Abstract
Clathrin is crucial for endocytosis and plays a recently described role in mitosis. Two clathrin heavy chains (CHCs) are found in humans: the ubiquitous CHC17 and CHC22, a CHC that is enriched in skeletal muscle. Functional differences have been proposed for these clathrins despite high sequence homology. Here, we compared each paralogue in functional assays of endocytosis and mitosis. We find that CHC17 and CHC22 are functionally equivalent. We also describe how previous work on CHC22 has involved a splice variant that is not usually expressed in cells.
Introduction
Clathrin is a triskelion comprising three clathrin heavy chains (CHCs) each with an associated light chain (Kirchhausen, 2000). Clathrin is crucial for membrane trafficking, particularly for endocytosis at the plasma membrane. It is also proposed to have a role in stabilising fibres of the spindle apparatus during mitosis (Royle, 2006). In humans there are two isoforms of CHC encoded by separate genes (Wakeham et al., 2005). The first, on chromosome 17 (CLTC) encodes a clathrin heavy chain (CHC17) which is ubiquitously expressed and has been intensively studied. The second, on chromosome 22 (CLTCL1) codes for CHC22, a homologue of CHC17 whose function is unclear. CHC22 is expressed at very low levels in most tissues but has a higher expression in skeletal muscle (Gong et al., 1996; Kedra et al., 1996; Lindsay et al., 1996; Long et al., 1996; Sirotkin et al., 1996; Liu et al., 2001). This expression is upregulated during myogenesis, suggesting a role for CHC22 in muscle repair and regeneration (Liu et al., 2001; Towler et al., 2004b).
CHC17 and CHC22 are very similar, with 85% identity (97% similarity) at the amino acid level. However, recent work has suggested that the functions of CHC17 and CHC22 are different. CHC22 appears to be unable to bind AP2, the main adaptor protein for clathrin-mediated endocytosis, but is able to bind other APs (Liu et al., 2001). This has led to the suggestion that CHC22 is not involved clathrin-mediated endocytosis but in other forms of intracellular traffic (Towler et al., 2004a). In apparent disagreement with this idea, a recent study has found that CHC22 is a component of clathrin-coated vesicles (Borner et al., 2006). Furthermore, biochemical work has shown that CHC22 cannot bind clathrin light chains, unlike CHC17 (Liu et al., 2001). However, an interaction between clathrin light chain b (LCb) and both CHCs was detected by yeast 2-hybrid (Towler et al., 2004b). On balance, the published results represent some confusion over what differences, if any, exist between CHC17 and CHC22.
During mitosis, clathrin becomes associated with the microtubules of the mitotic spindle (Maro et al., 1985; Okamoto et al., 2000; Royle et al., 2005), where it apparently stabilises kinetochore fibres. This has been well documented for CHC17 (Royle et al., 2005; Royle and Lagnado, 2006), but it is untested whether CHC22 also binds to the spindle.
We set out to compare the function of CHC17 and CHC22 in endocytosis and mitosis in mammalian cells. Our strategy was to express each protein on a background of CHC17-depletion. We find that CHC17 and CHC22 are functionally equivalent. During the course of this work we also established that some of the previous work on CHC22 has been carried out using an incorrect clone that is non-functional in the assays we have used.
Results and discussion
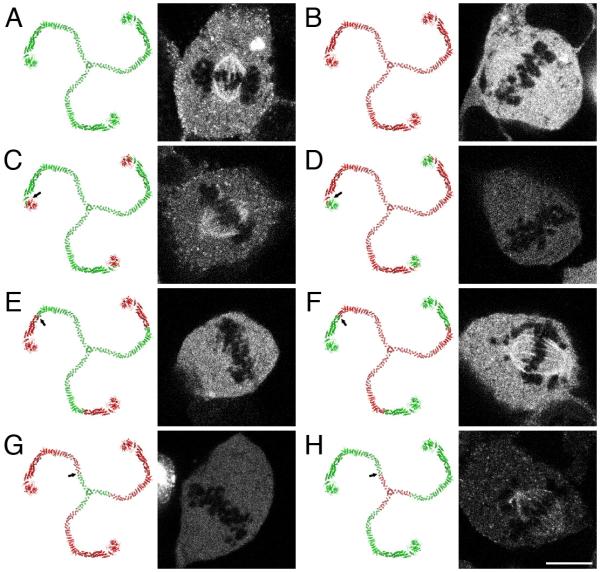
The subcellular localisation of GFP-tagged CHC22 was examined in mitotic cells. We used a CHC22 cDNA that is the basis for much of the previous work on CHC22 (Liu et al., 2001; Towler et al., 2004b). When expressed in HEK293 cells, this GFP-CHC22 construct, unlike GFP-CHC17, did not associate with the mitotic spindle (Fig 1). This difference between CHC22 and CHC17 was unexpected given the extensive similarity between the two proteins. In order to determine the region responsible for this difference, we generated six chimeric clathrins using CHC17 and CHC22 (Fig 1C-H). To our surprise we found that the region of non-equivalence was located between residues 291-728. This was puzzling due to our previous observation that the N-terminal domain (residues 1-330) of CHC17 is required for spindle-binding (Royle et al., 2005). The chimeric clathrins in Fig 1C & D, show that the N-terminal domain of CHC17 was not sufficient to confer spindle-binding to CHC22, yet the N-terminal domain from CHC22 could be substituted for that of CHC17 and the resultant chimera was competent for binding. One possible explanation for these results is that the non-equivalence of residues 291-728 is due to an ‘inhibitory sequence’ in CHC22 that prevents triskelia containing this region from binding to the mitotic spindle.
