;96:3–18. doi: 10.1016/j.pbiomolbio.2007.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang NC, Lee MH, Ohara T, Okuyama Y, Fishbein GA, Lin SF, Karagueuzian HS, Chen PS. Optical mapping of ventricular defibrillation in isolated swine right ventricles: demonstration of a postshock isoelectric window after near-threshold defibrillation shocks. Circulation. 2001;104:227–233. doi: 10.1161/01.cir.104.2.227. [DOI] [PubMed] [Google Scholar]
- Wiedmann RT, Tan RC, Joyner RW. Discontinuous conduction at Purkinje–ventricular muscle junction. Am J Physiol Heart Circ Physiol. 1996;271:H1507–H1516. doi: 10.1152/ajpheart.1996.271.4.H1507. [DOI] [PubMed] [Google Scholar]
- Wu EX, Wu Y, Nicholls JM, Wang J, Liao S, Zhu S, Lau CP, Tse HF. MR diffusion tensor imaging study of postinfarct myocardium structural remodeling in a porcine model. Magn Reson Med. 2007;58:687–695. doi: 10.1002/mrm.21350. [DOI] [PubMed] [Google Scholar]
- Wu M-T, Tseng W-YI, Su M-YM, Liu C-P, Chiou K-R, Wedeen VJ, Reese TG, Yang C-F. Diffusion tensor magnetic resonance imaging mapping the fiber architecture remodeling in human myocardium after infarction: correlation with viability and wall motion. Circulation. 2006;114:1036–1045. doi: 10.1161/CIRCULATIONAHA.105.545863. [DOI] [PubMed] [Google Scholar]
- Xie F, Qu Z, Yang J, Baher A, Weiss JN, Garfinkel A. A simulation study of the effects of cardiac anatomy in ventricular fibrillation. J Clin Invest. 2004;113:686–693. doi: 10.1172/JCI17341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan A, Shayne A, Brown K, Gupta S, Chan C, Luu T, Di Carli M, Reynolds G, Stevenson W, Kwong R. Characterization of the peri-infarct zone by contrast-enhanced cardiac magnetic resonance imaging is a powerful predictor of post-myocardial infarction mortality. Circulation. 2006;114:32–39. doi: 10.1161/CIRCULATIONAHA.106.613414. [DOI] [PubMed] [Google Scholar]
- Yao JA, Hussain W, Patel P, Peters NS, Boyden PA, Wit AL. Remodeling of gap junctional channel function in epicardial border zone of healing canine infarcts. Circ Res. 2003;92:437–443. doi: 10.1161/01.RES.0000059301.81035.06. [DOI] [PubMed] [Google Scholar]
- Young PG, Beresford-West TBH, Coward SRL, Notarberardino B, Walker B, Abdul-Aziz A. An efficient approach to converting three-dimensional image data into highly accurate computational models. Philos Trans R Soc A Math Phys Eng Sci. 2008;366:3155–3173. doi: 10.1098/rsta.2008.0090. [DOI] [PubMed] [Google Scholar]