Abstract
The pathway for base pair opening within a B-DNA duplex is investigated by theoretical molecular modeling. The results show that the disruption of a single base pair is energetically compatible with the deductions made from hydrogen exchange measurements. In addition, it is found that the opening process is greatly facilitated by DNA bending and that, conversely, once a base pair is disrupted, DNA can bend very easily. It appears that the energetic coupling between these two processes may play an important role in many biological reactions involving nucleic acid distortion.
Full text
PDF
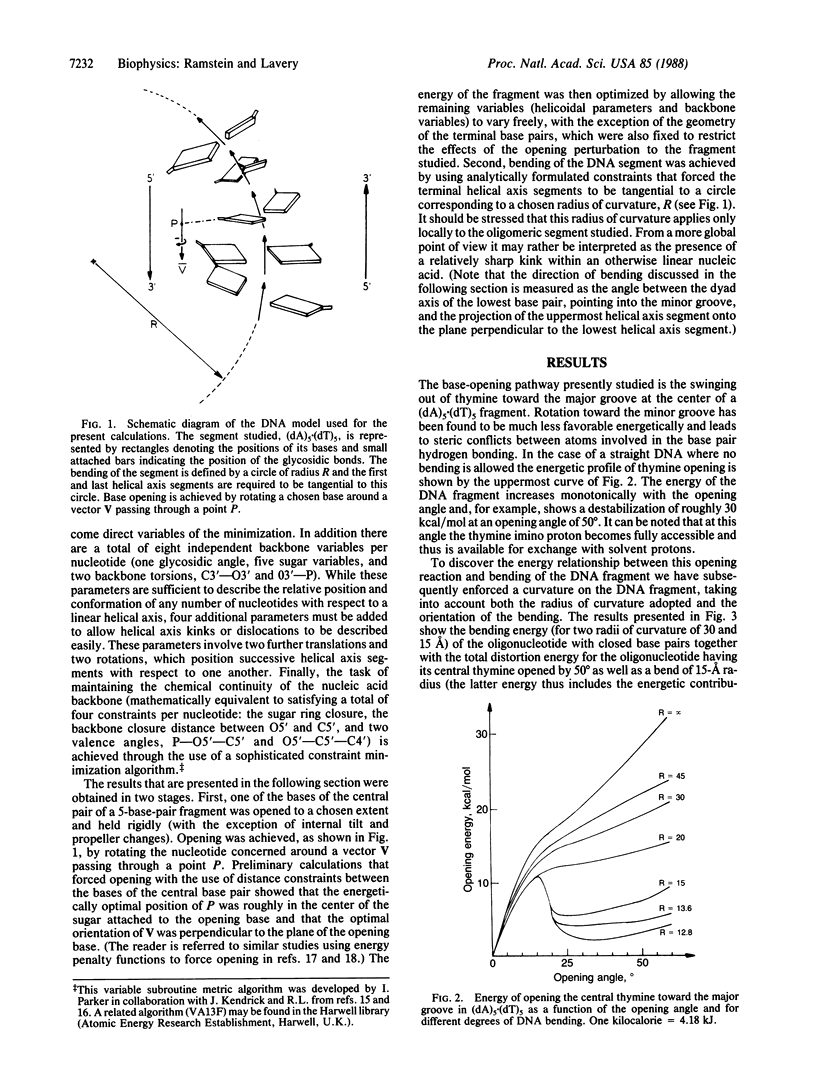
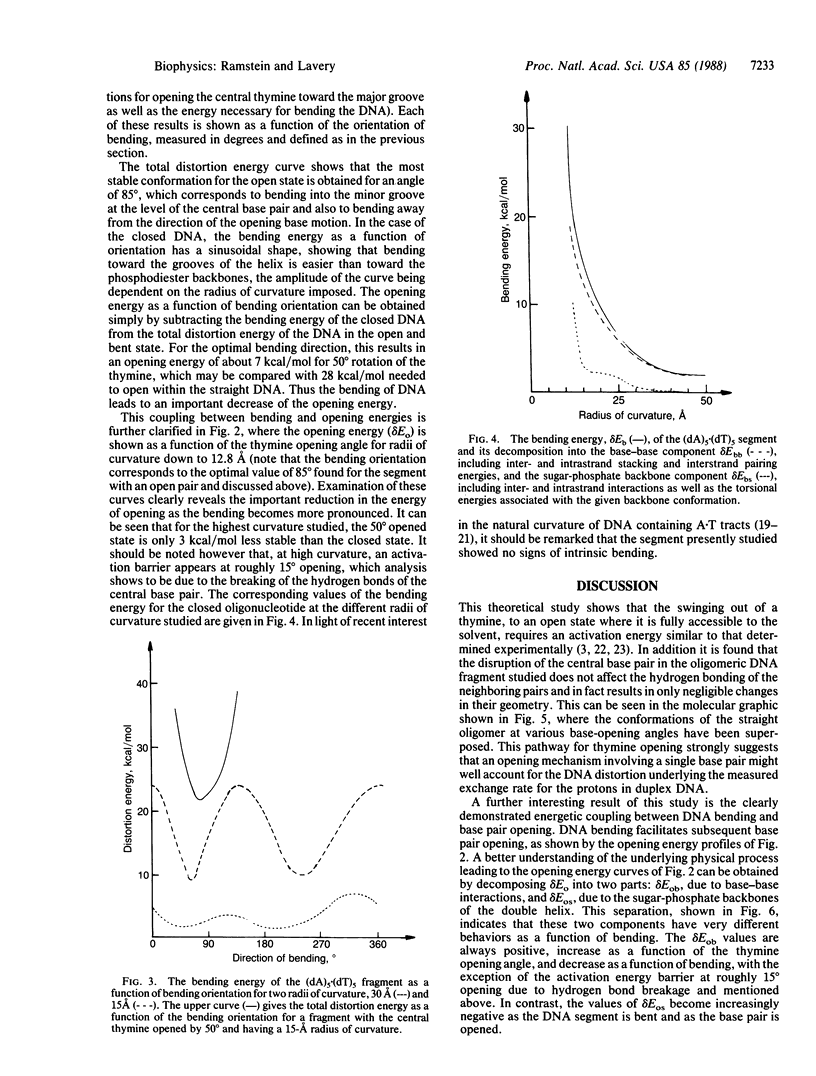
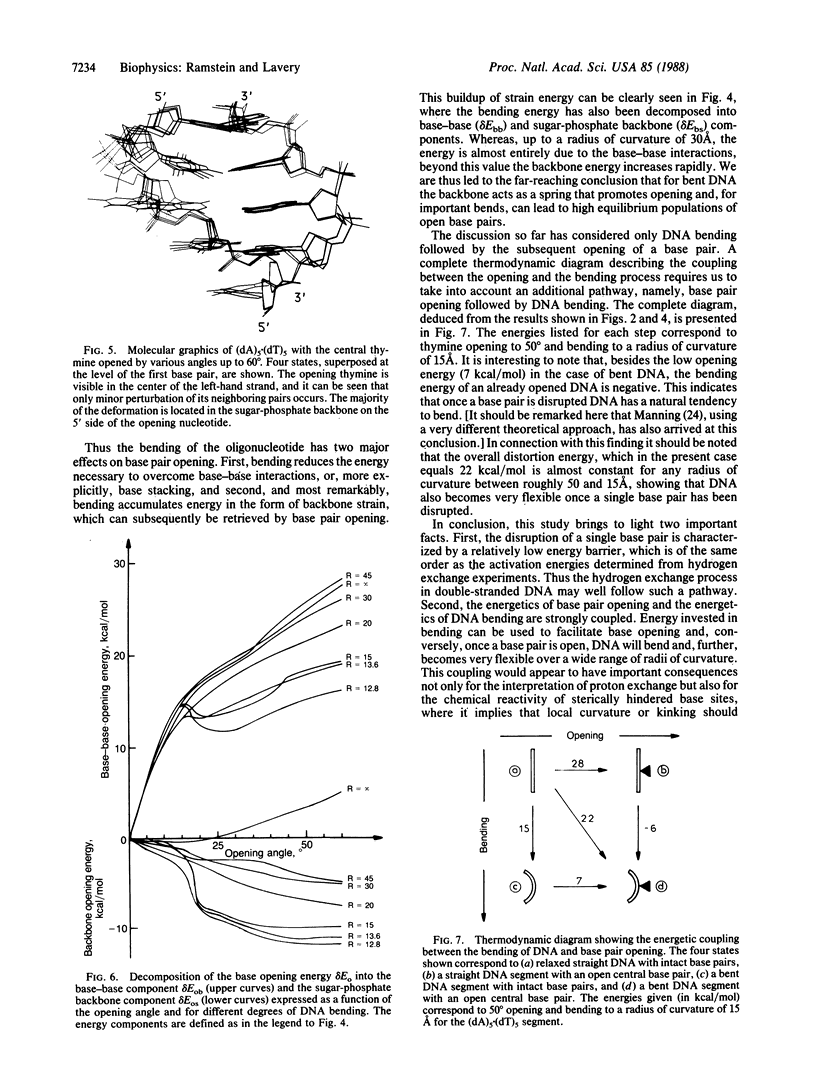

Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Englander S. W., Kallenbach N. R. Hydrogen exchange and structural dynamics of proteins and nucleic acids. Q Rev Biophys. 1983 Nov;16(4):521–655. doi: 10.1017/s0033583500005217. [DOI] [PubMed] [Google Scholar]
- Etchebest C., Pullman A. The gramicidin A channel: energetics and structural characteristics of the progression of a sodium ion in the presence of water. J Biomol Struct Dyn. 1986 Feb;3(4):805–825. doi: 10.1080/07391102.1986.10508463. [DOI] [PubMed] [Google Scholar]
- Guéron M., Kochoyan M., Leroy J. L. A single mode of DNA base-pair opening drives imino proton exchange. Nature. 1987 Jul 2;328(6125):89–92. doi: 10.1038/328089a0. [DOI] [PubMed] [Google Scholar]
- Hagerman P. J. Evidence for the existence of stable curvature of DNA in solution. Proc Natl Acad Sci U S A. 1984 Aug;81(15):4632–4636. doi: 10.1073/pnas.81.15.4632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartmann B., Leng M., Ramstein J. Poly(dA-dT).poly(dA-dT) two-pathway proton exchange mechanism. Effect of general and specific base catalysis on deuteration rates. Biochemistry. 1986 Jun 3;25(11):3073–3077. doi: 10.1021/bi00359a001. [DOI] [PubMed] [Google Scholar]