Fig 1.
Recruitment of chimeric clathrin constructs to the mitotic spindle
Representative confocal micrographs of HEK293 cells expressing GFP-tagged clathrin constructs. Schematic diagrams (left) show the arrangement of CHC17 (green) and CHC22 (red); arrows in C-F indicate changeover points. A, GFP-CHC17(1-1675); B, GFP-CHC22(1-1640); C. GFP-CHC22(1-290)CHC17(291-1675); D. GFP-CHC17(1-291)CHC22(292-1640); E, GFP-CHC22(1-728)CHC17(729-1675); F. GFP-CHC17(1-728)CHC22(729-1640); G, GFP-CHC22(1-1393)CHC17(1394-1675); H. GFP-CHC17(1-1393)CHC22(1394-1640). The GFP-CHC22 clone used in these experiments was a variant lacking exon9 (CHC22ΔExon9). The lack of exon 9 correlated with a lack of spindle recruitment (C, D, E and F). Scale bar = 10 μm
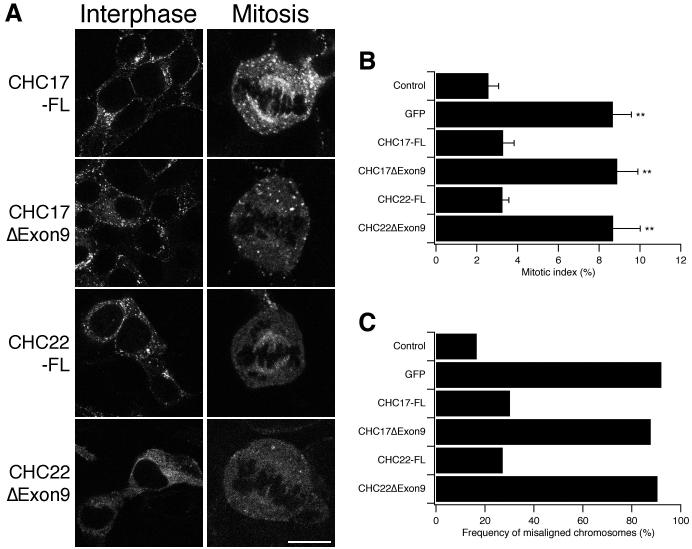
Upon further work to narrow down the region of non-equivalence we found that the CHC22 cDNA was a variant that lacked exon 9. Exon 9 is a 153 bp exon that codes for residues 457-507. These amino acids correspond to helices e-h in CHCR0 (Supplementary Fig S1). As the exon 9 residues are within the region of non-equivalence that we had identified, we postulated that the lack of these residues caused the inhibition of spindle-binding. To test this idea, we generated a full-length, exon 9-containing GFP-tagged CHC22 and expressed it in HEK293 cells. Fig 2A shows the subcellular distributions of the full-length and exon-skipped forms of CHC22. The full-length form had a subcellular distribution that was similar to that of CHC17. Full-length CHC22 was able to bind to the mitotic spindle and, in interphase cells, had a distribution typical of clathrin: numerous coated pits and vesicles throughout the cell with a perinuclear accumulation around the Golgi apparatus (compare CHC22-FL with CHC17-FL). By contrast the exon-skipped form (CHC22ΔExon9), which was not recruited to the mitotic spindle, had a diffuse, non-punctate distribution in interphase cells. In addition, we artificially removed exon 9 from CHC17 (CHC17ΔExon9) in order to test the effect of removing these residues in a better-studied clathrin. Fig 2A shows that CHC17ΔExon9, like CHC22ΔExon9, was no longer recruited to the spindle; but in contrast, CHC17ΔExon9 was still found in an interphase distribution that was similar to CHC17-FL and CHC22-FL. These results demonstrate that residues encoded by exon 9 are required for localisation of CHCs to the mitotic spindle.
Fig 2.
Rescue of normal mitosis in CHC17-depleted cells by full-length CHC17 or CHC22
A, Residues 457-507 (exon 9) are necessary for recruitment of CHC17 or CHC22 to the mitotic spindle. Representative confocal micrographs of CHC17-depleted HEK293 cells expressing GFP-tagged clathrin constructs in either interphase (left) or mitosis (right). Scale bar = 20 μm for interphase cells, 10 μm for mitotic cells. B, Histogram to show mean ± s.d. mitotic index, n = 3, ** = p < 0.01. C, Histogram to show the mean frequency of misaligned chromosomes in metaphase-like cells. Results are shown from a typical experiment, n = 1. ‘Control’ is GFP expressed on a control RNAi background. The remainder express the indicated GFP-tagged construct on a CHC17 RNAi background.
