Abstract
Background
Human promoter polymorphisms in the chemokine co-receptor 5 gene (CCR5) have been noted for association with mother-to-child transmission of HIV (HIV MTCT) as well as reduced receptor expression in vitro, but have not been clearly associated with CCR5 expression in vivo. Placental expression of CCR5 may be influenced by such polymorphisms as well as other in vivo regulatory factors.
Methodology/Principal Findings
We evaluated the associations between infant CCR5 polymorphisms, measures of maternal infection, and placental expression of CCR5 among mother-infant pairs in Blantyre, Malawi. RNA was extracted from placental tissue and used in multiplex real-time PCR to quantify gene expression. Through linear regression, we observed that CCR5-2554T (β = −0.67, 95% CI = −1.23, −0.11) and -2132T (β = −0.75, 95% CI = −0.131, −0.18) were significantly associated with reduced placental expression of CCR5. An incremental increase in CCR5 expression was observed for incremental increases in expression of two heparan sulfate genes involved in viral infection, HS3ST3A1 (β = 0.27, 95% CI = 0.18, 0.35) and HS3ST3B1 (β = 0.11, 95% CI = 0.06, 0.18). Among HIV infected mothers, an incremental increase in maternal HIV viral load was also associated with higher CCR5 expression (β = 0.76, 95% CI = 0.12, 1.39). Maternal HIV status had no overall effect (β = 0.072, 95% CI = −0.57, −0.72). Higher CCR5 expression was observed for mothers with malaria but was not statistically significant (β = 0.37, 95% CI = −0.43, 1.18).
Conclusions/Significance
These results provide in vivo evidence for genetic and environmental factors involved in the regulation of CCR5 expression in the placenta. Our findings also suggest that the measurement of placental expression of CCR5 alone is not an adequate indicator of the risk of mother-to-child transmission of HIV.
Introduction
In sub-Saharan Africa, over 1,300,000 pregnant women were living with HIV in 2007, and more than 300,000 children are newly infected with HIV each year, primarily through mother-to-child transmission (HIV MTCT) [1]. HIV MTCT can occur during pregnancy (intrauterine transmission), during labor and delivery (intrapartum transmission), or through breastfeeding (postpartum transmission).
The chemokine (CC motif) receptor 5 (CCR5), a co-receptor of the CD4 receptor, is used by macrophage-tropic (R5) HIV-1 for cell entry [2] and is genetically regulated by the CCR5 gene [3]. It has been demonstrated that HIV infection and progression is inhibited by competitive ligands (i.e. the chemokine RANTES) binding with the CCR5 receptor [4], [5]. Genetic variants of the CCR5 gene such as the 32-basepair deletion in the open reading frame (CCR5 Δ32) and promoter polymorphisms are also associated with human susceptibility to infection and/or progression of HIV-1 [2], [6], [7], [8], [9], [10], [11], [12], [13]. This is likely explained by variable receptor expression resulting from mutation [14], [15], [16].
Upregulation of CCR5 in the placenta has been noted to increase the risk of HIV MTCT [17]. However, CCR5 expression may be altered not only by variants in the CCR5 gene but also by environmental factors such as maternal infection. Variability in such factors may limit the external validity of in vivo findings. Thus, one aim of this study was to evaluate the affects of infant CCR5 promoter polymorphisms, maternal HIV infection, maternal HIV viral load, and maternal malaria, on CCR5 expression in placental tissue from mother-infant pairs in Malawi.
In cells lacking the CD4 receptor, such as brain microvascular endothelial cells (BMECs), or primary genital epithelial cells (PGECs), alternative routes for HIV-1 attachment and cell entry have been suggested, including the use of heparan sulfate proteoglycans (HSPGs) [18], [19]. HSPGs are one type of proteoglycan, which is composed of a core protein (i.e. syndecan) and one or more covalently attached glycosaminoglycan (GAG) chains. HSPGs are proteoglycans with heparan sulfate (HS) attached, a highly sulfated polysaccharide made up of glucosamine and glucuronic/iduronic acid repeating disaccharide units [20]. The binding properties and thereby function of HSPGs are determined by the structure and sequence of the disaccharide units, consequential of HS biosynthesis. Various HS subtypes are produced through HS biosynthesis, which involves genetically regulated biosynthetic enzymes. One example subtype is 3-O-sulfated HS, synthesized by 3-O-sulfotransferase, which is encoded by the genes, heparan sulfate (glucosamine) 3-O-sulfotransferase 3A1 (HS3ST3A1) and B1 (HS3ST3A1). The 3-O-sulfated HS subtype has been shown to play a key role in susceptibility to herpes simplex virus -1 (HSV-1) infection in vitro [21], [22], [23] but has not been evaluated in the context of HIV-1 infection.
Although not specific to HS subtype, it is known that HSPGs can facilitate internalization of HIV-1 transactivator protein, Tat [24], which can induce cytokine activity and bind to heparan [25]. Treatment of cells bearing HSPGs with heparinase diminishes HIV-1 attachment and infectivity for CD4+ HeLa cells and macrophages [26], [27], an effect that was shown to differ between HIV viruses using CCR5 as a coreceptor compared to the CXCR4 coreceptor [28]. Furthermore, chemokines such as RANTES, MIP-1α, and MIP-1ß, bind not only with the CCR5 coreceptor but also with glycossaminoglycans (GAGs) bearing heparin, heparan sulfate, or chondroitin sulfate A or C [29], [30], [31]. GAGs can also strongly influence the activity of chemokines [32], [33]. Specifically, chemokines can be stored and released from T lymphocytes cytolotic granules complexed to GAGs [34], and binding with GAGs can influence chemokine structure and lead to aggregation, possibly protecting chemokines from degradation [35]. One study demonstrated that CCL5 (RANTES)-CCR5 binding-mediated apoptosis was dependent on cell-surface GAG binding and that the addition of exogenous heparin or chondroitin sulfate plus GAG digestion protected cells from apoptosis [36]. More recent studies have suggested that GAGs facilitate chemokine binding with receptors [35], [37], perhaps through electrostatic interactions [38].
