Abstract
The diversity of functional and life-history traits of organisms depends on adaptation as well as the legacy of shared ancestry. Although the evolution of traits in macro-organisms is well studied, relatively little is known about character evolution in micro-organisms. Here, we surveyed an ancient and ecologically important group of microbial plant symbionts, the arbuscular mycorrhizal (AM) fungi, and tested hypotheses about the evolution of functional and life-history traits. Variation in the extent of root and soil colonization by AM fungi is constrained to a few nodes basal to the most diverse groups within the phylum, with relatively little variation associated with recent divergences. We found no evidence for a trade-off in biomass allocated to root versus soil colonization in three published glasshouse experiments; rather these traits were positively correlated. Partial support was observed for correlated evolution between fungal colonization strategies and functional benefits of the symbiosis to host plants. The evolution of increased soil colonization was positively correlated with total plant biomass and shoot phosphorus content. Although the effect of AM fungi on infection by root pathogens was phylogenetically conserved, there was no evidence for correlated evolution between the extent of AM fungal root colonization and pathogen infection. Variability in colonization strategies evolved early in the diversification of AM fungi, and we propose that these strategies were influenced by functional interactions with host plants, resulting in an evolutionary stasis resembling trait conservatism.
Keywords: arbuscular mycorrhizal fungi, functional traits, symbiosis, phylogenetic trait conservatism
1. Introduction
The functional characteristics of organisms are influenced by adaptation to environmental conditions and by shared ancestry. Because functional traits contribute to the acquisition of resources and define the niches that organisms occupy, the process of character evolution can have strong implications for the assembly and functioning of ecosystems (Webb et al. 2002; Wiens & Graham 2005; Cadotte et al. 2008). Although it has long been recognized that closely related species share functional characteristics (Darwin 1859), broad patterns of trait evolution have only been recently quantified in plant and animal lineages (Peterson et al. 1999; Prinzing et al. 2001; Losos et al. 2003; Cavender-Bares et al. 2004; Swenson & Enquist 2007).
Despite progress in understanding character evolution in macro-organisms, patterns of character evolution in micro-organisms have been more difficult to assess because of challenges associated with phenotyping and genotyping individuals (Green et al. 2008). Nevertheless, micro-organisms have important ecological roles in communities and ecosystems. In particular, arbuscular mycorrhizal (AM) fungi engage in functionally important symbioses with species from many terrestrial plant families and are completely reliant on their hosts to obtain photosynthetically fixed carbon (Smith & Read 2008). In exchange, they may enhance host fitness via a variety of mechanisms, including enhanced efficiency of nutrient uptake (Ames et al. 1983; Harrison & Buuren 1995) and protection from pathogenic micro-organisms (Borowicz 2001). Understanding how the functional and life-history traits of AM fungi have evolved can reveal the mechanisms of community assembly in these groups as well as predict the functional consequences of AM fungal diversity for ecosystems (Maherali & Klironomos 2007; Cadotte et al. 2008).
Theories of the evolution of AM fungi, a lineage that is more than 400 million years old (Berbee & Taylor 2000; Redecker 2002), suggest phylogenetic conservatism and trade-offs among multiple functions (Koide 2000; Hart & Reader 2002). The AM fungal phylum Glomeromycota is currently divided into four orders, with most described species belonging to the Glomerales and Diversisporales (Schüßler et al. 2001). Three families have been described within the Diversisporales; of these, the Acaulosporaceae and Gigasporaceae contain the greatest number of described species. The Glomerales consists of a single family (the Glomeraceae) and genus (Glomus), although these taxa group into two distinct clades (Schwarzott et al. 2001). Hart & Reader (2002) conducted a broad survey of colonization strategies expressed within the Glomeromycota. Generally, members of the Gigasporaceae tended to extensively colonize soil while exhibiting slow and limited colonization of roots. Conversely, members of the Glomeraceae rapidly and extensively colonized host root systems but produced limited biomass in soil. Members of the Acaulosporaceae tended to be poor colonizers of both soil and roots relative to species of Gigasporaceae and Glomeraceae, respectively. The dramatic differences in root and soil colonization between the Gigasporaceae and the Glomeraceae have raised the hypothesis that there is a functional trade-off in allocating biomass to colonize plant roots versus colonizing soil (Hart & Reader 2002) that has contributed to the divergence between these lineages.
The functional differences among AM fungal lineages in their colonization strategy could be related to specific mechanisms for promoting host plant growth (Fitter 2005; Maherali & Klironomos 2007). The extensive root colonization of the Glomeraceae in comparison with other AM fungal families is hypothesized to reduce root infection by soil pathogens (Newsham et al. 1995). By contrast, the high level of hyphal growth within the soil by the Gigasporaceae compared with other AM fungal families is hypothesized to enhance nutrient and water uptake (Newsham et al. 1995; Maherali & Klironomos 2007; Van der Heijden & Scheublin 2007).
In this study, we combined information on the phylogeny of AM fungi with trait data to examine patterns of character evolution and test for the evolution of trade-offs between functional traits. First, we determined whether AM fungal species that share a common evolutionary history also share certain traits and elicit common symbiotic responses (trait conservatism). We then quantified the extent of trait conservatism, identifying the nodes at which important divergences are likely to have occurred. We then addressed two hypotheses that could explain the existence of distinct colonization strategies in AM fungi. In particular, we asked (i) is there evidence for a trade-off in biomass being allocated to growth in one environment (roots) over the other (soil)? and (ii) is there evidence for correlated evolution between colonization strategies and the benefits obtained by host plants via the symbiosis?
2. Material and methods
(a). Study designs and trait data collection
To examine the functional traits of AM fungi and the benefits they provide to plant hosts, we have grown a variety of AM fungal species with several plant hosts derived from a single old-field site in Guelph, Ontario, Canada (Hart & Reader 2002, 2004, 2005; Maherali & Klironomos 2007). In the present study, we analysed these previously published data to specifically evaluate patterns of character evolution and functional trade-offs using comparative phylogenetic methods.