We next wanted to determine the cellular functions of these clathrins. In order to gain a fair comparison of protein function, we expressed each clathrin in HEK293 cells depleted of CHC17. This allowed us to compare the functions of CHC17 and CHC22 directly, without the complication of endogenous CHC17.
We first tested the mitotic function of these clathrins. RNAi of CHC17 causes defects in mitosis characterised by an increase in the frequency of misaligned chromosomes that results in prolonged mitosis, which can be measured as an increase in the mitotic index (Royle et al., 2005). We tested for functional rescue of normal mitosis by expressing the clathrins on a background of CHC17 RNAi. We found that CHC17-FL and CHC22-FL were able to fully rescue normal mitosis in CHC17-depleted cells (Fig 2B,C). CHC17ΔExon9 and CHC22ΔExon9 were unable to rescue normal mitosis, as assessed by no significant change in the mitotic index and the frequency of misaligned chromosomes in metaphase-like cells compared with the control RNAi condition (GFP, Fig 2B,C). These results are consistent with the observation that the exon-skipped forms of CHC17 and CHC22 were unable to bind to the mitotic spindle.
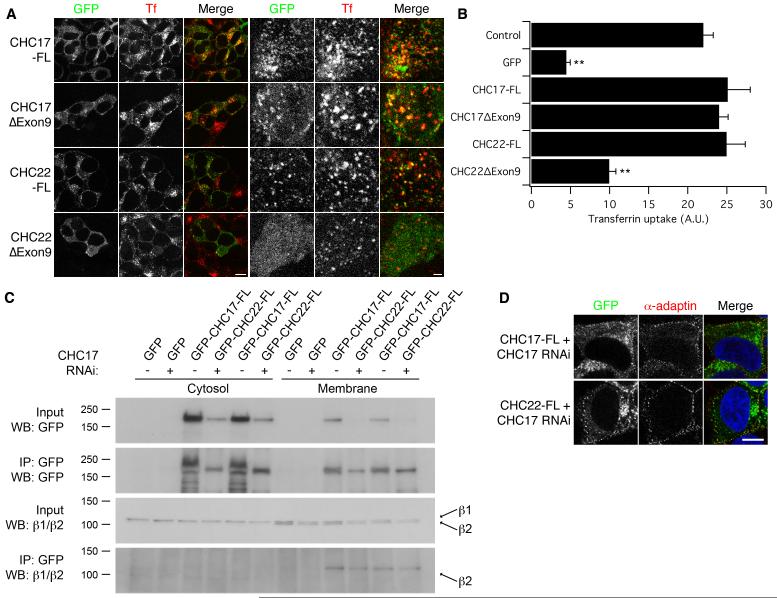
We next tested the function of these clathrin constructs in endocytosis (Fig 3). We found that CHC17-FL and CHC22-FL were able to rescue transferrin uptake in CHC17-depleted cells to levels similar to that seen in the control RNAi condition (GFP, Fig 3B). We saw very little transferrin uptake with CHC22ΔExon9, however CHC17ΔExon9 was able to mediate endocytosis (Fig 3A,B). The ventral surface of CHC17-depleted cells expressing these clathrins was imaged by confocal microscopy (Fig 3A). This allowed us to visualise individual transferrin-Alexa546 puncta that co-localise with these clathrins. We could clearly visualise transferrin in CHC22-positive spots, presumably CHC22-coated vesicles, whilst little or no association between CHC22ΔExon9 and the reduced number of transferrin puncta was found (Fig 3A).
Fig 3.
Functional test for endocytosis in CHC17-depleted HEK293 cells expressing clathrin constructs
A, Representative confocal micrographs to show relative distribution of GFP-tagged clathrin constructs (GFP, green in merge) and transferrin-Alexa546 (Tf, red in merge). Left panels give an indication of the rescue of clathrin-mediated endocytosis, right panels are confocal sections taken adjacent to the coverslip to visualise co-localisation of clathrin and transferrin spots. Scale bars = 10 μm (left) 2 μm (right). B, Histogram to show mean ± s.e.m. transferrin uptake in CHC17-depleted HEK293 cells. n = 3, ** = p < 0.01. C, Co-immunoprecipitation (co-IP) of β2-adaptin with GFP-CHC17-FL and GFP-CHC22-FL. HEK293 cells transfected as described and immunoprecipitations of either the cytosolic or membrane-enriched fractions were done using anti-GFP. Samples of the inputs and IPs were analysed by western blotting for GFP or β1/β2 adaptin. Under the conditions of our experiments, equivalent amounts of β1- and β2-adaptin are present in the membrane-enriched fraction, but only β2-adaptin co-IPd with CHCs. Western blots were typical of 3 experiments. Molecular weights (kDa) are as indicated to the left. D, Representative confocal micrographs to show co-localisation of GFP-CHC22-FL or GFP-CHC17-FL (green) with α-adaptin detected with AP.6/A546 (red). Cells were depleted of endogenous CHC17 by RNAi, DAPI/DNA is blue in the merged image. Scale bar = 10 μm.
