Abstract
The spontaneously hypertensive rat (SHR) is the most widely used animal model of essential hypertension and associated metabolic disturbances. Multiple quantitative trait loci associated with hemodynamic and metabolic parameters have been mapped in the SHR. Recently, it has become possible to identify some of the specific quantitative trait gene (QTG) variants that underlie quantitative trait loci linked to complex cardiovascular and metabolic traits in SHR related strains. Recombinant inbred strains derived from SHR and Brown Norway progenitors, together with SHR congenic and transgenic strains, have proven useful for establishing the identity of several QTGs in SHR models. It is anticipated that the combined use of linkage analyses and gene expression profiles, together with the recently available genome sequences of both the SHR and Brown Norway strains and new methods for manipulating the rat genome, will soon accelerate progress in identifying QTGs for complex traits in SHR-related strains.
Keywords: Spontaneously hypertensive rat, Quantitative trait genes, Genetics
Introduction
Genome-wide association studies (GWAS) represent the most widely used approach for genetic analyses of common human diseases with multifactorial inheritance [1•]. The identification of genetic variants associated with essential hypertension has turned out to be particularly challenging. The original GWAS (with 14,000 cases and 3000 controls), The Wellcome Trust Case Control Consortium, revealed no statistically significant associations between any of the approximately 500,000 single nucleotide polymorphisms (SNPs) and the dichotomous hypertension trait [2]. Recently, the results of two GWAS were reported, Global Blood Pressure Genetics (Global BP Gen) Consortium [3] and Cohorts for Heart and Aging Research in Genomic Epidemiology (CHARGE) Consortium [4], including respective meta-analyses of 34,433 and 29,136 individuals of European ancestry, with measurements of systolic and diastolic blood pressure. Both studies identified several SNPs in the proximity of known genes that were significantly associated with effects on systolic or diastolic blood pressures of less than 1 mm Hg. Notwithstanding the small effect sizes on blood pressure, results of this kind theoretically could point to interesting new candidate genes that may merit further study from both mechanistic and therapeutic perspectives. To date, however, GWAS explain only a small portion of total heritability, and very few publications involving GWAS have gone beyond statistical findings to include direct experimental testing of the SNPs to investigate their functional impact at the cellular level or to confirm putative effects on blood pressure in vivo [5•, 6, 7]. Given the dramatic advances occurring in next-generation sequencing technology [8], together with increasing interest in the rare-variant hypothesis of hypertension [9], it is possible that direct sequence analysis of thousands of hypertensive patients and controls may someday ultimately supplant the use of GWAS in hypertension.
The Spontaneously Hypertensive Rat
Although results obtained in animal models of human diseases must be interpreted with caution because of important differences between animals and humans in pathophysiologic processes, studies in animal models can provide useful information about genes and pathways underlying complex pathophysiologic traits in humans. The spontaneously hypertensive rat (SHR) is the most widely used animal model of essential hypertension. Under specific environmental conditions, such as when fed a high-fructose diet, the SHR also develops additional features of the metabolic syndrome, including insulin resistance and dyslipidemia [10]. As in humans, spontaneous hypertension and associated metabolic disturbances in the SHR represent complex, polygenic traits. Identification of responsible genes in the SHR could be used to study the role of human orthologous genes in complex cardiovascular and metabolic diseases and to identify new pathways and targets for pharmaceutical interventions [11].
Linkage analyses of crosses between the SHR and various control strains have revealed multiple quantitative trait loci (QTL) associated with blood pressure variation and with parameters of lipid and glucose metabolism on practically all rat chromosomes (see the Rat Genome Database [12] and additional online resources [13]). Based on stringent criteria for identifying QTL at the molecular level, including studies in co-isogenic strains [14], only a few of the responsible quantitative trait genes (QTGs) underlying these QTL for blood pressure and metabolic phenotypes have been confirmed in SHR-related models: 1) a deletion variant of Cd36 (fatty acid translocase) has been confirmed as a genetic determinant of multiple features of the metabolic syndrome including dyslipidemia, insulin resistance, and hypertension [15–17, 18•]; 2) a mutated form of Ogn (osteoglycin) has been confirmed as a gene predisposing to cardiac hypertrophy [19•]; and 3) a variant in Srebf1 (sterol regulatory element binding factor 1c isoform) has been confirmed to exert effects on hepatic lipid levels [20]. Despite many years of intense international effort, only one gene, Cd36, has been confirmed, using both transgenic and congenic rescue studies, to represent a specific molecular determinant influencing hypertension in a model derived from the SHR [18•]. This illustrates the inherent difficulty of identifying causative QTGs even in animal models in which the effects of genetic background and environmental variables can be strictly controlled.