In each experiment, plants were grown individually in media inoculated with a single AM fungal species, cultures of which were initiated from single fungal spores. The number of plant and fungal species used and the responses estimated varied among the studies, though there was considerable overlap (table S1 in the electronic supplementary material). Maherali & Klironomos (2007) (‘study 1’) quantified plant and fungal variables after 1 year of growth with Plantago lanceolata in 20 cm diameter pots containing sterilized field soil. Individual AM fungal species were maintained in culture with P. lanceolata as the host plant for 1 year, and to initiate the study, 50 g of AM fungal inocula from these cultures was added to each pot along with a germinated seedling. The series of papers by Hart & Reader (2002, 2004, 2005) were based on a single study and quantified plant and fungal variables in a group of AM fungal species grown with four plant species (P. lanceolata, Plantago major, Poa annua and Poa pratensis). Plants and fungi were grown in 4 cm diameter ‘Conetainers’ containing a sand- and potting soil-based growth medium. Individual AM fungal species in these containers were cultured for 30 days with a surrogate host species (Allium porrum) prior to planting with the four target species. In one experiment (‘study 2’), the 30-day-old mycelial network was disrupted by sieving soil through a 250 µm mesh prior to sowing seedlings of the plant host. In a second experiment, the 30-day-old mycelial network was left intact prior to sowing (‘study 3’). In studies 2 and 3, host plants were harvested after 12 weeks of growth.
In each study, root (percentage of root length infected; McGonigle et al. 1990) and soil (m hyphae g−1 soil; Newman 1966; Miller et al. 1995) colonization by fungi were measured. In studies 2 and 3, the rate of root colonization (days to initial colonization) and the rate of soil colonization (maximum growth rate, days to reach maximum growth rate) were measured. The rate of soil colonization was determined by fitting time-series data (Hart & Reader 2005) to a sigmoidal model using the ‘nlme’ package (Pinheiro et al. 2006) in R v. 2.8 (R Development Core Team 2008). To estimate the benefit that host plants received from AM fungi, total plant biomass and the phosphorus (P) concentration of leaf tissue were measured. In study 1, the degree to which hosts benefited from pathogen protection was determined by quantifying the per cent root length infected by pathogenic fungi (Fusarium oxysporum, Pythium spp.).
(b). Phylogenetic relationships
To obtain a phylogeny of the AM fungal species used in our experiments, we used the Glomeromycota phylogeny by Redecker & Raab (2006) as a guide for sequence and locus selection. We queried GenBank and retrieved all 18S ribosomal RNA gene sequences used to generate the phylogeny published by Redecker & Raab (2006) and also retrieved representative 18S sequences for additional taxa that were present in our dataset.
Sequence data were available for all of our additional taxa of interest except Glomus aggregatum and Glomus microaggregatum. For these two taxa, 18S sequence data were generated from spores derived from two isolates (G. aggregatum FL312D, G. microaggregatum UT126B), obtained from the International Culture Collection of (Vesicular) Arbuscular Mycorrhizal Fungi (West Virginia University, Morgantown, WV, USA). DNA was extracted from spores following the protocol of Redecker et al. (1997). PCR amplification of 18S rRNA gene sequences was performed using primers SR6.1 (Parrent & Vilgalys in press) and SR1R (Vilgalys & Hester 1990). PCR products were ligated into the PCR 2.1-TOPO cloning kit (Invitrogen, Carlsbad, CA, USA), sequenced with primers SR1R, SR6.1, NS1.5 (James et al. 2000) and NS4 (White et al. 1990) using Big Dye chemistry v. 3.1 and visualized on an ABI3730 automated sequencer (Applied Biosystems, Foster City, CA, USA).
To construct the phylogenetic tree, we first aligned the sequences using Muscle (Edgar 2004). The alignment was manually edited and ambiguous regions were excluded from the analysis. The resulting alignment (available upon request) was analysed using MrModeltest v. 2.3 (Nylander 2004) to determine the best-fitting model of nucleotide evolution (a general time-reversible model with invariant sites and gamma-distributed substitution rates). The phylogeny was estimated with MrBayes v. 3.1.2 (Ronquist & Huelsenbeck 2003) using two independent Markov chain Monte Carlo runs of 10 million generations; equilibrium was reached within the first two million generations, which were not included (burn-in) when generating the consensus phylogeny.
The phylogenetic analysis recovered a topology consistent with that presented by Redecker & Raab (2006) (figure S1 in the electronic supplementary material). All taxa except those for which trait data are available were deleted from the phylogeny with branch lengths retained using PAUP v. 4.01b (Swofford 2002). The resulting phylogeny was used for all comparative analyses.
(c). Statistical analyses
Because each of the studies differed in methodology (container size, experimental duration) we analysed trait evolution separately for each study. Data were log- or arcsine square root-transformed, where appropriate, to realize normality of error distributions.
We used command-line software (Phylocom v. 3.41; Webb et al. 2007) to test for trait conservatism, calculate the root mean square deviation in trait values between daughter nodes at each node (divergence, D), and estimate the contribution of these nodal divergences to overall variation (contribution index). Statistical significance of D was determined against a distribution of 1000 simulated estimates derived from randomization of trait values across the tree tips (Blomberg et al. 2003; Moles et al. 2005; Webb et al. 2007); a significant reduction in the tree-wide average for D is indicative of trait conservatism, while the statistical significance of individual divergences was evaluated at each node. The contribution index at a particular node is the product of (i) the trait variance at that node relative to variance among descended tips and (ii) the variance within the clade descended from that node relative to all clades descended from the tree root. A high value indicates that divergence at that node accounts for a large proportion of trait variation in the phylogeny (Moles et al. 2005; Webb et al. 2007).
Correlations among AM fungal traits and between AM fungal traits and host responses, after correcting for shared evolutionary histories (phylogenetically independent contrasts (PICs); Felsenstein 1985), were determined using the ‘ape’ package (v. 2.2-2; Paradis et al. 2004) in R. PICs account for covariance in trait values for closely related taxa by estimating differences in trait values between lineages and standardizing this difference by the estimated time since the two lineages diverged. Analyses were performed on an ultrametricized tree (figure S2 in the electronic supplementary material) obtained using the non-parametric rate smoothing algorithm of Sanderson (1997). PIC correlations might be subject to bias associated with non-random sampling of taxa (Ackerly 2000); however, we argue that this bias is probably minimal because the taxa were sampled in such a way to represent described AM fungal families and known taxonomic richness within these families, not with regard to character state divergence among these families and taxa. We obtained data for two isolates of Acaulospora spinosa and used the median value of each response in all subsequent analyses.
Because we analysed multiple studies and taxon sampling differed among studies, we obtained multiple PICs for many nodes. To allow us to use all PICs, and to account for the hierarchical structure (PICs estimated within each study) and lack of balance in the data (variable numbers of PICs between studies), we used linear mixed-effects models and likelihood ratio (LR) tests to determine statistical significance, as implemented in the ‘lme4’ package (v. 0.999375-28; Bates et al. 2008) in R. Observed LRs were compared with a simulated distribution of 1000 randomly generated LRs to assess the statistical significance of correlations (Faraway 2006). Tree-wide correlations were estimated as well as correlations specific to the Gigasporaceae and the Glomeraceae.