The CHC22-mediated endocytosis described here is in agreement with other work demonstrating that CHC22 is a component of CCVs (Borner et al., 2006). However previous work had suggested that endogenous CHC22 immunoprecipitated from HeLa cells did not associate with AP2, unlike CHC17 (Liu et al., 2001). The mode of AP-binding by the CHC17 N-terminal domain is well-defined (Dell’Angelica et al., 1998; ter Haar et al., 2000; Drake and Traub, 2001; Miele et al., 2004) and all of the residues involved in the interaction between CHC17 and proteins containing clathrin-box motifs are completely conserved in CHC22; so we would predict that such interactions are therefore equivalent. Our functional data is strong evidence that CHC22 can interact with AP2, since transferrin receptor endocytosis is AP2-dependent (Jing et al., 1990; Nesterov et al., 1999; Hinrichsen et al., 2003; Motley et al., 2003). To confirm that this was indeed the case we tested for such an interaction biochemically by immunoprecipitation (IP) of GFP-tagged CHCs from membrane-enriched fractions and western blotting to detect β1- and β2-adaptin using a monoclonal antibody, 100/1 (Fig 3C). Under the conditions of our experiments we found that β2-adaptin was co-IPd by GFP-CHC17-FL and by GFP-CHC22-FL. The interaction was evident irrespective of depletion of endogenous CHC17 (Fig 3C). In support of these results, cells expressing GFP-CHC22-FL but depleted of CHC17, that were co-stained for α-adaptin, had a subset of GFP spots that co-localised with α-adaptin puncta. The co-localisation was similar to that seen for GFP-CHC17-FL under the same conditions (Fig 3D). These observations indicate that CHC22-mediated endocytosis is similar to that mediated by CHC17; and that CHC22, like CHC17 can indeed interact with AP2. We interpret the earlier report of a lack of interaction between AP2 and CHC22 in HeLa cells as an out-competition of binding by the high levels of CHC17 relative to CHC22.
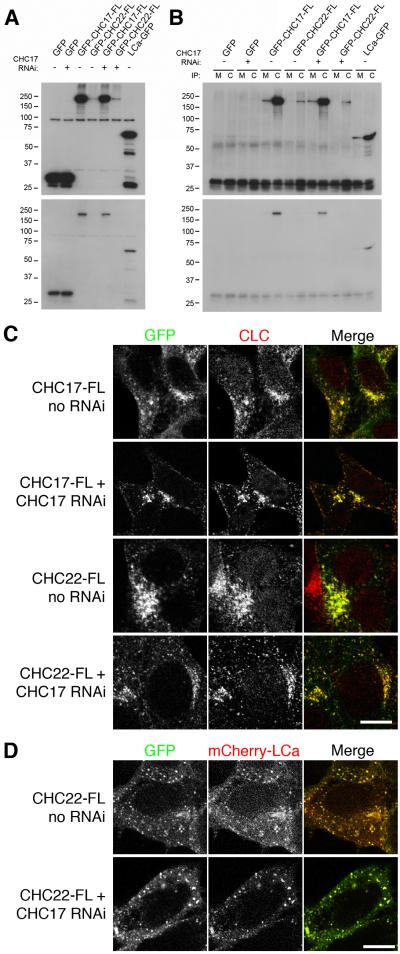
We finally set out to determine whether or not CHC22-FL can associate with clathrin light chains, as there have been conflicting reports on this issue (Liu et al., 2001; Towler et al., 2004b). To analyse binding between endogenous clathrin light chains and GFP-tagged CHCs, co-IP experiments were performed on HEK293 cells expressing GFP-CHC17-FL or GFP-CHC22-FL, using a monoclonal clathrin light chain antibody, CON.1 (Fig 4A-B). Analysis of the lysates used for IPs indicated that there was a large difference in expression between GFP-CHC17-FL and GFP-CHC22-FL (Fig 4A), reflecting the lower transfection rate and expression of CHC22-FL compared to CHC17-FL. Nonetheless, we detected specific co-IP of GFP-CHC22-FL with clathrin light chains (Fig 4B). This indicated a biochemical association between CHC22 and clathrin light chains. We next observed by confocal microscopy good co-localisation between GFP-CHC22-FL and endogenous clathrin light chains. The co-localisation was similar to that for GFP-CHC17-FL and also did not noticeably differ with or without CHC17 RNAi (Fig 4C). We also observed virtually complete co-localisation of GFP-CHC22-FL co-expressed with mCherry-tagged clathrin light chain a (LCa). The co-localisation of puncta was again independent of CHC17 levels (Fig 4D). These results suggest that CHC22, like CHC17, can indeed associate with clathrin light chains in cells.
Fig 4.