It is likely that both CCR5 and HS play a role in HIV MTCT, possibly through interactions within placental tissue. Because CCR5 and HS are genetically regulated, evaluation of pertinent gene expression may provide clues for what takes place at the protein level. Thus, in addition to an evaluation of CCR5 expression in the placenta, we quantified the expression of two key HS genes highly expressed in the placenta [39]: HS3ST3A1 and HS3ST3B1, responsible for the synthesis of 3-O-sulfated HS. HS gene expression was evaluated for association with CCR5 expression as well as with susceptibility to HIV MTCT in Malawi. The overall aim of this work was to describe how genetic and environmental factors may regulate CCR5 expression in the placenta.
Methods
Ethics Statement
This research was approved by the University of North Carolina Institutional Review Board and the University of Malawi College of Medicine Research Ethics Committee (COMREC). Written informed consent was obtained from all study participants at the time of recruitment. Consent forms were in both English and Chichewa languages.
Study Population
The participants were a subset of a larger cohort study of malaria and HIV in pregnancy (MHP), previously described [40], [41]. Fresh placental tissue samples from the MHP cohort were obtained from consenting study participants at delivery and immediately frozen at −80 degrees Celsius (°C). Placental tissue samples from 723 HIV positive mothers and 419 HIV negative mothers were transported to the University of North Carolina at Chapel Hill. Of the HIV positive mothers, a total of 411 samples had data on transmission status of the infant.
There were five transmission groups of interest: 1) HIV negative mother/negative infant, 2) HIV positive mother/negative infant at all visits, 3) HIV positive mother/intrauterine transmission to the infant, 4) HIV positive mother/intrapartum transmission to the infant, and 5) HIV positive mother/postpartum transmission to the infant. Power analyses indicated that a sample size of 200 would provide 80% power to detect a difference in r2 of 0.03 across groups. To obtain a slightly larger sample size of 250, a sample of 50 individuals was randomly selected from each of the five transmission categories, where possible. Only 47, 49, and 17 mother-infant pairs were available for intrauterine, intrapartum, or postpartum transmission events, respectively, giving a total sample size of N = 213.
Gene Expression and Genotyping
RNA was extracted from frozen placental tissue of the 213 mother-infant pairs using RNeasy mini kit (Qiagen, Valencia, CA). In order to quantify gene expression for each RNA sample, multiplex real-time PCR was run on 96-well plates using ABI 7700 Sequence Detector (PE Biosystems) according to methods previously described [42]. A total volume of 30 µl was used, which included 10 µl of RNA and 20 µl reaction mixture [42]. The cycle conditions were 30 min at 48°C for the RT reaction, 10 min at 94°C, and then 40 temperature cycles (15 sec at 94°C and 1 min at 60°C). Relative quantification was performed where each 96-well plate was normalized to an endogenous placental RNA control sample. Negative control samples (water) were used to assess the presence of genomic DNA contamination. The difference in cycle threshold (Ct) value between a control gene, GAPDH, and target gene (HS3ST3A1, HS3ST3B1, or CCR5) was obtained for each sample (ΔCt). That value was subtracted from the ΔCt value of the endogenous control sample (ΔΔCt) and then transformed to a percent change in gene expression between GAPDH and the target gene for each sample. Infant genotyping of CCR2-64I and CCR5 promoter polymorphisms were determined using a multiplex ligase detection reaction (LDR) based method with flow cytometric technology, previously described [43], [44]. Briefly, the CCR5 promoter region containing the seven promoter SNPs and the CCR2 open reading frame were PCR-amplified. The amplicon was probed with an upstream allele specific primer with a unique 24 nucleotide FlexMAP™TAG sequence extension (Luminex® Corporation, Austin, TX) and a downstream 5′ phosphorylated/3′ biotinylated conserved sequence primer. After allele specific hybridization, the primers were ligated, ligation products were hybridized with fluorescent bead-labeled anti-TAG probes, and the 3′ biotin group was labeled with phycoerythrin (PE). To determine genotypes, the mean fluorescence intensity of the allele-specific LDR:bead-labeled anti-TAG hybrid complexes was read on a BioPlex array reader (Bio-Rad Laboratories, Hercules, CA) into allele specific channels [43], [44].
Statistical Analysis
The percent change in CCR5 expression was log-transformed in order to approximate a normal distribution. Infant CCR2/CCR5 SNPs were categorized into haplotypes (Table S1), based on phylogeny as previously described [45]. In order to evaluate the association between SNP/haplotype and CCR5 expression in the placenta, linear regression was performed using log-transformed CCR5 expression as the continuous outcome. Logistic regression was also employed to evaluate the association between SNP/haplotype and high vs. low CCR5 expression, dichotomized at the median value. Due to small cell sizes for some polymorphisms, SNPs and haplotypes were categorized as carriers of the variant/haplotype compared to non-carriers and analyzed according to a dominant genetic model. Because the SNPs were not completely independent, exhibiting variable pairwise linkage disequilibrium (Table S2), Bonferroni correction was not employed to adjust for multiple comparisons.