3. Results
(a). Tree-wide trait conservatism and nodal contributions
As expected, the main data clusters in the dataset were grouped by the different studies (figures 1 and 2). Nonetheless, we were able to identify general patterns that, in most cases, were qualitatively similar across the different studies.
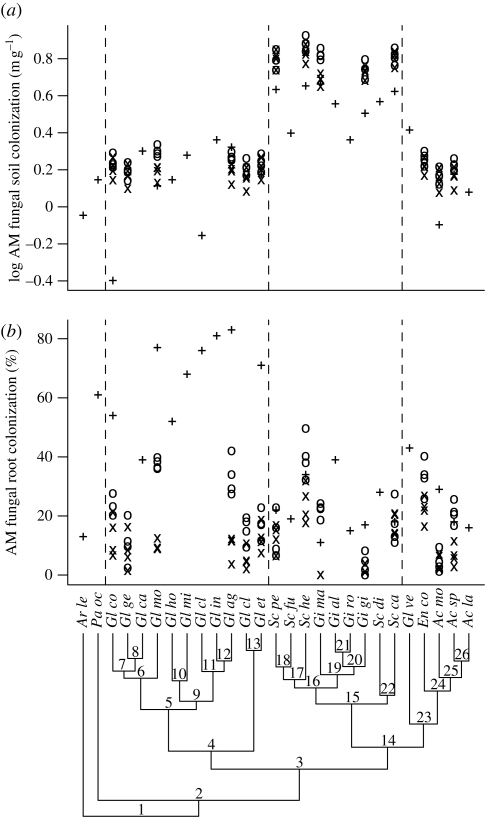
Figure 1.
Phylogenetic distribution of traits associated with the extent of AM fungal growth in the (a) soil and (b) root environments. Data were collected from three studies (plus symbols, study 1; × symbols, study 2; open circles, study 3). These parameters were estimated for four host species in studies 2 and 3.
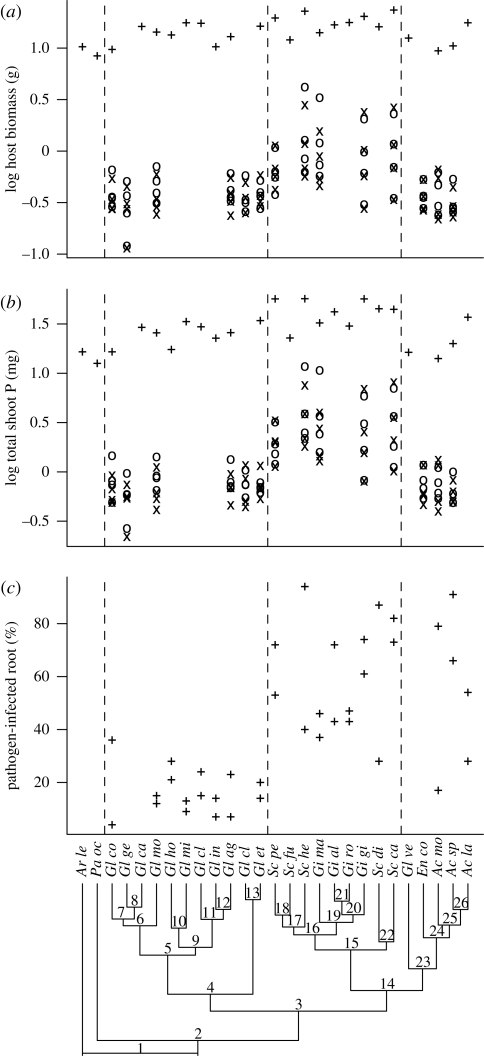
Figure 2.
Phylogenetic distribution of putative benefits associated with AM fungal symbiotic functioning: (a) Total host biomass and (b) P uptake in host shoots were estimated in three studies (plus symbols, study 1; × symbols, study 2; open circles, study 3) and these parameters were estimated for four host species in studies 2 and 3. (c) Extent of infection by two fungal pathogens (F. oxysporum, Pythium spp.) was estimated in study 1.
We found that the extent of soil colonization (soil hyphal length) was conserved within the Glomeromycota, a result that varied in magnitude but was statistically significant across all studies (figure 1a and table 1). Divergence estimates for this trait were significantly higher than expected by chance for the split between the Gigasporaceae and the rest of the Diversisporales and (marginally) for the Glomerales–Diversisporales split, and these nodes provided the largest contributions to variance in this trait across the phylum (table 2). Beyond these early divergences, the extent of soil colonization was conserved within lineages, with more recent nodes contributing little to the overall variation across the tree. In addition, divergence estimates were often small for ancestral nodes within the Glomeraceae and Gigasporaceae (nodes 4, 5 and 15; figure 1a; table S2 in the electronic supplementary material).
Table 1.
Average tree-wide divergences (D) for fungal traits and functions. (Asterisks indicate that responses were significantly conserved. n.a., response not estimated. ***p < 0.001, **p < 0.01, *p < 0.05.)
| response | study 1a |
study 2a |
study 3a |
||||||
|---|---|---|---|---|---|---|---|---|---|
| Plb | Pl | Pmb | Pab | Ppb | Pl | Pm | Pa | Pp | |
| trait | |||||||||
| AM fungal root colonization | 0.11*** | 0.08* | 0.08 | 0.08 | 0.09 | 0.11 | 0.15 | 0.12 | 0.10 |
| time to root colonization | n.a. | 15.33 | 12.33 | 17.56 | 14.11 | 17.65 | 20.86 | 17.65 | 11.30 |
| AM fungal soil hyphal length | 0.14*** | 0.07*** | 0.07*** | 0.07*** | 0.07*** | 0.08*** | 0.08** | 0.08** | 0.07*** |
| maximum rate of soil colonization | n.a. | 0.04 | 0.05 | 0.04 | 0.07 | 0.06 | 0.04 | 0.06 | 0.09 |
| time to maximum rate in soil | n.a. | 0.04 | 0.04 | 0.05 | 0.07 | 0.13 | 0.10 | 0.10 | 0.16 |
| symbiotic function | |||||||||
| host biomass | 0.08 | 0.06*** | 0.05*** | 0.23* | 0.07** | 0.06*** | 0.05*** | 0.24* | 0.07* |
| host shoot P | 0.11* | 0.09** | 0.08*** | 0.25* | 0.10** | 0.10*** | 0.09*** | 0.24** | 0.09** |
| Fusarium root infection | 0.12** | n.a. | n.a. | n.a. | n.a. | n.a. | n.a. | n.a. | n.a. |
| Pythium root infection | 0.13*** | n.a. | n.a. | n.a. | n.a. | n.a. | n.a. | n.a. | n.a. |
aStudy from which the data were collected (§2).
bHost plant species: Pl, P. lanceolata; Pm, P. major; Pa, P. annua; Pp, P. pratensis.