Traditional genetic analyses in the SHR have been based on mapping QTL in genetically segregating populations, followed by genetic isolation of QTL in SHR congenic strains and by positional mapping in congenic sublines. This is a time-consuming process in which QTL are isolated in so-called minimal congenic strains within relatively small differential chromosome segments (usually less than 2 Mbp). However, even minimal congenic strains can harbor multiple genes in their differential chromosome segments, and it is challenging to identify the responsible QTGs within the chromosome regions of interest. For instance, a 1.4 Mbp differential segment genetically isolated within the SHR.PD-D8Rat42/D8Arb23 congenic strain that differs from SHR controls in blood pressure and sensitivity to insulin action still carries 14 genes [21]; on the other hand, a 321 kbp segment of the SS.LEW congenic strain with decreased blood pressure carries no evident candidate gene [22]. Recently, however, new tools and approaches have become available, which may help accelerate identification of responsible gene variants that underlie QTL isolated in SHR congenic strains. These include 1) a combination of gene expression profiling of tissues relevant to the pathogenesis of traits of interest together with linkage and correlation analyses [23]; 2) availability of the complete genome sequence of the SHR [24••]; and 3) availability of new methods for manipulating the rat genome, including more effective transgenesis using Sleeping Beauty transposon vectors [25] and the possibility of producing knockout models using embryo microinjection of zinc-finger nucleases [26] as well as techniques for generating rat embryonic stem cells [27].
The BXH/HXB Recombinant Inbred Strains
Recombinant inbred (RI) strains are derived from F2 individuals obtained by crossing of two highly inbred strains. Randomly chosen F2 pairs are brother-sister mated for more than 20 generations to become genetically fixed. Individual RI strains have unique combinations of loci derived by segregation and recombination of the alleles present in the progenitor strains. An important feature of RI strains is that, because they are inbred, repeated assays can be made so that phenotypes of each strain can be more precisely estimated using average results obtained from measurements in multiple rats per each RI strain. A precise estimation of phenotype-genotype relationship is especially important when studying highly variable traits such as blood pressure or behavior. Moreover, because data are cumulative across assays, studies, and research groups, accumulated data can be analyzed and trait relationships studied in a manner not possible with conventional, genetically segregating populations. This is a major advantage for the analysis of complex pathophysiologic traits.
The BXH/HXB RI strains were derived from reciprocal crosses of the SHR/Ola and BN-Lx/Cub (Brown Norway) progenitors [28]. At present, 20 HXB RI strains and 10 BXH RI strains are available (F>80). The RI strains were genotyped in over 13,000 SNPs that bin into 1,200 strain distribution patterns [29]. In addition, over 130 copy number variants were determined in the RI strains [30]. More than 200 physiological phenotypes determined in RI strains are available online in the GeneNetwork database for genetic analyses [31]. The GeneNetwork database is a public resource that combines genomic and phenotypic information, including gene expression profiles, with fast software for linkage and correlation analyses [32].
The BXH/HXB RI strains were used to map the genetic determinants of gene expression in key tissues relevant to pathophysiology of hypertension and the metabolic syndrome, including kidney, retroperitoneal fat, adrenals, left ventricles, whole brains, livers, soleus muscles, aorta, and hypocampus [23, 33–35] (Hubner, Williams, Pravenec, et al., unpublished results). After assessment of genome-wide significance and accounting for multiple testing using false discovery rates, over 1000 expression QTL (eQTL) were found in these tissues. Linkage analyses of gene expression profiles identified cis- and trans- acting eQTL. These eQTL, especially cis-regulated eQTL in the vicinity of physiological QTL (pQTL), represent a large source of attractive candidate genes for pQTL. In addition, gene expression profiles in relevant tissues can be correlated with physiological traits to reveal quantitative trait transcripts (QTT) and thus prioritize specific genes as candidates for discovery of specific QTGs. Using this strategy, Ogn and Cd36 genes have been identified as QTGs predisposing to cardiac hypertrophy and hypertension, respectively [18•, 19•]. Similarly, Ephx2 (epoxide hydrolase 2, cytoplasmic) has been identified as a genetic determinant predisposing SHHF (spontaneously hypertensive heart failure) rats to heart failure [36]. Gene expression profiles thus represent useful intermediate phenotypes between variability at the DNA level and complex physiological traits. Additional intermediate phenotypes, including microRNA expression, quantitative proteome, metabonome, or epigenetic modifications (epigenome), will be determined in the BXH/HXB RI strains within the new integrated project EURATRANS, funded by the European Union [37]. These new intermediate traits will provide new levels of ever-increasing complexity, which will enable investigators to build gene regulatory networks and associate these with complex physiological traits.