- Keepers J. W., Kollman P. A., Weiner P. K., James T. L. Molecular mechanical studies of DNA flexibility: coupled backbone torsion angles and base-pair openings. Proc Natl Acad Sci U S A. 1982 Sep;79(18):5537–5541. doi: 10.1073/pnas.79.18.5537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keepers J., Kollman P. A., James T. L. Molecular mechanical studies of base-pair opening in d(CGCGC):d(GCGCG), dG5 . dC5, d(TATAT):d(ATATA), and dA5 . dT5 in the B and Z forms of DNA. Biopolymers. 1984 Nov;23(11 Pt 2):2499–2511. doi: 10.1002/bip.360231124. [DOI] [PubMed] [Google Scholar]
- Lavery R., Parker I., Kendrick J. A general approach to the optimization of the conformation of ring molecules with an application to valinomycin. J Biomol Struct Dyn. 1986 Dec;4(3):443–462. doi: 10.1080/07391102.1986.10506361. [DOI] [PubMed] [Google Scholar]
- Lavery R., Sklenar H., Zakrzewska K., Pullman B. The flexibility of the nucleic acids: (II). The calculation of internal energy and applications to mononucleotide repeat DNA. J Biomol Struct Dyn. 1986 Apr;3(5):989–1014. doi: 10.1080/07391102.1986.10508478. [DOI] [PubMed] [Google Scholar]
- Lavery R., Zakrzewska K., Pullman B. Binding of non-intercalating antibiotics to B-DNA: a theoretical study taking into account nucleic acid flexibility. J Biomol Struct Dyn. 1986 Jun;3(6):1155–1170. doi: 10.1080/07391102.1986.10508492. [DOI] [PubMed] [Google Scholar]