Association between CHC22 and clathrin light chains
A-B, IPs were performed using either anti-myc (M) or anti-clathrin light chain (C) from lysates of HEK293 cells transfected as indicated. Samples of the lysates (A) or the IP (B) were analysed by blotting for GFP. Long (above) and short (below) exposures are shown to visualise the signal for GFP-CHC22-FL, these blots are typical of three experiments. C, Representative confocal micrographs of cells expressing GFP-CHC17-FL or GFP-CHC22-FL (green) in the presence (no RNAi) or near absence (CHC17 RNAi) of endogenous CHC17. Cells were stained for endogenous clathrin light chains using CON.1/A546 (CLC, red). D, Representative confocal micrographs of cells expressing GFP-CHC22-FL (left, green in merge) and mCherry-LCa (middle, red in merge) in the presence (no RNAi) or near absence (CHC17 RNAi) of endogenous CHC17. Scale bar = 10 μm.
The observation of functional equivalence between CHC17-FL and CHC22-FL reported here is only of significance if CHC22-FL is actually expressed endogenously. To investigate which form of CHC22 is expressed, we cloned CHC22 from human skeletal muscle cDNA and from HeLa cDNA (Supplementary Fig S2). We found expression of full-length CHC22 only, i.e. the exon 9-containing form. A CHC22 transcript that lacked exon 9 is expressed, but only in a truncated form that terminates downstream of exon 10 and upstream of exon 27. This means that if translation does occur, this variant would not form triskelia and would be unlikely to function as anything other than a dominant-negative. We also found no evidence for splicing of exon 9 in CHC17 (Supplementary Fig S2). The implication is that the CHC22 form that has previously been studied (Liu et al., 2001; Towler et al., 2004b) is a variant that is not normally found in skeletal muscle or HeLa cells. This is further compounded by the observation that CHC22ΔExon9 is non-functional in the assays used here.
One potential concern with our experimental strategy is that the functional rescue that we have observed is due to CHCs that are non-functional associating with the trace amounts of endogenous CHC17 left in cells following RNAi. We believe this is unlikely as CHC22 does not appear to co-trimerise with CHC17 (Liu et al., 2001) and even transfected CHC17 does not co-trimerise with endogenous CHC17 (Liu et al., 1998; Wilde et al., 1999). In addition, we have expressed under the same RNAi conditions a CHC17 lacking the N-terminal domain necessary for binding to AP2 and found no functional rescue of endocytosis (Royle and Lagnado, 2006). This indicates that in our experimental system the CHCs that are transfected can form triskelia that operate independently of trace amounts of CHC17.
In summary, when expressed under similar conditions, CHC17 and CHC22 are equally functional in endocytosis and mitosis. In most cells in the body, the level of CHC22 expression is apparently too low for a significant contribution to endocytosis and to mitosis and this explains why endocytic and mitotic defects are actually observed upon CHC17-depletion (Hinrichsen et al., 2003; Motley et al., 2003; Royle et al., 2005). The ability of CHC22 to functionally substitute for CHC17 is likely to be relevant in cells expressing higher levels of CHC22, such as in skeletal muscle, especially during myogenesis when CHC22 expression is further enhanced (Liu et al., 2001; Towler et al., 2004b). We also do not rule out the possibility that CHC22 has unique functions. For example, CHC22, unlike CHC17, can bind to sorting nexin 5 (SNX5) a membrane trafficking protein that is enriched in skeletal muscle (Towler et al., 2004b).
The present work provides further support to the idea that the mitotic function of clathrin is separate from the function of clathrin in endocytosis. We have previously showed that (i) a dominant-negative construct that interferes with endocytosis does not cause mitotic defects as a secondary effect (Royle et al., 2005), (ii) mitosis can be rescued partially in CHC17-depleted cells by a construct that is not competent for endocytosis (Stunted in (Royle and Lagnado, 2006)). Here we found that CHC17ΔExon9 is a construct that can rescue endocytosis but not mitosis in CHC17-depleted cells. This further suggests that these two functions for clathrin are separable. Another interesting observation from this work is that removal of exon 9 from either CHC17 or CHC22 resulted in a reduction in spindle-binding. This raises the possibility that the residues encoded by exon 9, in addition to the N-terminal domain, constitute a binding site for microtubules or microtubule-associated proteins of spindle fibres. Alternatively, the removal of CHCR0 helices e-h, may result in a conformational change which results in the N-terminal domains no longer being available for spindle-binding (Supplementary Fig. S1). These two models are currently being tested in our laboratory.
Materials and Methods
Molecular biology
DNA constructs
A plasmid containing human CHC22ΔExon9 (in pEGFP-N3) was a gift from Frances Brodsky (UCSF) (Towler et al., 2004b). In order to make GFP-CHC22ΔExon9, a Hind III-Xma I fragment from the pEGFP-N3 plasmid was subcloned into pEGFP-C1. This gave GFP-CHC22ΔExon9 with an extra 7 amino acids (GPSTGSR) before the stop codon. Chimeric clathrin constructs were made by first inserting restriction sites that are present in CHC17 into GFP-CHC22ΔExon9 by site-directed mutagenesis and then swapping fragments between these plasmids and human GFP-CHC17-FL (available from previous work).