Genome Sequence of the SHR/Ola and BN-Lx/Cub Progenitor Strains
The BN/Mcwi strain, closely related to the BN-Lx progenitor of the BXH/HXB RI strains, was the first completely sequenced rat strain [38]. The other progenitor of the RI strain data set, the SHR/Ola strain, was recently sequenced at 10.7-fold coverage using Illumina high-throughput sequencing technology (Illumina, Inc., San Diego, CA). Comparison of the BN/Mcwi versus SHR/Ola sequences revealed thousands of nonsynonymous SNPs and hundreds of stop codons, frameshift mutations, and copy number variations. All sequence variants are available in the SHRBase database [24••]. Recently, the sequence of the BN-Lx progenitor strain has been determined at greater than 10-fold coverage [39]. The availability of genome sequences of both progenitors of the BXH/HXB RI strains as well as of the SHR.BN-congenic strains that are available for most rat chromosomes or that can be derived within a year using “supersonic“ breeding methods [40] will provide unprecedented opportunities to begin systematically identifying the most promising candidates for in vivo functional analyses and QTG isolation at the molecular level.
Advanced Resources for Manipulating the Rat Genome
Development of Transgenic Rats Using Sleeping Beauty Transposon Vectors
Sleeping Beauty transposon vectors have proven to be useful tools for transgenesis. Recently, a highly active Sleeping Beauty transposase (SB100X) was developed and was demonstrated to be very efficient in mouse transgenesis (about 45%) [25]. SB100X also enables a rate of transgenesis of approximately 25% in the SHR [41]. Transgenic founders showed no apparent signs of mosaic transgene expression and carried a few copies of the transgene. Figure 1 shows transgenic SHRs expressing green fluorescent protein derived with the Sleeping Beauty transposon system and segregation of insertional sites in transgenic lines. Heritable and stable gene knockdown in rats has been also achieved using RNA interference with the help of lentiviral-mediated transgenesis [42].
Fig. 1.
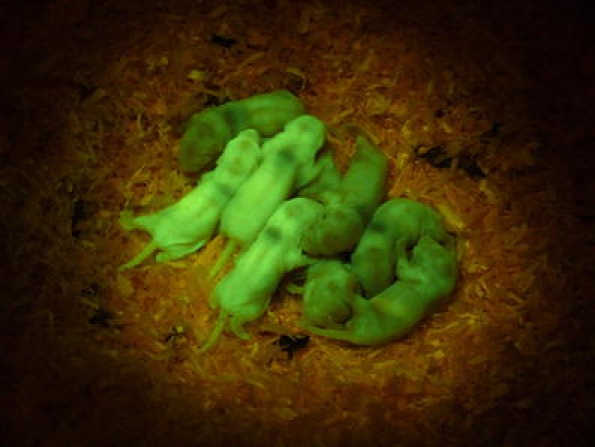
Transgenic spontaneously hypertensive rats (SHRs) derived with the Sleeping Beauty RMCE/Venus transposon vectors. F1 rats were obtained by mating the RMCE/Venus SHR line 80 with three insertional sites on chromosomes 2, 4, and 16 with the SHR progenitor. The different shades of green reflect segregation of the transgenes in F1 rats (Izsvak, Mates, Landa, Pravenec, unpublished results)
Development of New Techniques for Knocking out Specific Rat Genes
Recently, new tools for rat knockouts have become available. It is now possible to inactivate genes using zinc finger nucleases [26], and embryonic stem cells have been derived from rat blastocysts [27]. These new methods should prove useful for in vivo functional studies to analyze QTG candidates for complex traits in the SHR and other rat models.