The percent change in HS3ST3A1 expression and HS3ST3B1 expression were also log-transformed and individually evaluated for association with CCR5 expression through linear regression, using CCR5 expression as the continuous outcome. Measures of maternal infection including maternal HIV, maternal HIV viral load (MVL), and maternal malaria were uniquely evaluated for association with CCR5 expression, also through linear regression.
Finally, gene expression of CCR5, HS3ST3A1, and HS3ST3B1 was investigated for association with HIV MTCT through logistic regression. HIV MTCT was coded as 1 (transmission occurred) vs. 0 (no transmission). Different transmission time points (IU, IP, and PP) were evaluated in independent logistic regression models. Because gene expression did not meet the assumption of linearity of the logit, it was categorized into tertiles for this model.
Because MVL is known to be a strong predictor of HIV MTCT, it was evaluated for effect measure modification of the association between gene expression and HIV MTCT by calculating the Mantel-Haenszel test of homogeneity of the odds ratio (OR) across MVL quartiles. It was also evaluated for confounding by using the percent change in estimate criterion: |ln(CoRR)|*100>10%, where |ln(CoRR)| = |ln(crude OR–adjusted OR)|*100 [46].
Results
Study Population
A total of 212 mother-infant pairs evaluated for gene expression had previously been genotyped for CCR2/CCR5 polymorphisms [43]. This included 154 HIV positive mothers, 103 (67%) of which had infants who became HIV positive by 12 weeks postpartum and 51 (33%) of which had HIV negative infants. Following quality control, CCR5 SNP genotypes and placental expression was available for 196 mother-infant pairs. This included 44 (22%) HIV negative infants of HIV positive mothers, 98 (50%) HIV positive infants of HIV positive mothers, and 54 (28%) HIV negative infants of HIV negative mothers. A total of 160 mother-infant pairs also had data on HIV maternal viral load.
CCR5 Gene Expression and CCR5 Variants
The overall mean and standard deviation (SD) of the cycle threshold values were 26.99 (SD = 4.00), 27.93 (SD = 3.92), 24.56 (2.27), and 24.51 (SD = 3.50) for GAPDH, HS3ST3A1, HS3ST3B1, and CCR5, respectively. The mean and SD of the log %change in gene expression relative to GAPDH expression was 5.83 (SD = 1.39), 4.63 (SD = 0.83), and 5.74 (SD = 2.02), for HS3ST3A1, HS3ST3B1, and CCR5, respectively. The CCR5-2132C→T variant was significantly associated with variable placental expression of CCR5 (Table 1), where carriers of the T variant displayed lower placental expression of CCR5 (mean log %change = 5.53, range = −1.61, 9.40) compared to non carriers (mean log %change = 5.87, range = 1.95, 14.55). The CCR5-2554 G→T variant was also significantly associated with a lower expression of CCR5 (mean log %change for T allele = 5.69, range = −1.61, 9.91; mean log %change for G allele = 5.77, range = 1.95, 14.55), although this finding was not statistically significant in the analysis of high vs. low expression (Table 1). The minor allele frequency for CCR5-2132T and -2554T was 0.23 and 0.29, respectively. CCR5 SNPs -2459G, -2135T, and -1835T corresponded to a lower risk of HIV MTCT, but these results were not statistically significant (Table 1).
Table 1. Frequencies and mean CCR5 expression by CCR5 SNP/haplotype category.
| SNP/Haplotype | Genotype/# Copies£ | N | Log % Change by GenotypeN, Mean (Range) | β (95% CI)† | p | OR (95% CI) ‡ | p |
| CCR2-64V→I | VVVIII | 1426210 | 131, 5,82 (−1.61, 14.55)57, 5.59 (0.69, 12.33)10, 5.36 (2.30, 7.15) | −0.27 (−0.87, 0.32) | 0.374 | 0.69 (0.39, 1.24) | 0.213 |
| CCR5-2733A→G | AAAGGG | 190211 | 177, 5.74 (−1.61, 14.55)19, 5.75 (2.56, 9.33)0, NA | 0.01 (−0.95, 0.97) | 0.981 | 0.97 (0.38, 2.51) | 0.957 |
| CCR5-2554G→T | GGGTTT | 1029515 | 94, 6.09 (2.30, 14.55)89, 5.40 (−1.61, 9.91)13, 5.52 (2.77, 9.43) | −0.67 (−1.23, −0.11) | 0.019 | 0.61 (0.35, 1.08) | 0.091 |
| CCR5-2459A→G | AAAGGG | 5711342 | 53, 5.97 (−1.61, 14.55)105, 5.62 (0.69, 12.33)38, 5.76 (2.30, 9.25) | −0.32 (−0.96, 0.32) | 0.324 | 0.62 (0.33, 1.17) | 0.142 |
| CCR5-2135C→T | CCCTTT | 5811242 | 54, 6.00 (−1.61, 14.55)104, 5.62 (0.69, 12.33)38, 5.70 (2.30, 9.25) | −0.36 (−0.99, 0.28) | 0.269 | 0.59 (0.31, 1.11) | 0.104 |
| CCR5-2132C→T | CCCTTT | 1277114 | 114, 6.05 (2.30, 14.55)68, 5.09 (−1.61, 9.40)14, 6.33 (2.77, 9.43) | −0.75 (−0.131, −0.18) | 0.010 | 0.45 (0.25, 0.81) | 0.007 |
| CCR5-2086A→G | AAAGGG | 182283 | 171, 5.70 (−1.61, 14.55)25, 6.15 (3.49, 9.91)1, 3.37 (3.37, 3.37) | 0.34 (−0.49, 1.18) | 0.418 | 1.86 (0.80, 4.33) | 0.149 |
| CCR5-1835C→T | CCCTTT | 1307112 | 120, 5.93 (−1.61, 14.55)65, 5.41 (0.69, 12.32)12, 5.67 (2.30, 9.25) | −0.48 (−1.05, 0.099) | 0.104 | 0.59 (0.33, 1.05) | 0.074 |
Linear regression for the association between CCR5 expression and CCR5 SNP/haplotype: Continuous outcome of placental expression. β: Beta coefficient, 95% CI: 95% Confidence Interval for the Beta, p: p-value.