Table 2.
Nodal divergences (D) and contributions to tree-wide variation (contribution index, CI) in fungal traits. (Only nodes representing significantly high divergence estimates are presented. Asterisks indicate the level of statistical significance for the estimate of D. ***p < 0.001, **p < 0.01, *p < 0.05.)
| trait | study numbera | node numberb | hostc | D | CI |
|---|---|---|---|---|---|
| AM fungal root colonization | |||||
| 1 | 3 | Pl | 0.31** | 0.68 | |
| 2 | 16 | Pl | 0.31** | 0.61 | |
| 2 | 16 | Pm | 0.21* | 0.41 | |
| 2 | 16 | Pa | 0.22* | 0.46 | |
| 2 | 16 | Pp | 0.32** | 0.59 | |
| time to root colonization | |||||
| 2 | 6 | Pm | 44.97* | 0.43 | |
| 3 | 16 | Pp | 27.36* | 0.32 | |
| AM fungal soil hyphal length | |||||
| 1 | 14 | Pl | 0.27* | 0.22 | |
| 2 | 14 | Pl | 0.43*** | 0.59 | |
| 2 | 14 | Pm | 0.43** | 0.58 | |
| 2 | 14 | Pa | 0.42** | 0.59 | |
| 2 | 14 | Pp | 0.41** | 0.58 | |
| 3 | 14 | Pl | 0.42** | 0.60 | |
| 3 | 14 | Pm | 0.41** | 0.60 | |
| 3 | 14 | Pa | 0.42** | 0.60 | |
| 3 | 14 | Pp | 0.42** | 0.61 | |
| maximum rate of soil colonization | |||||
| 3 | 4 | Pl | 0.10* | 0.25 | |
| 3 | 19 | Pm | 0.25* | 0.70 | |
| time to maximum rate in soil | |||||
| 2 | 6 | Pa | 0.15** | 0.41 | |
| 3 | 19 | Pm | 0.30* | 0.38 | |
aStudy from which the data were collected (§2).
bAs labelled in figure 1.
cHost plant species: Pl, P. lanceolata; Pm, P. major; Pa, P. annua; Pp, P. pratensis.
Estimates of trait divergence for the extent of AM fungal root colonization at each node were more variable among the studies (figure 1b, and table 1), but we still detected evidence of trait conservatism. In study 1, there was little variation in this trait after the divergence of the Diversisporales and the Glomerales, which explained a majority of variation across the phylum (figure 2b and table 2). A large divergence was observed at node 16 in study 2 for all host species, but not for any other node in studies 2 or 3 (table 2). Although AM fungal species differed in the rate of root or soil colonization, these traits were highly variable throughout the AM fungal phylogeny (figure S3 in the electronic supplementary material and table 1).
With the exception of host biomass in study 1, we detected trait conservatism for many of the putative benefits that AM fungi provide to their plant hosts (figure 2 and table 1). Significant divergences were always associated with the Glomerales–Diversisporales split (pathogen colonization) and the split between the Gigasporaceae and the rest of the Diversisporales (host biomass, total P uptake), and these nodes taken together explained almost all of the variation in these responses across the phylum (table 3).
Table 3.
Nodal divergences (D) and contributions to tree-wide variation (contribution index, CI) in fungal symbiotic functions. (Only nodes representing significantly high divergence estimates are presented. Asterisks indicate the level of statistical significance for the estimate of D. ***p < 0.001, **p < 0.01, *p < 0.05.)
| symbiotic function | study numbera | node numberb | hostc | D | CI |
|---|---|---|---|---|---|
| host biomass | |||||
| 1 | 2 | Pl | 0.19* | 0.25 | |
| 1 | 14 | Pl | 0.13* | 0.19 | |
| 2 | 14 | Pl | 0.24** | 0.60 | |
| 2 | 14 | Pm | 0.28*** | 0.81 | |
| 2 | 3 | Pa | 0.36* | 0.39 | |
| 2 | 14 | Pp | 0.17* | 0.45 | |
| 3 | 14 | Pl | 0.25*** | 0.68 | |
| 3 | 14 | Pm | 0.25** | 0.68 | |
| 3 | 14 | Pa | 0.49* | 0.33 | |
| 3 | 14 | Pp | 0.16* | 0.44 | |
| host shoot P | |||||
| 1 | 14 | Pl | 0.25** | 0.28 | |
| 2 | 14 | Pl | 0.35** | 0.51 | |
| 2 | 14 | Pm | 0.39** | 0.67 | |
| 2 | 3 | Pa | 0.42* | 0.40 | |
| 2 | 14 | Pa | 0.57* | 0.33 | |
| 2 | 14 | Pp | 0.32** | 0.55 | |
| 3 | 14 | Pl | 0.39** | 0.62 | |
| 3 | 14 | Pm | 0.35** | 0.64 | |
| 3 | 14 | Pa | 0.65* | 0.42 | |
| 3 | 14 | Pp | 0.27* | 0.47 | |
| Fusarium root infection | |||||
| 1 | 3 | Pl | 0.25** | 0.55 | |
| Pythium root infection | |||||
| 1 | 3 | Pl | 0.42*** | 0.78 | |
aStudy from which the data were collected (§2).
bAs labelled in figure 2.
cHost plant species: Pl, P. lanceolata; Pm, P. major; Pa, P. annua; Pp, P. pratensis.