Variants in the Mitochondrial Genome Affect Glucose and Lipid Metabolism
The relationship of mitochondrial DNA variants to metabolic risk factors for diabetes and other common diseases has begun to attract increasing attention. However, progress in this area has been limited because the phenotypic effects of variation in the mitochondrial genome are difficult to isolate owing to confounding variation in the nuclear genome, imprinting phenomena, and environmental factors. Substitution of different mitochondrial genomes on the same nuclear genetic background in conplastic strains provides a way to unambiguously isolate effects of the mitochondrial genome on complex traits. The SHR/Ola and SHR/Ola-mtBN/Crl conplastic strains have identical nuclear genomes but divergent mitochondrial genomes that encode amino acid differences in seven proteins of oxidative phosphorylation, including a unique phenylalanine-to-leucine substitution in mitochondrial cytochrome c oxidase subunit 1 (mt-Co1), as well as variants in five transfer RNA (tRNA) genes [43]. Compared with the SHR, the conplastic strain exhibited impaired glucose tolerance, lower skeletal muscle glycogen and adenosine triphosphate (ATP) levels, as well as decreases in both the activity and content of cytochrome c oxidase in liver mitochondria. Genotyping of 43 rat inbred strains identified four haplotype groups represented by the BN, F344, LEW, and SHR mitochondrial genomes [43]. Additional conplastic strains with these haplotypes of mitochondrial DNA, including SHR/Ola-mtF344 and SHR/Ola-mtLEW, also exhibited dyslipidemia and decreased sensitivity of skeletal muscles to insulin action when compared with SHR controls (Pravenec et al., unpublished observations). These findings provide some of the first direct evidence linking naturally occurring variation in the mitochondrial genome (independent of variation in the nuclear genome and other confounding factors) to inherited variation in known risk factors for type 2 diabetes. They also establish that spontaneous variation in the mitochondrial genome per se can promote systemic metabolic disturbances relevant to the pathogenesis of common diseases.
Conclusions
During the past several years, major advances have been made in rat genome technology, and new resources have become available, particularly for studies in the SHR and other rat models of hypertension [44••]. The recent availability of the SHR genome sequence, one of the first nonhuman mammalian genomes sequenced by short-read sequencing, should help to further accelerate analyses of molecular mechanisms underlying spontaneous hypertension and related metabolic phenotypes. It is hoped that these new tools and others will enable investigators to gain better insight into the specific genes and mechanisms that influence blood pressure and other complex phenotypes in the SHR and that such results will ultimately prove relevant to our understanding of hypertension and related cardiovascular and metabolic disorders in humans.
Acknowledgments
This work was supported by the Grant Agency of the Ministry of Health of the Czech Republic (NS9754 and NS10036); the Ministry of Education of the Czech Republic (AV0Z 50110509, 1M0520); the Grant Agency of the Czech Republic (301/08/0166); Fondation Leducq (06 CVD 03); the European Commission within the Sixth Framework Programme through the Integrated Project EURATools (contract No. LSHG-CT-2005-019015); and the National Heart, Lung and Blood Institute of the US National Institutes of Health. Dr. Pravenec is an international research scholar of the Howard Hughes Medical Institute.
Disclosure
No potential conflicts of interest relevant to this article have been reported.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Contributor Information
Michal Pravenec, Email: pravenec@biomed.cas.cz.
Theodore W. Kurtz, Email: KurtzT@Labmed2.ucsf.edu
References
Papers of particular interest, published recently, have been highlighted as: • Of importance •• Of major importance
- 1.Altshuler D, Daly MJ, Lander ES. Genetic mapping in human disease. Science. 2008;322:881–888. doi: 10.1126/science.1156409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wellcome Trust Case Control Consortium Genome-wide association study of 14, 000 cases of seven common diseases and 3, 000 shared controls. Nature. 2007;447:661–678. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Newton-Cheh C, Johnson T, Gateva V, et al. Genome-wide association study identifies eight loci associated with blood pressure. Nat Genet. 2009;41:666–676. doi: 10.1038/ng.361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Levy D, Ehret GB, Rice K, et al. Genome-wide association study of blood pressure and hypertension. Nat Genet. 2009;41:677–687. doi: 10.1038/ng.384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Maher B. Personal genomes: the case of the missing heritability. Nature. 2008;456:18–21. doi: 10.1038/456018a. [DOI] [PubMed] [Google Scholar]
- 6.Manolio TA, Collins FS, Cox NJ, et al. Finding the missing heritability of complex diseases. Nature. 2009;461:747–753. doi: 10.1038/nature08494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang Y, O’Connell JR, McArdle PF, et al. Whole-genome association study identifies STK39 as a hypertension susceptibility gene. Proc Natl Acad Sci U S A. 2009;106:226–231. doi: 10.1073/pnas.0808358106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mardis ER. The impact of next-generation sequencing technology on genetics. Trends Genet. 2008;24:133–141. doi: 10.1016/j.tig.2007.12.007. [DOI] [PubMed] [Google Scholar]
- 9.Ji W, Foo JN, O’Roak BJ, et al. Rare independent mutations in renal salt handling genes contribute to blood pressure variation. Nat Genet. 2008;40:592–599. doi: 10.1038/ng.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pravenec M, Zidek V, Simakova M, et al. Genetics of Cd36 and the clustering of multiple cardiovascular risk factors in spontaneous hypertension. J Clin Invest. 1999;103:1651–1657. doi: 10.1172/JCI6691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pravenec M, Kurtz TW. Molecular genetics of experimental hypertension and the metabolic syndrome: from gene pathways to new therapies. Hypertension. 2007;49:941–952. doi: 10.1161/HYPERTENSIONAHA.107.086900. [DOI] [PubMed] [Google Scholar]
- 12.Rat Genome Database Web Site, Medical College of Wisconsin, Milwaukee, Wisconsin. Available at http://rgd.mcw.edu/.