Logistic regression for the association between CCR5 expression and CCR5 SNP/haplotype: Dichotomous outcome of high vs. low placental expression of CCR5 dichotomized at the median value. OR: Odds Ratio, 95% CI: 95% Confidence Interval for the Odds Ratio.
£ # Copies: Number of copies of haplotype: 0, 1, or 2 copies possible per subject. SNPs and haplotypes categorized as having one or more copies of the variant allele or haplotype compared to zero copies.
The A haplotype, which contains the infant alleles CCR5 -2132C and -2554G (Table S1), was associated with higher expression of CCR5 in the placenta (Table 1). A statistically significant association in the opposite direction (lower expression of CCR5) was observed for haplotype D (Table 1), which contains CCR5 -2132T and -2554T. All other alleles were the same across haplotypes A and D. Associations between CCR5 expression and other haplotypes were not statistically significant (Table 1).
Gene Expression Interplay in the Placenta
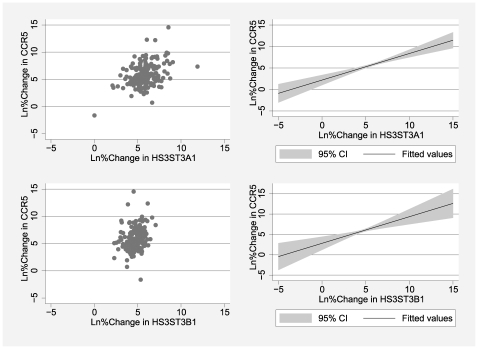
An interesting interplay between placental expression of HS3ST3A1, HS3ST3B1, and CCR5 was observed in this study. An incremental increase in placental expression of HS3ST3A1 or HS3ST3B1 corresponded to an incremental increase in placental expression of CCR5 (Figure 1). The positive association between placental expression of HS3ST3A1 or HS3ST3B1and CCR5 expression was statistically significant (Table 2).
Figure 1. Pattern of association between placental expression of heparan sulfate genes and CCR5.
Scatter Plots (left) and Predicted Linear Plots (right).
Table 2. Linear regression for the associations between gene expression variables.
| Comparison† | β (95% CI) | p |
| HS3ST3A1 vs. CCR5 expression (N = 197) | 0.27 (0.18, 0.35) | <0.0001 |
| HS3ST3B1 vs. CCR5 expression (N = 180) | 0.11 (0.06, 0.18) | <0.0001 |
Continuous placental gene expression variables compared via linear regression; β: Beta coefficient; 95% CI: 95% Confidence Interval for the Beta; p: p-value.
CCR5 Expression and Measures of Maternal Infection
Maternal HIV infection was not associated with CCR5 expression in the placenta (β = 0.072, 95% CI = −0.57, −0.72, p = 0.826, N = 194). However, among the HIV infected mothers, maternal HIV viral load was associated with CCR5 expression, where an incremental increase in viral load corresponded to an incremental increase in CCR5 expression (β = 0.76, 95% CI = 0.12, 1.39; p = 0.020, N = 92). Maternal malaria infection also corresponded to a higher placental expression of CCR5, but the association was not statistically significant (β = 0.37, 95% CI = −0.43, 1.18, p = 0.362, N = 170).
Gene Expression and Risk of HIV MTCT
A general trend of increasing risk of HIV MTCT with increasing expression of CCR5 was observed but these findings were not statistically significant (Medium vs. low tertile OR = 1.16, 95% CI = 0.49, 2.73; High vs. low tertile OR = 1.26, 95% CI = 0.52, 3.03). Log maternal viral load did not modify the association between HIV MTCT and placental expression of CCR5 (Mantel-Haenszel OR = 0.22, p = 0.683). In addition, MVL was not a confounder in the association based on a less than 10% change in the effect estimates after adjustment for MVL (%ln(CoRR) = 6.7%) and because MVL was not associated with CCR5 expression (β = 0.02, 95% CI = −0.35, 0.39). Although the estimates of the association between CCR5 expression and each transmission time point were not very precise, a similar increased risk of transmission was observed for higher expression of CCR5 at all time points (Table 3). No significant association between the heparan sulfate genes and HIV MTCT was observed (data not shown).