(b). Trade-offs in colonization strategies
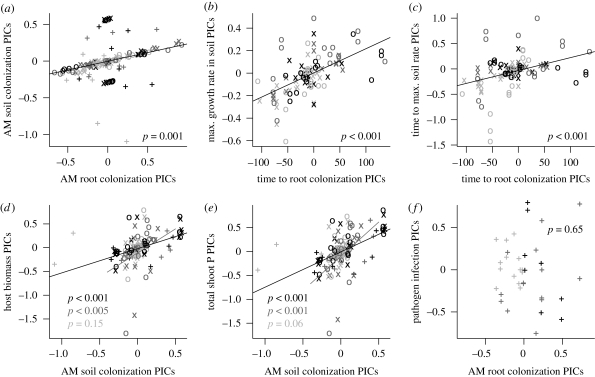
After accounting for shared evolutionary histories, we did not find that the extent of fungal soil colonization was negatively correlated with colonization within roots. On the contrary, soil-colonization PICs were positively associated with PICs of root colonization (LR = 13.4, p = 0.001) (figure 3a). This pattern was also observed when analysing the Gigasporaceae separately (LR = 32.8, p < 0.001). Within the Glomeraceae, the positive correlation was only significant for the traits estimated in studies 2 and 3 (LR = 79.5, p < 0.001; all data: LR = 1.3, p = 0.31). We also observed positive correlations between PICs for the rates at which the root and soil environments were colonized. This relationship was observed using either the estimated maximum rate of hyphal growth (LR = 39.0, p < 0.001; figure 3b) or the number of days to reach the maximum hyphal growth rate (LR = 14.4, p < 0.001; figure 3c) as a surrogate of soil colonization rate, and was largely consistent between the Glomeraceae (max. rate: LR = 14.5, p = 0.001; days to max. rate: LR = 16.5, p < 0.001) and the Gigasporaceae (max. rate: LR = 11.2, p = 0.001; days to max. rate: LR = 3.8, p = 0.06).
Figure 3.
PICs of AM fungal traits and putative symbiotic functions. Positive evolutionary correlations were observed between the (a) extents and (b,c) rates of root and soil colonization. PICs for the extent of AM fungal colonization were significantly correlated with those for (d) total host biomass and (e) total P in host shoots, but not for (f) pathogen infection. Symbols refer to the studies from which the data were collected (plus symbols, study 1; × symbols, study 2; open circles, study 3). Shading of points, lines and significance levels refer to the family-level affiliation of the nodes where contrasts were calculated (light grey, Glomeraceae; dark grey, Gigasporaceae; black points refer to other nodes, black lines refer to the model fit including all points).
(c). Fungal colonization and host benefit
AM fungal colonization of soil was positively correlated with plant performance. Soil colonization PICs were positively correlated with those for host biomass (LR = 20.5, p < 0.001; figure 3d) and total shoot P (LR = 34.9, p < 0.001; figure 3e). The effect coefficients and LRs were larger and highly significant in the Gigasporaceae (biomass: LR = 8, p = 0.005; total P: LR = 15.6, p < 0.001), but smaller and marginally or not significant in the Glomeraceae (biomass: LR = 2.2, p = 0.15; total P: LR = 3.6, p = 0.06). However, different patterns between these families were largely influenced by a few data points (nodes 7 and 11 with P. lanceolata in study 1, node 17 with P. annua as the host in studies 2 and 3); removal of these data points resulted in significant correlations with similar coefficients for both the Gigasporaceae and the Glomeraceae.
AM fungal root colonization PICs were also positively correlated with those of host biomass (LR = 9.5, p = 0.003) and P uptake (LR = 11.4, p < 0.001). However, this was largely owing to the confounding effects of the positive evolutionary correlation between the extent of root and soil colonization. Treating soil colonization extent as a covariate resulted in root colonization having marginal effects on plant performance (biomass: LR = 3.3, p = 0.072; total P: LR = 3.4, p = 0.065). PIC correlations between soil colonization extent and host benefit remained significant even when including root colonization as a covariate (biomass: LR = 13.3, p < 0.001; total P: LR = 24.5, p < 0.001), confirming the greater importance for colonization of the soil environment for host benefit.
AM fungal root colonization PICs were not significantly correlated with PICs for pathogen infection when assessing the relationship across the Glomeromycota (LR = 0.2, p = 0.65) or within the two AM fungal families (Glomeraceae: LR = 2.2, p = 0.22; Gigasporaceae: LR = 2.7, p = 0.12) (figure 3f).
4. Discussion
We found strong evidence for phylogenetic conservation of functional traits and host benefits of AM fungi in three glasshouse experiments, regardless of the identity of the host plant species. Though the magnitude of the nodal contributions for these traits and benefits varied among studies, much of the variation was associated with nodes representing early divergences within the Glomeromycota that occurred prior to the diversification of the Glomeraceae and Gigasporaceae, which presently harbour the greatest species richness within the phylum. Trait conservation is also supported by the observation that functional traits and host benefits were largely invariant within descendent lineages relative to variation between lineages.
The differences in nodal contributions to soil and root colonization and host benefit among studies may have been caused by differences in methodology. All traits were measured after one year in study 1 and after three months in studies 2 and 3. The differences between studies in the length of the experiments could have affected fungal traits and host benefit in several ways. For example, the shorter time period of studies 2 and 3 could have constrained the extent of root colonization in the Glomeraceae relative to study 1 (figure 1b). This would have reduced the root colonization differences between the Glomeraceae and other lineages, resulting in a lower contribution index score for the Glomerales–Diversisporales split in studies 2 and 3 (table 2). More generally, our results highlight the importance of developmental stage in characterizing the phenotypes of AM fungi and their effects on hosts.
Although there was evidence of phylogenetic conservatism for the extents of soil and root colonization, the rates of soil and root colonization were not conserved. Estimates of these traits varied in all lineages and among host plant species, suggesting that AM fungal–host interactions are important in determining the expression of these traits. However, further study on the evolutionary history of colonization rates at finer phylogenetic scales may require different methods from those used in the present study. We used fungal species that could be established by growing a single host plant in field soil to obtain spores (‘trap culture’) and inoculating individual spores onto a new host plant (Klironomos 2003), an approach that may be biased towards sampling rapid colonizers in all lineages. For example, Sýkorová et al. (2007) found that the likelihood of observing specific Glomus taxa depended on whether they were sampled directly from field-collected roots or by ‘baiting’ the soil by growing and harvesting a new host plant; many taxa present in the field-collected roots were lost in the process of baiting the soil. Brundrett et al. (1999a,b) found that using a trap culture method similar to the method used here resulted in a paucity of species from the Acaulosporaceae and Gigasporaceae when compared with direct isolation of spores from field soil, and also detected temporal variation in the abundance of AM fungal taxa recovered using trap cultures. Nevertheless, we were able to obtain several taxa from all of these lineages, and this cultivation bias probably did not influence our analyses excessively.