GFP-CHC22-FL was made by ‘repairing’ GFP-CHC22ΔExon9 by inserting a ScaI-BspEI fragment digested from an internal PCR from human testis cDNA library (Clontech). GFP-CHC17ΔExon9 was made by overlap extension PCR using GFP-CHC17(1-1675)KDP as template and Acc65 I-Sal I sites.
pBrain-GFP-CHC(1-1675)KDP-CHC4 was available from previous work. We made pBrain-GFP-CHC17ΔExon9-CHC4 by inserting a BglII-EcoRV fragment from GFP-CHC17ΔExon9 into pBrain-GFP-CHC(1-1675)KDP-CHC4. The pBrain-GFP-CHC22FL-CHC4 and pBrain-GFP-CHC22ΔExon9-CHC4 were made by inserting an ApaLI-AgeI fragment from pBrain-GFP-CHC4 into GFP-CHC22FL and GFP-CHC22ΔExon9. Any constructs that contained the sequence coding for CHC17 residues 60-66 were knockdown-proof (Royle et al., 2005). CHC22 has four changes from the CHC17 shRNA target sequence and was therefore not targeted by RNAi. All constructs were verified by restriction digest and any that involved PCR were further verified by automated DNA sequencing (Geneservice, UK).
Amplification and analysis of CHC22 and CHC17 transcripts
Total RNA from HeLa and HEK293 cells was extracted using Trizol (Invitrogen), human skeletal muscle total RNA (No. 540029) was from Stratagene. In each case cDNA was obtained from 5 μg total RNA using an oligo dT15 primer and amplified with Superscript II reverse transcriptase (Invitrogen). Following first strand cDNA synthesis, the hybridised mRNA was degraded using RNAse H. Amplification from these cDNA templates was by PCR using Pfu turbo (Stratagene). Details of primers used are given in Supplementary Table 1.
Cell biology
HEK293 cell culture, transfection, transferrin uptake and immunocytochemistry were carried out as described previously (Royle and Lagnado, 2006). The RNAi conditions for rescue experiments have been described elsewhere (Royle and Lagnado, 2006). In summary, we typically get >90 % reduction of CHC17 compared to controls at 72 h and re-expression levels of GFP-CHC17 are approximately two-thirds of endogenous CHC17 levels. Here, the fluorescence intensity of GFP-CHC22-FL was less than that of GFP-CHC17-FL and the transfection rate of GFP-CHC22-FL was also consistently lower than GFP-CHC17-FL. Monoclonal antibodies: anti-c-myc (9E10, Zymed laboratories, 13-2500), anti-CHC17 (TD.1 Covance, MMS-427P), anti-clathrin light chain (CON.1, Sigma Aldrich, C1985), anti-β1/β2 adaptin (100/1, AbCam, ab11327) and anti-α-adaptin (AP.6, Cambridge Bioscience, MA1-064). Polyclonal sheep anti-GFP was a generous gift of Francis Barr (University of Liverpool). For immunoblotting, antibody concentrations were as follows: 49.5 μg/ml 100/1, 1 μg/ml TD.1, 1:1000 anti-GFP. Anti-mouse HRP-conjugated IgG (GE Healthcare, 1:10000) and anti-sheep HRP-conjugated IgG (Cambridge Bioscience, 1:10000) were use for detection. For immunocytochemistry: 5 μg/ml CON1, 25 μg/ml AP6. Anti-mouse Alexa546-conjugated IgG (Invitrogen, 1:500) was used for detection.
Biochemistry
HEK293 cells transfected with GFP-clathrin constructs or control plasmids were harvested 72 hours post-transfection. The fractionation protocol was as described elsewhere (McCullough et al., 2006). Briefly, cells were homogenised in homogenisation buffer (10 mM Hepes, pH 7.2, 3 mM Imidazole, 250 mM sucrose, protease inhibitor cocktail) by passing through a 23G needle 6-8 times. Nuclei were spun out of the homogenate at 1900 × g, and the post-nuclear supernatant spun at 100000 × g in a Beckman TLA120.2 rotor to separate the cytosolic supernatant from the membrane-enriched pellet.
For whole cell lysates, transfected HEK293 cells were trypsinised and resuspended in lysis buffer A (20 mM Tris, pH 7.5, 150 mM NaCl, 1% IGEPAL, 30 mM NaF, 5 mM Na3VO4, 15 μg/ml DNase I, 100 nM okadaic acid, protease inhibitor cocktail), incubated on ice for 30 min, then spun at 20000 × g, 4°C, 15 min. The supernatant was retained as cell lysate. For immunoprecipiations, between 1 mg lysate per IP was pre-cleared with 10 μl protein G beads 1 h, 4°C, rotating, then incubated with either 3 μl GFP antibody or 2 μg CON.1 or 9E10 antibody, 1 hr, 4°C, rotating. Then 15 μl protein G beads were added and incubated for a further 1.5 h, 4° C, rotating in the case of CON.1 IPs, or overnight, 4° C, rotating for GFP IPs. Beads were washed once with 1 ml lysis buffer B (20 mM Tris, pH 7.5, 150 mM NaCl, 1% IGEPAL, protease inhibitor cocktail), and 4 times with 1 ml wash buffer (20 mM Tris, pH 7.5, 150 mM NaCl, 0.1% IGEPAL, protease inhibitor cocktail). Beads were resuspended in Laemmli buffer.