Table 3. Association between CCR5 placental expression and the risk of HIV MTCT at different time points.
| Log %Change in Gene Expression | Intrauterine Transmission | Intrapartum Transmission | Postpartum Transmission | |||
| OR (95% CI)† | p | OR (95% CI) | p | OR (95% CI) | p | |
| Medium low vs. Low | 1.06 (0.37, 3.04) | 0.907 | 1.11 (0.36, 3.42) | 0.862 | 1.15 (0.25, 5.22) | 0.853 |
| Medium high vs. Low | 1.02 (0.34, 3.02) | 0.974 | 1.88 (0.60, 5.96) | 0.280 | 0.21 (0.02, 2.18) | 0.190 |
| High vs. Low | 1.25 (0.42, 3.76) | 0.691 | 1.21 (0.36, 4.08) | 0.757 | 1.39 (0.28, 6.83) | 0.686 |
Logistic regression for association between CCR5 expression tertile and HIV MTCT (transmission vs. no transmission); OR: Odds Ratio; 95% CI: 95% Confidence Interval for the Odds Ratio; p: p-value.
CCR5 Variants and HIV MTCT
The findings of Pedersen et al. [44] were replicated in this subset of the original cohort from Malawi, with regards to the direction of association between each CCR5 SNP and HIV MTCT. One exception was the association between CCR5-1835T and the risk of HIV MTCT, which varied slightly in direction compared to previous findings (OR = 1.06 vs. RR = 0.84) [44]. Some findings also had variable statistical significance which may be a reflection of sample size (Table S3).
Discussion
This study evaluated the regulation of CCR5 expression in the placenta by genetic and environmental factors involved in the risk of HIV MTCT. Infant CCR5 promoter polymorphisms -2132T and -2554T were associated with lower expression of CCR5 in the placenta, as was the infant haplotype D, which is tagged by these alleles. Infant haplotype A was associated with significantly higher CCR5 expression in the placenta and differs from infant haplotype D in that it contains the wild type alleles, -2132C and -2554G (Table S1) [44]. These findings provide in vivo evidence for CCR5-2554 G→T and CCR5 -2132C→T related down regulation of CCR5 expression in the placenta.
We expected the SNPs associated with lower CCR5 expression to also be associated with a lower risk of HIV MTCT, and for SNPs associated with higher CCR5 expression to be associated with a higher risk of HIV MTCT. Referring to the association between infant CCR2/CCR5 SNPs and HIV MTCT previously described [44] and based on our replicate analyses, we found that this was not always the case. Notably, CCR5 -2733G was associated with higher CCR5 expression compared to a lower risk of HIV MTCT, although the association with expression was not statistically significant (Table S3). CCR5 SNPs -2554T, -2132T, and -2086G also showed discrepant associations for CCR5 expression and HIV MTCT, with variable statistical significance. The only discrepant finding that was statistically significant for both associations was observed for the D haplotype, which was associated with lower CCR5 expression but a higher risk of HIV MTCT (Table S3). Haplotype D varies from all other haplotypes with regards to the -2132T allele, which displayed similar results (Table S3). It is possible that these discrepancies reflect smaller sample sizes or the fact that we are comparing infant genotypes/haplotypes with a combined measurement of infant and maternal placental gene expression. More expensive techniques were required to separate mother and infant tissue and we were unable to pursue this in our study. Despite this limitation, the SNP/haplotype associations suggest that predictors of CCR5 expression do not directly correlate to the prediction of HIV MTCT and that these outcomes should be considered independently.
The discrepant SNP/haplotype associations with CCR5 expression and HIV MTCT were partnered with the finding that CCR5 placental expression was not associated with HIV MTCT. To obtain the best power, this association was first evaluated by using the cumulative transmission status of the infant (occurring at birth, 6 weeks, or 12 weeks postpartum), and showed no significant association. Because placenta samples were obtained at delivery, the measured CCR5 expression was viewed to be most representative of the expression occurring during labor and delivery and thus, most relevant to the risk of IP transmission. For both cumulative and IP transmission, although the direction of effect was consistent with previous findings [17], where an increase in CCR5 expression contributed to an increase in the risk of HIV MTCT, the association was not statistically significant. It is likely that other factors played a stronger role in predicting HIV MTCT in this study population.
Disease severity appeared to be an important regulator of placental expression of CCR5. Among HIV infected mothers, higher maternal HIV viral load was significantly associated with higher CCR5 placental expression. Thus, the presence of HIV infection alone may not make as great of an impact on CCR5 expression as does the severity of HIV disease or viral burden experienced by the individual.
Unlike maternal HIV viral load, maternal malaria was not a significant predictor of CCR5 expression. We limited this analysis to only HIV positive mothers and found that maternal malaria did increase CCR5 expression in the placenta but that it was not statistically significant (data not shown). Thus, we could not make any broad conclusions from the analyses with malaria. Furthermore, MVL and maternal malaria did not confound or act as effect measure modifiers in the associations between CCR5 expression and HIV MTCT, suggesting that accounting for a key co-infection or severity of HIV infection did not explain the lack of a significant association between CCR5 expression and HIV MTCT.
One of the most important findings from this study was the revelation of a possible interaction between CCR5 and heparan sulfate at the genetic level. Up-regulation of CCR5 expression in the placenta was observed at higher expression levels of two genes involved in the biosynthesis of 3-O-sulfated heparan sulfate, HS3ST3A1 and HS3ST3B1. These findings were statistically significant and to our knowledge, are novel in vivo findings. It is possible that each heparan sulfate gene interacts with CCR5 in the placenta, causing up-regulation, or that the genes share transcription regulatory regions or factors. As previously noted, heparan sulfate has been shown to interact with chemokines which bind to CCR5, such as RANTES, but has not been evaluated for any interaction with CCR5 or related factors at the genetic level. Our findings press the importance of additional research on heparan sulfate and CCR5–related factors that may individually or cooperatively contribute to viral infection in human populations.