There was no support for the hypothesis that trait conservation was caused by a trade-off in allocation of biomass to root versus soil colonization. Instead, we found evidence for positive correlated evolution between root and soil colonization. As a result, fungi have probably not evolved specialization in root colonization at the expense of soil colonization. The expectation that this type of life-history trade-off is common depends on the assumption that carbon resources available for growth are constant and limiting (i.e. Van Noordwijk & deJong 1986). However, whether carbon resource availability limits fungal growth is uncertain because AM fungi can increase host photosynthetic rate because of increased respiratory demands below ground (Wright et al. 1998) as well as through increased plant access to limiting nutrients such as nitrogen and P. In addition, roots of individual plants are often simultaneously colonized by both the Gigasporaceae and the Glomeraceae (e.g. Lekberg et al. 2007; Maherali & Klironomos 2007), suggesting that plants can support the carbon demands associated with both increased root and soil colonization. If carbon resources are variable and/or not limiting, differences in root and soil colonization may simply reflect variation in resource acquisition and growth rate among AM fungal lineages (Van Noordwijk & deJong 1986). In this case, a positive correlation between root and soil colonization would be expected because each trait reflects fungal growth. Nevertheless, it is also possible that the positive correlation we observed was caused by growing fungi with single plants in pots, where resource limitation is weaker than in the field. Competition among plants in the wild might reduce carbon availability to fungi, which could result in a trade-off between root and soil colonization or simply reduce overall growth.
One hypothesis for the origin and evolution of the mycorrhizal symbiosis with early terrestrial plants is that hyphae and absorptive mycelia of ancient fungi could provide phosphate ions for their hosts, which lacked true roots (Pirozynski & Malloch 1975). Our results support this hypothesis indirectly because there was correlated evolution between increased soil colonization by AM fungal hyphae and host plant benefit, in terms of increased plant biomass and shoot P concentration. However, the fact that correlated evolution between soil colonization and host benefit was observed in all AM fungal lineages raises the question of why the extensive soil colonization trait only evolved in the Gigasporaceae, a question for which we currently have no firm explanation.
We did not observe direct evidence of correlated evolution between the extent of root colonization and host benefit, but we cannot reject the hypothesis that selective feedbacks between fungi and their hosts did not exist. Extensive root colonization is probably a derived trait in the Glomeraceae, based on the current model of AM fungal evolution (Schüßler et al. 2001; Redecker & Raab 2006). The evolution of extensive root colonization in the Glomeraceae probably occurred following their divergence from the low root-colonizing Gigasporaceae and Acaulosporaceae lineages. There was strong evidence for trait conservation in root infection by pathogenic fungi, with this trait being expressed at much higher levels in the Glomeraceae. Fossil evidence suggests that endophytic fungi were common in early root-like structures (Taylor & Osborn 1996) prior to the estimated divergence between the Glomerales and Diversisporales lineages (Simon et al. 1993). Others have suggested that antagonism of pathogenic fungi may have been a function of primitive AM fungi (Brundrett 2002). Despite these lines of evidence, we did not observe correlated evolution between pathogen infection and the extent of AM fungal root colonization in pots by members of the Glomeraceae, suggesting that pathogen protection may not have been the primary cause of the evolution of increased root colonization.
Extensive root colonization in the Glomeraceae may have evolved as an early strategy for enhancing nutrient uptake by the host or a way to enhance carbon acquisition from the host, especially if these processes were limited by the available surface area or by the efficiency of mass flow across the root–fungal interface. High root colonization may have also evolved in concert with the appearance of extensive root systems in seed plants, which would have opened a spatial niche for the Glomeraceae that was not being exploited by the Gigasporaceae (Maherali & Klironomos 2007). Regardless of the evolutionary cause, increased protection against pathogen infection of host roots could explain why host plants maintained symbiosis with the Glomeraceae, even though the Gigasporaceae could provide greater P uptake benefits.
In plant and animal lineages, strong trait conservatism has typically been found at relatively recent divergences in comparisons of co-occurring species (e.g. Ackerly 2004; Cavender-Bares et al. 2004) or in regional and global species datasets that include multiple phyla or divisions (Prinzing et al. 2001; Moles et al. 2005; Swenson & Enquist 2007). The strong functional trait conservatism originating early in the ancient AM fungal lineage may be unique or possibly a common feature of microbial and/or asexual lineages, but these hypotheses still need to be tested. This trait conservatism has likely ecological consequences for the local assembly of AM fungal communities, in that confamilial AM fungal species may be less likely to co-occur because niche overlap within a lineage would lead to competitive exclusion and phylogenetic overdispersion (Maherali & Klironomos 2007). On an evolutionary time-scale, competitive exclusion may cause divergence within lineages that currently express a common colonization strategy. Although we have focused on traits associated with spatial colonization and host benefit, traits associated with environmental tolerances and phenology also have potential to explain the existing richness and distribution of AM fungal taxa (Van der Heijden & Scheublin 2007; Helgason & Fitter 2009).
Acknowledgements
This work was supported by funding from the Natural Sciences and Engineering Research Council of Canada and the Alexander von Humboldt Foundation. We thank Jinzhong Fu and Robert Hanner for helpful discussions.
References