Imaging and data analysis
Confocal imaging was done using a Leica confocal microscope SP2 with a 63x (1.4 NA) oil-immersion objective. GFP, Alexa546 and mCherry were excited using an Ar/Kr 488 nm laser, a He/Ne 543 nm laser or He/Ne 543/594 nm laser lines, respectively. H33342 or DAPI were excited using a multiphoton laser. Excitation and collection of emission were performed separately and sequentially. For quantitative experiments, identical laser power and acquisition settings were used. For analysis of transferrin uptake, images were imported into ImageJ (NIH), the outline of the cell was manually drawn using the GFP channel as a guide and this ROI was transferred to the red channel. The image was thresholded and the transferrin-Alexa546 puncta quantified. Experiments to determine the mitotic index were quantified by counting the number of GFP-positive cells with mitotic figures as a proportion of the total number of GFP-positive cells within a 1.215 mm2 area. The number of GFP-positive metaphase-like cells that had misaligned chromosomes was counted. For mitotic index measurements between 1026 and 2113 cells per condition were counted. For transferrin uptake measurements between 78 and 99 cells were analysed for each condition. All experiments were preformed three or more times. Normally distributed data were analysed by ANOVA with Dunnet’s post-hoc test and binomial data were analysed by Chi-squared approximation using Instat (Graphpad). Figures were assembled using Igor Pro (Wavemetrics), PyMOL (DeLano Scientific) and Adobe Photoshop.
Supplementary Material
Acknowledgements
We thank Hannah Dunford for technical assistance during the early stages of this study; Frances Brodsky for the gift of CHC22ΔExon9-GFP; Francis Barr and Ulrike Gruneberg for helpful suggestions and useful reagents. We thank Sylvie Urbé for help with preparing membrane-enriched fractions. This work was supported by a Career Establishment Award from Cancer Research UK.
References
- Borner GH, Harbour M, Hester S, Lilley KS, Robinson MS. Comparative proteomics of clathrin-coated vesicles. J. Cell Biol. 2006;175:571–578. doi: 10.1083/jcb.200607164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dell’Angelica EC, Klumperman J, Stoorvogel W, Bonifacino JS. Association of the AP-3 adaptor complex with clathrin. Science. 1998;280:431–434. doi: 10.1126/science.280.5362.431. [DOI] [PubMed] [Google Scholar]
- Drake MT, Traub LM. Interaction of two structurally distinct sequence types with the clathrin terminal domain beta-propeller. J. Biol. Chem. 2001;276:28700–28709. doi: 10.1074/jbc.M104226200. [DOI] [PubMed] [Google Scholar]
- Fotin A, Cheng Y, Sliz P, Grigorieff N, Harrison SC, Kirchhausen T, Walz T. Molecular model for a complete clathrin lattice from electron cryomicroscopy. Nature. 2004;432:573–579. doi: 10.1038/nature03079. [DOI] [PubMed] [Google Scholar]
- Gong W, Emanuel BS, Collins J, Kim DH, Wang Z, Chen F, Zhang G, Roe B, Budarf ML. A transcription map of the DiGeorge and velo-cardio-facial syndrome minimal critical region on 22q11. Hum. Mol. Genet. 1996;5:789–800. doi: 10.1093/hmg/5.6.789. [DOI] [PubMed] [Google Scholar]
- Hinrichsen L, Harborth J, Andrees L, Weber K, Ungewickell EJ. Effect of clathrin heavy chain- and alpha-adaptin-specific small inhibitory RNAs on endocytic accessory proteins and receptor trafficking in HeLa cells. J. Biol. Chem. 2003;278:45160–45170. doi: 10.1074/jbc.M307290200. [DOI] [PubMed] [Google Scholar]
- Jing SQ, Spencer T, Miller K, Hopkins C, Trowbridge IS. Role of the human transferrin receptor cytoplasmic domain in endocytosis: localization of a specific signal sequence for internalization. J. Cell Biol. 1990;110:283–294. doi: 10.1083/jcb.110.2.283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kedra D, Peyrard M, Fransson I, Collins JE, Dunham I, Roe BA, Dumanski JP. Characterization of a second human clathrin heavy chain polypeptide gene (CLH-22) from chromosome 22q11. Hum. Mol. Genet. 1996;5:625–631. doi: 10.1093/hmg/5.5.625. [DOI] [PubMed] [Google Scholar]
- Kirchhausen T. Clathrin. Annu. Rev. Biochem. 2000;69:699–727. doi: 10.1146/annurev.biochem.69.1.699. [DOI] [PubMed] [Google Scholar]
- Lindsay EA, Rizzu P, Antonacci R, Jurecic V, Delmas-Mata J, Lee CC, Kim UJ, Scambler PJ, Baldini A. A transcription map in the CATCH22 critical region: identification, mapping, and ordering of four novel transcripts expressed in heart. Genomics. 1996;32:104–112. doi: 10.1006/geno.1996.0082. [DOI] [PubMed] [Google Scholar]