Overall, this study demonstrated the complexity of predicting HIV MTCT in human populations and offers new insights into regulatory factors of CCR5 expression in the placenta. Additional epidemiological investigations are warranted in order to more clearly elucidate how CCR5 and heparan sulfate genes may interact in vivo and whether combined genetic and environmental factors contribute to the risk of HIV MTCT in other populations.
Supporting Information
CCR2/CCR5 haplotypes.
(0.04 MB DOC)
Pairwise linkage disequilibrium for CCR2-64I and CCR5 promoter polymorphisms.
(0.04 MB DOC)
CCR5 SNP associations with CCR5 placental expression compared to CCR5 SNP associations with HIV MTCT.
(0.06 MB DOC)
Acknowledgments
We wish to thank Dr. Hyungsuk Kim for the gene expression work conducted at the Animal Clinical Chemistry and Gene Expression Laboratories, University of North Carolina at Chapel Hill, and Dr. Jian Liu from the University of North Carolina at Chapel Hill School of Pharmacy, for his intellectual contributions.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This research was supported by the Centers for Disease Control and Prevention Dissertation Award (PAR 07-231, 2008) and the NIH Virology Training Grant (T32 AI007419, 2007). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.UNAIDS. Geneva: Joint United Nations programme on HIV/IADS (UNAIDS) and the World Health Organization; 2008. Report on the Global AIDS Epidemic: 2008. [Google Scholar]
- 2.Dragic T, Litwin V, Allaway GP, Martin SR, Huang Y, et al. HIV-1 entry into CD4+ cells is mediated by the chemokine receptor CC-CKR-5. Nature. 1996;381:667–673. doi: 10.1038/381667a0. [DOI] [PubMed] [Google Scholar]
- 3.Samson M, Labbe O, Mollereau C, Vassart G, Parmentier M. Molecular cloning and functional expression of a new human CC-chemokine receptor gene. Biochemistry. 1996;35:3362–3367. doi: 10.1021/bi952950g. [DOI] [PubMed] [Google Scholar]
- 4.Burns JC, Shimizu C, Gonzalez E, Kulkarni H, Patel S, et al. Genetic variations in the receptor-ligand pair CCR5 and CCL3L1 are important determinants of susceptibility to Kawasaki disease. J Infect Dis. 2005;192:344–349. doi: 10.1086/430953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fernandez EJ, Lolis E. Structural studies of chemokines that inhibit HIV-1 entry. Antivir Chem Chemother. 2001;12(Suppl 1):43–49. [PubMed] [Google Scholar]
- 6.Dean M, Carrington M, Winkler C, Huttley GA, Smith MW, et al. Genetic restriction of HIV-1 infection and progression to AIDS by a deletion allele of the CKR5 structural gene. Hemophilia Growth and Development Study, Multicenter AIDS Cohort Study, Multicenter Hemophilia Cohort Study, San Francisco City Cohort, ALIVE Study. Science. 1996;273:1856–1862. doi: 10.1126/science.273.5283.1856. [DOI] [PubMed] [Google Scholar]
- 7.Deng H, Liu R, Ellmeier W, Choe S, Unutmaz D, et al. Identification of a major co-receptor for primary isolates of HIV-1. Nature. 1996;381:661–666. doi: 10.1038/381661a0. [DOI] [PubMed] [Google Scholar]
- 8.Doranz BJ, Rucker J, Yi Y, Smyth RJ, Samson M, et al. A dual-tropic primary HIV-1 isolate that uses fusin and the beta-chemokine receptors CKR-5, CKR-3, and CKR-2b as fusion cofactors. Cell. 1996;85:1149–1158. doi: 10.1016/s0092-8674(00)81314-8. [DOI] [PubMed] [Google Scholar]
- 9.Zimmerman PA, Buckler-White A, Alkhatib G, Spalding T, Kubofcik J, et al. Inherited resistance to HIV-1 conferred by an inactivating mutation in CC chemokine receptor 5: studies in populations with contrasting clinical phenotypes, defined racial background, and quantified risk. Mol Med. 1997;3:23–36. [PMC free article] [PubMed] [Google Scholar]
- 10.Clegg AO, Ashton LJ, Biti RA, Badhwar P, Williamson P, et al. CCR5 promoter polymorphisms, CCR5 59029A and CCR5 59353C, are under represented in HIV-1-infected long-term non-progressors. The Australian Long-Term Non-Progressor Study Group. Aids. 2000;14:103–108. doi: 10.1097/00002030-200001280-00004. [DOI] [PubMed] [Google Scholar]
- 11.Knudsen TB, Kristiansen TB, Katzenstein TL, Eugen-Olsen J. Adverse effect of the CCR5 promoter -2459A allele on HIV-1 disease progression. J Med Virol. 2001;65:441–444. [PubMed] [Google Scholar]
- 12.Kostrikis LG, Neumann AU, Thomson B, Korber BT, McHardy P, et al. A polymorphism in the regulatory region of the CC-chemokine receptor 5 gene influences perinatal transmission of human immunodeficiency virus type 1 to African-American infants. J Virol. 1999;73:10264–10271. doi: 10.1128/jvi.73.12.10264-10271.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McDermott DH, Zimmerman PA, Guignard F, Kleeberger CA, Leitman SF, et al. CCR5 promoter polymorphism and HIV-1 disease progression. Multicenter AIDS Cohort Study (MACS). Lancet. 1998;352:866–870. doi: 10.1016/s0140-6736(98)04158-0. [DOI] [PubMed] [Google Scholar]