- 13.Twigger SN, Pruitt KD, Fernández-Suárez XM, et al. What everybody should know about the rat genome and its online resources. Nat Genet. 2008;40:523–527. doi: 10.1038/ng0508-523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Glazier AM, Nadeau JH, Aitman TJ. Finding genes that underlie complex traits. Science. 2002;298:2345–2349. doi: 10.1126/science.1076641. [DOI] [PubMed] [Google Scholar]
- 15.Aitman TJ, Gotoda T, Evans AL, et al. Quantitative trait loci for cellular defects in glucose and fatty acid metabolism in hypertensive rats. Nat Genet. 1997;16:197–201. doi: 10.1038/ng0697-197. [DOI] [PubMed] [Google Scholar]
- 16.Aitman TJ, Glazier AM, Wallace CA, et al. Identification of Cd36 (Fat) as an insulin-resistance gene causing defective fatty acid and glucose metabolism in hypertensive rats. Nat Genet. 1999;21:76–83. doi: 10.1038/5013. [DOI] [PubMed] [Google Scholar]
- 17.Pravenec M, Landa V, Zidek V, et al. Transgenic rescue of defective Cd36 ameliorates insulin resistance in spontaneously hypertensive rats. Nat Genet. 2001;27:156–158. doi: 10.1038/84777. [DOI] [PubMed] [Google Scholar]
- 18.Pravenec M, Churchill PC, Churchill MC, et al. Identification of renal Cd36 as a determinant of blood pressure and risk for hypertension. Nat Genet. 2008;40:952–954. doi: 10.1038/ng.164. [DOI] [PubMed] [Google Scholar]
- 19.Petretto E, Sarwar R, Grieve I, et al. Integrated genomic approaches implicate osteoglycin (Ogn) in the regulation of left ventricular mass. Nat Genet. 2008;40:546–552. doi: 10.1038/ng.134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pravenec M, Kazdova L, Landa V, et al. Identification of mutated Srebf1 as a QTL influencing risk for hepatic steatosis in the spontaneously hypertensive rat. Hypertension. 2008;51:148–153. doi: 10.1161/HYPERTENSIONAHA.107.100743. [DOI] [PubMed] [Google Scholar]
- 21.Seda O, Liska F, Sedová L, et al. A 14-gene region of rat chromosome 8 in SHR-derived polydactylous congenic substrain affects muscle-specific insulin resistance, dyslipidaemia and visceral adiposity. Folia Biol (Praha) 2005;51:53–61. [PubMed] [Google Scholar]
- 22.Gopalakrishnan K, Kumarasamy S, Thangavel J, et al.: Alleles of a S.LEW congenic rat spanning 320.6 KB further increase the BP of the hypertensive Dahl S rat—Evidence for Rffl and/or two miRNAs as potential QTL effectors [abstract]. Presented at the Rat Genomics & Models meeting. Cold Spring Harbor, NY, USA; December 2–5, 2009.
- 23.Hubner N, Wallace CA, Zimdahl H, et al. Integrated transcriptional profiling and linkage analysis for identification of genes underlying disease. Nat Genet. 2005;37:243–253. doi: 10.1038/ng1522. [DOI] [PubMed] [Google Scholar]
- 24.•• SHR Base: a SHR genomic resource. Available at http://shr.csc.mrc.ac.uk/index.cgi. Accessed December 2009. The first report of the full genome sequence of the SHR, including comparative analysis with the genome sequence of the Brown Norway (BN/Mcwi) rat.