- Ackerly D. D.2000Taxon sampling, correlated evolution, and independent contrasts. Evolution 54, 1480–1492 (doi:10.1554/0014-3820(2000)054[1480:TSCEAI]2.0.CO;2) [DOI] [PubMed] [Google Scholar]
- Ackerly D. D.2004Adaptation, niche conservatism, and convergence: comparative studies of leaf evolution in the California chaparral. Am. Nat 163, 654–671 (doi:10.1086/383062) [DOI] [PubMed] [Google Scholar]
- Ames R. N., Reid C. P. P., Porter L. K., Cambardella C.1983Hyphal uptake and transport of nitrogen from two 15N-labelled sources by Glomus mosseae, a vesicular-arbuscular mycorrhizal fungus. New Phytol. 95, 381–396 (doi:10.1111/j.1469-8137.1983.tb03506.x) [Google Scholar]
- Bates D., Maechler M., Dai B.2008lme4: linear mixed-effects models using S4 classes, R package version 0.999375-28. See http://www.r-project.org [Google Scholar]
- Berbee M. L., Taylor J. W.2000The Mycota, volume VIIB. Systematics and evolution New York, NY: Springer-Verlag [Google Scholar]
- Blomberg S. P., Garland T., Ives A. R.2003Testing for phylogenetic signal in comparative data: behavioral traits are more labile. Evolution 57, 717–745 (doi:10.1111/j.0014-3820.2003.tb00285.x) [DOI] [PubMed] [Google Scholar]
- Borowicz V. A.2001Do arbuscular mycorrhizal fungi alter plant–pathogen relations? Ecology 82, 3057–3068 (doi:10.1890/0012-9658(2001)082[3057:DAMFAP]2.0.CO;2) [Google Scholar]
- Brundrett M. C.2002Coevolution of roots and mycorrhizas of land plants. New Phytol. 154, 275–304 (doi:10.1046/j.1469-8137.2002.00397.x) [DOI] [PubMed] [Google Scholar]
- Brundrett M., Abbott L., Jasper D.1999aGlomalean fungi from tropical Australia. I. Comparison of the effectiveness of isolation procedures. Mycorrhiza 8, 305–314 (doi:10.1007/s005720050251) [Google Scholar]
- Brundrett M. C., Jasper D. A., Ashwath N.1999bGlomalean mycorrhizal fungi from tropical Australia. II. The effect of nutrient levels and host species on the isolation of fungi. Mycorrhiza 8, 315–321 (doi:10.1007/s005720050252) [Google Scholar]
- Cadotte M. W., Cardinale B. J., Oakley T. H.2008Evolutionary history and the effect of biodiversity on plant productivity. Proc. Natl Acad. Sci. USA 105, 17 012–17 017 (doi:10.1073/pnas.0805962105) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavender-Bares J., Ackerly D. D., Baum D. A., Bazzaz F. A.2004Phylogenetic overdispersion in Floridian oak communities. Am. Nat. 163, 823–843 (doi:10.1086/386375) [DOI] [PubMed] [Google Scholar]
- Darwin C.1859On the origin of species Cambridge, MA: Harvard University Press [Google Scholar]
- Edgar R. C.2004Muscle: multiple sequence alignment with high accuracy and high throughput. Nucl. Acids Res. 32, 1792–1797 (doi:10.1093/nar/gkh340) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faraway J. J.2006Extending the linear model with R Boca Raton, FL: Chapman & Hall/CRC [Google Scholar]
- Felsenstein J.1985Phylogenies and the comparative method. Am. Nat. 125, 1–15 (doi:10.1086/284325) [Google Scholar]
- Fitter A. H.2005Darkness visible: reflections on underground ecology. J. Ecol. 93, 231–243 (doi:10.1111/j.0022-0477.2005.00990.x) [Google Scholar]
- Green J. L., Bohannan B. J. M., Whitaker R. J.2008Microbial biogeography: from taxonomy to traits. Science 320, 1039–1043 (doi:10.1126/science.1153475) [DOI] [PubMed] [Google Scholar]
- Harrison M. J., Buuren M. L. V.1995A phosphate transporter from the mycorrhizal fungus Glomus versiforme. Nature 378, 626–629 (doi:10.1038/378626a0) [DOI] [PubMed] [Google Scholar]
- Hart M. M., Reader R. J.2002Taxonomic basis for variation in the colonization strategy of arbuscular mycorrhizal fungi. New Phytol. 153, 335–344 (doi:10.1046/j.0028-646X.2001.00312.x) [Google Scholar]
- Hart M. M., Reader R. J.2004Do arbuscular mycorrhizal fungi recover from soil disturbance differently? Trop. Ecol. 45, 97–111 [Google Scholar]
- Hart M. M., Reader R. J.2005The role of the external mycelium in early colonization for three arbuscular mycorrhizal fungal species with different colonization strategies. Pedobiologia 49, 269–279 (doi:10.1016/j.pedobi.2004.12.001) [Google Scholar]
- Helgason T., Fitter A. H.2009Natural selection and the evolutionary ecology of the arbuscular mycorrhizal fungi (Phylum Glomeromycota). J. Exp. Bot. 60, 2465–2480 (doi:10.1093/jxb/erp144) [DOI] [PubMed] [Google Scholar]
- James T. Y., Porter D., Leander C. A., Vilgalys R., Longcore J. E.2000Molecular phylogenetics of the Chytridiomycota supports the utility of ultrastructural data in chytrid systematics. Can. J. Bot. 78, 336–350 (doi:10.1139/cjb-78-3-336) [Google Scholar]
- Klironomos J. N.2003Variation in plant response to native and exotic arbuscular mycorrhizal fungi. Ecology 84, 2292–2301 (doi:10.1890/02-0413) [Google Scholar]
- Koide R. T.2000Functional complementarity in the arbuscular mycorrhizal symbiosis. New Phytol. 147, 233–235 (doi:10.1111/j.1469-8137.2000.00710.x) [Google Scholar]
- Lekberg Y., Koide R., Rohr J., Aldrich-Wolfe L., Morton J.2007Role of niche restrictions and dispersal in the composition of arbuscular mycorrhizal fungal communities. J. Ecol. 95, 95–105 (doi:10.1111/j.1365-2745.2006.01193.x) [Google Scholar]
- Losos J. B., Leal M., Glor R. E., de Queiroz K., Hertz P. E., Rodriguez Schettino L., Chamizo Lara A., Jackman T. R., Larson A.2003Niche lability in the evolution of a Caribbean lizard community. Nature 424, 542–545 (doi:10.1038/nature01814) [DOI] [PubMed] [Google Scholar]
- Maherali H., Klironomos J. N.2007Influence of phylogeny on fungal community assembly and ecosystem functioning. Science 316, 1746–1748 (doi:10.1126/science.1143082) [DOI] [PubMed] [Google Scholar]
- McGonigle T. P., Miller M. H., Evans D. G., Fairchild G. L., Swan J. A.1990A new method which gives an objective measure of colonization of roots by vesicular–arbuscular mycorrhizal fungi. New Phytol. 115, 495–501 (doi:10.1111/j.1469-8137.1990.tb00476.x) [DOI] [PubMed] [Google Scholar]
- Miller R. M., Jastrow J. D., Reinhardt D. R.1995External hyphal production of vesicular–arbuscular mycorrhizal fungi in pasture and tallgrass prairie communities. Oecologia 103, 17–23 (doi:10.1007/BF00328420) [DOI] [PubMed] [Google Scholar]
- Moles A. T., Ackerly D. D., Webb C. O., Tweddle J. C., Dickie J. B., Westoby M.2005A brief history of seed size. Science 307, 576–580 (doi:10.1126/science.1104863) [DOI] [PubMed] [Google Scholar]
- Newman E. I.1966A method of estimating the total length of root in a sample. J. Appl. Ecol. 3, 139–145 (doi:10.2307/2401670) [Google Scholar]