- 14.Benkirane M, Jin DY, Chun RF, Koup RA, Jeang KT. Mechanism of transdominant inhibition of CCR5-mediated HIV-1 infection by ccr5delta32. J Biol Chem. 1997;272:30603–30606. doi: 10.1074/jbc.272.49.30603. [DOI] [PubMed] [Google Scholar]
- 15.Hladik F, Liu H, Speelmon E, Livingston-Rosanoff D, Wilson S, et al. Combined effect of CCR5-Delta32 heterozygosity and the CCR5 promoter polymorphism -2459 A/G on CCR5 expression and resistance to human immunodeficiency virus type 1 transmission. J Virol. 2005;79:11677–11684. doi: 10.1128/JVI.79.18.11677-11684.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Salkowitz JR, Bruse SE, Meyerson H, Valdez H, Mosier DE, et al. CCR5 promoter polymorphism determines macrophage CCR5 density and magnitude of HIV-1 propagation in vitro. Clin Immunol. 2003;108:234–240. doi: 10.1016/s1521-6616(03)00147-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Behbahani H, Popek E, Garcia P, Andersson J, Spetz AL, et al. Up-regulation of CCR5 expression in the placenta is associated with human immunodeficiency virus-1 vertical transmission. Am J Pathol. 2000;157:1811–1818. doi: 10.1016/S0002-9440(10)64819-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bobardt MD, Salmon P, Wang L, Esko JD, Gabuzda D, et al. Contribution of proteoglycans to human immunodeficiency virus type 1 brain invasion. J Virol. 2004;78:6567–6584. doi: 10.1128/JVI.78.12.6567-6584.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Argyris EG, Acheampong E, Nunnari G, Mukhtar M, Williams KJ, et al. Human immunodeficiency virus type 1 enters primary human brain microvascular endothelial cells by a mechanism involving cell surface proteoglycans independent of lipid rafts. J Virol. 2003;77:12140–12151. doi: 10.1128/JVI.77.22.12140-12151.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gallagher JT, Lyon M, Steward WP. Structure and function of heparan sulphate proteoglycans. Biochem J. 1986;236:313–325. doi: 10.1042/bj2360313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Geraghty RJ, Krummenacher C, Cohen GH, Eisenberg RJ, Spear PG. Entry of alphaherpesviruses mediated by poliovirus receptor-related protein 1 and poliovirus receptor. Science. 1998;280:1618–1620. doi: 10.1126/science.280.5369.1618. [DOI] [PubMed] [Google Scholar]
- 22.Shukla D, Liu J, Blaiklock P, Shworak NW, Bai X, et al. A novel role for 3-O-sulfated heparan sulfate in herpes simplex virus 1 entry. Cell. 1999;99:13–22. doi: 10.1016/s0092-8674(00)80058-6. [DOI] [PubMed] [Google Scholar]
- 23.Warner MS, Geraghty RJ, Martinez WM, Montgomery RI, Whitbeck JC, et al. A cell surface protein with herpesvirus entry activity (HveB) confers susceptibility to infection by mutants of herpes simplex virus type 1, herpes simplex virus type 2, and pseudorabies virus. Virology. 1998;246:179–189. doi: 10.1006/viro.1998.9218. [DOI] [PubMed] [Google Scholar]
- 24.Cladera J, Martin I, O'Shea P. The fusion domain of HIV gp41 interacts specifically with heparan sulfate on the T-lymphocyte cell surface. Embo J. 2001;20:19–26. doi: 10.1093/emboj/20.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gatignol A, Jeang KT. Tat as a transcriptional activator and a potential therapeutic target for HIV-1. Adv Pharmacol. 2000;48:209–227. doi: 10.1016/s1054-3589(00)48007-5. [DOI] [PubMed] [Google Scholar]
- 26.Mondor I, Ugolini S, Sattentau QJ. Human immunodeficiency virus type 1 attachment to HeLa CD4 cells is CD4 independent and gp120 dependent and requires cell surface heparans. J Virol. 1998;72:3623–3634. doi: 10.1128/jvi.72.5.3623-3634.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Saphire AC, Bobardt MD, Zhang Z, David G, Gallay PA. Syndecans serve as attachment receptors for human immunodeficiency virus type 1 on macrophages. J Virol. 2001;75:9187–9200. doi: 10.1128/JVI.75.19.9187-9200.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ugolini S, Mondor I, Sattentau QJ. HIV-1 attachment: another look. Trends Microbiol. 1999;7:144–149. doi: 10.1016/s0966-842x(99)01474-2. [DOI] [PubMed] [Google Scholar]
- 29.Slimani H, Charnaux N, Mbemba E, Saffar L, Vassy R, et al. Interaction of RANTES with syndecan-1 and syndecan-4 expressed by human primary macrophages. Biochim Biophys Acta. 2003;1617:80–88. doi: 10.1016/j.bbamem.2003.09.006. [DOI] [PubMed] [Google Scholar]