- 25.Mátés L, Chuah MK, Belay E, et al. Molecular evolution of a novel hyperactive Sleeping Beauty transposase enables robust stable gene transfer in vertebrates. Nat Genet. 2009;41:753–761. doi: 10.1038/ng.343. [DOI] [PubMed] [Google Scholar]
- 26.Geurts AM, Cost GJ, Freyvert Y, et al. Knockout rats via embryo microinjection of zinc-finger nucleases. Science. 2009;325:433. doi: 10.1126/science.1172447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Buehr M, Meek S, Blair K, et al. Capture of authentic embryonic stem cells from rat blastocysts. Cell. 2008;135:1287–1298. doi: 10.1016/j.cell.2008.12.007. [DOI] [PubMed] [Google Scholar]
- 28.Pravenec M, Klír P, Kren V, et al. An analysis of spontaneous hypertension in spontaneously hypertensive rats by means of new recombinant inbred strains. J Hypertens. 1989;7:217–221. doi: 10.1097/00004872-198903000-00008. [DOI] [PubMed] [Google Scholar]
- 29.Saar K, Beck A, Bihoreau MT, et al. SNP and haplotype mapping for genetic analysis in the rat. Nat Genet. 2008;40:560–566. doi: 10.1038/ng.124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Guryev V, Saar K, Adamovic T, et al. Distribution and functional impact of DNA copy number variation in the rat. Nat Genet. 2008;40:538–545. doi: 10.1038/ng.141. [DOI] [PubMed] [Google Scholar]
- 31.GeneNetwork. University of Tennessee: http://www.genenetwork.org.
- 32.Wang J, Williams RW, Manly KF. WebQTL: web-based complex trait analysis. Neuroinformatics. 2003;1:299–308. doi: 10.1385/NI:1:4:299. [DOI] [PubMed] [Google Scholar]
- 33.Grieve IC, Dickens NJ, Pravenec M, et al. Genome-wide co-expression analysis in multiple tissues. PLoS One. 2008;3:e4033. doi: 10.1371/journal.pone.0004033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tabakoff B, Saba L, Printz M, et al. Genetical genomic determinants of alcohol consumption in rats and humans. BMC Biol. 2009;7:70. doi: 10.1186/1741-7007-7-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Petretto E, Mangion J, Dickens NJ, et al. Heritability and tissue specificity of expression quantitative trait loci. PLoS Genet. 2006;2:e172. doi: 10.1371/journal.pgen.0020172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Monti J, Fischer J, Paskas S, et al. Soluble epoxide hydrolase is a susceptibility factor for heart failure in a rat model of human disease. Nat Genet. 2008;40:529–537. doi: 10.1038/ng.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Abbott A. Return of the rat. Nature. 2009;460:788. doi: 10.1038/460788a. [DOI] [PubMed] [Google Scholar]
- 38.Gibbs RA, Weinstock GM, Metzker ML, et al. Genome sequence of the Brown Norway rat yields insights into mammalian evolution. Nature. 2004;428:493–521. doi: 10.1038/nature02426. [DOI] [PubMed] [Google Scholar]
- 39.Simonis M, de Bruijn E, Guryev V, et al.: Genomic and RNA analysis of recombinant inbred panel founder strain [abstract]. Presented at the Rat Genomics & Models meeting. Cold Spring Harbor, NY, USA; December 2–5, 2009.
- 40.Landa V, Zidek V, Pravenec M. Generation of rat “supersonic” congenic/conplastic strains using superovulation and embryo transfer. Methods Mol Biol. 2010;597:267–275. doi: 10.1007/978-1-60327-389-3_18. [DOI] [PubMed] [Google Scholar]
- 41.Mátés L, Landa V, Zidek V, et al.: Harnessing of the hyperactive Sleeping Beauty transposase, SB100X, for genome manipulation in rat [abstract]. Presented at the Rat Genomics & Models meeting. Cold Spring Harbor, NY, USA; December 2–5, 2009.
- 42.Dann CT, Alvarado AL, Hammer RE, et al. Heritable and stable gene knockdown in rats. Proc Natl Acad Sci U S A. 2006;103:11246–11251. doi: 10.1073/pnas.0604657103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pravenec M, Hyakukoku M, Houstek J, et al. Direct linkage of mitochondrial genome variation to risk factors for type 2 diabetes in conplastic strains. Genome Res. 2007;17:1319–1326. doi: 10.1101/gr.6548207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Aitman TJ, Critser JK, Cuppen E, et al. Progress and prospects in rat genetics: a community view. Nat Genet. 2008;40:516–522. doi: 10.1038/ng.147. [DOI] [PubMed] [Google Scholar]