- Newsham K. K., Fitter A. H., Watkinson A. R.1995Multi-functionality and biodiversity in arbuscular mycorrhizas. Trends Ecol. Evol. 10, 407–411 (doi:10.1016/S0169-5347(00)89157-0) [DOI] [PubMed] [Google Scholar]
- Nylander J. A. MrModeltest, version 2.3. 2004. Distributed by the author. See http://www.abc.se/~nylander/ [Google Scholar]
- Paradis E., Claude J., Strimmer K.2004APE: analyses of phylogenetics and evolution in R language. Bioinformatics 20, 289–290 (doi:10.1093/bioinformatics/btg412) [DOI] [PubMed] [Google Scholar]
- Parrent J. L., Vilgalys R.In press Expression of genes involved in symbiotic carbon and nitrogen transport in Pinus taeda mycorrhizal roots exposed to CO2 enrichment and nitrogen fertilization. Mycorrhiza (doi:10.1007/s00572-009-0250-5) [DOI] [PubMed] [Google Scholar]
- Peterson A. T., Soberón J., Sánchez-Cordero V.1999Conservatism of ecological niches in evolutionary time. Science 285, 1265–1267 (doi:10.1126/science.285.5431.1265) [DOI] [PubMed] [Google Scholar]
- Pinheiro J., Bates D., DebRoy S., Sarkar D. & R Core Development Team 2006nlme: linear and nonlinear mixed effects models, R package version 3.1-89. See http://www.r-project.org [Google Scholar]
- Pirozynski K. A., Malloch D. W.1975The origin of land plants: a matter of mycotrophism. BioSystems 6, 153–164 [DOI] [PubMed] [Google Scholar]
- Prinzing A., Durka W., Klotz S., Brandl R.2001The niche of higher plants: evidence for phylogenetic conservatism. Proc. R. Soc. Lond. B 268, 2383–2389 (doi:10.1098/rspb.2001.1801) [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Development Core Team 2008R: a language and environment for statistical computing Vienna, Austria: R Foundation for Statistical Computing; See http://www.R-project.org [Google Scholar]
- Redecker D.2002New views on fungal evolution based on DNA markers and the fossil record. Res. Microbiol. 153, 125–130 (doi:10.1016/S0923-2508(02)01297-4) [DOI] [PubMed] [Google Scholar]
- Redecker D., Raab P.2006Phylogeny of the Glomeromycota (arbuscular mycorrhizal fungi): recent developments and new gene markers. Mycologia 98, 885–895 (doi:10.3852/mycologia.98.6.885) [DOI] [PubMed] [Google Scholar]
- Redecker D., Thierfelder H., Walker C., Werner D.1997Restriction analysis of PCR-amplified internal transcribed spacers of ribosomal DNA as a tool for species identification in different genera of the order Glomales. Appl. Environ. Microbiol. 63, 1756–1761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronquist F., Huelsenbeck J. P.2003MrBayes 3: bayesian phylogenetic inference under mixed models. Bioinformatics 19, 1572–1574 (doi:10.1093/bioinformatics/btg180) [DOI] [PubMed] [Google Scholar]
- Sanderson M. J.1997A nonparametric approach to estimating divergence times in the absence of rate constancy. Mol. Biol. Evol. 14, 1218 [Google Scholar]
- Schüßler A., Schwarzott D., Walker C.2001A new fungal phylum, the Glomeromycota: phylogeny and evolution. Mycol. Res. 105, 1413–1421 (doi:10.1017/S0953756201005196) [Google Scholar]
- Schwarzott D., Walker C., Schüßler A.2001Glomus, the largest genus of the arbuscular mycorrhizal fungi (Glomales), is nonmonophyletic. Mol. Phylogenet. Evol. 21, 190–197 (doi:10.1006/mpev.2001.1007) [DOI] [PubMed] [Google Scholar]
- Simon L., Bousquet J., Levesque R. C., Lalonde M.1993Origin and diversification of endomycorrhizal fungi and coincidence with vascular land plants. Nature 363, 67–69 (doi:10.1038/363067a0) [Google Scholar]
- Smith S. E., Read D. J.2008Mycorrhizal symbiosis, 3rd edn London, UK: Academic Press [Google Scholar]
- Swenson N. G., Enquist B. J.2007Ecological and evolutionary determinants of a key plant functional trait: wood density and its community-wide variation across latitude and elevation. Am. J. Bot. 94, 451–459 (doi:10.3732/ajb.94.3.451) [DOI] [PubMed] [Google Scholar]
- Swofford D. L.2002PAUP*: phylogenetic analysis using parsimony (* and other methods), version 4.0b10. Sunderland, MA: Sinauer Associates [Google Scholar]
- Sýkorová Z., Ineichen K., Wiemken A., Redecker D.2007The cultivation bias: different communities of arbuscular mycorrhizal fungi detected in roots from the field, from bait plants transplanted to the field, and from a greenhouse trap experiment. Mycorrhiza 18, 1–14 (doi:10.1007/s00572-007-0147-0) [DOI] [PubMed] [Google Scholar]
- Taylor T. N., Osborn J. M.1996The importance of fungi in shaping the paleoecosystem. Rev. Palaeobot. Palynol. 90, 249–262 (doi:10.1016/0034-6667(95)00086-0) [Google Scholar]
- Van der Heijden M. G. A., Scheublin T. R.2007Functional traits in mycorrhizal ecology: their use for predicting the impact of arbuscular mycorrhizal fungal communities on plant growth and ecosystem functioning. New Phytol. 174, 244–250 (doi:10.1111/j.1469-8137.2007.02041.x) [DOI] [PubMed] [Google Scholar]
- Van Noordwijk A. J., de Jong G.1986Acquisition and allocation of resources: their influence on variation in life history tactics. Am. Nat. 128, 137–142 (doi:10.1086/284547) [Google Scholar]
- Vilgalys R., Hester M.1990Rapid genetic identification and mapping of enzymatically amplified ribosomal DNA from several Cryptococcus species. J. Bacteriol. 172, 4238–4246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webb C. O., Ackerly D. D., McPeek M. A., Donoghue M. J.2002Phylogenies and community ecology. Ann. Rev. Ecol. Syst. 33, 475–505 (doi:10.1146/annurev.ecolsys.33.010802.150448) [Google Scholar]
- Webb C. O., Ackerly D. D., Kembel S. W. Phylocom: software for the analysis of community phylogenetic structure and trait evolution, version 3.41. 2007. See http://www.phylodiversity.net/phylocom/ [DOI] [PubMed] [Google Scholar]
- White T. J., Bruns T., Lee S., Taylor J.1990Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In PCR protocols: a guide to methods and applications (eds Innis M., Gelfand D., Sninsky J., White T.), pp. 315–322 San Diego, CA: Academic Press [Google Scholar]
- Wiens J. J., Graham C. H.2005Niche conservatism: integrating evolution, ecology, and conservation biology. Ann. Rev. Ecol. Evol. Syst. 36, 519–539 (doi:10.1146/annurev.ecolsys.36.102803.095431) [Google Scholar]
- Wright D. P., Scholes J. D., Read D. J.1998Effects of VA mycorrhizal colonization on photosynthesis and biomass production of Trifolium repens L. Plant Cell Environ. 21, 209–216 (doi:10.1046/j.1365-3040.1998.00280.x) [Google Scholar]