- 30.Slimani H, Charnaux N, Mbemba E, Saffar L, Vassy R, et al. Binding of the CC-chemokine RANTES to syndecan-1 and syndecan-4 expressed on HeLa cells. Glycobiology. 2003;13:623–634. doi: 10.1093/glycob/cwg083. [DOI] [PubMed] [Google Scholar]
- 31.Tkachenko E, Rhodes JM, Simons M. Syndecans: new kids on the signaling block. Circ Res. 2005;96:488–500. doi: 10.1161/01.RES.0000159708.71142.c8. [DOI] [PubMed] [Google Scholar]
- 32.Middleton J, Neil S, Wintle J, Clark-Lewis I, Moore H, et al. Transcytosis and surface presentation of IL-8 by venular endothelial cells. Cell. 1997;91:385–395. doi: 10.1016/s0092-8674(00)80422-5. [DOI] [PubMed] [Google Scholar]
- 33.Witt DP, Lander AD. Differential binding of chemokines to glycosaminoglycan subpopulations. Curr Biol. 1994;4:394–400. doi: 10.1016/s0960-9822(00)00088-9. [DOI] [PubMed] [Google Scholar]
- 34.Wagner L, Yang OO, Garcia-Zepeda EA, Ge Y, Kalams SA, et al. Beta-chemokines are released from HIV-1-specific cytolytic T-cell granules complexed to proteoglycans. Nature. 1998;391:908–911. doi: 10.1038/36129. [DOI] [PubMed] [Google Scholar]
- 35.Hoogewerf AJ, Kuschert GS, Proudfoot AE, Borlat F, Clark-Lewis I, et al. Glycosaminoglycans mediate cell surface oligomerization of chemokines. Biochemistry. 1997;36:13570–13578. doi: 10.1021/bi971125s. [DOI] [PubMed] [Google Scholar]
- 36.Murooka TT, Wong MM, Rahbar R, Majchrzak-Kita B, Proudfoot AE, et al. CCL5-CCR5-mediated apoptosis in T cells: Requirement for glycosaminoglycan binding and CCL5 aggregation. J Biol Chem. 2006;281:25184–25194. doi: 10.1074/jbc.M603912200. [DOI] [PubMed] [Google Scholar]
- 37.Martin L, Blanpain C, Garnier P, Wittamer V, Parmentier M, et al. Structural and functional analysis of the RANTES-glycosaminoglycans interactions. Biochemistry. 2001;40:6303–6318. doi: 10.1021/bi002670n. [DOI] [PubMed] [Google Scholar]
- 38.Maione TE, Gray GS, Hunt AJ, Sharpe RJ. Inhibition of tumor growth in mice by an analogue of platelet factor 4 that lacks affinity for heparin and retains potent angiostatic activity. Cancer Res. 1991;51:2077–2083. [PubMed] [Google Scholar]
- 39.Razi N, Lindahl U. Biosynthesis of heparin/heparan sulfate. The D-glucosaminyl 3-O-sulfotransferase reaction: target and inhibitor saccharides. J Biol Chem. 1995;270:11267–11275. doi: 10.1074/jbc.270.19.11267. [DOI] [PubMed] [Google Scholar]
- 40.Mwapasa V, Rogerson SJ, Kwiek JJ, Wilson PE, Milner D, et al. Maternal syphilis infection is associated with increased risk of mother-to-child transmission of HIV in Malawi. Aids. 2006;20:1869–1877. doi: 10.1097/01.aids.0000244206.41500.27. [DOI] [PubMed] [Google Scholar]
- 41.Mwapasa V, Rogerson SJ, Molyneux ME, Abrams ET, Kamwendo DD, et al. The effect of Plasmodium falciparum malaria on peripheral and placental HIV-1 RNA concentrations in pregnant Malawian women. Aids. 2004;18:1051–1059. doi: 10.1097/00002030-200404300-00014. [DOI] [PubMed] [Google Scholar]
- 42.Kim HS, Lee G, John SW, Maeda N, Smithies O. Molecular phenotyping for analyzing subtle genetic effects in mice: application to an angiotensinogen gene titration. Proc Natl Acad Sci U S A. 2002;99:4602–4607. doi: 10.1073/pnas.072083799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bruse S, Blood M, Zimmerman P. Multiplex Ligase Detection Reaction-based Genotyping Analysis of CCR2-CCR5 Polymorphism Improves Large-scale Population Studies of HIV Co-receptors.: Case Western Reserve University Rutgers University. 2006.
- 44.Pedersen BR, Kamwendo D, Blood M, Mwapasa V, Molyneux M, et al. CCR5 haplotypes and mother-to-child HIV transmission in Malawi. PLoS ONE. 2007;2:e838. doi: 10.1371/journal.pone.0000838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mummidi S, Bamshad M, Ahuja SS, Gonzalez E, Feuillet PM, et al. Evolution of human and non-human primate CC chemokine receptor 5 gene and mRNA. Potential roles for haplotype and mRNA diversity, differential haplotype-specific transcriptional activity, and altered transcription factor binding to polymorphic nucleotides in the pathogenesis of HIV-1 and simian immunodeficiency virus. J Biol Chem. 2000;275:18946–18961. doi: 10.1074/jbc.M000169200. [DOI] [PubMed] [Google Scholar]
- 46.Rothman K GS. Philapdelphia: Lippincott-Raven Publishers, Inc; 1998. Modern Epidemiology. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
CCR2/CCR5 haplotypes.
(0.04 MB DOC)
Pairwise linkage disequilibrium for CCR2-64I and CCR5 promoter polymorphisms.
(0.04 MB DOC)
CCR5 SNP associations with CCR5 placental expression compared to CCR5 SNP associations with HIV MTCT.
(0.06 MB DOC)