- Liu SH, Marks MS, Brodsky FM. A dominant-negative clathrin mutant differentially affects trafficking of molecules with distinct sorting motifs in the class II major histocompatibility complex (MHC) pathway. J. Cell Biol. 1998;140:1023–1037. doi: 10.1083/jcb.140.5.1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu SH, Towler MC, Chen E, Chen CY, Song W, Apodaca G, Brodsky FM. A novel clathrin homolog that co-distributes with cytoskeletal components functions in the trans-Golgi network. EMBO J. 2001;20:272–284. doi: 10.1093/emboj/20.1.272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long KR, Trofatter JA, Ramesh V, McCormick MK, Buckler AJ. Cloning and characterization of a novel human clathrin heavy chain gene (CLTCL) Genomics. 1996;35:466–472. doi: 10.1006/geno.1996.0386. [DOI] [PubMed] [Google Scholar]
- Maro B, Johnson MH, Pickering SJ, Louvard D. Changes in the distribution of membranous organelles during mouse early development. J. Embryol. Exp. Morphol. 1985;90:287–309. [PubMed] [Google Scholar]
- McCullough J, Row PE, Lorenzo O, Doherty M, Beynon R, Clague MJ, Urbe S. Activation of the endosome-associated ubiquitin isopeptidase AMSH by STAM, a component of the multivesicular body-sorting machinery. Curr. Biol. 2006;16:160–165. doi: 10.1016/j.cub.2005.11.073. [DOI] [PubMed] [Google Scholar]
- Miele AE, Watson PJ, Evans PR, Traub LM, Owen DJ. Two distinct interaction motifs in amphiphysin bind two independent sites on the clathrin terminal domain beta-propeller. Nat. Struct. Mol. Biol. 2004;11:242–248. doi: 10.1038/nsmb736. [DOI] [PubMed] [Google Scholar]
- Motley A, Bright NA, Seaman MN, Robinson MS. Clathrin-mediated endocytosis in AP-2-depleted cells. J. Cell Biol. 2003;162:909–918. doi: 10.1083/jcb.200305145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nesterov A, Carter RE, Sorkina T, Gill GN, Sorkin A. Inhibition of the receptor-binding function of clathrin adaptor protein AP-2 by dominant-negative mutant mu2 subunit and its effects on endocytosis. EMBO J. 1999;18:2489–2499. doi: 10.1093/emboj/18.9.2489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamoto CT, McKinney J, Jeng YY. Clathrin in mitotic spindles. Am. J. Physiol. Cell Physiol. 2000;279:C369–374. doi: 10.1152/ajpcell.2000.279.2.C369. [DOI] [PubMed] [Google Scholar]
- Royle SJ. The cellular functions of clathrin. Cell Mol Life Sci. 2006;63:1823–1832. doi: 10.1007/s00018-005-5587-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Royle SJ, Lagnado L. Trimerisation is important for the function of clathrin at the mitotic spindle. J. Cell Sci. 2006;119:4071–4078. doi: 10.1242/jcs.03192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Royle SJ, Bright NA, Lagnado L. Clathrin is required for the function of the mitotic spindle. Nature. 2005;434:1152–1157. doi: 10.1038/nature03502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sirotkin H, Morrow B, DasGupta R, Goldberg R, Patanjali SR, Shi G, Cannizzaro L, Shprintzen R, Weissman SM, Kucherlapati R. Isolation of a new clathrin heavy chain gene with muscle-specific expression from the region commonly deleted in velo-cardio-facial syndrome. Hum. Mol. Genet. 1996;5:617–624. doi: 10.1093/hmg/5.5.617. [DOI] [PubMed] [Google Scholar]
- ter Haar E, Harrison SC, Kirchhausen T. Peptide-in-groove interactions link target proteins to the beta-propeller of clathrin. Proc. Natl. Acad. Sci. U.S.A. 2000;97:1096–1100. doi: 10.1073/pnas.97.3.1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Towler MC, Kaufman SJ, Brodsky FM. Membrane traffic in skeletal muscle. Traffic. 2004a;5:129–139. doi: 10.1111/j.1600-0854.2003.00164.x. [DOI] [PubMed] [Google Scholar]
- Towler MC, Gleeson PA, Hoshino S, Rahkila P, Manalo V, Ohkoshi N, Ordahl C, Parton RG, Brodsky FM. Clathrin isoform CHC22, a component of neuromuscular and myotendinous junctions, binds sorting nexin 5 and has increased expression during myogenesis and muscle regeneration. Mol. Biol. Cell. 2004b;15:3181–3195. doi: 10.1091/mbc.E04-03-0249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakeham DE, Abi-Rached L, Towler MC, Wilbur JD, Parham P, Brodsky FM. Clathrin heavy and light chain isoforms originated by independent mechanisms of gene duplication during chordate evolution. Proc. Natl. Acad. Sci. U.S.A. 2005;102:7209–7214. doi: 10.1073/pnas.0502058102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilde A, Beattie EC, Lem L, Riethof DA, Liu SH, Mobley WC, Soriano P, Brodsky FM. EGF receptor signaling stimulates SRC kinase phosphorylation of clathrin, influencing clathrin redistribution and EGF uptake. Cell. 1999;96:677–687. doi: 10.1016/s0092-8674(00)80578-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.