- Mandal C., Kallenbach N. R., Englander S. W. Base-pair opening and closing reactions in the double helix. A stopped-flow hydrogen exchange study in poly(rA).poly(rU). J Mol Biol. 1979 Dec 5;135(2):391–411. doi: 10.1016/0022-2836(79)90443-1. [DOI] [PubMed] [Google Scholar]
- Manning G. S. Breathing and bending fluctuations in DNA modeled by an open-base-pair kink coupled to axial compression. Biopolymers. 1983 Feb;22(2):689–729. doi: 10.1002/bip.360220211. [DOI] [PubMed] [Google Scholar]
- Marini J. C., Levene S. D., Crothers D. M., Englund P. T. Bent helical structure in kinetoplast DNA. Proc Natl Acad Sci U S A. 1982 Dec;79(24):7664–7668. doi: 10.1073/pnas.79.24.7664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakanishi M., Mitane Y., Tsuboi M. A hydrogen exchange study of the open segment in a DNA double helix. Biochim Biophys Acta. 1984 Mar 22;798(1):46–52. doi: 10.1016/0304-4165(84)90008-4. [DOI] [PubMed] [Google Scholar]
- Takashima H., Nakanishi M., Tsuboi M. Determination of the kinetics of deuteration of DNA.RNA hybrids by ultraviolet spectroscopy. Biochemistry. 1985 Aug 27;24(18):4823–4825. doi: 10.1021/bi00339a016. [DOI] [PubMed] [Google Scholar]
- Ulanovsky L. E., Trifonov E. N. Estimation of wedge components in curved DNA. Nature. 1987 Apr 16;326(6114):720–722. doi: 10.1038/326720a0. [DOI] [PubMed] [Google Scholar]
- Zakrzewska K., Pullman B. Sequence selectivity, a test of the nature of the covalent adduct formed between benzo[a]pyrene and DNA. J Biomol Struct Dyn. 1987 Apr;4(5):845–858. doi: 10.1080/07391102.1987.10507682. [DOI] [PubMed] [Google Scholar]
- Zhurkin V. B., Poltev V. I., Florent'ev V. L. Atom-atomnye potentsial'nye funktsii dlia konformatsionnykh raschetov nukleinovykh kislot. Mol Biol (Mosk) 1980 Sep-Oct;14(5):1116–1130. [PubMed] [Google Scholar]



